Large-scale gene trapping in C57BL/6N mouse embryonic stem cells (original) (raw)
Abstract
We report the construction and analysis of a mouse gene trap mutant resource created in the C57BL/6N genetic background containing more than 350,000 sequence-tagged embryonic stem (ES) cell clones. We also demonstrate the ability of these ES cell clones to contribute to the germline and produce knockout mice. Each mutant clone is identified by a genomic sequence tag representing the exact insertion location, allowing accurate prediction of mutagenicity and enabling direct genotyping of mutant alleles. Mutations have been identified in more than 10,000 genes and show a bias toward the first intron. The trapped ES cell lines, which can be requested from the Texas A&M Institute for Genomic Medicine, are readily available to the scientific community.
It has been appreciated for some time that humans and mice share sufficiently similar genomes and physiology to allow the prediction of human gene function using the mouse as a model system. One of the most scalable genetic technologies available for the study of gene function in mice is “gene trapping,” a method of random mutagenesis in which the insertion of a synthetic DNA element into endogenous genes leads to their transcriptional disruption. In its most common form, a gene trapping construct consists of a splice acceptor, a selectable marker gene, and a polyadenylation signal that is placed within a retroviral genome such that it can be packaged into retroviral particles and used to infect cells (for review, see Abuin et al. 2007). When insertions occur within transcriptionally active regions, the marker gene is expressed and translated, allowing selection of mutant clones. Gene disruption is accomplished most often through the capture of endogenous gene transcription by the splice acceptor element within the trapping construct, or alternatively, by direct gene disruption as a result of insertion within an exon. Gene trapping is inherently amenable to high-throughput, cost-effective mutant clone production and mutation identification. A single gene trapping vector can be used to produce thousands of mutations and associated sequence tags, over the course of only a few weeks. In contrast, gene targeting via homologous recombination, while aided by the availability of complete genome sequences, still requires a unique construct for every mutation as well as subsequent clone screening to find the desired targeted mutation. The efficiency of homologous recombination is dependent on the characteristics of the targeting construct (extent of homology, positive/negative selection schemes, etc.) and the characteristics of each unique locus. A third method, chemical mutagenesis, produces basepair mutations that, while of value for understanding protein function, cannot be identified directly and thus necessitate complex genetic screens and mapping procedures.
Transcript-based technologies such as RACE (rapid amplification of cDNA ends) have been historically used to identify genes disrupted by gene trapping, as they allow amplification of fusion transcripts that are produced by splicing between endogenous gene exons and the gene trap construct, also known as “transcriptional tagging.” These technologies do not require extensive knowledge of gene structure or sequence; therefore, they were the ideal methodologies for mutation identification prior to completion of the mouse genome sequencing efforts. Ready access to the essentially complete sequence of the mouse genome now provides the basis for precise mapping of retroviral insertion mutations using genomic sequence tags. Direct genomic-based insertion site amplification, sequencing, and mapping obviate the problems associated with transcript-based sequence acquisition (e.g., variable RNA expression levels, effects of insertion site proximity to the 5′- and 3′-ends of the transcribed gene, and RNA stability). In addition, desirable mutation classes that cannot be identified through transcriptional tagging, such as those in single exon genes, can be detected readily from genomic insertion site sequence data. Furthermore, genomic-based insertion site sequence data permit the study of retroviral insertion patterns, genome and chromatin structure, and transcriptional activity in embryonic stem (ES) cells, in addition to producing a greater proportion of confirmed sequence-tagged clones in the resulting library.
The Knockout Mouse Project, initiated by the NIH, emphasized the generally acknowledged utility of a new resource of knockout mice in a non-hybrid C57 background (Austin et al. 2004). Even though C57-derived ES cell lines have been available for nearly two decades, the robust performance of 129 lines in cell culture and mouse production has led to their nearly exclusive use in knockouts to date. Germline-transmission breeding of 129-derived chimeras with C57 animals produces F1 hybrid heterozygotes and subsequent generations of individuals with variable background inheritance. Making knockout mice using mutated C57 ES cells would alleviate doubts about the effects of hybrid backgrounds on phenotypic expression and would eliminate the delays and costs associated with isogenization breeding. We report here the construction and analysis of a library consisting of more than 350,000 genomically tagged gene trapped ES cell clones of C57BL/6N origin. The creation of this fully public resource was supported by the state of Texas through the Texas Enterprise Fund and serves as the principal genetic resource of the Texas A&M Institute for Genomic Medicine (Collins et al. 2007). We have phenotyped more than 2000 lines of mutant mice derived from OmniBank, a gene trap library of transcription-tagged 129-derived ES cells (Zambrowicz et al. 1998, 2003), and we are using these data to identify medically relevant genes to aid drug discovery (Rice et al. 2004; Powell et al. 2005; Desai et al. 2007; Brommage et al. 2008). Compared to existing gene trap resources such as OmniBank, this new effort represents advances in both the strain of origin of the ES cells and in the mode of acquisition and mapping of mutation sequence tags. Furthermore, we describe the use of this library for the generation of C57BL/6 knockout mice.
Results
Generation of the OmniBankII gene trap library
We have used high-throughput gene trapping with retroviral vectors in mouse C57BL/6N ES cells to generate OmniBankII, a library of 481,152 mutated ES cell clones that are cryopreserved in duplicate in vapor-phase liquid nitrogen. Using an automated, genomic-based inverse-PCR (IPCR) sequencing and annotation protocol (Supplemental Fig. 1), we have produced a total of 532,829 insertion site sequence tags (ISTs) derived from 352,402 (or 73%) of these clones. For inclusion into the OmniBankII sequence data set, ISTs are required to show significant contiguous sequence identity with mouse genome sequence (NCBI build 36). ISTs range in length from 25 to 790 bp with an average length of 267 bp (Supplemental Fig. 2) and contain an average of 4.2% sequence ambiguities.
Our sequence acquisition methodology requires fragmentation of the gene trapped ES cell DNA prior to circularization and amplification such that target templates are within a size range amenable to high-throughput PCR. We used restriction endonuclease cleavage to achieve this fragmentation; however, this approach can limit access to insertions that lie at a distance from the restriction enzyme recognition sequence. Therefore, to maximize the insertions mapped through this method, we processed the entire library in duplicate using two different multienzyme digestion reactions. The insertion site sequence and genome map locations of 33% of OmniBankII clones have been confirmed by two or more independent ISTs. Twelve percent of clones show disparate mapping information, owing either to ambiguous mapping of insertion site sequence or to a mixture of gene trapped cell populations within the sample. A total of 396 OmniBankII clones have been thawed, expanded, and resequenced to determine the accuracy of the automated analysis. Ninety-six percent of expanded clones were confirmed to match the automated analysis. In 3% of cases, only one of two different annotated mutations assigned to the same well was identified in the sample following clone expansion. This suggests that clone loss from a mixed population could be one factor limiting the overall confirmation rate. Following expansion, 7% of clones have been shown to contain mixed populations of gene trap mutations.
OmniBankII gene coverage
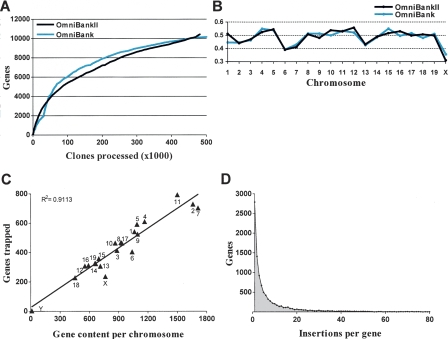
To identify the genes disrupted in the OmniBankII library, gene trap vector insertion site positions were compared with a collection of 28,907 gene and transcript entries (including, but not limited to, NCBI Refseq genes). These entries were derived from NCBI’s gene annotation database, Entrez Gene, for which corresponding mouse genome map locations (NCBI build 36) were available. Figure 1A shows the gene acquisition rate over the course of OmniBankII production. The rate is comparable to that observed for OmniBank (Zambrowicz et al. 2003), despite the fact that the methodologies for sequence acquisition and gene identification differ. Trapped genes were distributed among all chromosomes (Fig. 1B), and there was a linear correlation between chromosomal gene content and gene acquisition (Fig. 1C; Supplemental Fig. 3). A total of 10,433 unique genes have been trapped one or more times in the OmniBankII library. In agreement with previous studies (Nord et al. 2007), gene acquisition increased with increasing gene span; 63% of genes with a chromosomal span of ≥20 kb were trapped in OmniBankII, whereas less than 35% of genes with a span of <20 kb were trapped. Analysis of the density of insertions at each gene locus revealed that although many genes have only a single associated gene trap clone (2793), the majority (73%) of genes that have been trapped in the OmniBankII library have more than one mutant clone available for mouse production (Fig. 1D).
Figure 1.
OmniBankII gene acquisition and chromosomal distribution. (A) Gene acquisition rate over the course of OmniBankII production, shown as a function of the number of clones processed for sequence acquisition. For comparison, a plot of OmniBank gene acquisition (Zambrowicz et al. 2003) is shown. (B) Frequency of gene trap acquisition across the mouse genome. Frequency is calculated as a percentage of RefSeq genes trapped in each library. (C) Correlation between chromosomal gene content and gene acquisition for OmniBankII. RefSeq genes mapped to the random contig set were excluded from analysis. (D) Density of gene trap mutations per gene for the OmniBankII library.
Novel mutation classes
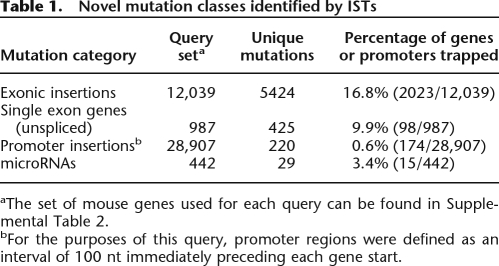
We next evaluated the frequency of three specific mutation classes: exonic insertions, single exon gene insertions, and insertions within gene promoters. To date, these mutation classes have not been effectively harvested from gene trap screens because the previously used transcriptional tagging methodologies are not accurate predictors of the exact location of retroviral integration. We first examined the frequency of exonic insertions within the OmniBankII data set. Using a subset of our gene query table for which we had complete exon position information (see Supplemental Table 2), we queried for insertions that occurred within exons. Our analysis showed that 16.8% of the genes queried have one or more exonic insertion in OmniBankII (Table 1). Next we examined gene trap insertions that occurred within single exon genes. Single exon genes do not undergo splicing; therefore, they should not be particularly amenable to gene trapping screens that select for functional splicing between endogenous exons and the selectable marker within the vector construct. We found that 9.9% of single exon genes were disrupted by direct insertion (Table 1). Finally, we looked for insertions occurring within an arbitrary but small distance (100 bp) immediately preceding each gene’s transcriptional start site (TSS) within the genome in an effort to identify mutations that could disrupt gene transcription by physical disruption of promoter elements. Only 174 genes were found to carry insertions within the 100 bp preceding the TSS (Table 1). Eleven percent of these genes are not otherwise represented in the OmniBankII library. Therefore, although very few genes may carry potentially useful promoter mutations, a small number of additional mutations useful for studying gene function could be identified by expanding the gene coordinates to include gene promoter elements.
Table 1.
Novel mutation classes identified by ISTs
OmniBankII mutations in microRNAs
To identify gene trap insertion mutations that might be useful for defining the function of microRNAs (miRNAs), a class of noncoding RNA genes, we obtained the most recent set of mouse miRNA sequences from the Sanger Center miRNA database miRBASE (release 10.0). This data set (Griffiths-Jones 2006; Griffiths-Jones et al. 2006) contains 442 mouse miRNAs, of which 423 can be mapped to discrete locations within the NCBI mouse genome assembly (build 36). Because the primary transcripts, TSSs, and promoter elements for most miRNAs have not yet been described in sufficient detail for accurate gene trap mutation prediction, we limited our analysis to mutations that directly disrupted the characteristic stem–loop sequence (called a pre-mir) of the miRNA. Comparisons of these pre-mir genome positions with the OmniBankII gene trap insertion site data set revealed 26 gene trap mutations that occur within a total of 12 miRNA genes (Supplemental Table 3). The full set of miRNA sequences was also used to search the OmniBankII gene trap library sequence tag data set directly using BLAST. This search identified three additional gene trap mutations within miRNAs that are not present in genome build 36, but can be found in earlier versions of the mouse genome assemblies. Together these mutations disrupt 15 unique miRNAs, representing 3% of the mouse miRNA genes (Table 1). These 15 miRNA genes show a proportional distribution between genic and intergenic locations, implying that our selection scheme had no particular bias for miRNAs located within larger spliced transcriptional units. However, we found a strong bias for a single miRNA located on mouse chromosome 17, accounting for 39% of all OmniBankII miRNA gene trap clones (Supplemental Table 3). The greater genomic context for this particular miRNA showed frequent trapping in both OmniBank and OmniBankII insertion site data sets, suggesting that Mirn715 (mmu-mir-715) is a frequent target for gene trapping, and may be highly expressed in embryonic stem cells.
OmniBankII insertions show a first intron bias
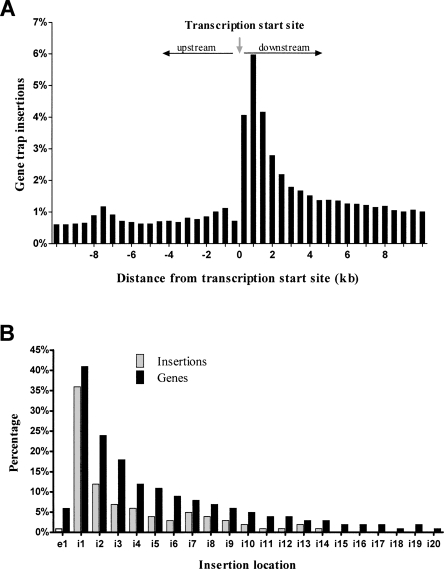
When no selection scheme is used, murine leukemia retroviruses (MLV) exhibit a nondirectional preference for inserting near the TSSs of genes (Wu et al. 2003). This bias toward TSSs is unique to MLV viruses, as opposed to other classes of retroviruses, and is reported to be the most important factor influencing MLV integration site preference (Mitchell et al. 2004). This bias has been reported to be coincident with genomic distance, where insertion density is highest within 1 kb of the TSS (Wu et al. 2003). Under a gene trap selection scheme, insertions within a transcriptional unit are highly favored; therefore, one would expect a directional bias toward the TSS, within the transcriptional unit. Our previous gene trap library showed a strong bias toward the 5′-end of genes, as measured by 3′-RACE tagging (Zambrowicz et al. 2003); however, the exact distribution of insertions with respect to the TSS could not be determined from a transcript-tagged data set. Therefore, we calculated the exact physical distance of each OmniBankII gene trap from its associated TSS and plotted the percentage of insertions found within 500-bp intervals on either side of the TSS as a percentage of total insertions (Fig. 2A). This analysis showed that insertion density was highest near the TSS, and decreased as physical distance from the TSS increased. The overall density was greatest within a 2-kb interval downstream of the gene’s TSS. A similar pattern was observed when plotting insertion frequency with respect to CpG islands (Supplemental Fig. 4). We subsequently plotted the distribution of insertions with respect to intron rather than by physical distance as a secondary measure of TSS bias. This distribution revealed a strong preference for insertions in intron 1, where 36% of all OmniBankII gene traps are located within the first intron of the trapped gene (Fig. 2B).
Figure 2.
OmniBankII gene trap insertions show first intron bias. (A) All insertions occurring within 10 kb upstream or downstream of a TSS were identified and used to plot the percentage of insertions occurring within 500-bp intervals on either side of the TSS. (B) Graph showing the percentage of total genic insertions occurring within exon 1 (e1) or introns 1 through 20 (i1–i20). The percentage was calculated as a function of total genic insertions, or as a function of genes containing insertions in each category.
OmniBankII mutations display insertion site sequence preferences that vary according to the offset of strand cleavage
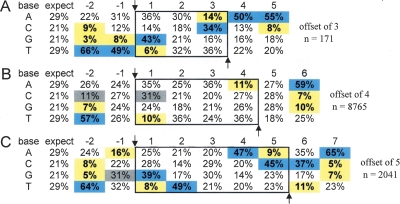
Most regions of the genome can be accessed by retroviral elements; however, retroviral insertion is essentially nonrandom. Target site selection is influenced by many factors including the sequence of the insertion site itself. Although our insertion data set is biased in that we used a selection scheme to enrich for insertions within transcriptional units, we sought to determine whether we could identify a significant consensus sequence for insertion within our large gene trap data set, and if so, whether it was similar to the sequences reported for MuLV target sites in human cells where no selection was used (Holman and Coffin 2005; Wu et al. 2005). Surprisingly, our initial observation revealed that the mutations in OmniBankII showed a variable length of duplicated target sequence at the insertion site, rather than a consistent 4-base duplication as has been described for MuLV retroviruses (Varmus 1983). This prompted us to query our data set to determine the fidelity of the described 4-base duplication. We analyzed 10,977 gene trap insertion sites from which we obtained high-quality sequence from both the upstream and downstream proviral/genomic junction. Sequence overlaps ranged from 3 to 5 bp, with 79.8% displaying a 4-bp duplication, 18.6% displaying a 5-bp duplication, and 1.6% displaying a 3-bp duplication. We hypothesized that the insertion site signature might differ according to the distance of the cleavage offset, as insertion is thought to be governed by spatial and energy requirements for the interaction of the integration complex with the host DNA strand (Wu et al. 2005). Therefore, we calculated the frequencies of A, C, G, and T at each position around the insertion site separately for insertion sites with each of the three different cleavage offsets. The values were compared to the expected nucleotide frequency based on the average frequency of each nucleotide within the mouse genome (Waterston et al. 2002). Significant base preferences were identified near the insertion site for each cleavage class (Fig. 3A–C). The base preferences for insertions showing a 4-bp cleavage offset were consistent with those reported previously. Base preferences for insertions showing a 3-bp or 5-bp cleavage offset, however, showed obvious differences, yet maintained an overall symmetry that appeared to scale according to the cleavage offset (Fig. 3A–C). For all insertion classes, the strongest base preferences were observed 2 nt distal to each cleavage position (position −2 [preference T] and positions 5, 6, or 7 [preference A], respectively).
Figure 3.
Symmetrical sequence preferences at OmniBankII gene trap insertion sites. (A–C) Base compositions surrounding each gene trap insertion site are shown according to chromosomal cleavage offset. Insertion occurs between position −1 and 1 on the top strand (with respect to the orientation of the retroviral backbone rather than the gene trap construct components). Confirmed duplicated junction sequences are shown within the solid box. (Arrows) Base positions for DNA strand transfer. Positions showing statistically different base frequencies are highlighted. (Gray) Differences of 10%; differences of >10% (blue, increased frequency; or yellow, decreased frequency). Additional discussion and an expanded version of this table can be found in the Supplemental material and Supplemental Figure 7.
Qualification of clones for mouse production
Isolation and qualification of C57BL/6N ES cell lines, initial characterization for germline transmissibility, and tissue culture for high-throughput gene trapping were performed using standard methods (described in Supplemental material). We used quantitative PCR to measure the proportions of neo and the Sry gene (located on the Y chromosome) in the ES cell population as a quality control procedure to approve or reject clones for mouse production. A neo gene copy number of 1 indicated that there were not multiple integrations per cell (value > 1) and that the clone was not mixed with wild-type cells (value < 1). An Sry copy number equal to 1 is a useful indicator of the ability of the mutated ES cells to convert female embryos to male and for the ability of all male chimeras to pass the mutation through the germline. Clones with low Sry values (<0.5) typically produce a high percentage of female chimeras. In cases where Sry or neo QPCR values (either or both) were low, we performed clonal isolations and additional G418 selection to generate subclones that carry the Y chromosome and the gene trap construct in a high proportion of the cell population. Likewise, cultures that were determined to be mixtures of two different mutation populations (by IST sequences and IPCR confirmation) were subjected to subculture for isolation of clonal populations of each mutation. To date, all such mixed clones have been successfully separated and confirmed by both IPCR and QPCR. No individual clones bearing more than one insertion per genome have been identified in OmniBankII or the original 129S5/SvEvBrd-derived OmniBank library, although multiple retroviral insertions per ES cell clone have been reported for other library resources (Gragerov et al. 2007).
The rate of loss of the Y chromosome and the neomycin-resistance marker may reflect genomic stability of the ES cells during in vitro culture. We compared post-expansion neo and Sry QPCR results (16–18 passages) for the C57BL/6N clones with results from 129S5/SvEvBrd clones expanded during the same time period. Both clone types were expanded in the absence of antibiotic selection for neo. The number of clones that failed to meet standards for microinjection was compared statistically using χ2 analysis. No significant difference was found for loss of the markers Sry (χ2, P = 0.2990) or neo (χ2, P = 0.0753), suggesting that stability of the Y chromosome and autosomes is comparable during standard expansion in culture (Supplemental Table 4). For C57BL/6N clones, an increased rate of loss of the Y chromosome was observed only after subjecting cultures to 10 additional passages (data not shown).
Chimera production by blastocyst microinjection
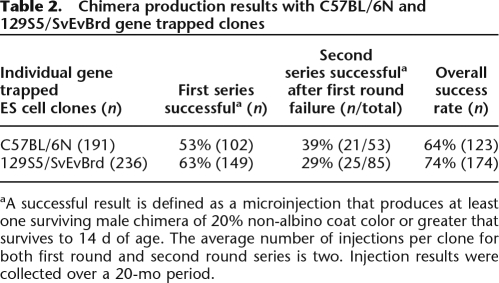
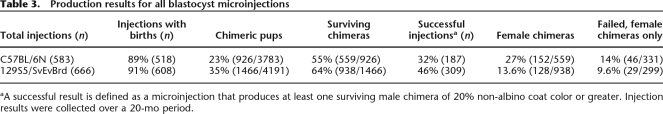
We examined performance of the C57BL/6N clones in our standardized high-throughput knockout mouse production operation (see Supplemental material). Results of chimera production over a 20-mo period are shown in Tables 2 and 3. We compared the success rates in successive injection rounds for individual gene trapped clones of 129S5/SvEvBrd or C57BL/6N origin. Our criterion for injection success was based on our experience with 129S5/SvEvBrd cells and was defined as one that produced at least one male chimera of 20% or greater non-albino fur that survives to 14 d of age. On a per-clone basis, as described in Table 2, the C57BL/6N clones showed a lower overall success rate after two injection rounds, as compared to the 129S5/SvEvBrd clones. The difference is statistically significant (χ2, P = 0.0481). The injection success rate of individual clones of the OmniBankII library (64%) is consistent with the rate observed in our pilot experiment (see Supplemental material). Our overall production statistics for all microinjections performed using gene trapped clones from the C57BL/6N and 129S5 libraries are presented in Table 3. The proportion of black-eyed chimeras born from C57BL/6N ES cell microinjections is significantly lower than that produced from 129S5/SvEvBrd ES cells (P < 0.0001).
Table 2.
Chimera production results with C57BL/6N and 129S5/SvEvBrd gene trapped clones
Table 3.
Production results for all blastocyst microinjections
Other factors used to define success, such as survival of chimeras and extent of chimerism, have a smaller but still detectable effect on the success rate. A recognizable difference between the two types of ES cells, which appears to contribute to the differential success rate for chimera production, is seen in the proportion of female chimera births (Table 3). The relative ability of the C57BL/6N and 129S5/SvEvBrd cells to convert female blastocysts (50% of all blastocyst hosts are expected to be female) to male chimeras is dependent on the extent of contribution of the male ES cells to the chimera and requires the presence of the Sry gene on the Y chromosome. The female chimeras from C57BL/6N cells tend to occur in a greater range of coat color contribution than those from the 129S5/SvEvBrd ES cells (C57, n = 152, median = 35%; 129 n = 128, median = 20%) (see Supplemental Fig. 5).
Chimera breeding for germline transmission
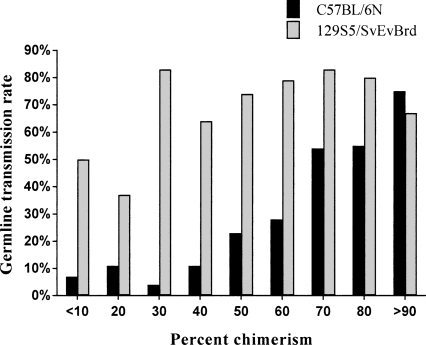
A germline transmission event was scored when any chimera produced progeny with coat color. These progeny were analyzed for transmission of the mutation. For C57BL/6N chimeras, the rate of germline transmission was related to coat color contribution as the estimated contribution increased. As shown in Figure 4, even for C57BL/6N-derived chimeras of 20% or less black coat color, germline transmission was obtained 10% of the time. The C57BL/6N-derived chimeras approached transmission rates of 129S5/SvEvBrd chimeras only at the highest rates of chimerism. Sterile chimeras of both strains were not included in the analysis; the percentage of sterile chimeras was stable and apparently unrelated to the percent chimerism (data not shown).
Figure 4.
Germline transmission rates as a function of percent chimerism. The C57BL/6N derived chimeras approach those of 129S5/SvEvBrd chimeras only at the highest percent contribution.
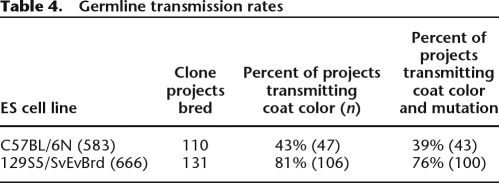
On a clone-by-clone basis, the average germline transmission rate achieved from each of the C57BL/6N clone projects was 43% for coat color transmission, roughly half the rate 129S5/SvEvBrd lines at 81% (Table 4). Looking at the rate of mutation transmission (heterozygotes) versus coat color transmission, there was no apparent difference in loss of the mutation in C57BL/6N as compared to 129S5/SvEvBrd, suggesting autosome stability (χ2, P = 0.4973). Significantly, the coat color transmission rate by C57BL/6N OmniBankII clone projects (43%) approached the result established by the pilot experiment (55%) with no statistically significant difference observed between the two sets of results (χ2, P = 0.1406). Additional chimera production and breeding of these projects will likely increase this overall success rate for OmniBankII.
Table 4.
Germline transmission rates
Analysis of OmniBankII mouse lines
The first OmniBankII gene trap clone from which we obtained homozygous progeny carries a mutation in the solute carrier protein family member Slc25a40. The insertion in Slc25a40 was mapped to intron 2 of this 14-exon gene, within the 5′-untranslated region (Supplemental Fig. 6A). The insertion was flanked by two alternately spliced exons, potentially locating the splice acceptor of the gene trap vector within an intronic context favoring less effective capture of the endogenous transcription by the vector trapping cassette (Jarvik et al. 1996). To determine the efficiency of transcript capture by the splice acceptor of the gene trap vector, we isolated RNA from the spleen and kidney of both wild-type and homozygous mutant animals and subjected it to RT-PCR using primers that flanked both the insertion site and the alternately spliced exons. No endogenous splicing of any of the Slc25a40 splice forms was detected in the homozygous mutant animals (Supplemental Fig. 6B). To verify that the insertion produced a null allele, a second RT-PCR assay directed at the coding region of the gene was performed, showing that the Slc25a40 transcript was, indeed, absent in the mutant. Mutant mice were born in the expected Mendelian ratios and appeared overtly normal in a battery of physiological, metabolic, and behavioral assays (Abuin et al. 2002; Beltrandelrio et al. 2003).
We have since bred 18 OmniBankII mouse lines to homozygosity and have analyzed all nonlethal lines by RT-PCR. Endogenous gene expression was eliminated or drastically reduced in all lines. Unlike the results reported by Gragerov et al. (2007), we did not observe incomplete gene inactivation for insertions located within the 5′-untranslated region of trapped genes (six of 18 lines). This is probably because all available transcript data present in public databases are reviewed to select insertions located downstream from alternate promoters/TSSs.
Discussion
OmniBankII represents the first large-scale use of a C57BL/6 ES cell line in the production of a library of mutagenized ES cell clones for generating knockout mice. Increased interest in deriving new C57 ES cell lines and improving methodology for using existing lines is demonstrated by multiple recent publications (Cheng et al. 2004; Seong et al. 2004; Shimizukawa et al. 2005; Collins et al. 2007; Hughes et al. 2007; Keskintepe et al. 2007; Mishina and Sakimura 2007). Although these reports provide valuable assessments of performance of the C57 lines, ultimately they are limited by the scale of the characterization effort reported, as methodology and definitions of success have not been standardized. Any study including relatively few injection and chimera breeding attempts could present results biased either in favor of, or against, a particular clone or cell line. Because 129 cells are in use worldwide as a standard line for knockout mouse production, they provide an important benchmark for establishing production methods using other lines. Our direct comparison of 191 OmniBankII C57BL/6N clones with OmniBank 129S5/SvEvBrd clones over 20 mo of production in our core facility is the most comprehensive report of performance of mutant C57BL/6 ES cells to date.
A library of insertion site tags has limited value if the cells that carry the mutations cannot be used successfully to generate mutant mice. C57BL/6 ES cell lines have typically been considered to be less robust in cell culture, that is, more sensitive to less than optimal handling (Auerbach et al. 2000; Seong et al. 2004). Our results demonstrate that when cultured with attention to regular media changes and subculturing as required to resist differentiation that is promoted by overcrowded or clumpy growth, OmniBankII clones are of sufficient quality for use in large-scale knockout mouse production. Importantly, we found that blastocyst microinjection success and germline transmission rates of these C57BL/6N clones met expectations established in a small-scale pilot experiment. This is a strong indicator that techniques for tissue culture, quality control, and quality assurance were well maintained during the high-throughput phase of production of the library. Furthermore, the uniformity of this library and the scale of our tests allow us to provide realistic guidance to investigators regarding expectations for achieving germline transmission from any particular mutant cell line obtained from OmniBankII.
For blastocyst microinjection, we did not investigate alternative host strains to find a combination that would favor growth of the injected C57 ES cells, preferring instead to assess performance in our established operation, using C57BL/6J-Tyrc-Brd albino blastocyst recipients. This approach provided a consistent background of technical and environmental effects for direct comparison of the OmniBankII cells to the laboratory standard 129S5/SvEvBrd cells. Our results for germline transmission rates for 110 unique gene trapped clone projects show that even when chimera matings are set up without bias for high coat color contribution, overall, better than one in three clones (39%) will transmit the desired mutation to the F1 generation, a rate about half that of the 129S5/SvEvBrd lines.
As expected, the extent of coat color contribution and the probability of germline transmission are linked for both 129 and C57 clones. The correlation between chimerism and germline transmission for 129S5/SvEvBrd lines is clearly better than 1:1 and is pronounced at percentages of 30% or less. Our definition of success in microinjection for chimera production is based on the observation that, for 129 clones, we routinely obtain germline transmission from ∼50% of chimeras of 20% agouti fur. In contrast, for OmniBankII cells, our germline transmission results suggest that microinjection should be pursued until chimeras of 70% or greater black coat color contribution are obtained. The reduced correlation of chimerism to germline transmission rate for OmniBankII may be an indication of comparative defects of C57BL/6 cell lines. For instance, in vitro characterizations indicate that C57 lines exhibit altered differentiation potential and decreased expression of pluripotency markers as compared to 129 ES cells (Hughes et al. 2007; Sharova et al. 2007). A common approach to improve germline transmission potential is to subclone isolates from a mutant cell line to eliminate aneuploid cells. In cases where microinjection fails to produce high percentage chimeras, it may be advantageous to take these extra steps when alternate gene trap clones are not available. The results may also be a demonstration that 129S5/SvEvBrd-derived cells have a growth advantage in the germline of the C57BL/6J host that is not shared by isogenic cells.
We expect that an aggressive program of microinjection of two mutant ES cell clones (that meet our quality criteria) will nearly always provide sufficient chimeras for successful germline transmission. We have used the same C57BL/6N cell line described here for creation of knockout mice by gene-targeting (data not shown). To date, 36 of 106 targeted clones from 49 homologous recombination projects have transmitted through the germline (34%), an outcome comparable to that of the OmniBankII clones. The overall germline transmission rate, with an average of two clones per targeted knockout project, is 67% (33 of 49 projects to date). Performance expectations for C57BL/6 cell lines, as established for the NIH Knockout Mouse Project, were set at one in four clones per project (25%) giving germline transmission. Our C57BL/6N OmniBankII clones clearly exceed that minimum standard.
Based on the most recent studies of gene number in the mammalian genome, the OmniBankII mutation collection is estimated to contain mutations in roughly one-half of the protein-coding genes in the mouse (Clamp et al. 2007). The trapped gene set shows significant (74%) overlap with genes trapped in our original OmniBank library (Zambrowicz et al. 2003). A notable exception to consistency of gene capture between these two libraries is provided by the Nod1 gene (previously known as Card4). This gene was not captured in OmniBank; however, it was frequently trapped in OmniBankII (0 events vs. 94 events). This is unlikely to be explained by bias or deficiencies in the RACE sequence-tagging methodology used for OmniBank, as >10% of the OmniBank clone set has also been sequenced using IPCR. The exclusivity of Nod1 trapping in OmniBankII is likely due to increased expression of this gene in C57BL/6N ES cells, as suggested by a recent study characterizing differences in gene expression between 129 and C57 ES cell lines (Sharova et al. 2007). The Nod1 gene was identified as a C57 “signature gene,” being one of the most differentially expressed genes between these two genetic backgrounds. Several other differentially expressed genes from that study also show correspondingly differential rates of trapping in our two libraries, for example, Hectd1, Dhrs7, and Ostf1 for C57, and Casc4, Fxyd6, and Necab1 for 129. Nonetheless, the majority of differentially expressed genes show roughly equivalent rates of trapping. Gene expression differences, therefore, play a minor role in differences between OmniBank and OmniBankII. Access to mutations inaccessible through RACE, and improvements in the accuracy of our mutation prediction as a consequence of our switch to genomic-based sequence tagging also contributed to the observed differences in gene acquisition and trap frequency between these libraries.
Our analysis of miRNA capture through gene trapping revealed a small number of mutations in this gene class. It is important to note that we limited our search to only those mutations that directly disrupted the 75–100-nt pre-mir interval that contains the characteristic stem–loop sequence (Kim 2005). Any insertion that disrupts production of the primary transcript concomitantly abrogates production of the miRNA; however, the primary transcription unit for most miRNAs is not yet known. Therefore, it is reasonable to assume that we have underestimated this mutation class.
Retroviral transfer of gene trapping constructs into cells provides a key advantage over other DNA delivery methods, most importantly the consistent generation of discrete insertion events not associated with deletions or rearrangements of host chromosomal DNA. This is advantageous for insertion site tagging and high-throughput gene identification, but also essential for the direct and consistent interpretation of the mutagenicity of a gene trap event. Concomitantly, retrovirus based gene trapping outcomes are constrained by underlying integration site preferences or biases. In the absence of selective screens, MLV retroviruses display a local, nondirectional preference for insertion near CpG islands, gene TSSs, and DNase I-hypersensitive sites (Wu et al. 2003; Mitchell et al. 2004; Lewinski and Bushman 2005; Lewinski et al. 2006). We also observed increased percentages of insertions near CpG islands and TSSs; however, owing to our selection scheme, insertions showed a unidirectional bias where insertions within the transcriptional unit were highly favored. These insertion biases translate into a collection of mutations that are frequently located within the first intron of trapped genes, a particular advantage for effective gene disruption.
In addition to transcription-focused biases, this analysis also revealed variability in the action of the retroviral integrase enzyme that executes the cleavage and strand transfer that produces near-perfect insertion mutation events. The MLV-derived retroviral integrase typically cleaves chromosomal DNA with an offset of 4 bp; however, we observed a range of chromosomal cleavage offsets, each showing a unique but weak insertion site sequence preference. While the insertion site sequence preferences limit the randomness of retroviral integration in this system, the observed flexibility of this enzyme in its interaction with the host chromosome reduces the extent of the overall effect.
In the context of gene trapping, the characteristic biases of retroviral insertion combined with the requirement for sufficient locus expression to allow mutant clone survival through the selection process ultimately limit gene acquisition by favoring particular loci. In an attempt to overcome these limitations, a recent study produced 10 million clones, and potentially 22.7 million retroviral insertion mutations in an unsequenced but screenable format (Gragerov et al. 2007). Based on analysis of capture rate of 403 GPCR and nuclear receptor genes, the mutation collection was estimated to contain 90% coverage of mouse genes, with a mutation density per gene equivalent to that achieved in the present study. In a significant change from standard gene trapping approaches, selective requirements for splicing with endogenous gene exons were removed. Whether this change was necessary to achieve an acquisition rate of 90% is not clear, and a bias toward expressed genes is still in evidence. As demonstrated by the present study, standard splice-acceptor gene trapping approaches can yield insertions in unspliced genes and poorly expressed genes, albeit at a low rate of acquisition. Rather than eliminating the advantage provided by screening schemes that require genic insertion for mutation selection, it might instead be useful to explore changes at the level of the retroviral integrase to increase the efficiency and extent of gene capture. Other classes of retroviral proteins show increased rates of genic insertion in the absence of selection, and hybrid integrase proteins have been created whereby biases of one integrase can be shifted toward another (Lewinski et al. 2006). Modifying the underlying mutation profile of the retroviral-mediated DNA transfer seems an unexplored avenue for increasing acquisition of useful mutations and may ultimately aid in the creation of near-complete gene mutation libraries with individually isolated and sequenced clones available for widespread use.
In conclusion, the OmniBankII resource described here represents a large pool of germline-competent ES cell clones in the novel, desirable C57 genetic background. The genome-based sequence data collected for each mutation allow more accurate prediction of mutagenicity and more straightforward genotyping. OmniBankII clones can be accessed through TIGM, at www.tigm.org.
Methods
ES cell line derivation, cell culture for gene trapping, chimera production, QPCR, and RT-PCR were performed using standard methods and are described in the Supplemental material, along with descriptions of pilot studies for qualification of cell line performance for germline transmission.
Generation of insertion site sequence tags (ISTs)
Genomic DNA was isolated in 96-well format from confluent G418-resistant ES cell clones. Cells were rinsed twice with PBS and frozen at −80°C prior to processing. ES cell clone samples were treated with 50 μL of lysis buffer (50 mM Tris at pH 7.5, 50 mM EDTA at pH 8.0, 100 mM NaCl, 1% SDS, and 3 mg/mL Proteinase K) and were incubated overnight at 65°C. DNA was precipitated and washed with ethanol and resuspended in TE buffer. Templates for inverse PCR (IPCR) were prepared by digesting ∼0.5 μg of each DNA sample in parallel with either BamHI and BglII or HincII and MscI according to the manufacturer’s recommendations (NEB). Digestion reactions were prepared in 384-well format and incubated in an air incubator at 37°C. Completed reactions were heat-inactivated and then ligated in a final volume of 75 μL using 100 U of T4 DNA ligase (NEB). Two rounds of PCR using nested primers complementary to the gene trapping vector (Supplemental Table 1) were used to amplify vector-genomic junction sites. Reactions were prepared with 1 M betaine, 0.2 mM dNTPs, 1.5 mM MgCl2, 1.25 U of Taq polymerase, 2.5 pmol of each primer, and 5 μL of ligation template for round 1 (or 1 μL of first round PCR template for round two). Cycling conditions consisted of 1 min at 94°C followed by 30 cycles of 30 sec at 94°C, 1 min at 60°C, and 2 min at 72°C; followed by a 2-min hold at 72°C, Mastercycler Ep384 (Eppendorf). The number of cycles was increased to 40 for the second round of PCR. The PCR product yield was determined using Picogreen fluorescence. Products were purified using size-exclusion filtration in 384-well MultiScreen PCR purification plates (Millipore). Purified products were cycle-sequenced using a primer (Supplemental Table 1) complementary to the retroviral vector LTR (long terminal repeat). The sequencing primer was extended in length with nontemplate-derived nucleotides to improve sequence resolution within the first 50 nt of the sequence trace. Sequence reactions were prepared with 0.5 μL of BigDye Terminator v1.1 premix (Applied Biosystems), ∼1–10 ng of PCR template, and 1.6 pmol of sequencing primer in a 10-μL total volume. Unincorporated dye terminators were removed from completed reactions using either size-exclusion filtration (Millipore) or the BigDye Xterminator Purification Kit (Applied Biosystems). The Xterminator reactions were maintained at 4°C during the entire 30 min vortex step to prevent irreversible binding of low-molecular-weight products to the resin. Sequence data were collected with a 3730 capillary sequencer (Applied Biosystems). Liquid handling was performed in 96- and 384-well formats using a Sciclone ALH 3000 (Caliper Life Sciences). Disposable polypropylene tips for liquid handling were cleaned for reuse during method runs using a 384-well plasma tipcharger (Cerionx).
Bioinformatics processing and genomic mapping of ISTs
ISTs were defined as IPCR sequences showing a significant, contiguous alignment with the mouse genome. IPCR sequence reads were compared with the NCBI Build 36, UCSC mm8 assembly mouse genome assembly using BLAT (Kent 2002) after masking retroviral LTR sequence derived from the gene trap vector. Alignments shorter than 25 nt or with <90% nucleotide identity were excluded from analysis. The remaining alignments were ranked according to contiguous hit length; the longest alignment beginning coincident with the end of the provirus was selected for annotation. The sequence terminus was truncated at the end of the genome match after merging all adjacent alignments showing a gap of ≤6 nt. The annotation process was then reiterated for all sequences that failed to produce a valid IST, substituting a BLAST analysis (Altschul et al. 1997) for the previous BLAT search (Supplemental Fig. 1). ISTs were then tailored to retain a maximum of 30 nt of LTR sequence (in lowercase) preceding the contiguous genome match identified by BLAT or BLAST. A total of 37,080 IST annotation entries have been reviewed manually to verify the LTR-genomic junction and mapping assignments. Additionally, manual curation was used to annotate IST entries where the IPCR sequence tag produced an alignment with an earlier genome sequence build rather than the current build (NCBI Build 34). Clones with multiple valid ISTs were categorized as “confirmed” if two or more independent IST annotations produced the same chromosomal map position. If disparate map positions were produced, the clone was categorized as “mixed.” Each IST was then assigned an accession that contains four elements: (1) a sample well identifier, (2) a designation for the enzyme combination used for IPCR template preparation, (3) a designation for sequence orientation (F for upstream junctions, R for downstream junctions), and (4) a final number that serves to count the number of uniquely mapped insertions associated with each individual clone. A minimum of two independent IPCR experiments were attempted for each ES cell clone.
Identification of trapped genes
Gene disruption was defined as any gene trap vector insertion event occurring within the exons or introns of a gene. The genome position of each mapped insertion site was compared with the mouse genome map positions of a comprehensive list of genes derived from the NCBI and UCSC gene data tables (Supplemental Table 2). All gene entries have a valid Entrez gene identification number (GeneID) and a defined genome map location reported for NCBI Build 36. To more accurately annotate the full genomic span of each gene, gene start and stop positions were adjusted as necessary from the positions reported by NCBI to accommodate alternate exons and transcription start sites defined by transcripts annotated in the UCSC Build 36 data set. The complete list includes 28,907 entries.
Acknowledgments
We thank L. Johns, C. Poteet, P. Whitted, M. Vega, M. Gonzalez, T. Perry, L. Jump, and W. Jarman for excellent technical assistance and J. Shaw for careful reading of the manuscript.
Footnotes
References
- Abuin A., Holt K.H., Platt K.A., Sands A.T., Zambrowicz B.P., Holt K.H., Platt K.A., Sands A.T., Zambrowicz B.P., Platt K.A., Sands A.T., Zambrowicz B.P., Sands A.T., Zambrowicz B.P., Zambrowicz B.P. Full-speed mammalian genetics: In vivo target validation in the drug discovery process. Trends Biotechnol. 2002;20:36–42. doi: 10.1016/s0167-7799(01)01843-1. [DOI] [PubMed] [Google Scholar]
- Abuin A., Hansen G.M., Zambrowicz B., Hansen G.M., Zambrowicz B., Zambrowicz B. Gene trap mutagenesis. Handb. Exp. Pharmacol. 2007;2007:129–147. doi: 10.1007/978-3-540-35109-2_6. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach W., Dunmore J.H., Fairchild-Huntress V., Fang Q., Auerbach A.B., Huszar D., Joyner A.L., Dunmore J.H., Fairchild-Huntress V., Fang Q., Auerbach A.B., Huszar D., Joyner A.L., Fairchild-Huntress V., Fang Q., Auerbach A.B., Huszar D., Joyner A.L., Fang Q., Auerbach A.B., Huszar D., Joyner A.L., Auerbach A.B., Huszar D., Joyner A.L., Huszar D., Joyner A.L., Joyner A.L. Establishment and chimera analysis of 129/SvEv- and C57BL/6-derived mouse embryonic stem cell lines. Biotechniques. 2000;29:1024–1032. doi: 10.2144/00295st04. [DOI] [PubMed] [Google Scholar]
- Austin C.P., Battey J.F., Bradley A., Bucan M., Capecchi M., Collins F.S., Dove W.F., Duyk G., Dymecki S., Eppig J.T., Battey J.F., Bradley A., Bucan M., Capecchi M., Collins F.S., Dove W.F., Duyk G., Dymecki S., Eppig J.T., Bradley A., Bucan M., Capecchi M., Collins F.S., Dove W.F., Duyk G., Dymecki S., Eppig J.T., Bucan M., Capecchi M., Collins F.S., Dove W.F., Duyk G., Dymecki S., Eppig J.T., Capecchi M., Collins F.S., Dove W.F., Duyk G., Dymecki S., Eppig J.T., Collins F.S., Dove W.F., Duyk G., Dymecki S., Eppig J.T., Dove W.F., Duyk G., Dymecki S., Eppig J.T., Duyk G., Dymecki S., Eppig J.T., Dymecki S., Eppig J.T., Eppig J.T., et al. The knockout mouse project. Nat. Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrandelrio H., Kern F., Lanthorn T., Oravecz T., Piggot J., Powell D., Ramirez-Solis R., Sands A.T., Zambrowicz B., Kern F., Lanthorn T., Oravecz T., Piggot J., Powell D., Ramirez-Solis R., Sands A.T., Zambrowicz B., Lanthorn T., Oravecz T., Piggot J., Powell D., Ramirez-Solis R., Sands A.T., Zambrowicz B., Oravecz T., Piggot J., Powell D., Ramirez-Solis R., Sands A.T., Zambrowicz B., Piggot J., Powell D., Ramirez-Solis R., Sands A.T., Zambrowicz B., Powell D., Ramirez-Solis R., Sands A.T., Zambrowicz B., Ramirez-Solis R., Sands A.T., Zambrowicz B., Sands A.T., Zambrowicz B., Zambrowicz B. Saturation screening of the druggable mammalian genome. In: Carroll P.M., Fitzgerald K., Fitzgerald K., editors. Model organisms in drug discovery. John Wiley & Sons, Ltd.; Chichester, UK: 2003. pp. 251–278. [Google Scholar]
- Brommage R., Desai U., Revelli J.-P., Donoviel D., Fontenot G., DaCosta C., Smith D., Kirkpatrick L.L., Coker K.J., Donoviel M., Desai U., Revelli J.-P., Donoviel D., Fontenot G., DaCosta C., Smith D., Kirkpatrick L.L., Coker K.J., Donoviel M., Revelli J.-P., Donoviel D., Fontenot G., DaCosta C., Smith D., Kirkpatrick L.L., Coker K.J., Donoviel M., Donoviel D., Fontenot G., DaCosta C., Smith D., Kirkpatrick L.L., Coker K.J., Donoviel M., Fontenot G., DaCosta C., Smith D., Kirkpatrick L.L., Coker K.J., Donoviel M., DaCosta C., Smith D., Kirkpatrick L.L., Coker K.J., Donoviel M., Smith D., Kirkpatrick L.L., Coker K.J., Donoviel M., Kirkpatrick L.L., Coker K.J., Donoviel M., Coker K.J., Donoviel M., Donoviel M., et al. High-throughput screening of mouse knockout lines identifies true lean and obese phenotypes. Obesity. 2008 doi: 10.1038/oby.2008.361. in press. [DOI] [PubMed] [Google Scholar]
- Cheng J., Dutra A., Takesono A., Garrett-Beal L., Schwartzberg P.L., Dutra A., Takesono A., Garrett-Beal L., Schwartzberg P.L., Takesono A., Garrett-Beal L., Schwartzberg P.L., Garrett-Beal L., Schwartzberg P.L., Schwartzberg P.L. Improved generation of C57BL/6J mouse embryonic stem cells in a defined serum-free media. Genesis. 2004;39:100–104. doi: 10.1002/gene.20031. [DOI] [PubMed] [Google Scholar]
- Clamp M., Fry B., Kamal M., Xie X., Cuff J., Lin M.F., Kellis M., Lindblad-Toh K., Lander E.S., Fry B., Kamal M., Xie X., Cuff J., Lin M.F., Kellis M., Lindblad-Toh K., Lander E.S., Kamal M., Xie X., Cuff J., Lin M.F., Kellis M., Lindblad-Toh K., Lander E.S., Xie X., Cuff J., Lin M.F., Kellis M., Lindblad-Toh K., Lander E.S., Cuff J., Lin M.F., Kellis M., Lindblad-Toh K., Lander E.S., Lin M.F., Kellis M., Lindblad-Toh K., Lander E.S., Kellis M., Lindblad-Toh K., Lander E.S., Lindblad-Toh K., Lander E.S., Lander E.S. Distinguishing protein-coding and noncoding genes in the human genome. Proc. Natl. Acad. Sci. 2007;104:19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F.S., Finnell R.H., Rossant J., Wurst W., Finnell R.H., Rossant J., Wurst W., Rossant J., Wurst W., Wurst W. A new partner for the International Knockout Mouse Consortium. Cell. 2007;129:235. doi: 10.1016/j.cell.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Desai U., Lee E.C., Chung K., Gao C., Gay J., Key B., Hansen G., Machajewski D., Platt K.A., Sands A.T., Lee E.C., Chung K., Gao C., Gay J., Key B., Hansen G., Machajewski D., Platt K.A., Sands A.T., Chung K., Gao C., Gay J., Key B., Hansen G., Machajewski D., Platt K.A., Sands A.T., Gao C., Gay J., Key B., Hansen G., Machajewski D., Platt K.A., Sands A.T., Gay J., Key B., Hansen G., Machajewski D., Platt K.A., Sands A.T., Key B., Hansen G., Machajewski D., Platt K.A., Sands A.T., Hansen G., Machajewski D., Platt K.A., Sands A.T., Machajewski D., Platt K.A., Sands A.T., Platt K.A., Sands A.T., Sands A.T., et al. Lipid-lowering effects of anti-angiopoietin-like 4 antibody recapitulate the lipid phenotype found in angiopoietin-like 4 knockout mice. Proc. Natl. Acad. Sci. 2007;104:11766–11771. doi: 10.1073/pnas.0705041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gragerov A., Horie K., Pavlova M., Madisen L., Zeng H., Gragerova G., Rhode A., Dolka I., Roth P., Ebbert A., Horie K., Pavlova M., Madisen L., Zeng H., Gragerova G., Rhode A., Dolka I., Roth P., Ebbert A., Pavlova M., Madisen L., Zeng H., Gragerova G., Rhode A., Dolka I., Roth P., Ebbert A., Madisen L., Zeng H., Gragerova G., Rhode A., Dolka I., Roth P., Ebbert A., Zeng H., Gragerova G., Rhode A., Dolka I., Roth P., Ebbert A., Gragerova G., Rhode A., Dolka I., Roth P., Ebbert A., Rhode A., Dolka I., Roth P., Ebbert A., Dolka I., Roth P., Ebbert A., Roth P., Ebbert A., Ebbert A., et al. Large-scale, saturating insertional mutagenesis of the mouse genome. Proc. Natl. Acad. Sci. 2007;104:14406–14411. doi: 10.1073/pnas.0700608104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. miRBase: The microRNA sequence database. Methods Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J., Grocock R.J., van Dongen S., Bateman A., Enright A.J., van Dongen S., Bateman A., Enright A.J., Bateman A., Enright A.J., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman A.G., Coffin J.M., Coffin J.M. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc. Natl. Acad. Sci. 2005;102:6103–6107. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes E.D., Qu Y.Y., Genik S.J., Lyons R.H., Pacheco C.D., Lieberman A.P., Samuelson L.C., Nasonkin I.O., Camper S.A., Van Keuren M.L., Qu Y.Y., Genik S.J., Lyons R.H., Pacheco C.D., Lieberman A.P., Samuelson L.C., Nasonkin I.O., Camper S.A., Van Keuren M.L., Genik S.J., Lyons R.H., Pacheco C.D., Lieberman A.P., Samuelson L.C., Nasonkin I.O., Camper S.A., Van Keuren M.L., Lyons R.H., Pacheco C.D., Lieberman A.P., Samuelson L.C., Nasonkin I.O., Camper S.A., Van Keuren M.L., Pacheco C.D., Lieberman A.P., Samuelson L.C., Nasonkin I.O., Camper S.A., Van Keuren M.L., Lieberman A.P., Samuelson L.C., Nasonkin I.O., Camper S.A., Van Keuren M.L., Samuelson L.C., Nasonkin I.O., Camper S.A., Van Keuren M.L., Nasonkin I.O., Camper S.A., Van Keuren M.L., Camper S.A., Van Keuren M.L., Van Keuren M.L., et al. Genetic variation in C57BL/6 ES cell lines and genetic instability in the Bruce4 C57BL/6 ES cell line. Mamm. Genome. 2007;18:549–558. doi: 10.1007/s00335-007-9054-0. [DOI] [PubMed] [Google Scholar]
- Jarvik J.W., Adler S.A., Telmer C.A., Subramaniam V., Lopez A.J., Adler S.A., Telmer C.A., Subramaniam V., Lopez A.J., Telmer C.A., Subramaniam V., Lopez A.J., Subramaniam V., Lopez A.J., Lopez A.J. CD-tagging: A new approach to gene and protein discovery and analysis. Biotechniques. 1996;20:896–904. doi: 10.2144/96205rr03. [DOI] [PubMed] [Google Scholar]
- Kent W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskintepe L., Norris K., Pacholczyk G., Dederscheck S.M., Eroglu A., Norris K., Pacholczyk G., Dederscheck S.M., Eroglu A., Pacholczyk G., Dederscheck S.M., Eroglu A., Dederscheck S.M., Eroglu A., Eroglu A. Derivation and comparison of C57BL/6 embryonic stem cells to a widely used 129 embryonic stem cell line. Transgenic Res. 2007;16:751–758. doi: 10.1007/s11248-007-9125-8. [DOI] [PubMed] [Google Scholar]
- Kim V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Lewinski M.K., Bushman F.D., Bushman F.D. Retroviral DNA integration–mechanism and consequences. Adv. Genet. 2005;55: 147–181. doi: 10.1016/S0065-2660(05)55005-3. [DOI] [PubMed] [Google Scholar]
- Lewinski M.K., Yamashita M., Emerman M., Ciuffi A., Marshall H., Crawford G., Collins F., Shinn P., Leipzig J., Hannenhalli S., Yamashita M., Emerman M., Ciuffi A., Marshall H., Crawford G., Collins F., Shinn P., Leipzig J., Hannenhalli S., Emerman M., Ciuffi A., Marshall H., Crawford G., Collins F., Shinn P., Leipzig J., Hannenhalli S., Ciuffi A., Marshall H., Crawford G., Collins F., Shinn P., Leipzig J., Hannenhalli S., Marshall H., Crawford G., Collins F., Shinn P., Leipzig J., Hannenhalli S., Crawford G., Collins F., Shinn P., Leipzig J., Hannenhalli S., Collins F., Shinn P., Leipzig J., Hannenhalli S., Shinn P., Leipzig J., Hannenhalli S., Leipzig J., Hannenhalli S., Hannenhalli S., et al. Retroviral DNA integration: Viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina M., Sakimura K., Sakimura K. Conditional gene targeting on the pure C57BL/6 genetic background. Neurosci. Res. 2007;58:105–112. doi: 10.1016/j.neures.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Mitchell R.S., Beitzel B.F., Schroder A.R., Shinn P., Chen H., Berry C.C., Ecker J.R., Bushman F.D., Beitzel B.F., Schroder A.R., Shinn P., Chen H., Berry C.C., Ecker J.R., Bushman F.D., Schroder A.R., Shinn P., Chen H., Berry C.C., Ecker J.R., Bushman F.D., Shinn P., Chen H., Berry C.C., Ecker J.R., Bushman F.D., Chen H., Berry C.C., Ecker J.R., Bushman F.D., Berry C.C., Ecker J.R., Bushman F.D., Ecker J.R., Bushman F.D., Bushman F.D. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:e234. doi: 10.1371/journal.pbio. 0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord A.S., Vranizan K., Tingley W., Zambon A.C., Hanspers K., Fong L.G., Hu Y., Bacchetti P., Ferrin T.E., Babbitt P.C., Vranizan K., Tingley W., Zambon A.C., Hanspers K., Fong L.G., Hu Y., Bacchetti P., Ferrin T.E., Babbitt P.C., Tingley W., Zambon A.C., Hanspers K., Fong L.G., Hu Y., Bacchetti P., Ferrin T.E., Babbitt P.C., Zambon A.C., Hanspers K., Fong L.G., Hu Y., Bacchetti P., Ferrin T.E., Babbitt P.C., Hanspers K., Fong L.G., Hu Y., Bacchetti P., Ferrin T.E., Babbitt P.C., Fong L.G., Hu Y., Bacchetti P., Ferrin T.E., Babbitt P.C., Hu Y., Bacchetti P., Ferrin T.E., Babbitt P.C., Bacchetti P., Ferrin T.E., Babbitt P.C., Ferrin T.E., Babbitt P.C., Babbitt P.C., et al. Modeling insertional mutagenesis using gene length and expression in murine embryonic stem cells. PLoS One. 2007;2:e617. doi: 10.1371/journal.pone.0000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell D.R., Desai U., Sparks M.J., Hansen G., Gay J., Schrick J., Shi Z.Z., Hicks J., Vogel P., Desai U., Sparks M.J., Hansen G., Gay J., Schrick J., Shi Z.Z., Hicks J., Vogel P., Sparks M.J., Hansen G., Gay J., Schrick J., Shi Z.Z., Hicks J., Vogel P., Hansen G., Gay J., Schrick J., Shi Z.Z., Hicks J., Vogel P., Gay J., Schrick J., Shi Z.Z., Hicks J., Vogel P., Schrick J., Shi Z.Z., Hicks J., Vogel P., Shi Z.Z., Hicks J., Vogel P., Hicks J., Vogel P., Vogel P. Rapid development of glomerular injury and renal failure in mice lacking p53R2. Pediatr. Nephrol. 2005;20:432–440. doi: 10.1007/s00467-004-1696-5. [DOI] [PubMed] [Google Scholar]
- Rice D.S., Huang W., Jones H.A., Hansen G., Ye G.L., Xu N., Wilson E.A., Troughton K., Vaddi K., Newton R.C., Huang W., Jones H.A., Hansen G., Ye G.L., Xu N., Wilson E.A., Troughton K., Vaddi K., Newton R.C., Jones H.A., Hansen G., Ye G.L., Xu N., Wilson E.A., Troughton K., Vaddi K., Newton R.C., Hansen G., Ye G.L., Xu N., Wilson E.A., Troughton K., Vaddi K., Newton R.C., Ye G.L., Xu N., Wilson E.A., Troughton K., Vaddi K., Newton R.C., Xu N., Wilson E.A., Troughton K., Vaddi K., Newton R.C., Wilson E.A., Troughton K., Vaddi K., Newton R.C., Troughton K., Vaddi K., Newton R.C., Vaddi K., Newton R.C., Newton R.C., et al. Severe retinal degeneration associated with disruption of semaphorin 4A. Invest. Ophthalmol. Vis. Sci. 2004;45:2767–2777. doi: 10.1167/iovs.04-0020. [DOI] [PubMed] [Google Scholar]
- Seong E., Saunders T.L., Stewart C.L., Burmeister M., Saunders T.L., Stewart C.L., Burmeister M., Stewart C.L., Burmeister M., Burmeister M. To knockout in 129 or in C57BL/6: That is the question. Trends Genet. 2004;20:59–62. doi: 10.1016/j.tig.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Sharova L.V., Sharov A.A., Piao Y., Shaik N., Sullivan T., Stewart C.L., Hogan B.L., Ko M.S., Sharov A.A., Piao Y., Shaik N., Sullivan T., Stewart C.L., Hogan B.L., Ko M.S., Piao Y., Shaik N., Sullivan T., Stewart C.L., Hogan B.L., Ko M.S., Shaik N., Sullivan T., Stewart C.L., Hogan B.L., Ko M.S., Sullivan T., Stewart C.L., Hogan B.L., Ko M.S., Stewart C.L., Hogan B.L., Ko M.S., Hogan B.L., Ko M.S., Ko M.S. Global gene expression profiling reveals similarities and differences among mouse pluripotent stem cells of different origins and strains. Dev. Biol. 2007;307:446–459. doi: 10.1016/j.ydbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizukawa R., Sakata A., Hirose M., Takahashi A., Iseki H., Liu Y., Kunita S., Sugiyama F., Yagami K., Sakata A., Hirose M., Takahashi A., Iseki H., Liu Y., Kunita S., Sugiyama F., Yagami K., Hirose M., Takahashi A., Iseki H., Liu Y., Kunita S., Sugiyama F., Yagami K., Takahashi A., Iseki H., Liu Y., Kunita S., Sugiyama F., Yagami K., Iseki H., Liu Y., Kunita S., Sugiyama F., Yagami K., Liu Y., Kunita S., Sugiyama F., Yagami K., Kunita S., Sugiyama F., Yagami K., Sugiyama F., Yagami K., Yagami K. Establishment of a new embryonic stem cell line derived from C57BL/6 mouse expressing EGFP ubiquitously. Genesis. 2005;42:47–52. doi: 10.1002/gene.20122. [DOI] [PubMed] [Google Scholar]
- Varmus H.E. Retroviruses. Academic Press; New York: 1983. [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wu X., Li Y., Crise B., Burgess S.M., Li Y., Crise B., Burgess S.M., Crise B., Burgess S.M., Burgess S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Wu X., Li Y., Crise B., Burgess S.M., Munroe D.J., Li Y., Crise B., Burgess S.M., Munroe D.J., Crise B., Burgess S.M., Munroe D.J., Burgess S.M., Munroe D.J., Munroe D.J. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J. Virol. 2005;79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz B.P., Friedrich G.A., Buxton E.C., Lilleberg S.L., Person C., Sands A.T., Friedrich G.A., Buxton E.C., Lilleberg S.L., Person C., Sands A.T., Buxton E.C., Lilleberg S.L., Person C., Sands A.T., Lilleberg S.L., Person C., Sands A.T., Person C., Sands A.T., Sands A.T. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature. 1998;392:608–611. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- Zambrowicz B.P., Abuin A., Ramirez-Solis R., Richter L.J., Piggott J., Beltrandelrio H., Buxton E.C., Edwards J., Finch R.A., Friddle C.J., Abuin A., Ramirez-Solis R., Richter L.J., Piggott J., Beltrandelrio H., Buxton E.C., Edwards J., Finch R.A., Friddle C.J., Ramirez-Solis R., Richter L.J., Piggott J., Beltrandelrio H., Buxton E.C., Edwards J., Finch R.A., Friddle C.J., Richter L.J., Piggott J., Beltrandelrio H., Buxton E.C., Edwards J., Finch R.A., Friddle C.J., Piggott J., Beltrandelrio H., Buxton E.C., Edwards J., Finch R.A., Friddle C.J., Beltrandelrio H., Buxton E.C., Edwards J., Finch R.A., Friddle C.J., Buxton E.C., Edwards J., Finch R.A., Friddle C.J., Edwards J., Finch R.A., Friddle C.J., Finch R.A., Friddle C.J., Friddle C.J., et al. Wnk1 kinase deficiency lowers blood pressure in mice: A gene-trap screen to identify potential targets for therapeutic intervention. Proc. Natl. Acad. Sci. 2003;100:14109–14114. doi: 10.1073/pnas.2336103100. [DOI] [PMC free article] [PubMed] [Google Scholar]