Architecture of the SWI/SNF-Nucleosome Complex (original) (raw)
Abstract
The SWI/SNF complex disrupts and mobilizes chromatin in an ATP-dependent manner. SWI/SNF interactions with nucleosomes were mapped by DNA footprinting and site-directed DNA and protein cross-linking when SWI/SNF was recruited by a transcription activator. SWI/SNF was found by DNA footprinting to contact tightly around one gyre of DNA spanning ∼50 bp from the nucleosomal entry site to near the dyad axis. The DNA footprint is consistent with nucleosomes binding to an asymmetric trough of SWI/SNF that was revealed by the improved imaging of free SWI/SNF. The DNA site-directed cross-linking revealed that the catalytic subunit Swi2/Snf2 is associated with nucleosomes two helical turns from the dyad axis and that the Snf6 subunit is proximal to the transcription factor recruiting SWI/SNF. The highly conserved Snf5 subunit associates with the histone octamer and not with nucleosomal DNA. The model of the binding trough of SWI/SNF illustrates how nucleosomal DNA can be mobilized while SWI/SNF remains bound.
The organization of DNA into tightly packed nucleosomal and supranucleosomal structures plays a central role in many aspects of cellular biology and development in eukaryotes. Cellular processes regulated by chromatin structure range from transcription to DNA repair, replication, recombination, and genetic stability (13, 34, 48). Cells have developed a variety of mechanisms to overcome this barrier by altering chromatin structure and controlling the accessibility of DNA.
One of these mechanisms is ATP-dependent chromatin remodeling, which is catalyzed by a large family of multisubunit chromatin remodeling complexes that alter DNA-histone interactions (2). Yeast SWI/SNF (1.14 MDa from STEM or 1.15 MDa from stoichiometry data), a prototype of this family of chromatin remodeling complexes, is composed of 12 different subunits whose homologs have been identified in organisms ranging from other fungi to mammals (2, 14). SWI/SNF disrupts the nucleosome structure, increases the binding of transcription factors to nucleosomes, mobilizes histone octamers along DNA in cis, transfers histone octamers to different DNA fragments, displaces histone H2A/H2B dimers, generates superhelical torsion in DNA, and binds preferentially to four-way DNA (5, 16, 35). SWI/SNF is required for the transcriptional regulation of about 5% of the total yeast genome (17, 44). SWI/SNF binds to DNA and nucleosomes without any DNA sequence specificity. The complex is found in a low abundance in the cell, suggesting that it is not a general component of chromatin (10). In vitro and in vivo data demonstrate a direct interaction between SWI/SNF and transcription activators or repressors, showing that SWI/SNF is recruited to target genes by transcription factors (2, 17, 29, 31, 52).
Most of the SWI/SNF subunits are required to maintain its common functions in vivo, but our knowledge of the distinct roles of each subunit is limited. The ATPase activity of the Swi2/Snf2 subunit is stimulated by double-stranded DNA and is critical for the transcriptional activation and chromatin remodeling functions of the SWI/SNF complex. The yeast Swi2, Arp7, and Arp9 subunits form a core complex that can remodel nucleosomes (50), and similarly, the human homologs of Swi2/Snf2, Swi3, Snf5, and Swp73 form a core complex that can remodel nucleosomes (36). Swi2/Snf2, Swi1, Swp73, Snf5, and Snf6 are indispensable for the transcription activation of a subset of genes mediated by Gcn4 (29, 45). Snf5, Snf6, and Swi3 are required for the optimal recruitment of SWI/SNF by Gcn4. Swi1 contains an ARID domain that likely has a lower affinity for DNA than do ARID domains in other proteins. Arp7 and Arp9 are two actin-related proteins that are also part of the RSC complex, another member of the Snf2 subfamily in yeast (2, 14). In addition to being a component of the SWI/SNF complex, Swp29 is a subunit of the yeast TFIIF and TFIID complexes (7). Snf11 was demonstrated to interact with the Swi2/Snf2 subunit; however, the deletion of the Snf11 subunit has no detectable effect on SWI/SNF-dependent gene expression (47). The elucidation of the role of subunits and domains of the complex in its interaction with the nucleosome is critical for an understanding of the mechanism of remodeling.
Electron microscopy and three-dimensional reconstruction of three members of the SWI/SNF family of remodelers have been reported. The yeast RSC and human PBAF complexes show some features in common (1, 22, 23); however, yeast SWI/SNF has marked differences from them (42). The RSC and PBAF reconstructions feature a large central cavity, which can accommodate mononucleosomes, whereas yeast SWI/SNF revealed a shallow depression, which could serve as a nucleosome binding site. Structural models have been proposed for RSC- and PBAF-nucleosome complexes based on the electron microscopy structure and limited footprinting data (1, 22, 23). Specifics of how the remodelers interact with nucleosomal substrates and the mechanistic implications of these interactions remain unclear due to limitations in resolving nucleosome-bound structures by electron microscopy methods.
To better determine the nucleosomal and extranucleosomal regions of the nucleosome bound by SWI/SNF, we have taken advantage of SWI/SNF recruitment by acidic transcription activators to orient SWI/SNF in one preferred direction on the nucleosome. Data from DNA footprinting and site-directed DNA cross-linking have identified the regions of DNA bound by SWI/SNF and the subunits of SWI/SNF that were bound at these different sites, improving on and greatly expanding previous studies with 5S ribosomal DNA (rDNA) nucleosomes (39). We have also developed a protein cross-linking approach to attach photoreactive moieties at specific sites within the histone octamer and thereby map the interactions of SWI/SNF with the histone octamer face of the nucleosome. The combination of these approaches has provided an unprecedented view of the structure of an ATP-dependent chromatin remodeler and its interaction with a nucleosomal substrate.
MATERIALS AND METHODS
Plasmids.
Oligonucleotides 5′-CGGAGGACTGTCCTCCGA-3′ and 5′-AGCTTCGGAGGACAGTCCTCCGTGCA-3′ were annealed to form a single Gal4 binding site, and 5′-CGGAGTACTGTCCTCCGAGCGGAGTACTGTCCTCCGCA-3′ and 5′-AGCTTGCGGAGGACAGTACTCCGCTCGGAGGACAGTACTCCGTGCA-3′ annealed to form two Gal4 binding sites in a DNA duplex with flanking PstI and HindIII sites. Plasmids with the Gal4 binding site(s) placed 27 bp from the edge of the 601 sequence, p159-1G-27 and p159-1-2G2-27, were formed by ligating the short double-stranded DNA duplex to p159 cut with PstI and HindIII. After ligation, the recognition site for HindIII was recovered and that of PstI was destroyed, and the constructs were confirmed by DNA sequencing (54).
Standard site-directed mutagenesis was used to introduce mutations into Xenopus laevis core histone proteins using the pET-histone expression plasmid reported previously (26), and the cysteine at residue 110 of H3 was changed to alanine.
Gal4-VP16 purification.
Gal4-VP16 was overexpressed in Escherichia coli Xa-90 and purified as described previously, with some minor changes (40). Briefly, a Q Sepharose column was used instead of a phenyl-Sepharose column, and a MonoS column was added with a linear gradient of 250 to 450 mM NaCl. The purified protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with anti-Gal4 antibody.
Purification of histone proteins and octamer refolding.
Recombinant histones were expressed in Escherichia coli BL21 cells with the pET-histone expression vector. The four histones were purified by Sephacryl S-200 gel filtration and MonoS cation-exchange chromatography as described previously (26). Octamers were refolded with the substitution of one of the four histones containing a single cysteine. The refolded octamer was purified by Superdex 200 size exclusion chromatography.
Purification of SWI/SNF.
Yeast SWI/SNF was purified from Saccharomyces cerevisiae strains YJW426 (30) and YBB001. YBB001 (BY4742 _mat_α _his3_Δ_1 leu2_Δ_0 lys2_Δ_0 ura3_Δ_0 swi3_-HA-V5-His 6::_kanMX4 snf6_-FLAG::LEU2) was derived from BY4742 by tagging the C terminus of Snf6 with FLAG and Swi3 with HA-V5-His6. Yeast cells were grown in yeast extract-peptone-dextrose medium to an optical density at 600 of ca. 5 to 6. Harvested cells were frozen and ground under liquid N2 using a mortar and pestle. Cell extract was prepared by mixing yeast cell powder with buffer containing 20 mM Na-HEPES, 350 mM NaCl, 20% glycerol, 0.1% Tween 20, and protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, 1 μg/ml pepstatin, and 2 μg/ml aprotinin) and spinning at 100,000 × g for 1 h. SWI/SNF was purified from the soluble whole-cell extract by Biorex70, Ni-nitrilotriacetic acid, and M2 agarose affinity chromatography. In some cases, SWI/SNF was purified only by M2 agarose affinity chromatography of the whole-cell extract. The one-step affinity purification of SWI/SNF had a higher yield (∼5 μg/liter of culture at an optical density at 600 nm of 5 to 6) and had a few contaminating proteins that did not adversely affect the binding or remodeling activities of SWI/SNF or the cross-linking pattern of SWI/SNF.
DNA probes, nucleosome reconstitution, DNA footprinting, and photoaffinity labeling.
A 555-bp NdeI-HindIII DNA fragment derived from plasmid p159-1 with one or two Gal4 binding sites was used as a template to synthesize photoaffinity DNA probes. Solid-phase DNA probe synthesis was performed as described previously (33) except that the probe was cleaned with a PCR purification kit instead of by phenol-chloroform extraction and ethanol precipitation. The probe was removed from the beads by EcoRI digestion of a 242-bp DNA that assembles into nucleosomes with extranucleosomal DNA of 34 bp and 61 bp on the other side containing the two Gal4 binding sites.
The DNA used for homogenous nucleosome reconstitution was synthesized by PCR using p159-1-1Gal4-27 as a template and oligonucleotides 5′-biotin/TCCCCAGTCACGACGTTGTAAAAC-3′ and 5′-ACCATGATTACGCCAAGCTTCGG-3′ of the upper and lower strands, respectively. A 276-bp DNA, biotinylated at one end with a Gal4 binding site at the opposite side, was generated, and the 601 positioning DNA sequence was flanked by 69 bp (biotinylated) and 59 bp (with the Gal4 binding site) of linker DNA.
Mononucleosomes were reconstituted at 37°C with a solution containing 3.8 ng/μl DNA probe, 1.5 μg/μl salmon sperm DNA, and 1 μg/μl histone octamer in 2 M NaCl in 10 μl using the rapid salt dilution method. Homogenous mononucleosomes were reconstituted using a solution containing 0.6 μg/μl biotinylated 276-bp DNA, 1 μg/μl histone octamer, and 2 M NaCl, and DNA footprinting and photoaffinity labeling were done as described previously (19).
Modification of nucleosomes and histone cross-linking.
After the removal of reducing agents by dialysis, nucleosomes (20 pmol) were modified with 3 nmol _N_-[(2-pyridyldithio)ethyl]-4-azidosalicylamide (PEAS) (Molecular Probes) for 1 h at room temperature. Excess PEAS reagent was removed by dialysis, and the PEAS-conjugated nucleosomes were radiolabeled by indirect iodination (9) using Iodo-Gen precoated iodination tubes (Pierce). The cysteine mutant nucleosomes modified with [125I]PEAS were immobilized on magnetic beads (streptavidin-coated M-280 Dynabeads). Five picomoles of the nucleosomes was incubated with 200 μg of equilibrated beads with vigorous mixing at room temperature for 45 min. After the removal of unbound nucleosomes by washing, 6 nM SWI/SNF was bound to the immobilized 4 nM nucleosome in the presence or absence of 6.4 nM Gal4-VP16 and cross-linked by UV irradiation. Gal4-VP16-dependent SWI/SNF binding to nucleosomes was achieved by including 50 ng sheared salmon DNA in the binding reaction mixture. Unbound free SWI/SNF was removed from the beads, and the bound cross-linked SWI/SNF was resuspended in buffer with 100 mM dithiothreitol, 400 mM NaCl, and 10 mM Na-HEPES (pH 7.8) to break the disulfide bond connecting cross-linked nucleosomes to SWI/SNF and reverse SWI/SNF binding to immobilized nucleosomes. The samples were analyzed by Bis-Tris SDS-PAGE and phosphorimaging to identify the radiolabeled subunits.
Cryoelectron microscopy of SWI/SNF.
Tandem affinity purification-purified yeast SWI/SNF was prepared as previously described (43). Unfixed samples at a nominal concentration of 1 mg/ml were applied to perforated carbon films in 3-μl droplets in a humidity-controlled chamber maintained at 95% relative humidity. After 1 min, the droplet was blotted from both sides with Whatman 425 filter paper for 3 to 5 s and then plunged into liquefied ethane for vitrification. The same procedure was used for fixed samples, which were applied to perforated substrates backed with an ultrathin layer of carbon.
Vitrified samples were held in liquid nitrogen through transfer to a Gatan 626 cryoholder for observation with an FEI Tecnai12 electron microscope at 120 kV, and images were recorded at defocus values of between 600 and 1,000 Å on a 2,048-by-2,048 Tietz video and image processing system charge-coupled-device camera at a pixel size of 2.45 Å in the images.
Single-particle reconstruction was carried out as previously described (18, 42) with EMAN (25). All image data were truncated to include only up to the first zero of the contrast transfer function, and no phase correction was performed. Only images whose defocus value included data to 15 Å inside the first zero were used.
Frozen-hydrated samples of unfixed yeast SWI/SNF frozen over holes had a bimodal distribution caused by subunit loss when complexes came into contact with one of the air-liquid interfaces during freezing. The subset of particles fully contained in ice did not show a preferred orientation or any size or density changes. This was determined by comparison to data collected from fixed samples, which were known to retain their full mass and to have no preferred orientation on carbon film (42). Model bias was precluded by evaluating two different structures for refinement in the initial classification of particles, a featureless sphere of appropriate dimensions, and the previously calculated structure from negatively stained particles low-pass filtered to 50 Å. The size of classes and distribution of projections confirmed that good coverage of Euler angles was represented, and no preferred orientation was present. In both cases, the close agreement between projections and class averages was indicative of homogeneity of the data sets (25).
The two data sets (fixed and unfixed) were independently reconstructed and compared for convergence, judged by the absence of change in the Fourier shell correlation (FSC) of subsequent iterations. This occurred after five iterations, with a value of 28 Å for the FSC of 0.5 for the independent sets. At this point, the two data sets were merged, and convergence at eight iterations led to a reconstruction with values of 19 Å at an FSC of 0.3 and 23 Å at an FSC of 0.5. Volumes were constrained to the mass determined by STEM (43) of 1.14 MDa, assuming a protein density of 1.35 g/ml (0.81 Da Å−3), and Gaussian low-pass filtered to the threshold of an FSC value of 0.5. Figures have been filtered to 25 Å.
The three-dimensional reconstruction was visualized using Amira software (Visage Imaging, Carlsbad, CA) with the isosurface calculated to include 1.14 MDa. The crystal structure of the nucleosome (11) was modified by the addition of 15 bp of B-DNA to simulate linker DNA. The nucleosome was first positioned to maximize the number of bases identified by footprinting as contacting the SWI/SNF surface. The position was further refined by maximizing histone contacts. Contacts were prioritized in order of their distance from the surface, with all contacts within 9 Å (the length of the chemical cross-linker) being given highest priority and contacts greater than 12 Å being disallowed.
RESULTS
SWI/SNF binds 50 bp or more of nucleosomal DNA.
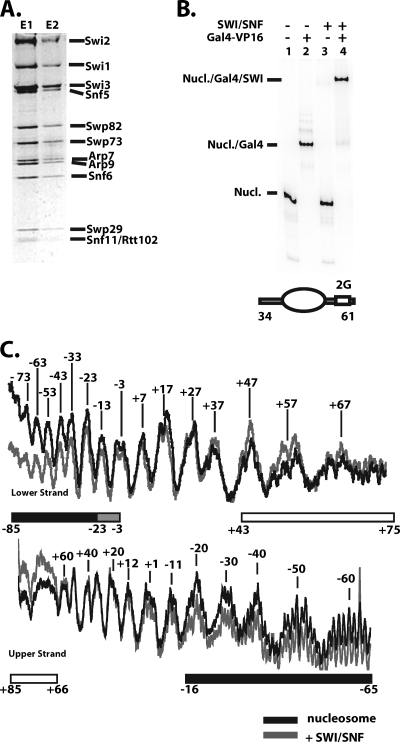
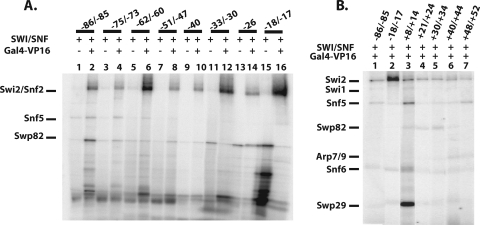
The 601 nucleosome positioning sequence described previously was used to construct nucleosomes with a common translational position (19, 24). Recruitment of SWI/SNF by the transcription activator Gal4-VP16 bound to extranucleosomal DNA ensured SWI/SNF binding to nucleosomes in one preferred orientation. SWI/SNF on its own would likely have two orientations, which it could bind nucleosomes due to their pseudo-twofold symmetry. Conditions were established using an excess of competitor DNA and nucleosomes to make SWI/SNF binding to the labeled nucleosomes dependent on Gal4-VP16 binding (Fig. 1B, compare lanes 3 and 4). Gal4-VP16 was observed to bind well to one or two Gal4-VP16 sites positioned 27 bp from the edge of the nucleosome (Fig. 1B, compare lanes 1 and 2, and data not shown).
FIG. 1.
DNA footprinting of SWI/SNF bound to nucleosomes after recruitment by Gal4-VP16. (A) Purified SWI/SNF was separated by 4 to 20% SDS-PAGE and stained with Coomassie blue. E1 and E2 are the first two fractions from the M2-agarose column. (B) Nucleosomes (Nucl.) were assembled with a radiolabeled 242-bp DNA probe (601 nucleosome positioning sequence flanked by 34 and 61 bp of extranucleosomal DNA). The positions of the nucleosome (oval), flanking DNA, and one or two Gal4 binding sites (2G) are shown. Nucleosomes (55 nM with 0.14 nM of the nucleosomes contains Gal4 binding sites) were incubated with 2.5 nM SWI/SNF in the presence or absence of Gal4-VP16 (75 nM) for 30 min at 30°C. Nucleosomes alone (lane 1) or with SWI/SNF (lanes 3 and 4) and/or Gal4-VP16 (lanes 2 and 4) added were separated on a native 4% polyacrylamide gel and visualized by phosphorimaging. (C) DNA footprinting with the hydroxyl radical was performed on complexes of the nucleosome plus Gal4-VP16 with (gray) or without (black) SWI/SNF. Quantification of the different lanes from a 6% denaturing polyacrylamide gel after normalization is overlaid for comparison. The region protected by SWI/SNF is indicated below by either a solid black (strongest) or gray (moderate) bar. The open bar indicates the region in which SWI/SNF binding enhances cutting, and the numbering refers to the number of base pairs from the right (+) or left (−) of the dyad axis.
The interactions of SWI/SNF with DNA in these nucleosomes were determined by DNA footprinting with hydroxyl radical (Fig. 1C). The strongest protection was on the side of the dyad axis opposite the Gal4 binding sites in the nucleosomal region starting ∼1 to 2 helical turns from the dyad axis and extending into the extranucleosomal DNA region (positions −85 to −3 and −16 to −65 in the upper and lower strands, respectively). Protection of nucleosomal DNA corresponded approximately to one turn or gyre of DNA around the nucleosome core (see Fig. 6A and B, shown in red). The other half of the nucleosomal DNA region had little or no protection, and there was enhanced DNA cutting at the extranucleosomal DNA region proximal to the Gal4 binding site. The enhanced cutting is suggestive of SWI/SNF binding altering the DNA structure in some manner to make it more readily cleaved by hydroxyl radicals and implies that SWI/SNF may be interacting at or close to this region. The asymmetry of the DNA footprint pattern confirmed that SWI/SNF is bound in one preferred orientation rather than two symmetrical positions about the nucleosome.
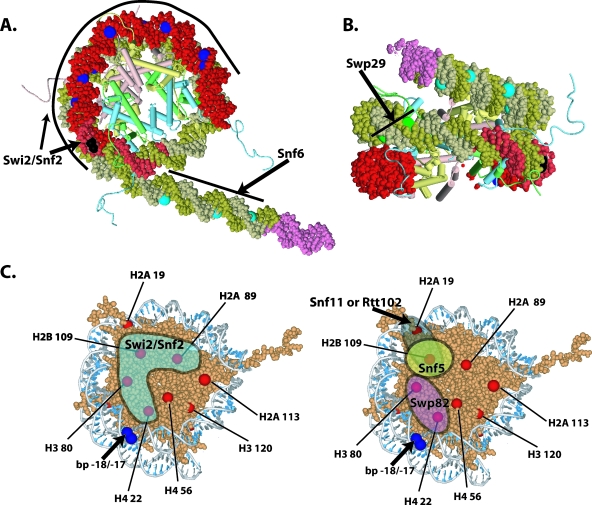
FIG. 6.
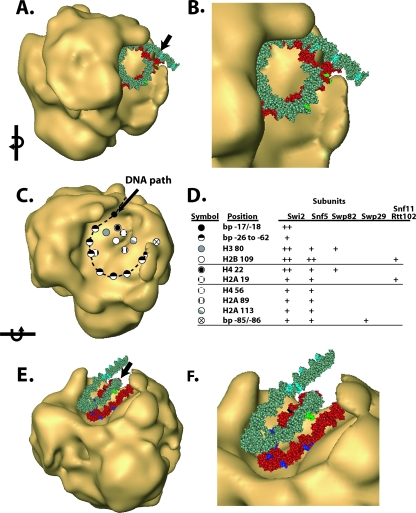
Display of the locations in DNA and histone octamer that were cross-linked to SWI/SNF subunits. (A and B) The DNA protected by SWI/SNF binding is highlighted in red in the nucleosome model, and the position of the Gal4 binding site is highlighted in pink. The black (strong) and dark blue (weak) spots on the DNA indicated the location to which Swi2/Snf2 was cross-linked. The light blue spots indicate the location of Snf6 cross-linked in the extranucleosomal DNA region. The location of Swp29 cross-linking is indicated by green spots (B). (C) The locations of the eight different cysteine sites are shown on the surface of the nucleosome core particle along with that of the DNA position to which Swi2/Snf2 is most efficiently cross-linked (bp −18/−17). Those sites that were >0.3 times as efficiently cross-linked to Swi2/Snf2 as residue 80 of histone H3 are shown in the shaded region (left). The locations of the sites that are most efficiently cross-linked to Snf5, Swp82, and Snf11 or Rtt102 are in the shaded regions depicted on the right.
SWI/SNF has one large asymmetric trough on its surface that matches the contour of the nucleosome.
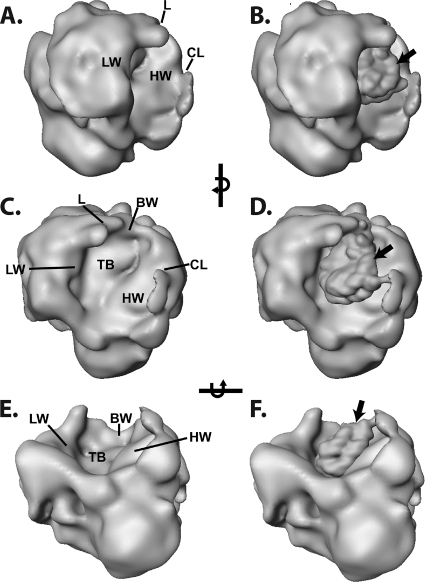
The DNA footprinting data demonstrate that the nucleosomal binding surface of SWI/SNF should accommodate one entire gyre of the nucleosome without making close contact with the adjacent gyre. To find if there are key structural features consistent with our DNA footprinting data, we have reexamined the structure of yeast SWI/SNF utilizing cryoelectron microscopy (cryoEM) to avoid potential artifacts inherent in previous studies due to negative staining and drying of the sample. The single-particle reconstruction of cryoEM data (resolution of 23 Å at an FSC of 0.5) (see Materials and Methods) revealed a shape closely related to the negative-stain reconstruction (42), with most of the differences being attributable to flattening in stain and indistinct boundaries between stain penetration and exclusion. The foremost feature of the reconstruction in ice is the deepened trough whose base closely matches the contour of a nucleosome. The trough has a base and three walls referred to as back, high, and low, with heights of 56 Å, 120 Å, and 85 Å, respectively, and with defined features at their upper edges identified as clamp and lip (Fig. 2A, C, and E).
FIG. 2.
cryoEM reconstruction of SWI/SNF and model of the SWI/SNF-nucleosome complex. Panels A, C, and E show three different views of the SWI/SNF structure obtained from cryoEM. Panels B, D, and F are the models of the SWI/SNF-nucleosome complex obtained by fitting the crystal structure of the nucleosome low pass filtered to 25 Å into the putative nucleosome binding surface of SWI/SNF. Features of the nucleosome binding face of SWI/SNF are a trough whose base (TB) is met by a high wall (HW), a low wall (LW), and a back wall (BW). The base slopes very gently up into the high wall and down to the nearly rimless front; junctions with the low and back walls are more pronounced. Along the rim of the trough are two prominent features, labeled lip (L) and clamp (CL). The dyad axis of the nucleosome is indicated by an arrow (B, D, and F).
The contours of the trough (Fig. 2C and E) suggest that a nucleosome could be bound on edge with limited rotational freedom about the dyad axis but with a large degree of freedom perpendicular to it. The DNA footprinting data suggest that the nucleosome is located inside this trough, abutting the high wall of SWI/SNF, thereby protecting one gyre of the nucleosome. The other gyre, facing the low wall, would be readily accessible (Fig. 2B, D, and F). Based on this model, the Gal4 binding site would be located in the outer surface of the back wall, and the dyad axis of the nucleosome would be exposed, facing out of the trough, rotated toward the back wall.
The Swi2/Snf2 subunit of SWI/SNF is closely associated with nucleosomal DNA two helical turns from the dyad axis.
The subunits of SWI/SNF bound near nucleosomal and extranucleosomal DNA were identified by site-directed DNA cross-linking. In these experiments, a photoreactive deoxyribonucleotide was enzymatically incorporated adjacent to a radioactive nucleotide(s) in DNA at 20 different sites throughout the nucleosome (Fig. 3A). These DNA modifications were shown previously to not interfere with SWI/SNF binding and remodeling of nucleosomes (38) (results not shown). Deoxyuridine and deoxycytidine nucleotide analogs contained either a photoreactive aryl azide or diazirine moiety. The aryl azide preferentially cross-links to nucleophilic side chains, while the diazirine has no amino acid side chain preference (46). The subunits of SWI/SNF bound proximal to these sites were covalently linked to DNA by UV irradiation and the DNA degraded by DNase I and S1 nuclease. The short radiolabeled DNA left cross-linked to the protein was previously shown to add only about 3 to 5 kDa to the apparent molecular mass of the protein (38). Gel shift assays were used to monitor the binding of SWI/SNF to nucleosomes, and SWI/SNF binding was found to be dependent on Gal4-VP16 under these conditions (Fig. 1B). Nucleosome assembly for all 20 positions was as high as 97%, indicating that the binding and cross-linking of SWI/SNF to free DNA were negligible.
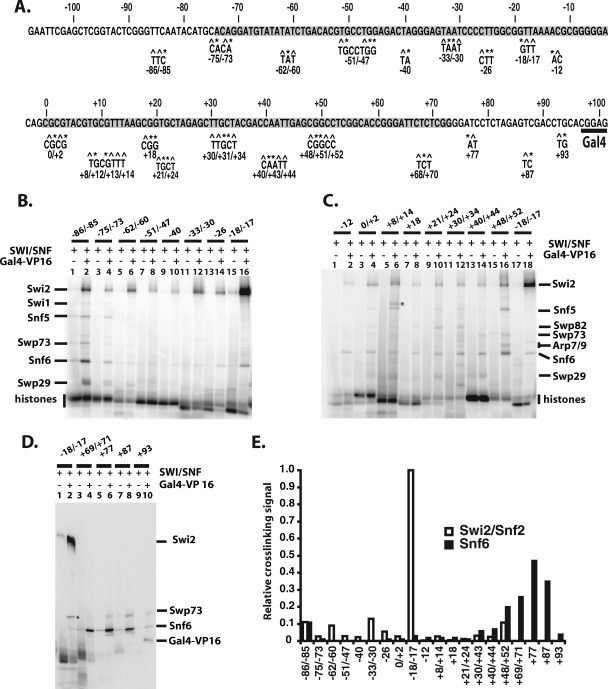
FIG. 3.
Swi2/Snf2 is cross-linked most extensively to the nucleosomal region, and Snf6 is cross-linked most extensively to the extranucleosomal region closest to the Gal4 binding site. (A) The 601 DNA sequence is shown, along with the 20 regions modified by the incorporation of a photoreactive nucleotide and radiolabel. The numbering is the same as that in Fig. 1, and the highlighted region is the core nucleosome region. Those stretches of sequence shown below have photoreactive nucleotides, indicated by “^,” and the incorporated α-32P-deoxynucleotide is indicated by “*.” Numbering refers to the location of the photoreactive nucleotide(s) relative to the dyad axis. (B to D) Binding of SWI/SNF to nucleosomes was done as indicated in the legend of Fig. 1. SWI/SNF-nucleosome complexes were cross-linked, DNA digested, and analyzed by 4 to 20% SDS-PAGE and phosphorimaging. Nucleosomes were reconstituted with different DNA photoaffinity probes and SWI/SNF with (even lanes) and without (odd lanes) Gal4-VP16 added. The * in panel C indicates a band that does not correlate to a SWI/SNF subunit, and the * in panel D indicates labeled bands due to trace amounts of undigested DNA that were not consistently observed. (E) The relative efficiency of cross-linking Swi2/Snf2 or Snf6 to DNA was obtained by dividing the labeling intensity of these two subunits at different positions with that of Swi2/Snf2 at bp −18/−17.
The region of nucleosomal DNA from positions −62 to −18/−17 bound by SWI/SNF was cross-linked almost exclusively to the Swi2/Snf2 subunit (Fig. 3B to D). The site of strongest cross-linking is at a region two helical turns from the dyad axis at bp −17 and −18 and corresponds to where the translocation of SWI/SNF along DNA was shown to be required for nucleosome mobilization (54). Swi2/Snf2 was reproducibly cross-linked at least 10-fold more efficiently at bp −18/−17 than at any other position examined (Fig. 3E). These data indicate that although the Swi2/Snf2 subunit makes extensive contact with nucleosomal DNA, its primary contact with the major groove of DNA is two helical turns from the dyad axis, consistent with its catalytic role in chromatin remodeling. Cross-linking of SWI/SNF was highly specific, as shown by its strong dependency on Gal4-VP16 (compare odd versus even lanes in Fig. 3B to D). Based on the model of nucleosome binding to the trough region of SWI/SNF, the strong cross-linking of Swi2/Snf2 at bp −18 and −17 would be located at the base of the trough next to the back wall (see Fig. 8C and D).
FIG. 8.
Subunit topology in the nucleosome binding pocket of SWI/SNF. The views in panels A, C, and E are in the same orientation as that shown in Fig. 2A, C, and E. Nucleosomal DNA is displayed with the same color scheme as that in Fig. 6A and B and refers to sites that cross-linked Swi2, Snf6, and Swp29. The histone octamer is not shown for better visualization of the proximity of DNA cross-linking sites to the surface of SWI/SNF. The DNA protected by SWI/SNF binding is shown in red, and that not protected is shown in gray (A to B and E and F). In this model, the linker trajectory is arbitrarily assigned. (C) The locations of particular DNA and histone sites are shown with the nucleosome removed and are based on the SWI/SNF-nucleosome model described above. The legend for the symbols used to mark these locations is shown (D), along with a summary of the SWI/SNF subunits cross-linked at these sites and their relative efficiencies, i.e., strong (++) and weak (+). The proposed path of nucleosomal DNA along the surface of the SWI/SNF trough is shown as a dashed line. Arrows in panels A and E indicate the dyad axis of the nucleosome.
The Swp73 and Swp82 subunits are less efficiently cross-linked to nucleosomal DNA than Swi2/Snf2 from bp +40 to +52 and bp +21 to +34, respectively. Snf5 weakly cross-linked DNA at three positions (bp +48 to +52 and −73 to −85 and near the dyad axis from bp 0 to +14) proximal to each other on the nucleosome surface. Although these contacts are weak, they appear to be specific and are not likely to be due to minor amounts of SWI/SNF bound to free DNA, as for the weak cross-linking of Snf6 that is observed throughout the nucleosome-bound region (see below).
The Snf6 subunit binds near the Gal4-VP16 activator protein, and the Swp29 subunit is associated close to the dyad axis.
The cross-linking of extranucleosomal DNA to SWI/SNF revealed the association of several other SWI/SNF subunits with the nucleosome. Extranucleosomal DNA closest to the Gal4-VP16 binding site efficiently cross-linked primarily the Snf6 subunit in a region spanning from the edge of the nucleosome (position +69/+71) to the side of the Gal4 binding site (position +87) as shown in Fig. 3D (compare lanes 4 to 8 to lane 2). Snf6 was most efficiently cross-linked at bp +77 and was only half as efficient as optimal Swi2/Snf2 cross-linking at bp −18/−17 (Fig. 3E). In contrast, extranucleosomal DNA at bp −85/−86 cross-linked the Swi2/Snf2, Snf6, and Swp29 subunits ∼10-fold less efficiently than did Swi2/Snf2 at bp −18/−17 (Fig. 3B, compare lane 2 to lane 16).
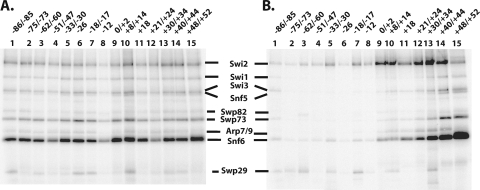
The Snf6 subunit was found to be most readily cross-linked to DNA independent of the base pair position in SWI/SNF-DNA complexes (Fig. 4A). In other experiments where the binding of SWI/SNF to DNA is restricted by Gal4-VP16 recruitment, DNA cross-linking revealed that Snf6 was preferentially cross-linked to sites nearest the Gal4-VP16 binding site (Fig. 4B). These data indicate that the recruitment of SWI/SNF to either free DNA or nucleosomes by Gal4-VP16 causes the placement of the Snf6 subunit proximal to Gal4-VP16 and that Snf6 likely interacts with Gal4-VP16. In vivo studies have shown that Snf6 along with Snf5 and Swi3 are required for the optimal recruitment of SWI/SNF by Gcn4 (51). Other cross-linking studies using photoreactive Gcn4 and Hap4 have found Snf5, Swi1, and Swi2/Snf2 instead of Snf6 to be associated with these two acidic transcription activators (30).
FIG. 4.
Snf6 is the subunit of SWI/SNF most proximal to the Gal4-VP16 binding site. The same DNA probes as those in Fig. 3 were used in binding reactions with SWI/SNF without (A) and with (B) Gal4-VP16 added but without prior assembly into nucleosomes. Samples were analyzed as described in the legend to Fig. 3, and the relative electrophoretic mobilities of the SWI/SNF subunits are shown.
The DNA cross-linking data reveal new details about SWI/SNF interactions with nucleosomal and extranucleosomal DNA that were not uncovered in our previous cross-linking studies using nucleosomes reconstituted with 5S rDNA (39). Previously, the Swi2/Snf2 and other subunits were not seen to be as well localized to regions of the nucleosome as observed in this study. The two reasons that the previous results likely failed to show the specificity of these contacts are that (i) 5S rDNA nucleosomes do not have a single nucleosomal translational position and (ii) the recruitment of SWI/SNF to nucleosomes eliminates the possibility of SWI/SNF docking nucleosomes in two equivalent and symmetrical orientations on nucleosomes.
Additional DNA photoaffinity experiments utilized DNA that contained the photoreactive diazirine group instead of the aryl azido group. An advantage of diazirine is that upon photolysis, it forms the more reactive and less selective carbene compared to the aryl nitrene formed upon the photolysis of aryl azide (46). The same positions on DNA were scanned using diazirine-modified DNA (Fig. 5). A key difference in the cross-linking pattern with diazirine-modified DNA was the strong and almost exclusive cross-linking of the Swp29 subunit at bp +8 to +14 (Fig. 5B, lane 3). Cross-linking of the Swp29 subunit places this subunit close to the dyad axis in a region not strongly protected by SWI/SNF binding (Fig. 6B). The association of the Swp29 subunit with this area of the nucleosome is consistent with the cross-linking of Swp29 observed in the extranucleosomal region at bp −85 and −86 (Fig. 3B, lane 2). The likely reason for Swp29 not being cross-linked well by aryl azide containing DNA at bp +8 to +14 is that the protein surface in close proximity to this DNA site is deficient in nucleophilic side chains (46). The cross-linking of Swi2/Snf2 was also different with DNA probes containing diazirine versus those with aryl azide. The Swi2/Snf2 subunit was most efficiently cross-linked 17 to 18, 30 to 33, and 60 to 62 bp from the dyad axis (Fig. 5A, lanes 6, 12, and 16), consistent with Swi2/Snf2 being the principal subunit of SWI/SNF that contacts nucleosomal DNA in the region protected from hydroxyl radical cleavage.
FIG. 5.
DNA cross-linking of SWI/SNF with photoreactive diazirine probes. DNA cross-linking was done as described in the legend to Fig. 3 with photoreactive nucleosomes using the more photoreactive diazirine-containing DNA probes. (A) Samples contained SWI/SNF with (even lanes) and without (odd lanes) Gal4-VP16. (B) All samples contained SWI/SNF and Gal4-VP16.
Swi2/Snf2 and Snf5 bind discrete regions of the histone octamer.
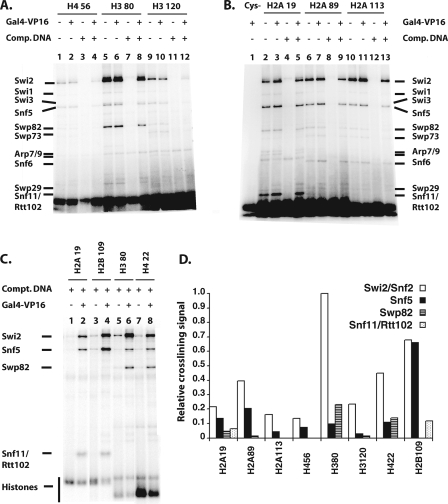
DNA cross-linking provided a limited view of the nucleosome interaction surface of SWI/SNF by focusing on the edge of the nucleosome involved in binding DNA. Site-directed modification of histones in the nucleosome was used to examine the interaction of SWI/SNF with the other two sides of the nucleosome comprising the histone octamer faces. Photoreactive aryl azides were conjugated to different sites in the histone octamer using recombinant histones engineered with unique cysteines (8). Residues in the histone proteins that were changed to cysteine were selected based on (i) surface accessibility, (ii) not being in secondary structures, and (iii) not being essential or well conserved. Histones were expressed and purified (26), and histone octamers were prepared by replacing one of the wild-type histones (cysteine minus) with a mutant histone protein (single cysteine). The introduction of cysteine at these sites and its subsequent modification did not affect nucleosome reconstitution or the ability of nucleosomes to be remodeled, as shown by gel shift and remodeling assays (data not shown). The cross-linking reagent used was PEAS, which can be radioiodinated and cleaved by disulfide reduction (12). PEAS contains a 2-thiopyridyl moiety for conjugation to cysteine, which creates a cleavable disulfide bond and has the photoreactive 4-azidosalicylamide moiety that can be radioiodinated. The specificity of PEAS conjugation to the mutant octamers was tested using 125I-labeled PEAS and analyzed by SDS-PAGE and phosphorimaging. Conjugation site specificity was confirmed using nucleosomes without cysteine as well as varying the histone containing the unique cysteine as controls, and cross-linking and label transfer were UV irradiation dependent (data not shown).
Histone cross-linking showed the association of Swi2/Snf2 with a part of the histone octamer. Swi2/Snf2 was most efficiently cross-linked to residue 80 of histone H3, while residues 22, 89, and 109 of histone H4, H2A, and H2B, respectively, were one-third, or more, as efficiently cross-linked (Fig. 7 and data not shown). These four residues cluster to a region of the histone octamer that coincides well with the part of the nucleosome shown by DNA footprinting to be bound to SWI/SNF (Fig. 6C). Also consistent with this idea is that residue 80 of histone H3 is proximal to the position in DNA that is cross-linked most efficiently to Swi2/Snf2. The Swi2/Snf2 obviously not only extensively interacts with nucleosomal DNA but also interacts with a large portion of one or both faces of the histone octamer not bound to DNA. For each mutant histone octamer, there are two symmetrical modification sites due to the dual copies of each histone in the octamer such that SWI/SNF could be cross-linked from either side of the histone octamer. The DNA footprinting data, however, indicate that one face of the nucleosome is tightly bound to SWI/SNF while the other face is fairly accessible. Given these data and the observation of the asymmetry of the putative nucleosome binding trough of SWI/SNF, most of the histone cross-linking will likely arise from just one face of the nucleosome.
FIG. 7.
Identification of SWI/SNF subunits bound to the histone octamer surface by site-directed histone cross-linking. (A to C). Nucleosomes reconstituted with different cysteine mutant octamers were modified with PEAS (see Materials and Methods). Modified nucleosomes (4 nM) were bound to SWI/SNF (6 nM) in the presence or absence of Gal4-VP16 (6.4 nM) and cross-linked by UV irradiation. After transfer of the radiolabel by disulfide reduction, the samples were analyzed by 4 to 12% (A and B) or 10% (C) Bis-Tris SDS-PAGE and phosphorimaging. The location of the modification is indicated above the lanes by referring to the histone protein and residue that was changed to cysteine. Gal4-VP16 and/or competitor DNA (2 ng/μl in panels A and B and 1 ng/μl in panel C) were added as indicated, and all reaction mixtures contained SWI/SNF. (D) The relative efficiency of histone cross-linking to SWI/SNF subunits was shown by normalizing the labeling intensity relative to Swi2/Snf2 cross-linking to residue 80 of histone H3. For each experiment, the relative labeling signal of Swi2/Snf2 and other subunits was obtained by dividing the signal to that of Swi2/Snf2 at residue 80 of histone H3. The data presented are the average data for two to three independent experiments for all sites except for H422 and H2B109. This comparison is shown for four primary SWI/SNF subunits (Swi2/Snf2, Snf5, Swp82, and Snf11 or Rtt102) shown to be cross-linked to the histone octamer face.
Although Snf5 does not appear to be bound stably near nucleosomal DNA, at residue 109 of H2B, it was found to be efficiently cross-linked and was comparable to Swi2 cross-linking at the same position (Fig. 6C and 7C, compare lane 4 to lanes 2, 6, and 8). The relative efficiency of Snf5 cross-linking progressively diminished at more distal positions. At the next most proximal positions at residues 89 of H2A and 19 of H2A, the relative efficiency is reduced ≥3.5-fold and even more extensively at more distal positions such as residues 113 of H2A and 56 of H4 (Fig. 7D). The Swp82 subunit was found to be located near residue 80 of histone H3 and residue 22 of histone H4 (Fig. 6C and 7C, lanes 6 and 8, and D). Likewise, one of the smallest subunits of SWI/SNF (Snf11 or Rtt102) was found to be discretely positioned at an opposite side of the nucleosome, near residue 19 of histone H2A and residue 109 of histone H2B (Fig. 6C and 7B, lanes 2 to 5, and C, lanes 2 and 4).
The specificity of histone cross-linking to SWI/SNF was demonstrated by (i) cross-linking requiring the presence of mutant cysteine histone protein, (ii) competition by free DNA eliminating SWI/SNF cross-linking, (iii) radiolabeling of SWI/SNF requiring UV irradiation, (iv) the cross-linking pattern varying in a position-dependent manner, and (v) SWI/SNF cross-linking requiring Gal4-VP16 under competitive conditions. The interaction of SWI/SNF with the histone octamer surface was not substantially different whether it bound nucleosomes directly or was recruited by Gal4-VP16. These data suggest that the interactions of SWI/SNF with the histone portion of the nucleosome, although facilitated by recruitment, do not appear to be altered by recruitment. Conditions for the Gal4-VP16-dependent binding of SWI/SNF required the addition of sufficient competitor DNA to avoid the direct binding of SWI/SNF to nucleosomes, as seen by gel shift assay (data not shown).
DISCUSSION
The structure of SWI/SNF and its interaction with nucleosomes has been studied using a combination of DNA footprinting, cross-linking to specific sites in DNA or histone proteins of the SWI/SNF-nucleosome complex, and cryoEM of free SWI/SNF. A salient feature of the SWI/SNF structure is an asymmetric trough region that has dimensions appropriate for the binding of a mononucleosome. Modeling of the nucleosome bound to this trough region of SWI/SNF coincides strikingly with the pattern of DNA footprinting observed when SWI/SNF is recruited to the nucleosome by Gal4-VP16 and protects almost one complete gyre of DNA. The dimensions of the trough are such that nucleosomes can bind only on edge between the two walls of different heights (Fig. 2). Our model for the nucleosome bound to the trough region of SWI/SNF accommodates not only the DNA footprinting information but also the DNA and histone cross-linking data such that all the observed cross-links are directly in contact with the surface of the trough or within the length of the cross-linker (≤9 Å) (Fig. 8A to C). Although the histone cross-linking could occur from either face of the nucleosome core particle, we suggest that it is primarily from the part of the nucleosome that is likely bound to the high wall of the trough. The proposed path of nucleosomal DNA along the base of the SWI/SNF trough is depicted in Fig. 8C, with locations of different subunits of SWI/SNF in the trough based on DNA or histone cross-linking results.
RSC is closely related to SWI/SNF, with several subunits shared between these two complexes (14); however, there are surprising differences between the cryoEM structures of the RSC and the SWI/SNF complex (27). RSC was shown to have a large central cavity surrounded by four regions of high density with dimensions that could accommodate a nucleosome inside the central cavity (1, 22, 23). The RSC-nucleosome model is devoid of the extensive contacts, with the nucleosome evident in the cross-linking data presented here. Further experiments will be required to determine to what extent the structural differences between the complexes are related to their respective subfamilies.
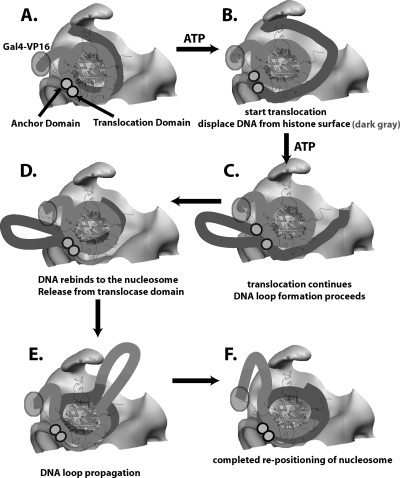
We present a new model for SWI/SNF remodeling based on mapping of the interactions of SWI/SNF with nucleosomes and other biochemical and biophysical data. Remodeling begins with the translocation of SWI/SNF along nucleosomal DNA at the edge of the trough region. The translocation of SWI/SNF along nucleosomal DNA ∼20 bp from the dyad axis was shown to be required for nucleosome remodeling (54) and is the basis for placing translocation at the back of the trough region. Consistent with previously reported data (54), DNA between the translocation site and the other side of the bound trough region in our model is displaced from the surface of the histone octamer in conjunction with the translocation, as shown in Fig. 9A and B. Since DNA loop formation appears to be a common feature of SWI/SNF and RSC remodeling (6, 28, 41, 49, 53), we postulate that a small DNA loop is initially formed between the translocation domain and an anchor domain. As SWI/SNF hydrolyzes more ATP, a larger DNA loop that is primarily free and accessible because of its location in the trough region of SWI/SNF is created (Fig. 9C). Subsequently, the translocation and anchor domain release their DNA contact. The DNA bulge migrates across the histone octamer away from the translocation site and away from the location in which DNA was initially displaced, with the eventual reassociation with the histone surface as shown (Fig. 9D). Propagation of the DNA loop in this direction is favored because it first proceeds in the region of the nucleosome facing away from the cleft, and as it proceeds around to the other side of the nucleosome, it is in the area of the cleft with the low wall, where there is sufficient space for the passage of a DNA loop. The loop will continue to propagate until it reaches the other entry/exit site and thereby complete the movement of DNA around the nucleosome (Fig. 9E and F). A key difference in this model compared to others is the extent of the remodeler's interaction with nucleosomes. RSC has been suggested to contact only a small region of nucleosomal DNA, whereas SWI/SNF makes extensive contact and could reasonably cause a larger disruption of the histone-DNA interface due to compensating interactions of SWI/SNF with nucleosomal DNA and thus move the nucleosome in larger steps (6, 37, 54).
FIG. 9.
Structure-based model for SWI/SNF remodeling. (A) A cutaway view of the SWI/SNF trough surface with a schematic view of the bound nucleosome is shown. The dark gray line represents the DNA in close contact with the high wall surface of SWI/SNF, and the light gray line is the DNA that is more accessible and is located next to the low wall. The position of the translocation and anchor domains of SWI/SNF is indicated by two gray spheres, and the approximate location of the Gal4-VP16 is indicated. (B and C) Remodeling starts with the translocation domain pulling DNA toward it and the concerted displacement of ∼50 bp of DNA from the surface of the nucleosome. The translocation domain continues to pull DNA toward itself and creates a large DNA loop by ratcheting DNA on one side, while the other part of DNA remains fixed to an adjacent site in SWI/SNF. (D and E) The DNA loop is released to move around the nucleosome in a wave-like process. (F) The movement of the DNA loop toward the other entry/exit site causes the movement of the histone octamer to a new position on DNA.
A natural outcome of this process will be the progressive reduction in the length of linker DNA at the side where the DNA is displaced and then pulled into the translocation domain of SWI/SNF. As DNA from this entry/exit site is pulled into SWI/SNF, proteins bound to neighboring DNA, such as adjacent nucleosomes or DNA-bound transcription factors, will be brought into closer proximity to the SWI/SNF-nucleosome complex. This process will ultimately cause a clash between any protein bound to the linker DNA and SWI/SNF with its bound nucleosome. Either the SWI/SNF-bound nucleosome may be displaced causing remodeling to terminate or the factor or nucleosome bound to the linker DNA may give way. Given this type of mechanism, it can be envisaged how SWI/SNF remodeling in a nucleosomal array might cause the displacement through nucleosome collision. In vivo studies provided evidence for nucleosome eviction by SWI/SNF at the PHO5 promoter, which would be consistent with the idea of eviction through collision (3, 4, 20, 21). Recent experiments have shown the stimulation of octamer transfer in vitro by an acidic activator, and previously reported data indicated that SWI/SNF can cause nucleosome displacement in nucleosomal arrays in vitro (15, 32). While there are other models of SWI/SNF remodeling of nucleosomes that could be based on our structural data, we favor the one presented, as it accounts well for the properties of SWI/SNF remodeling that have been observed to date.
Acknowledgments
We thank Brad Cairns for providing anti-Swp73 antibodies and members of the Bartholomew laboratory for their critical review and comments.
This work was supported by Public Health Service grant GM 48413 to B.B., GM 43786 to C.L.W., and GM 49650 to C.L.P.
Footnotes
▿
Published ahead of print on 21 July 2008.
REFERENCES
- 1.Asturias, F. J., W. H. Chung, R. D. Kornberg, and Y. Lorch. 2002. Structural analysis of the RSC chromatin-remodeling complex. Proc. Natl. Acad. Sci. USA 9913477-13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71247-273. [DOI] [PubMed] [Google Scholar]
- 3.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 111587-1598. [DOI] [PubMed] [Google Scholar]
- 4.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14667-673. [DOI] [PubMed] [Google Scholar]
- 5.Bruno, M., A. Flaus, C. Stockdale, C. Rencurel, H. Ferreira, and T. Owen-Hughes. 2003. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol. Cell 121599-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cairns, B. R. 2007. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat. Struct. Mol. Biol. 14989-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns, B. R., N. L. Henry, and R. D. Kornberg. 1996. TFG/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol. Cell. Biol. 163308-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, H. T., and S. Hahn. 2003. Binding of TFIIB to RNA polymerase II: mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol. Cell 12437-447. [DOI] [PubMed] [Google Scholar]
- 9.Chizzonite, R., T. Truitt, B. B. Desai, P. Nunes, F. J. Podlaski, A. S. Stern, and M. K. Gately. 1992. IL-12 receptor. I. Characterization of the receptor on phytohemagglutinin-activated human lymphoblasts. J. Immunol. 1483117-3124. [PubMed] [Google Scholar]
- 10.Cote, J., J. Quinn, J. L. Workman, and C. L. Peterson. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 26553-60. [DOI] [PubMed] [Google Scholar]
- 11.Davey, C. A., D. F. Sargent, K. Luger, A. W. Maeder, and T. J. Richmond. 2002. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 3191097-1113. [DOI] [PubMed] [Google Scholar]
- 12.Ebright, Y. W., Y. Chen, Y. Kim, and R. H. Ebright. 1996. S-[2-(4-Azidosalicylamido)ethylthio]-2-thiopyridine: radioiodinatable, cleavable, photoactivatible cross-linking agent. Bioconjug. Chem. 7380-384. [DOI] [PubMed] [Google Scholar]
- 13.Ehrenhofer-Murray, A. E. 2004. Chromatin dynamics at DNA replication, transcription and repair. Eur. J. Biochem. 2712335-2349. [DOI] [PubMed] [Google Scholar]
- 14.Gangaraju, V. K., and B. Bartholomew. 2007. Mechanisms of ATP dependent chromatin remodeling. Mutat. Res. 6183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez, J. L., M. Chandy, M. J. Carrozza, and J. L. Workman. 2007. Activation domains drive nucleosome eviction by SWI/SNF. EMBO J. 26730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havas, K., A. Flaus, M. Phelan, R. Kingston, P. A. Wade, D. M. Lilley, and T. Owen-Hughes. 2000. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell 1031133-1142. [DOI] [PubMed] [Google Scholar]
- 17.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95717-728. [DOI] [PubMed] [Google Scholar]
- 18.Horowitz-Scherer, R. A., and C. L. Woodcock. 2004. Visualization and 3D structure determination of defined sequence chromatin and chromatin remodeling complexes. Methods Enzymol. 37629-48. [DOI] [PubMed] [Google Scholar]
- 19.Kagalwala, M. N., B. J. Glaus, W. Dang, M. Zofall, and B. Bartholomew. 2004. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 232092-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korber, P., S. Barbaric, T. Luckenbach, A. Schmid, U. J. Schermer, D. Blaschke, and W. Horz. 2006. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J. Biol. Chem. 2815539-5545. [DOI] [PubMed] [Google Scholar]
- 21.Korber, P., T. Luckenbach, D. Blaschke, and W. Horz. 2004. Evidence for histone eviction in trans upon induction of the yeast PHO5 promoter. Mol. Cell. Biol. 2410965-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leschziner, A. E., B. Lemon, R. Tjian, and E. Nogales. 2005. Structural studies of the human PBAF chromatin-remodeling complex. Structure 13267-275. [DOI] [PubMed] [Google Scholar]
- 23.Leschziner, A. E., A. Saha, J. Wittmeyer, Y. Zhang, C. Bustamante, B. R. Cairns, and E. Nogales. 2007. Conformational flexibility in the chromatin remodeler RSC observed by electron microscopy and the orthogonal tilt reconstruction method. Proc. Natl. Acad. Sci. USA 1044913-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowary, P. T., and J. Widom. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 27619-42. [DOI] [PubMed] [Google Scholar]
- 25.Ludtke, S. J., D. H. Chen, J. L. Song, D. T. Chuang, and W. Chiu. 2004. Seeing GroEL at 6 A resolution by single particle electron cryomicroscopy. Structure 121129-1136. [DOI] [PubMed] [Google Scholar]
- 26.Luger, K., T. J. Rechsteiner, and T. J. Richmond. 1999. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 1191-16. [DOI] [PubMed] [Google Scholar]
- 27.Mohrmann, L., K. Langenberg, J. Krijgsveld, A. J. Kal, A. J. Heck, and C. P. Verrijzer. 2004. Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol. Cell. Biol. 243077-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narlikar, G. J., M. L. Phelan, and R. E. Kingston. 2001. Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol. Cell 81219-1230. [DOI] [PubMed] [Google Scholar]
- 29.Natarajan, K., B. M. Jackson, H. Zhou, F. Winston, and A. G. Hinnebusch. 1999. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol. Cell 4657-664. [DOI] [PubMed] [Google Scholar]
- 30.Neely, K. E., A. H. Hassan, C. E. Brown, L. Howe, and J. L. Workman. 2002. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 221615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neely, K. E., A. H. Hassan, A. E. Wallberg, D. J. Steger, B. R. Cairns, A. P. Wright, and J. L. Workman. 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 4649-655. [DOI] [PubMed] [Google Scholar]
- 32.Owen-Hughes, T., R. T. Utley, J. Cote, C. L. Peterson, and J. L. Workman. 1996. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science 273513-516. [DOI] [PubMed] [Google Scholar]
- 33.Persinger, J., and B. Bartholomew. 1996. Mapping the contacts of yeast TFIIIB and RNA polymerase III at various distances from the major groove of DNA by DNA photoaffinity labeling. J. Biol. Chem. 27133039-33046. [DOI] [PubMed] [Google Scholar]
- 34.Peterson, C. L., and J. Cote. 2004. Cellular machineries for chromosomal DNA repair. Genes Dev. 18602-616. [DOI] [PubMed] [Google Scholar]
- 35.Phelan, M. L., G. R. Schnitzler, and R. E. Kingston. 2000. Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases. Mol. Cell. Biol. 206380-6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phelan, M. L., S. Sif, G. J. Narlikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3247-253. [DOI] [PubMed] [Google Scholar]
- 37.Saha, A., J. Wittmeyer, and B. R. Cairns. 2005. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat. Struct. Mol. Biol. 12747-755. [DOI] [PubMed] [Google Scholar]
- 38.Sengupta, S. M., J. Persinger, B. Bartholomew, and C. L. Peterson. 1999. Use of DNA photoaffinity labeling to study nucleosome remodeling by SWI/SNF. Methods 19434-446. [DOI] [PubMed] [Google Scholar]
- 39.Sengupta, S. M., M. VanKanegan, J. Persinger, C. Logie, B. R. Cairns, C. L. Peterson, and B. Bartholomew. 2001. The interactions of yeast SWI/SNF and RSC with the nucleosome before and after chromatin remodeling. J. Biol. Chem. 27612636-12644. [DOI] [PubMed] [Google Scholar]
- 40.Shen, F., S. J. Triezenberg, P. Hensley, D. Porter, and J. R. Knutson. 1996. Critical amino acids in the transcriptional activation domain of the herpesvirus protein VP16 are solvent-exposed in highly mobile protein segments. An intrinsic fluorescence study. J. Biol. Chem. 2714819-4826. [DOI] [PubMed] [Google Scholar]
- 41.Shundrovsky, A., C. L. Smith, J. T. Lis, C. L. Peterson, and M. D. Wang. 2006. Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nat. Struct. Mol. Biol. 13549-554. [DOI] [PubMed] [Google Scholar]
- 42.Smith, C. L., R. Horowitz-Scherer, J. F. Flanagan, C. L. Woodcock, and C. L. Peterson. 2003. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat. Struct. Biol. 10141-145. [DOI] [PubMed] [Google Scholar]
- 43.Smith, C. L., and C. L. Peterson. 2003. Coupling tandem affinity purification and quantitative tyrosine iodination to determine subunit stoichiometry of protein complexes. Methods 31104-109. [DOI] [PubMed] [Google Scholar]
- 44.Sudarsanam, P., V. R. Iyer, P. O. Brown, and F. Winston. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 973364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swanson, M. J., H. Qiu, L. Sumibcay, A. Krueger, S. J. Kim, K. Natarajan, S. Yoon, and A. G. Hinnebusch. 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 232800-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tate, J. J., J. Persinger, and B. Bartholomew. 1998. Survey of four different photoreactive moieties for DNA photoaffinity labeling of yeast RNA polymerase III transcription complexes. Nucleic Acids Res. 261421-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Treich, I., B. R. Cairns, T. de los Santos, E. Brewster, and M. Carlson. 1995. SNF11, a new component of the yeast SNF-SWI complex that interacts with a conserved region of SNF2. Mol. Cell. Biol. 154240-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsukiyama, T., and C. Wu. 1997. Chromatin remodeling and transcription. Curr. Opin. Genet. Dev. 7182-191. [DOI] [PubMed] [Google Scholar]
- 49.Wang, H., R. Bash, S. M. Lindsay, and D. Lohr. 2005. Solution AFM studies of human Swi-Snf and its interactions with MMTV DNA and chromatin. Biophys. J. 893386-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, X., R. Zaurin, M. Beato, and C. L. Peterson. 2007. Swi3p controls SWI/SNF assembly and ATP-dependent H2A-H2B displacement. Nat. Struct. Mol. Biol. 14540-547. [DOI] [PubMed] [Google Scholar]
- 51.Yoon, S., H. Qiu, M. J. Swanson, and A. G. Hinnebusch. 2003. Recruitment of SWI/SNF by Gcn4p does not require Snf2p or Gcn5p but depends strongly on SWI/SNF integrity, SRB mediator, and SAGA. Mol. Cell. Biol. 238829-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yudkovsky, N., C. Logie, S. Hahn, and C. L. Peterson. 1999. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 132369-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, Y., C. L. Smith, A. Saha, S. W. Grill, S. Mihardja, S. B. Smith, B. R. Cairns, C. L. Peterson, and C. Bustamante. 2006. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol. Cell 24559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zofall, M., J. Persinger, S. R. Kassabov, and B. Bartholomew. 2006. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat. Struct. Mol. Biol. 13339-346. [DOI] [PubMed] [Google Scholar]