A system for Cre-regulated RNA interference in vivo (original) (raw)
Abstract
We report a system for Cre-regulated expression of RNA interference in vivo. Expression cassettes comprise selectable and FACS-sortable markers in tandem with additional marker genes and shRNAs in the antisense orientation. The cassettes are flanked by tandem LoxP sites arranged so that Cre expression inverts the marker–shRNA construct, allowing its regulated expression (and, at the same time, deletes the original selection/marker genes). The cassettes can be incorporated into retroviral or lentiviral vectors and delivered to cells in culture or used to generate transgenic mice. We describe cassettes incorporating various combinations of reporter genes, miRNA-based RNAi (including two shRNA constructs at once), and oncogenes and demonstrate the delivery of effective RNA interference in cells in culture, efficient transduction into hematopoietic stem cells with cell-type-specific knockdown in their progeny, and rapid generation of regulated shRNA knockdown in transgenic mice. These vector systems allow regulated combinatorial manipulation (both overexpression and loss of function) of gene expression in multiple systems in vitro and in vivo.
Keywords: lentivirus, retrovirus, FLIP vectors
RNAi has proven to be a valuable research tool for investigating biological processes. Various groups have devoted significant efforts toward developing genome-scale, vector-based RNAi libraries with the intent of probing cellular phenotypes in high-throughput assays in vitro. These efforts are beginning to offer insights into the complex network of interactions that potentiate the development of differentiated cell types or drive the transformed phenotypes of cancer cells (1, 2). Although these in vitro RNAi screens identify many gene “hits,” validating their roles in vivo has remained daunting. The two most vexing obstacles to realizing in vivo RNAi have been efficient delivery and effective regulation of expression. Vector-based RNAi can silence gene expression over long periods and allows the probing of developmental programs and the modeling of disease states. Early vector-based RNAi systems were composed of RNA polymerase III promoters expressing self-complementary shRNAs (3). Transgenic mice engineered to express these constructs exhibited long-term reductions in gene expression and can phenocopy gene-knockout animals (4). The introduction of shRNAs into a larger microRNA (miRNA) context that could be expressed from RNA polymerase II promoters and the discovery that these miRNAs could be placed in the 3′ untranslated regions of transgenes allowed the coupling of reporter and RNAi expression (5–7). Engineering these RNAi expression constructs into retroviral vectors provides a relatively rapid method for introducing RNAi into cell lines and primary cells.
Lentiviral vectors provide efficient gene delivery in vitro, can infect nondividing cells, and may be used to generate transgenic animals (8). However, variegated expression has been observed consistently in lentiviral transgenic mice. This variegation in vivo can be reduced by inclusion of DNA insulating elements (9). In a mouse model of diabetes, inclusion of two DNA insulating elements in the lentiviral vector, pLB, resulted in significantly improved penetrance and effective knockdown of a suspected diabetes-promoting gene. This approach was sufficient to delay significantly the onset of diabetes (10).
Besides transgenics, an attractive alternative to address gene function in vivo is bone marrow reconstitution, in which recipient mice are irradiated to eradicate hematopoietic stem cell (HSC) progenitors and then “reconstituted” with long-term-repopulating HSCs (LTR-HSCs) from donor bone marrow. The stem cells in donor marrow can be transduced with viral vectors in vitro to assess gene functions in vivo.
Retroviral vectors offer efficient gene delivery, but regulated expression of the RNAi construct has thus far proven daunting. Several regulated RNA polIII systems have been developed, but they lack markers to differentiate cells in both the “on” and “off” states (11, 12). Recently, miRNA-based designs have taken advantage of the RNA polII-driven tetracycline-regulated system. These strategies allow reversible “on–off” regulation of RNAi expression in vivo but rely on a limited (although expanding) set of tissue-specific tet-transactivator mice (13).
For in vivo regulation, the Cre-recombinase–loxP system has proven to be a robust method for genetic manipulations (14). The panoply of tissue-specific Cre mice provides a valuable resource from which to manipulate the mouse genome. To take advantage of this “Cre zoo,” we developed a retrovirus-based miRNA expression system that could be regulated by Cre recombinase.
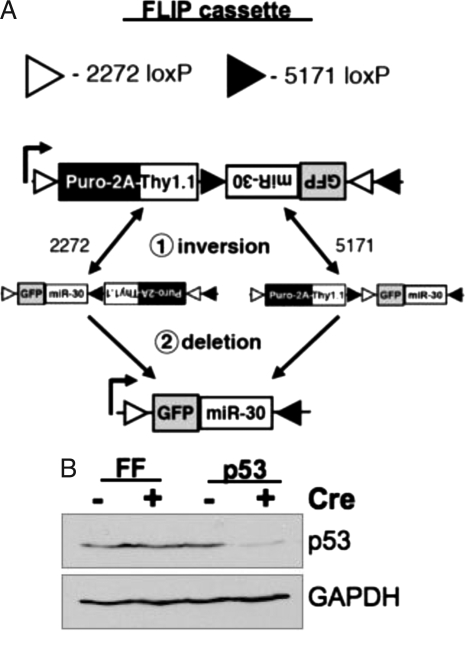
Cre recombinase mediates sequence-specific recombination and, depending on the relative orientation of the target loxP sites, produces either deletion or inversion of the intervening DNA sequence (15, 16). We use this dual activity of Cre recombinase to regulate miRNA processing. Using two sets of mutant loxP sites that cannot recombine with each other, we generated vectors that could mediate stable and irreversible inversion of a segment of DNA after Cre-mediated recombination (17, 18). This “flipping” of a segment of DNA containing the miRNA from the antisense to the sense orientation is sufficient to confer Cre-regulated expression of the shRNA. We have termed these “FLIP vectors” (Fig. 1A).
Fig. 1.
Cre-regulated RNAi. (A) Diagram showing recombination of the FLIP expression cassette with the promoter depicted by the arrow. The vector expresses puromycin resistance and the surface marker Thy1.1. After recombination, the puroR cassette is deleted and GFP and the miR-30 RNAi construct are expressed. (B) Primary MEFs were transduced with MSCV-FLIP targeting FF or the tumor suppressor p53. Cells were selected with puromycin and then infected with MSCV-Cre to induce recombination. Cells were treated with doxorubicin for 4 h, lysed, and immunoblotted for p53 and GAPDH (loading control) protein levels.
FLIP vectors may be used for inducible expression of multiple shRNA constructs in combination with transgenes. The versatility of the FLIP vectors should accelerate the pace of validating in vivo the results from high-throughput screens, and open new possibilities for manipulating the mouse genome. Incorporating FLIP vectors with tissue-specific Cre mouse lines will allow cell-type-specific expression of transgenes and RNAi to model human diseases and probe developmental programs.
Results
FLIP Cassette for Cre-Inducible miRNA Expression.
We set out to develop a system to regulate gene expression in vivo with Cre recombinase. We chose a vector design in which Cre-mediated recombination would stably and irreversibly invert a segment of DNA. The design consists of two pairs of mutant loxP sites (loxP-2272 and 5171) that can recombine with the identical sequence but not with the other mutant loxP site (Fig. 1A) (18). Irreversible Cre-mediated DNA inversion requires two sequential recombination steps. The first consists of a metastable inversion that may revert to the original configuration or proceed through a second productive recombination. The second recombination deletes the cognate loxP site and blocks reversion to the original configuration, ensuring permanent inversion.
Infected cells are selected by a dual selection cassette that consists of the puromycin-_N_-acetyl-transferase gene fused to a segment of the foot-and-mouth disease virus 2A peptide followed by the congenic surface protein Thy1.1 (19, 20). Before recombination, the GFP-miRNA is in the antisense orientation with regard to transcription from the viral LTR (for mouse stem cell virus [MSCV]) or internal promoter (for lentivirus), and the antisense transcript is not productively processed. Upon Cre-mediated recombination, the GFP-miRNA inverts to the sense orientation and is transcribed and properly translated and processed by the cellular miRNA machinery.
The FLIP vector may be used to confer Cre-regulated expression of transgenes and miR-30-based shRNA, as illustrated in Fig. 1B. Primary mouse embryo fibroblasts (MEFs) were transduced with MSCV-FLIP containing miR-30-based shRNA constructs targeting either Firefly luciferase (FF) or the tumor suppressor p53. MEFs were selected with puromycin and subsequently infected with a retrovirus expressing Cre recombinase (MSCV-Cre), resulting in FLIP vector recombination in approximately 80% of MEFs (as measured by GFP expression). Further infections with high-titer Cre virus resulted in >95% GFP+ cells. MEFs were treated for 4 h with the genotoxic agent doxorubicin to induce p53 expression and then analyzed by Western blot for levels of p53 protein. The MEFs transduced with MSCV-FLIP-p53 and MSCV-Cre showed a marked reduction in p53 protein expression. Importantly, p53 remains at wild-type levels in MEFs transduced with MSCV-FLIP-p53 in the absence of Cre expression, indicating that the shRNA is not processed productively when transcribed in the antisense orientation.
Lentiviral Transgenics.
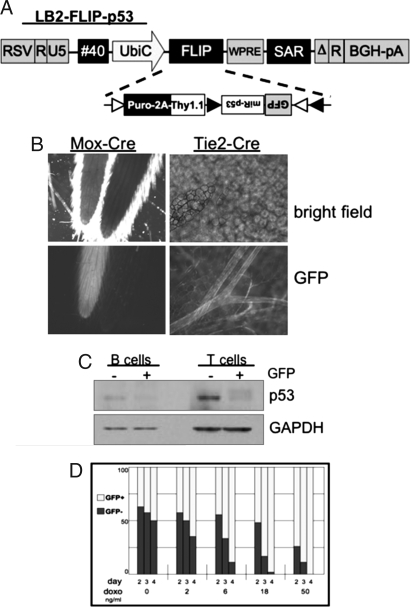
We incorporated the FLIP cassette into a lentiviral vector optimized for transgenesis by inclusion of two DNA insulators (element #40 and SAR), a modified 3′ LTR, and an internal UbiquitinC promoter, in addition to other modifications (Fig. 2A). This vector was used to generate transgenic mice by infection of ES cells. F1-ES cells were infected with LB2-FLIP-p53 virus, selected with puromycin, and chimeric mice were generated by tetraploid blastocyst complementation. Injection of ES cells into tetraploid blastocysts ensures that the chimeric animal is derived almost exclusively from the ES cells, because tetraploid cells can generate extra-embryonic tissue but do not significantly contribute to the embryo (21).
Fig. 2.
Cre-regulated RNAi and gene expression in lentiviral transgenic mice. (A) Schematic representation of the lentiviral transfer vector LB2-FLIP-p53. The UbiquitinC promoter drives the FLIP expression cassette and is flanked by the insulators #40 and SAR. (B) LB2-FLIP-p53 mice were crossed to Mox-Cre mice; Mox-Cre is expressed throughout the embryo. The tails of two littermates exhibit contrasting GFP expression: LB2+; Cre+ (left tail) compared with LB2+; Cre- (right tail). LB2-FLIP-p53 mice were crossed to the endothelial- and hematopoietic-specific Tie2-Cre. LB2+; Cre+ offspring exhibit GFP+ blood vessels throughout the small intestine. (C) Spleen cells from LB2-FLIP-p53+; Tie2-Cre+ mice were sorted by FACS for T or B cells with or without GFP, stimulated in vitro for 3 days, treated with doxorubicin for 4 h, then lysed and analyzed by immunoblot for levels of p53 and GAPDH. GFP+ (and hence Cre-recombined) cells show reduced levels of p53. (D) Spleen cells from LB2-FLIP-p53+; Tie2-Cre+ mice were sorted by FACS for B cells and cultured for several days with increasing concentrations of doxorubicin. The GFP+ (p53 knockdown) B cells are more resistant to the toxic effects of doxorubicin, as judged by the increased fraction of surviving cells.
To test the effectiveness of lenti-FLIP in vivo, tetraploid founder males were crossed to various Cre-transgenic females. LB2-FLIP-p53 males were crossed to Mox-Cre females, which express Cre in all embryonic tissues. Fig. 2B (left) is a picture of the tails of two littermates (bright field, top), but only the left tail exhibits GFP fluorescence (bottom). Whole embryos showed similar GFP expression [supporting information (SI) Fig. S1_A_]. LB2-FLIP-p53:Mox-Cre mice express GFP throughout embryonic and adult tissue.
To test expression in specific cell types, LB2-FLIP-p53 males were crossed to Tie2-Cre females, which express Cre in the endothelial and hematopoietic lineages. LB2-FLIP-p53;Tie2-Cre mice expressed GFP in peripheral blood lymphocytes (PBLs) and in blood vessels. Fig. 2B (right) shows a segment of small intestine (top), and blood vessels are clearly visible by GFP fluorescence (bottom). Analysis of several LB2-FLIP-p53:Tie2-Cre mice demonstrated in some mice a fraction of PBLs that lacked lentivirus vector expression (neither Thy1.1+ nor GFP+ cells), but we did not observe a significant fraction of Thy1.1+ cells, indicating that Tie2-Cre mediates efficient recombination.
To analyze the knockdown of p53, T and B cells from LB2-FLIP-p53;Tie2-Cre mice were sorted for GFP-negative and GFP-positive cells (from a mouse with mediocre penetrance of the lentiviral vector), stimulated through their antigen receptors, cultured in vitro for several days, and then treated with doxorubicin to induce expression of p53. Fig. 2C shows that GFP-positive T cells and B cells have markedly reduced levels of p53.
To test whether the p53 knockdown was protective, primary splenic B cells were stimulated and cultured in vitro for several days with increasing concentrations of doxorubicin, ranging from 2 ng/ml to 50 ng/ml. Fig. 2D demonstrates that GFP-positive B cells showed enhanced survival compared with GFP-negative B cells, indicating the FLIP-mediated reduction of endogenous p53 levels is sufficient to confer relative resistance to p53-dependent apoptosis.
FLIP Variants.
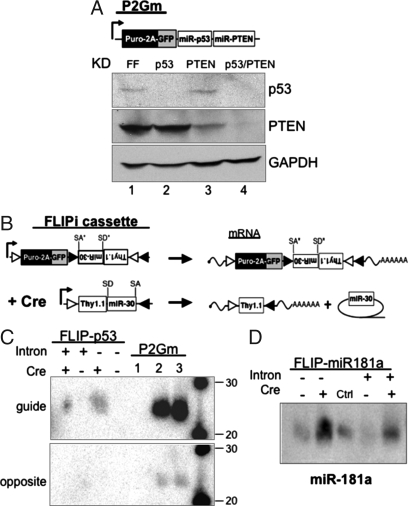
Some endogenous miRNAs are processed from single transcripts that contain multiple miRNA stem-loops (22). We wanted to investigate whether miR30-based shRNA constructs could be regulated similarly. The miR30 shRNA targeting p53 was cloned in front of the miR30 shRNA targeting the tumor suppressor PTEN in the 3′ UTR of the MSCV vector expressing _p_uro-_2_A-_G_FP _m_iR-30 (P2Gm; Fig. 3A). MEFs were transduced with P2Gm targeting FF, p53, PTEN, or both tumor suppressors, selected with puromycin, treated with doxorubicin to induce p53 expression or rosiglitazone to induce PTEN expression, and assayed by Western blot. Fig. 3A demonstrates efficient knockdown of both targeted tumor suppressors when two shRNA constructs are expressed from the same vector. The decreased level of PTEN in lane 4 compared with lane 3 is due to reduced levels of PTEN′s transcriptional transactivator p53 (23). In contrast to some reports, we did not observe any increased cell death in vitro or in vivo (see below) due to overexpression of any miR-30 shRNA constructs.
Fig. 3.
FLIP improvements and characterization. (A) Schematic representation of the expression cassette P2Gm, which expresses the puromycin-resistance gene, GFP, and the miR-30-based RNAi construct. The miR-30 constructs may be concatamerized to knock down multiple genes. Primary MEFs were transduced with MSCV-P2Gm targeting FF, p53, the tumor suppressor PTEN, or p53 and PTEN. MEFs were treated with doxorubicin (for p53 blot) or rosiglitazone (for PTEN blot), lysed, and analyzed by immunoblot for p53, PTEN, and GAPDH expression. (B) Schematic representation of FLIPi. After recombination, cells express Thy1.1 and therefore can be sorted by antibody labeling. The miR-30 RNAi construct is flanked by synthetic splice donor and splice acceptor sequences in an artificial intron. The splice sites are not recognized in the antisense orientation (denoted by *). After recombination, miR-30 is spliced from the transcript and processed separately from the mRNA. (C) 293T cells were transduced with MSCV-FLIP-p53 (with or without intron), selected with puromycin, then transfected with control or Cre-expressing plasmid. For positive and negative controls, cells were infected with MSCV-P2Gm targeting FF (lane 1), p53 (lane 2), or p53/PTEN (lane 3). Cell lysates were analyzed by Northern blot for the 21nt guide strand or opposite strand derived from the processed miR-30. The guide strand is only detected when the miR-30 has recombined to the sense orientation. (D) 293T cells were transduced with MSCV-FLIP-miR181a (with or without intron) that can express the endogenous miR-181a, selected with puromycin, and transfected with control or Cre-expressing plasmid. The middle control lane is 293T cells expressing miR-15b. Cells were lysed and analyzed by Northern blot for the presence of processed miR-181a. A significant increase in miR-181a expression over background is detected only when the vector has recombined.
Inclusion of the miR-30 construct in the 3′ UTR of the viral transcript can result in decreased GFP expression (approximately twofold), likely owing to the miRNA being cleaved from a fraction of the transcripts and those processed transcripts no longer contributing to the pool of translated GFP (data not shown). To remedy this effect, we wanted to separate the miRNA from the translated mRNA. Many naturally occurring miRNAs are present in introns (24), but introns placed in the same transcriptional direction as proviral transcription can be excised during proviral transmission. However, because a portion of the FLIP vector is in the antisense orientation, antisense splicing signals are not recognized and may be included. Consensus synthetic splice donor and splice acceptor sites were introduced flanking the miR-30 in the 3′ UTR of Thy1.1 and positioned near the stop codon to avoid nonsense-mediated decay of the transcript (Fig. 3B) (25). The configuration of reporter genes was reversed for the vector MSCV-FLIPi (FLIP _i_ntron) to facilitate recovery of “flipped” cells by bead-based sorting for surface Thy1.1. Viral titer was unaffected by the synthetic intron (data not shown). DNA and RNA were isolated from FLIP-, FLIPi-, and Cre- transduced cells and compared by PCR analysis for presence of the intron. The synthetic intron mediates proper splicing of the miR30 from the viral transcript (Fig. S2_A_). Knockdown of genes with the intronic miR-30 shRNA was unaffected by the flanking splice sites (data not shown).
Our analysis of p53 protein levels in FLIP-transduced cells was consistent with a lack of miRNA processing from the antisense strand and correct processing of the Cre-recombined sense form. We wanted to investigate this further by analyzing the small RNAs incorporated into RNA-induced silencing complexes that mediate silencing of target transcripts. We performed Northern blots for the small 21 nt RNA that silences p53 expression, the guide strand, and the opposite strand that is nonfunctional for silencing p53. 293FT cells were infected with MSCV-P2Gm targeting FF (control 1), p53 (control 2), or p53/PTEN (control 3) or MSCV-FLIP-p53. Transduced cells were selected with puromycin then transfected with a Cre-expressing vector or a control expression vector. Fig. 3C demonstrates that only cells that recombined the integrated FLIP vector (≈40% of total cells) express the guide strand, whether or not the miR-30 shRNA is included within a synthetic intron. The difference in intensity between the control and induced cells is because control cells contain multiple integrants per cell and hence express more of the guide strand RNA. Importantly, we did not detect processing of the antisense miR-30 shRNA to generate the guide strand or opposite strand species. Similar results were obtained for miR-30 RNAi targeting the PTEN tumor suppressor (Fig. S2_B_). When guide strand expression is normalized to the number of cells expressing the recombined FLIP vector, inclusion of the intron seems to cause a slight but reproducible reduction in expression of guide strand assayed by Northern blot. The intron did not seem to affect the efficacy of gene knockdown.
Because the miR-30 can be efficiently regulated by an antisense to sense reorientation, we wanted to determine whether this regulation may apply to endogenous miRNAs. Fig. 3D shows that processing of miR181a may be efficiently regulated by “flipping” from the antisense to sense orientation and that processing occurs with or without flanking splice sites. Similar data were obtained for miR-15b (Fig. S2_C_).
Temporal Regulation of RNAi in Vivo.
The aim of these experiments was to temporally regulate RNAi expression in immune cells. We isolated donor bone marrow cells, transduced them with MSCV-FLIP in vitro, and introduced them into lethally irradiated recipients to reconstitute the hematopoietic system. To increase infection and reconstitution efficiency, we incorporated an in vitro stem cell expansion protocol using standard HSC culture conditions with the cytokine Angiopoietin-like-2. LTR-HSCs expanded in vitro, allowing reconstitution of at least 20 mice from 1 donor. LTR-HSCs could be cultured for up to 10 days, providing enough time for multiple rounds of infection and selection with puromycin to enrich for transduced cells. This drug-selection protocol resulted in two- to threefold enhanced reconstitution efficiencies after bone marrow transplant (Fig. S3_A_). Recipient mice exhibited stable GFP expression in PBLs 8 months after reconstitution, indicating transduction of LTR-HSCs (Fig. S3_B_).
We wanted a system to investigate RNAi in primary immune cells in which the shRNA remained silent in the bone marrow stem cells and progenitor cells to prevent any disruption of proper development. To this end, we isolated bone marrow stem cells from Cre-ER mice, where Cre remains inactive until administration of the estrogen analogue tamoxifen. Stem cells were infected with MSCV-FLIPi targeting FF, p53, PTEN, or both tumor suppressors p53 and PTEN, selected with puromycin, and injected into lethally irradiated hosts.
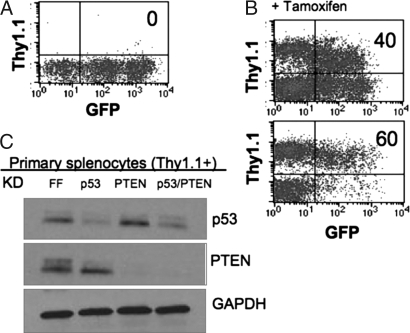
Six weeks after reconstitution, PBLs were isolated and analyzed by FACS for expression of GFP. FLIPi vectors express puromycin resistance and GFP before Cre activation and the cell-surface marker Thy1.1 after recombination. Reconstituted mice contained 30%–70% GFP+ PBLs, and Fig. 4A is a representative FACS plot. Mice were given tamoxifen, and PBLs were analyzed by FACS 1 week later. Fig. 4B contains two representative FACS plots that demonstrate Thy1.1 expression in a significant fraction of PBLs after tamoxifen (40 and 60%), indicating induction of Cre activity and recombination of the FLIPi vectors. Some cells were double-positive for Thy1.1 and GFP, perhaps owing to the long half-life of GFP or to multiple viral integrants and recombination in only a subset of these loci. When these mice were analyzed several weeks later, the majority of double-positive cells were no longer present.
Fig. 4.
Inducible knockdown in primary splenocytes in vivo. (A) HSCs were cultured for a total of 8 days in the presence of STIF and angiopoietin-like-2 (Angptl2). Cells were transduced twice with MSCV-FLIPi supernatants and selected with puromycin for 2 days, then injected into recipient mice. Six weeks after reconstitution, PBLs were analyzed for FLIP marker expression. A representative FACS plot shows GFP+ cells and no Thy1.1+ cells. (B) Reconstituted mice were administered tamoxifen on days 1, 3, and 5, and PBLs were analyzed on day 7 for FLIPi marker expression. Many cells have gained Thy1.1 expression and lost GFP expression. (C) Splenocytes were isolated from mice reconstituted with HSCs transduced with MSCV-FLIPi vectors targeting FF, p53, PTEN, or p53/PTEN. Unstimulated cells were sorted by MACS for Thy1.1+ cells, treated in vitro for 4 h with doxorubicin (for p53) or rosiglitazine (for PTEN), lysed, and analyzed by immunoblot for p53, PTEN, and GAPDH.
Spleens were isolated and sorted by magnetic bead separation for Thy1.1+ cells. Isolated Thy1.1+ spleen cells were cultured in vitro for 5 h with either doxorubicin or rosiglitazone to induce p53 or PTEN, respectively. Protein expression was then analyzed by Western blot. Fig. 4C demonstrates corresponding knockdown of p53, PTEN, or both tumor suppressors in the primary splenocytes expressing the recombined FLIPi vectors. Inducible Cre partnered with FLIP vectors thus allows development of hematopoietic cells in the absence of genetic perturbation followed by acute induction of RNAi to knock down genes in naïve splenocytes.
Spatial Regulation of RNAi in Vivo.
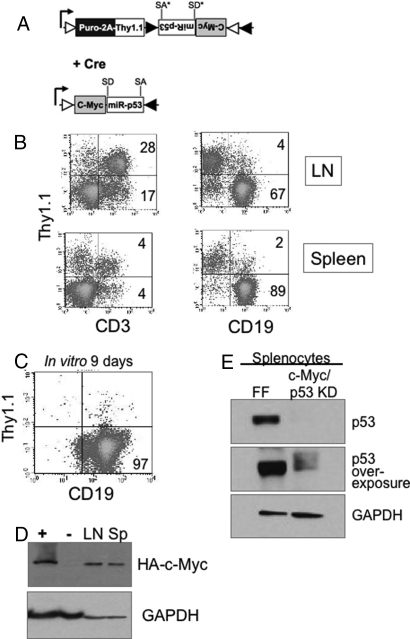
In addition to using a ligand-inducible Cre, we wanted to test the FLIP vectors in the presence of tissue-specific Cre expression. We incorporated an MSCV-FLIPi vector that expresses puromycin resistance and Thy1.1 before recombination and after Cre activity would express the proto-oncogene c-Myc and also knock down p53 (Fig. 5A). This FLIP vector was introduced into CD19-Cre (B-cell-specific Cre) bone marrow cells in culture. After puromycin selection, the transduced bone marrow cells were injected into lethally irradiated recipient mice. This design would allow us to produce oncogenic lesions only in a select cell type within a hematopoietic system derived from common stem cell precursors.
Fig. 5.
B cell-specific oncogene expression and tumor suppressor knockdown. (A) Schematic representation of MSCV-FLIPi-Myc. Upon Cre-recombination, the vector expresses c-Myc T38A and miR-30 targeting p53. (B) HSCs from CD19-Cre mice were cultured with STIF and Angptl2, transduced with MSCV-FLIPi-Myc, and injected into recipient mice. At 105 days after reconstitution, lymph node cells and spleen cells from a moribund mouse were analyzed by FACS for Thy1.1 expression in T cells (CD3) and B cell lineages (CD19). Note the expansion of the CD19+ cells and low level of Thy1.1 expression. (C) Spleen cells were cultured in vitro for 9 days (split 1:10 on days 3 and 6) and analyzed by FACS. (D) Spleen and lymph node cells from a moribund mouse were lysed and analyzed by immunoblot for expression of HA-tagged c-Myc and GAPDH. MEFs transduced with MSCV-Cre and MSCV-FLIPi-Myc (+) or MSCV-FLIP-FF (−) serve as controls.
Expression of c-Myc in the B cell compartment in conjunction with p53 inactivation results in lymphoma within approximately 100 days (26). In our MSCV-FLIPi-Myc/miRp53:CD19-Cre reconstitutions, mice became moribund (three of three) at approximately 100 days and were killed and analyzed. FACS plots in Fig. 5B show that at least half of CD3+ cells (T cells) from lymph node and spleen expressed Thy1.1, indicating an approximately 50% reconstitution efficiency. In contrast, very few CD19+ cells expressed Thy1.1, and the CD19+ population comprised two thirds of lymph node cells and 90% of spleen cells. This resulting expansion was consistent with dysregulated growth of CD19+ cells by the combined effects of c-Myc expression and loss of p53 function.
Lymph node and spleen cells were cultured in vitro for 9 days in the absence of B cell mitogens, and Fig. 5C shows that virtually all cells that survived for the culture period were CD19+. Spleen and lymph node cells were analyzed for c-Myc expression by detecting c-Myc's N-terminal HA tag on Western blot. Fig. 5D shows that both lymph node and spleen cells expressed HA-positive protein at the expected molecular weight. Fig. 5E shows significant reduction of p53 protein levels in spleen cells derived from MSCV-FLIPi-Myc/miRp53 compared with MSCV-FLIPi-miRFF-expressing splenocytes. In vitro expansion in the absence of specific mitogens confirms transformation of CD19+ cells, likely driven by expression of c-Myc and concomitant loss of p53.
Discussion
We present here a system for Cre-regulated RNAi and transgene expression. Before recombination, the vectors express a selection cassette composed of the puromycin-resistance gene followed by a marker suitable for FACS analysis. The Cre-induced genes and miRNA-based shRNA constructs are adjacent but in the antisense orientation and are silent. After recombination, the selection cassette is deleted, and the antisense portion of the vector permanently “flips” to the sense orientation and is properly expressed. This expression construct may be delivered by retroviral or lentiviral vectors, and we demonstrate its utility in lentiviral transgenics and bone marrow reconstitutions.
To generate transgenic mice, ES cells were infected with lentiviral FLIP constructs targeting the tumor suppressor p53, and chimeric mice were subsequently crossed to various Cre-expressing mice. Through this methodology, we generated knockdown mice for p53 in 4 months and demonstrated tissue-specific loss of function for p53 in <6 months. Although the level of p53 expression in shRNA-expressing cells is significantly reduced, we have not observed any increased frequency of diseases associated with p53 deficiency.
Infection of lineage-specific or inducible Cre-expressing bone marrow stem cells with retroviral FLIP constructs allows tight control of RNAi or transgene expression within the hematopoietic compartment. By combining MSCV-FLIP with bone marrow stem cells ubiquitously expressing the tamoxifen-regulatable Cre-ER, we were able to regulate RNAi expression inducibly and selectively in immune cells. We demonstrate knockdown of the tumor suppressors p53, PTEN, or both from a single vector in naïve primary splenocytes. This method permits hematopoietic development to progress from stem cell to fully differentiated progeny in the absence of genetic perturbations. Furthermore, induction of RNAi expression in relatively quiescent, lineage-committed cells allows analysis of the effects of RNAi in truly naïve immune cells, a scenario that previously has proven experimentally intractable.
In addition to expression of multiple shRNA constructs by concatamerizing the miR-30s to target two endogenous genes (or more; P.S., unpublished data), shRNA constructs can be combined with transgenes (such as oncogenes). By infecting bone marrow harboring B-cell-specific Cre with MSCV-FLIP expressing c-Myc and a p53 knockdown, we were able to transform B cells selectively in vivo. This strategy allows rapid generation of Cre-regulated gain-of-function and loss-of function activities from a single vector. This design allows knockdown of one or more functionally redundant gene(s) and to replace expression with mutant alleles to model disease, or with the human homologue to investigate drug efficacy in a humanized mouse model.
The FLIP vectors were designed to be compatible with integration into pre-existing Cre models and should significantly extend current transgenic efforts. Because many developmental and disease models in mice rely on Cre-mediated genetic modification, FLIP transgenic mice can be incorporated directly into these systems. In this way, genetic modifiers that exacerbate or suppress a phenotype may be rapidly assessed in vivo.
The FLIP vectors nicely complement the current set of regulated RNAi vectors. FLIP vectors offer Cre-regulated expression of one or more miRNA-based shRNA constructs, whereas pSICO uses a polIII-shRNA design to express a single construct (11). These Cre-regulated vectors take advantage of the “Cre zoo” that offers numerous strains for tissue-specific regulation of RNAi. The tet-regulated RNAi systems do not yet have the same depth of tissue-specific lines as Cre mice but do offer the prospect of reversible RNAi expression (13).
The FLIP vectors provide a relatively rapid and versatile means for introducing compound genetic modifications to a particular cell type or at a particular time in development. FLIP vectors combined with transgenic Cre lines should prove a powerful tool for investigating the multiplicity of interactions that comprise biological networks and a means for determining how perturbations in these networks can manifest as disease states.
Materials and Methods
Vectors.
LB2 and FLIP constructs were generated using standard molecular biology techniques. Maps, sequences, and cloning information are available on the Hynes laboratory web site (http://web.mit.edu/ccr/labs/hynes.htm). The HA-tagged c-Myc T38A construct was a kind gift from Michael Hemann, Massachusetts Institute of Technology (27). The FLAG-tagged angiopoietin-like-2 vector was a kind gift from Cheng Cheng Zhang, Whitehead Institute (28).
Mice.
All mice were housed and handled in accordance with Massachusetts Institute of Technology Division of Comparative Medicine protocols. All mice were C57BL6/J except tetraploid chimeric mice (see below). Cre-expressing Tie-2 (29), CD19 (30), and Cre-ER (31) transgenic mice are described elsewhere.
Cell Culture.
Primary MEFs, 293FT, T, and B cells were cultured under standard conditions and stimulated as indicated (see SI Text). Long-term-repopulating HSCs were cultured in Stemspan (Stemcell Technologies) supplemented with STIF (see SI Text). Angptl2 was purified on an anti-FLAG affinity column from the supernatant of transiently transfected 293FT cells.
Virus Production and Titer.
Production of lentivirus and infection was performed as described previously and in SI Text (10). Viral titers of the FLIP vectors were compared with MSCV-GFP, a retroviral vector expressing only GFP. MSCV-GFP and MSCV-FLIP titers were similar, all ≈1 × 106 infectious particles/ml. Lentivirus titers ranged from 1–6 × 105 infectious particle/ml and were not significantly affected by inclusion of the FLIP cassette.
LTR-HSC Culture, Infection, and Reconstitution.
HSCs were purified from bone marrow (20), cultured in STIF + Angptl2 (28), infected with retroviral constructs, and selected with puromycin over a period of 8 days (see SI Text). Cultured HSCs were injected into lethally irradiated recipients. Tamoxifen (Sigma) was dissolved in corn oil (10 mg/ml) and administered i.p. at 200 μg per mouse.
Derivation of 129S4/C57BL/6J F1 ES Cells.
F1 ES cells were generated as described in ref. 32.
Generation of F1 ES Cells Expressing FLIP.
ES cells (106) were infected with 105 infectious units of self-inactivating lentiviral vector pseudotyped with VSV-G and 8 μg/ml polybrene (Sigma) at room temperature for 1 h while rocking. ES cell medium containing 1 μg/ml of puromycin was added 72 h after infection. The surviving colonies were used in generation of tetraploid chimeras.
Tetraploid Chimeras.
Tetraploid chimeras were generated using ES cells expressing LB2-FLIP-p53 as described in ref. 32. Founder mice were screened by FACS of PBLs for expression of Thy1.1, and highest expressors were used for subsequent matings and experiments.
Northern Blot Analysis.
HEK-293FT cells were transduced with the appropriate MSCV-FLIP vectors, and each group was split in half and transfected with either MSCV-Cre or control MSCV vector to recombine the loxP sites and activate shRNA expression. After 48 h, total RNA was extracted and Northern blot performed (see SI Text).
Western Blot Analysis.
Equal numbers of cells were lysed in 70 μl 2× SDS sample buffer, boiled, and run on 4%–20% gradient Tris-Glycine gels (Invitrogen). Proteins were transferred to nitrocellulose and Western blots performed using mouse monoclonal anti-PTEN (Santa Cruz Biotechnology), mouse monoclonal anti-p53 (A. Ventura, Massachusetts Institute of Technology, unpublished), and mouse monoclonal anti-GAPDH (Chemicon).
Supplementary Material
Supporting Information
Acknowledgments.
We thank Lakshmi Balachandra and Adam Lacy-Hulbert for critical reading and discussions; Michael Hemann for lymphoma assistance; and Peimin Qi and the Massachusetts Institute of Technology transgenic facility for help in generating tetraploid and chimeric mice. This work was supported by the Integrative Cancer Biology Program Grant (U54-CA112967) from the National Institutes of Health (to R.O.H. and P.A.S.); the National Heart Lung and Blood Institute (PO1-HL066105) and the Tumor Microenvironment Grant (U54-CA126515) from the National Institutes of Health and by the Howard Hughes Medical Institute (R.O.H.); United States Public Health Service grants (RO1-GM34277 to P.A.S.); and partially by a Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute. R.O.H. is an investigator of the Howard Hughes Medical Institute.
Footnotes
References
- 1.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3:715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 2.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 3.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 4.Lickert H, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 5.McManus MT, Petersen CP, Haines BB, Chen J, Sharp PA. Gene silencing using micro-RNA designed hairpins. RNA. 2002;8:842–850. doi: 10.1017/s1355838202024032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paddison PJ, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 7.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci USA. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 9.Agarwal M, et al. Scaffold attachment region-mediated enhancement of retroviral vector expression in primary T cells. J Virol. 1998;72:3720–3728. doi: 10.1128/jvi.72.5.3720-3728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kissler S, et al. In vivo RNA interference demonstrates a role for Nramp1 in modifying susceptibility to type 1 diabetes. Nat Genet. 2006;38:479–483. doi: 10.1038/ng1766. [DOI] [PubMed] [Google Scholar]
- 11.Ventura A, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci USA. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin KJ, et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc Natl Acad Sci USA. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickins RA, et al. Tissue-specific and reversible RNA interference in transgenic mice. Nat Genet. 2007;39:914–921. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauer B, Henderson N. Cre-stimulated recombination at loxP-containing DNA sequences placed into the mammalian genome. Nucleic Acids Res. 1989;17:147–161. doi: 10.1093/nar/17.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kano M, Igarashi H, Saito I, Masuda M. Cre-loxP-mediated DNA flip-flop in mammalian cells leading to alternate expression of retrovirally transduced genes. Biochem Biophys Res Commun. 1998;248:806–811. doi: 10.1006/bbrc.1998.9011. [DOI] [PubMed] [Google Scholar]
- 16.Ungrin MD, Harrington L. Strict control of telomerase activation using Cre-mediated inversion. BMC Biotechnol. 2006;6:10. doi: 10.1186/1472-6750-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnutgen F, et al. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol. 2003;21:562–565. doi: 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- 18.Siegel RW, Jain R, Bradbury A. Using an in vivo phagemid system to identify non-compatible loxP sequences. FEBS Lett. 2001;499:147–153. doi: 10.1016/s0014-5793(01)02541-8. [DOI] [PubMed] [Google Scholar]
- 19.Donnelly ML, et al. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: A putative ribosomal ‘skip’. J Gen Virol. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- 20.Donnelly ML, et al. The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J Gen Virol. 2001;82:1027–1041. doi: 10.1099/0022-1317-82-5-1027. [DOI] [PubMed] [Google Scholar]
- 21.Eakin GS, Hadjantonakis AK, Papaioannou VE, Behringer RR. Developmental potential and behavior of tetraploid cells in the mouse embryo. Dev Biol. 2005;288:150–159. doi: 10.1016/j.ydbio.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 22.Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 23.Stambolic V, et al. Regulation of PTEN transcription by p53. Mol Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 24.Ying SY, Lin SL. Intron-derived microRNAs—fine tuning of gene functions. Gene. 2004;342:25–28. doi: 10.1016/j.gene.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 25.Lim LP, Burge CB. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci USA. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemann MT, et al. Probing tumor phenotypes using stable and regulated synthetic miRNA precursors. Nat Genet. 2003;33:396–400. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 27.Hemann MT, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang CC, et al. Angiopoietin-like proteins stimulate ex vivo expansion of hematopoietic stem cells. Nat Med. 2006;12:240–245. doi: 10.1038/nm1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kisanuki YY, et al. Tie2-Cre transgenic mice: A new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 30.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 32.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratories Press; 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information