A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb (original) (raw)
Abstract
Sonic hedgehog (Shh) signals via Gli transcription factors to direct digit number and identity in the vertebrate limb. We characterized the Gli-dependent _cis_-regulatory network through a combination of whole-genome chromatin immunoprecipitation (ChIP)-on-chip and transcriptional profiling of the developing mouse limb. These analyses identified ∼5000 high-quality Gli3-binding sites, including all known Gli-dependent enhancers. Discrete binding regions exhibit a higher-order clustering, highlighting the complexity of _cis_-regulatory interactions. Further, Gli3 binds inertly to previously identified neural-specific Gli enhancers, demonstrating the accessibility of their _cis_-regulatory elements. Intersection of DNA binding data with gene expression profiles predicted 205 putative limb target genes. A subset of putative _cis_-regulatory regions were analyzed in transgenic embryos, establishing Blimp1 as a direct Gli target and identifying Gli activator signaling in a direct, long-range regulation of the BMP antagonist Gremlin. In contrast, a long-range silencer cassette downstream from Hand2 likely mediates Gli3 repression in the anterior limb. These studies provide the first comprehensive characterization of the transcriptional output of a Shh-patterning process in the mammalian embryo and a framework for elaborating regulatory networks in the developing limb.
Keywords: Sonic hedgehog, limb, morphogen, gli, _cis_-regulatory network
The vertebrate limb is one of the best studied models of how morphogen signaling elaborates a complex pattern (for review, see McGlinn and Tabin 2006). Shh secreted by a discrete posterior organizing center, the zone of polarizing activity (ZPA), is thought to act as a long-range, concentration-dependent signal that regulates both the number and identity of digits that arise from the distal mesenchyme of the developing limb bud. Both the concentration and time of Shh signaling are critical, and growth couples with morphogen activity to give the final digit pattern (Yang et al. 1997; Harfe et al. 2004; Towers et al. 2008; Zhu et al. 2008). Shh actions are mediated through the Gli transcriptional effector family (Gli1-3). Of these, Gli3 appears to play a crucial role in regulating digit number; loss of Gli3 repressor leads to polydactyly and suppresses the loss of digits (2–5) observed in Shh mutants (Litingtung et al. 2002; te Welscher et al. 2002b). Interactions between the limb mesenchyme and the apical ectodermal ridge (AER) are critical for digit development.
The Shh pathway is thought to maintain the limb outgrowth-promoting role of AER produced FGFs through the regulation of a BMP antagonist, Gremlin (Zuniga et al. 1999; Khokha et al. 2003). In turn, AER signaling is essential for maintaining Shh expression (Laufer et al. 1994; Niswander et al. 1994). Genetic analyses have suggested that Shh-mediated loss of Gli3 repressor activity underlies the Shh → Gremlin → AER circuit, but whether this is a direct action of Gli repressor has not been addressed (Litingtung et al. 2002; te Welscher et al. 2002b).
How digits are regulated at the transcriptional level is less clear. Genetic studies have indicated a close interaction between members of the 5′HoxD complex, which have been implicated in the regulation of digit identity and Shh. Initially, 5′HoxD activity is required for the onset of Shh expression (Tarchini et al. 2006). However, as the digit field emerges, HoxD members become targets of Shh regulation. How is not clear, but the identification of a global control region (GCR) outside of the HoxD complex suggests an interaction with this distal regulatory element. Shh also regulates expression of Bmp2 within the distal, digit-forming limb mesenchyme, and experiments modulating BMP levels in the chick have suggested that Bmp2 may act as a secondary relay to regulate digit identity (Dahn and Fallon 2000; Drossopoulou et al. 2000). However, genetic studies in the mouse have not supported this view (Bandyopadhyay et al. 2006); consequently, the precise roles of BMPs are contentious.
We attempted to understand the roles of Shh signaling by identifying the targets of Gli3 action in the developing mouse limb. These studies provide a framework for the primary regulatory networks downstream from Shh signaling and identify new links between Shh and other signaling pathways in driving limb outgrowth. In addition to identifying targets of Gli repression regulating critical outputs in the developmental program, our work demonstrates that the Gli activator forms play a critical role in the limb patterning circuitry.
Results
In a previous study, we developed an approach to conditionally produce a Flag-tagged form of a Gli activator and examined Hedgehog (Hh) action in patterning neural progenitors in vitro (Vokes et al. 2007). Here, we introduced a cDNA encoding a Flag-epitope-tagged Gli3 repressor (Yuen et al. 2006) into the ubiquitous Rosa26 promoter to enable Cre-dependent conditional expression in Hh target regions in vivo (see the Materials and Methods; Supplemetnal Fig. S1A–C). When RosaGli3TFlag c/c mice were crossed to mice carrying the early limb mesenchyme-specific Prrx1∷Cre transgene (Logan et al. 2002), they produce Gli3TFlag at levels that are comparable with the endogenous protein (Supplemental Fig. S1C). Mice exhibited a variety of limb defects including a variable preaxial forelimb polydactyly, limb truncation, and reduced mineralization (Supplemental Fig. S1D). These phenotypes suggest that Gli3TFlag is active and modifies both Shh action in initial limb patterning and later Indian hedgehog action in growth and differentiation of the endochondral skeleton.
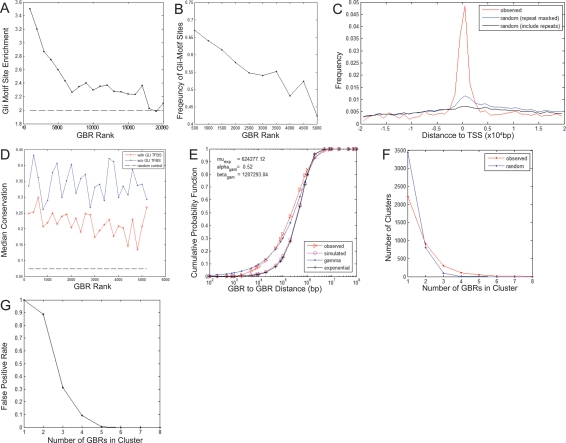
To focus our analysis on the period of Shh-mediated regulation of distal digit organization, we optimized a chromatin immunoprecipitation (ChIP) protocol to enable the direct identification of Gli3 targets in limb buds at embryonic day 11.5 (E11.5). A high level of enrichment of Gli target genes was obtained using ∼2.3 × 106 cells per ChIP. Three technical replicates comprising all limbs from a single litter (approximately nine embryos) provided sufficient sample for the interrogation of multiple tiling arrays required for a whole-genome hybridization. Data was processed using CisGenome software (http://www.biostat.jhsph.edu/~hji/cisgenome; H. Ji, H. Jiang, W. Ma, D.S. Johnson, R.M. Myers, and W.H. Wong, in prep.), identifying 20,587 potential Gli-binding regions (GBRs) with an estimated false discovery rate (FDR) of 9.8% (Supplemental Table S1). The GBRs were binned according to rank and the overall Gli enrichment level was obtained for each bin. Bins containing a Gli enrichment level of ≥2 were selected, reducing the list to the top 19,732 scoring regions (FDR < 5%) (Fig. 1A). A selected subset of the 19,732 regions were validated by qPCR. Within the top ranked 5274 GBRs qPCR validated Gli3-dependent enrichment was confirmed in >50%; 11of 17 shared an enrichment >4.39-fold (>three standard deviations from controls) (Supplemental Fig. S2A). This group of 5274 regions displayed a mean ChIP-binding region of 854 base pairs (bp) (Supplemental Fig. S2B) and a low FDR (FDR < 0.1%, SD ≥ 6). All subsequent computational and experimental analyses focused on this highly biologically significant data set of GBRs with high levels of enrichment (Supplemental Data Set 1) although regions identified in the remaining data set are nonetheless statistically significant (Fig. 1A; Supplemental Table S1).
Figure 1.
Gli3-binding region summary statistics. (A) Gli motif enrichment versus matched genomic controls indicates that all Gli3-binding sites are enriched for Gli motifs throughout all ranked regions. Note highest enrichment in the top 5000 regions, which are used for all subsequent anaysis. (B) The frequency of Gli motif sites in the 5274 binding regions is highest in the top ranking binding regions. (C) These regions are locally enriched around the TSS. (D) Binding regions that do not contain Gli motifs (blue) are significantly more conserved that are binding regions that contain Gli motifs (red). Conservation is measured by percentage of base pairs that have a phastCons score no less than the top 10% of the genome-wide scores. (E) Gli-containing binding regions are located significantly closer to each other than a random distribution; this peak–peak clustering occurs at the level of 100 kb. (F,G) The total number of binding regions (with and without Gli motifs) also exhibit higher-order clustering at 100 kb.
Genomic topography and binding site clustering
Analysis of ChIP products discovered a substantial enrichment in Gli motif sites within GBRs consistent with our expectations (Vokes et al. 2007). In all, 55% of these regions contain a high-quality Gli motif. Those GBRs exhibiting the highest levels of fold enrichment also contain the highest percentage of Gli motif enrichment (Fig. 1B). When compared with matched control genomic regions, we observed a significant enrichment of GBRs within ±2 kb of the transcriptional start site (TSS) (Fig. 1C). However, relatively few (594; 11%) of the GBRs were present within this proximal regulatory region, suggesting long-range interactions between promoter and distal regulatory sites (see later for functional evidence). GBRs were not significantly enriched within any other genomic feature (e.g., intergenic regions, intragenic regions, exons, introns, or untranslated regions) (Supplemental Table S2).
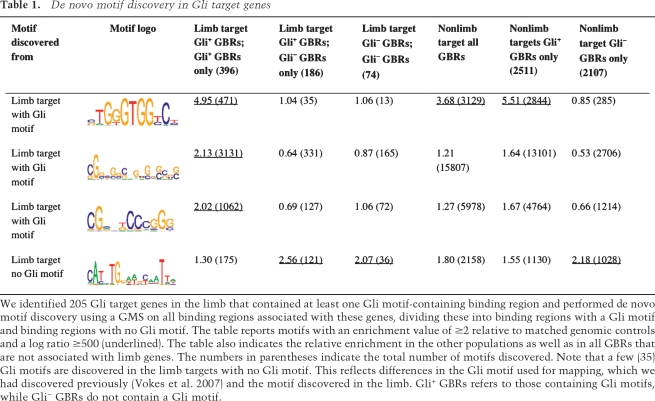
The high percentage of GBRs (45%) that did not contain a Gli-binding motif may reflect an indirect mechanism (e.g., Gli3-dependent protein–protein interactions with another DNA-binding protein or a nonphysiological DNA association). When the overall rate of sequence conservation was compared, GBRs without Gli motifs were, in fact, somewhat more conserved (Fig. 1D). Thus, as a population, these are unlikely to represent spurious binding sites. When compared with the entire data set, ChIP products containing Gli motifs were more significantly enriched around a TSS (16.3% of all binding regions) than those without Gli motifs (5.1% vs. 3.9% for controls) (Supplemental Table S2). A very small number of the binding regions without mapped Gli motifs do, in fact, contain a limb-specific variant of the Gli motif (Table 1), and a further subset of these binding sites could contain low-quality Gli motifs that were below the likelihood ratio cutoff of 500.
Table 1.
De novo motif discovery in Gli target genes
To determine whether GBRs containing Gli motifs cluster, we computed the pairwise distance between directly neighboring binding regions and compared this with randomly chosen Gli motifs from the genome (Ji et al. 2006). This analysis demonstrates that GBRs are not randomly distributed but cluster at the 100-kb scale (Fig. 1E). We next examined the clustering tendency of all ChIP-binding regions irrespective of a Gli motif and also detected a significant clustering of all peaks at the level of 100 kb (Fig. 1F,G). Clustering was even evident in 1–2-Mb windows (Supplemental Fig. S3); within a 1-Mb window, we observe significant clusters of seven or more binding regions accounting for 1917 Gli-binding sites (Supplemental Fig. S3C,D) that represent 36.35% of all Gli sites. Thus, long-range clustering of Gli3-binding sites is a significant overall property. However, we did not observe any correlation in the clustering between Gli motif-positive and -negative GBRs (see the Supplemental Material).
Gli3 recognizes homologous and heterologous enhancers
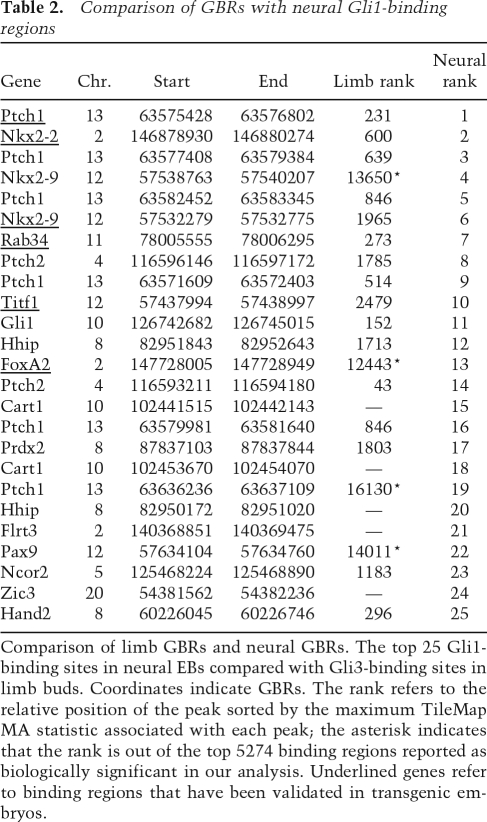
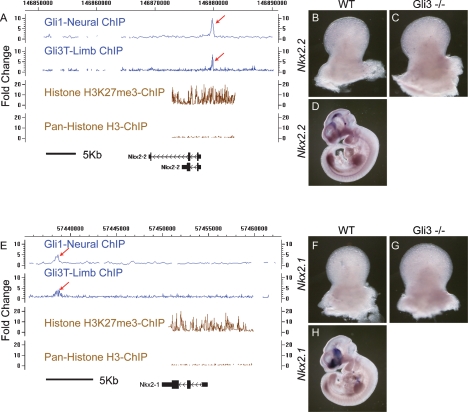
A previous analysis of Shh action on neural progenitors identified 25 biologically significant Gli1-binding sites (Vokes et al. 2007). Among these are neural specific Shh target genes (FoxA2, Nkx2-2, Nkx2-9, and Titf1/Nkx2.1) and more general targets of the pathway (Ptch1, Ptch2, Hhip, Rab34, and Gli1) that are also expressed in the limb. Gli3 bound to each of the general targets in the limb (with multiple inputs confirmed for Ptch1) to similar regions observed by Gli1 in neural progenitors (Table 2). In all, the set of 5274 GBRs included 16/25 neural Gli1-binding regions (four more were in the 19,732 GBR data set) with the surprising inclusion of neural-specific Gli1-binding regions. Thus, _cis_-binding regions mediating neural-specific regulation of a Gli program are accessible to Gli factors in the limb. However, the binding of Gli3 to the above characterized neural-specific Gli enhancers appears to be nonfunctional (Table 2; Fig. 2A,E; Supplemental Fig. S2A). FoxA2, Nkx2.1, and Nkx2.2 are not expressed in limbs of wild-type or Gli3 mutant embryos by in situ hybridization or transcriptional profiling analysis, arguing against a requirement for a direct Gli3-bound transcriptional silencing (Fig. 2B–D, F–H; data not shown). The analysis of proximal regulatory regions of these genes indicated high levels of histone H3 trimethylation (H3K27me3), indicative of global silencing (Fig. 2A,E; Lee et al. 2006). Thus, Gli3 is able to recognize and bind enhancer elements previously silenced by other mechanisms.
Table 2.
Comparison of GBRs with neural Gli1-binding regions
Figure 2.
Gli3 binds to heterologous neural Gli enhancers associated with silenced genes. (A) CisGenome visualization of neural Gli1 enrichment for Nkx2-2 using Agilent arrays reported in Vokes et al. (2007) shows that the binding region overlaps with that for Gli3 binding in the limb (red arrows). The transcript is enriched for Histone H3K27me3. (B,C) Nkx2.2 transcripts are not detectable by in situ hybridization in either wild-type or Gli3 mutant limb buds at E11.5. (D) Embryos processed in parallel show appropriate neural expression at E10.5. (E) Similar results are seen for Nkx2.1 ChIPs and these transcripts are also not detectable by in situ hybridization in wild-type or Gli3 mutant limb buds (shown in F,G). (H) Embryos processed in parallel show appropriate neural expression at E10.5. An artifact of the transmitted light generates a shadow in D and H. No signal is observed in dissected limb samples.
Identification of a core set of Shh-responsive limb gli target genes
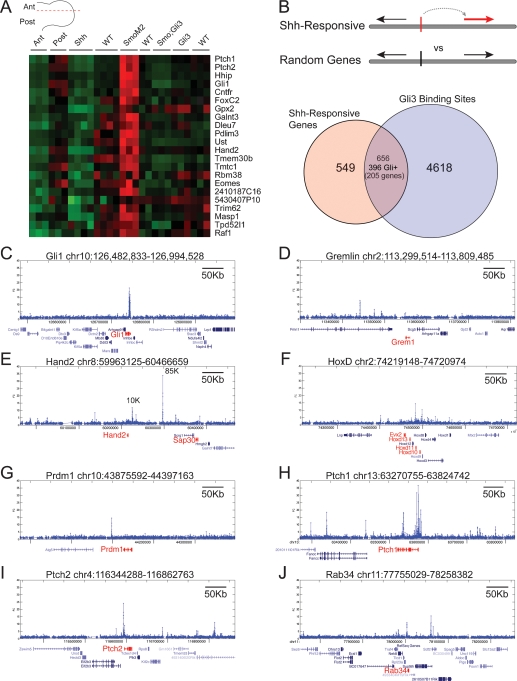
In order to define a core set of Shh-responsive direct target genes in the limb, we associated Gli3-binding sites with Shh-dependent transcriptional responses. Using exon arrays, we compared wild-type E11.5 forelimbs with forelimbs from Shh-null (constitutive Gli repressor), MhoxCre;R26SmoM2 (maximal Gli activation) (Jeong et al. 2004), Gli3-null (loss of major repressor), and Smo;Gli3 double mutant (loss of Gli activator and Gli repressor) backgrounds. In addition, we profiled E11.5 forelimbs microdissected into an anterior 1/3 compartment (Ant) or posterior 2/3-d compartment (Post); Gli1 expression in the posterior sample defines the active Shh signaling domain (Fig. 3A; Lewis et al. 2001). These lists were used to identify 753 genes whose expression profiles resembled known Shh-responsive genes and genes exhibiting pairwise changes in mutant scenarios or in Ant/Post fractions (Fig. 3B; Supplemental Data Set 2).
Figure 3.
(A) A range of Shh pathway genotypes were assayed for gene expression at E11.5, as well as limb buds dissected into anterior (Ant) and posterior (Post) compartments to define Hh-nonresponsive and Hh-responsive compartments, respectively. Gene expression indices derived from exon arrays were used in several pairwise combinations as well as multivariable combinations. The criteria shown in the heat map are Ant < Post and Shh < wild type < SmoM2 and Gli3 > SmoGli3 with an FDR < 10% and posterior probability cutoff of <25%. (B) By associating 753 genes differentially expressed in these conditions with GBRs (FDR < 50%), we are able to assign binding regions putative target genes. We obtain 656 binding regions, including 396 that contain Gli motifs; these are associated with 205 genes. (C–J) Examples of binding regions (∼500 kb) and the associated differentially expressed genes. The coordinates (mm8) are shown above each graph, while Ref_Seq transcripts are shown below. Genes and transcripts in red indicate differentially expressed genes.
To determine if a meaningful association would be made between GBRs and differentially expressed genes, we counted the number of GBRs associated with Shh-responsive genes and compared it with binding sites associated with randomly selected genes to define an FDR. Significantly more binding regions were associated with Shh-responsive genes (Supplemental Fig. S4A). Those GBRs located within a region 10 kb upstream of and 25 kb downstream from the TSS of differentially regulated genes showed a FDR of 50%. As an independent measure of the significance of binding regions associated with differentially expressed genes, we examined their conservation levels. GBRs associated with differentially expressed genes are significantly more conserved out to a distance of >1 Mb, albeit with a very high FDR (Supplemental Fig. S4B). Thus, our initial strategy likely underestimates the total number of associated GBRs. To further optimize this association, we noted that genes associated with development and transcription are enriched within gene deserts that are devoid of coding regions (Ovcharenko et al. 2005). Consistent with this, Gli3 peaks associated with differentially expressed genes have fewer intervening TSSs than randomly chosen genes (Supplemental Fig. S4C). We therefore incorporated information about the gene density into the final FDR determination (Supplemental Fig. S4D). We supplemented this list by searching for clustered binding regions and intersected these with differentially expressed genes (see the Materials and Methods).
The pooled data identified 656 GBR–gene pairs, involving 656 GBRs and 261 genes. We further restricted our criteria for direct target genes to those associated with the binding regions containing Gli motifs (Fig. 3B). These 396 binding regions correspond to 205 unique genes (Supplemental Data Set 3), representing the core set of candidates under direct Gli transcriptional control in the developing limb. The Gli target genes include known Hh pathway coponents such as Ptch1, Ptch2, Hhip, and Gli1, several of which are known to undergo Shh-dependent transcriptional feedback regulation. A surprising number of putative targets are themselves transcription factors; 48 of the 205 unique genes are associated with DNA-dependent regulation of transcription. These include multiple members of the HoxD family, and cofactors such as Pbx, Meis1, and Meis2 suggestive of extensive regulation of Hox transcriptional complexes. Several interactions are observed with Tbx transcription factors (Tbx2, Tbx3, and Tbx4). Interestingly, Tbx2 and Tbx3 are thought to play upstream roles in the activation of Shh (Nissim et al. 2007). The data suggest a reciprocal regulatory mechanism. Putative targets are highly enriched for GO categories involved in development and morphogenesis (Supplemental Fig. S5). Many of these genes, including Gremlin and Hand2 (Fig. 4C,J) and other predicted targets, exhibit limb expression patterns that are broader or only partially overlapping with the Shh-responsive region determined by Ptch1 and Gli1 expression. Thus, the observed gene expression patterns may reflect the integration of multiple regulatory inputs. That cross-regulatory interactions may engage other signaling pathways is evident in the significant enrichment for genes associated with the TGF-β family (_BMP_s), and Wnt pathway activity in addition to the Shh pathway components (Supplemental Table S5).
Figure 4.
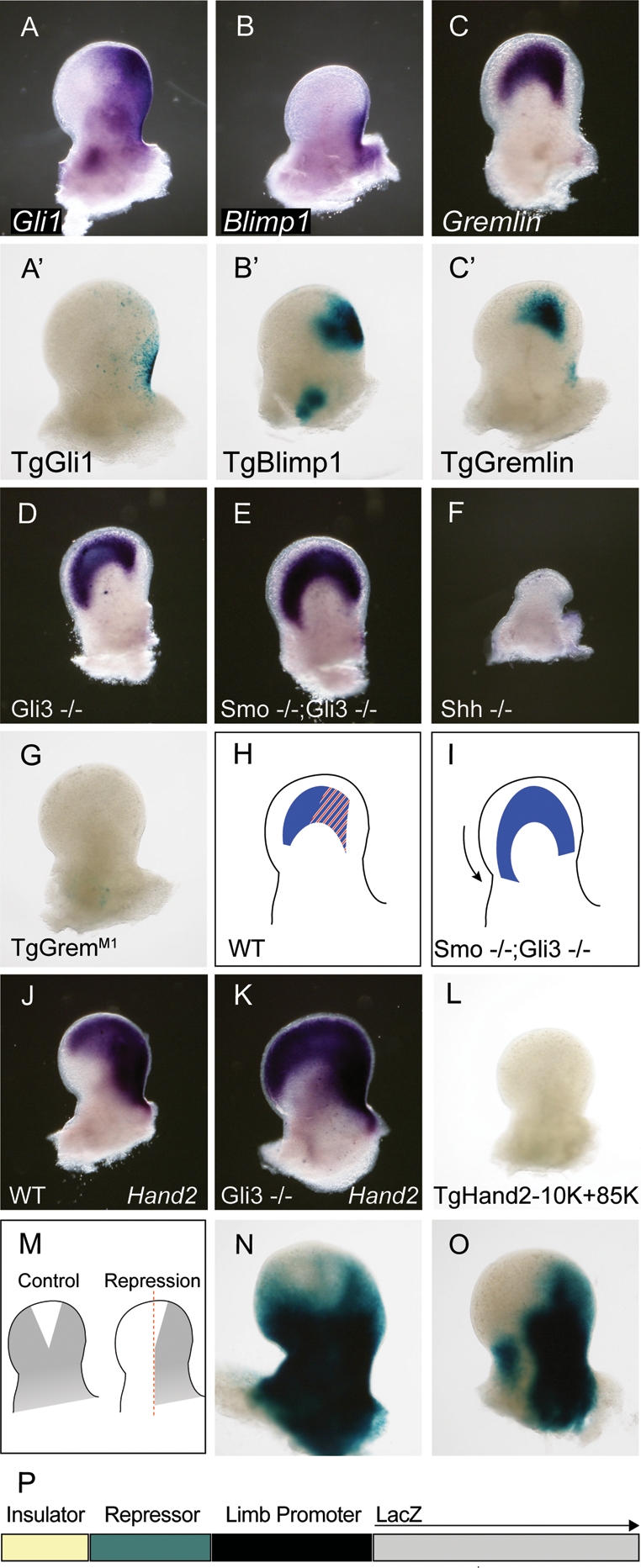
(A–C) In situ hybridization of E11.5 limb buds indicating gene expression patterns for Gli1, Blimp1, and Gremlin. (A′–C′) G0 transgenic analysis of putative Gli enhancer domains predicted on the basis of gene expression and binding, and stained for β-galactosidase activity. (D–F) In situ hybridization of Gremlin in Gli3 mutants, Smo;Gli3 double mutants and Shh mutants, respectively (cf. wild-type expression in C). (G) Mutation of the single Gli site in the construct in _C_′ results in a loss of all β-galactosidase activity. (H,I) The requirement for Gli-activity in enhancer expression contrasts with the persistence of Gremlin gene activity in Smo−/−;Gli3−/− mutants in E. This suggests that wild-type Gremlin gene expression is a composite of a Gli activator-responsive enhancer (red) and a second enhancer (blue) that does not require Gli activation but is nonetheless repressed by Gli3. (J,K) Hand2 expression is expanded anteriorly in Gli3 mutants (cf. J and K), but Gli3-binding sites in Hand2 do not exhibit enhancer activity (O). (M–O) To test these elements for silencing activity, we generated a control transgenic driving LacZ expression equally in the anterior and posterior domains (P) and compared this expression with transgenics in which both GBRs were placed upstream. Embryos containing Hand2 GBRs exhibit a loss of reporter activity in the anterior limb (cf. N and O).
Discovery of DNA motifs enriched in GBRs
To determine whether any additional transcription factor-binding sites were enriched in Gli-containing binding regions and to identify sites enriched in non-Gli-containing regions, we performed de novo motif analysis using a Gibbs motif sampler (GMS) as described previously (Ji et al. 2006; Vokes et al. 2007). In addition to a Gli motif, we recovered two G/C-rich motifs (Table 1); a similar sequence has been observed in other large-scale ChIP analyses (Ji et al. 2006). We also identified a composite motif specifically enriched in Gli-binding sites that do not contain Gli motifs. This motif contained an E-box indicative of basix helix–loop–helix (bHLH) homeodomain transcription factor binding fused to an ATTA motif, the core binding site for homeodomain transcription factors (FDR = 39%). However, as this motif was enriched within the larger subset of GBRs not associated with Shh modulated gene expression (Table 1), the significance to Gli3 regulation in the limbs is not clear. We also mapped all TRANSFAC human and mouse motifs to GBRs relative to matched genomic controls. Using this criteria, we found variants of an E-box enriched in limb GBRs with or without Gli motifs. Further, Mef2 and Chx10 motifs were specifically enriched in limb peaks devoid of Gli motifs (Supplemental Table S3). The core TAAT sequences in the Chx10 homeodomain factor is consistent with the composite E-box + Homeobox motif discussed above.
Functional characterization of Gli _cis_-regulatory elements
In order to explore the contributions of GBRs, we selected those associated with a number of genes that display distinct expression within the limb for functional transgenic analysis. Gli1 is a transcriptional activator and a global target of Gli activator response in all known Hh target fields (Bai et al. 2004). An unusually broad, bimodal GBR (3.7 kb) is observed in Gli1 intronic sequences that span the first noncoding exon (Fig. 3C). Six Gli motifs are identified within this domain; multiple sites have been noted in previous gel shift assays (Dai et al. 1999). A 3.4-kb fragment encompassing this region-directed β-galactosidase activity to the ventral CNS, consistent with our earlier report of Gli1 binding to this region in neural progenitors (Table 2) and a subregion of the Gli1 expression domain in the posterior limb (seven of eight embryos) (Fig. 4A, A′). The GBR contained two E-box motifs; when these were mutated, a lower percentage of transgenics exhibited posterior limb expression (one of four embryos). However, the expression domain was identical to the unmodified version (data not shown). Thus, these motifs are not essential for directing Gli1 expression to its normal limb domains.
Blimp1 (Prdm1) encodes a DNA-binding protein that recruits chromatin modifiers and plays a central role in the regulation of several stem/progenitor cell populations (Ohinata et al. 2005; Horsley et al. 2006). Blimp1 is expressed in the ZPA where its activity is required to maintain the ZPA and, consequently, normal Shh production (Robertson et al. 2007). A single GBR containing two Gli motifs was identified ∼27 kb downstream from the Blimp1 transcriptional unit (Fig. 3G). A 996-bp region spanning the binding region directed LacZ expression to the posterior limb in transgenic embryos, overlapping the normal Blimp1 domain (eight of 17 transgenics) (Fig. 4B, B′). Interestingly, the transgenic expression domain extends anterior to the domain of endogenous Blimp1 expression, mirroring the demonstrated anterior movement of ZPA cells (Harfe et al. 2004). Thus, the enhancer may lack sequences that normally repress Blimp1 outside of the ZPA. Alternatively, the perdurance of β-galactosidase activity may continue to label cells moving from the ZPA.
Gremlin encodes a BMP antagonist that is postulated to act downstream from Shh to maintain the AER. Consequently, Gremlin mutants lose Shh expression as the AER to ZPA feedback loop is broken and fail to develop distal limb structures (Zuniga et al. 1999; Khokha et al. 2003). In contrast to Blimp1, Gremlin expression is excluded from the ZPA but extends throughout most of the distal limb mesenchyme (Fig. 4C) (Zuniga et al. 1999; Scherz et al. 2004). We identified four GBRs (Fig. 3D) in a ∼70-kb region ∼40–110 kb downstream from Gremlin that was previously shown to drive reporter gene expression to the Gremlin limb domain (Zuniga et al. 2004). A 438-bp fragment representing the major binding site replicated the posterior Gremlin domain observed with a 70-kb fragment in the earlier study (eight of 12 transgenic embryos) (Figs. 4C, C′). As with the 70-kb element, expression is excluded from the anterior limb mesenchyme where Gremlin is normally present. Thus, distinct modes of regulation appear to govern Gremlin expression in different regions of the limb mesenchyme (see the Discussion).
Comparison of _Shh_−/− and _Shh_−/−; _Gli3_−/− compound mutants has suggested that Shh regulates Gremlin through the loss of Gli3 repressor rather than by a Gli activator function (Fig. 4D–F; Litingtung et al. 2002; te Welscher et al. 2002b). To determine if transgenic expression was Gli-dependent, we generated two independent mutations in this site (GremM1 and GremM2) and in one of these (GremM1) also nine additional mutations altering the sequence of even low-probability Gli sites (GremM3). Surprisingly, no appropriate reporter expression was observed in any of these regions (total of zero of 36 transgenics; Fig. 4G; data not shown). This data suggests that a direct Gli activator input governs Gremlin’s Shh-dependent limb expression and that the restoration of Gremlin expression in _Shh_−/−; _Gli3_−/− mutants reflects an indirect role of the removal of Gli3 repressor activity (see the Discussion).
Hand2 encodes a transcriptional regulator that acts genetically upstream of Shh to positively regulate Shh expression. In turn, Hand2 is restricted posteriorly by Gli3-mediated repression (Fig. 4J,K; Charité et al. 2000; te Welscher et al. 2002a). Thus, Shh-mediated derepression of Gli3 activity appears to govern the limb mesenchyme domain of Hand2. Two major GBRs were identified in phylogenetically conserved regions ∼10 kb and ∼85 kb downstream from the Hand2 transcript unit (Hand2-10K and Hand2-85K, respectively; Figure 3E). Neither a 433-bp region encompassing Hand2-10K or a composite of the 10K (433 bp) and 85K (850 bp) GBRs (Hand2-10K + 85K) displayed limb expression (zero of five embryos [data not shown] and zero of four embryos [Fig. 4L], respectively). The assayed regions therefore lack enhancer activity and either act as silencers or have no discernible function.
To address the former possibility, the 10K + 85K binding regions were attached to a Prrx1 regulatory cassette; this contains a strong limb enhancer that drives broad mesenchymal expression of transgenes in the limb (see schematic Fig. 4M; Martin and Olson 2000). While Prrx1∷lacZ expression is somewhat variable (nine of 14 transgenics exhibit limb expression), all embryos with limb expression showed approximately similar levels of β-galactosidase activity in both the anterior and posterior limb mesenchyme (Fig. 4N). In contrast, when the 10K + 85K binding regions were placed upstream of the Prrx1 LacZ cassette, transgene activity was markedly reduced specifically in the anterior mesenchyme of the limb (six of nine transgenics with limb expression) (Fig. 4O). This suggests that these GBRs may act as silencer elements in Gli3-mediated repression of Hand2 that result in appropriate restriction to the posterior half of the limb.
_cis_-regulatory mechanisms of Shh-dependent AP patterning in the limb bud
The 656 GBRs associated with 205 genes showing Shh/Gli-regulated expression changes provide a foundation for dissecting the Gli _cis_-regulatory circuitry in limb development. The Gli1, Blimp1, Gremlin, and Hand2 analyses functionally validate the data set. However, a full validation through transgenic experiments is not feasible. To shed further light on novel regulatory interactions, we examined a subset of genes that exhibit the most pronounced asymmetry in their anterior versus posterior expression through computational analysis of the gene expression data (see Supplemental Table S6). Whereas only 14% of randomly chosen genes are associated with GBRs, the posterior gene set contains 35 genes, 23 of which are closely associated with GBRs (Supplemental Table S6). The core Shh pathway components Gli1, Ptch1 and Ptch2 are all present. As these require Gli activator input, these data likely represent Gli activator targets where Shh signaling levels are highest and provide further evidence that Gli repressor and activator forms recognize common targets. In addition to Hand2 discussed above, the posterior set includes a large number of transcription factors with known roles in limb development: Hand2 (discussed above), Tbx2 (early role in initiating Shh expression) (Nissim et al. 2007), Hoxd13 (skeletal patterning) (Zakany and Duboule 1996), Sall1 (limb abnormality mirroring that seen in Townes-Brocks syndrome (Kiefer et al. 2003)), and Tbx4 (hindlimb outgrowth (Naiche and Papaioannou 2007). Further, the identification of BMP (Bmp2) and Notch (Dlk1) pathway components may point to cross-regulatory interactions in signaling pathways.
In contrast, only eight of the anterior gene set (31 genes) are associated with GBRs (Supplemental Table S6). The lower number is consistent with a largely repressive role of Gli3 in the anterior limb bud. Remarkably, of the 11 transcription factors in this set, six of them associated with GBRs: _Pax_1, Alx4, Pax9, Zic3, Irx3, and Dlx5. Alx4 mutants exhibit polydactyly (Qu et al. 1997), and Dlx5 is a candidate gene for split-hand/split-foot malformation (Robledo et al. 2002). Further, Alx4 and Pax9 expression is Gli3-dependent—both are down-regulated on loss of Gli3 activity (te Welscher et al. 2002a; McGlinn et al. 2005). This suggests that in certain cellular contexts, Gli3 binding to a regulatory region may indirectly promote anterior expression, perhaps by blocking the action of some other repressive function. However, analysis of one of these genes, Alx4, has identified a regulatory region that recapitulate Gli3-dependent expression and this region shows no GBRs in our data set (Kuijper et al. 2005).
Discussion
In this study we attempted to define the complete set of in vivo binding sites for a mammalian transcription factor in the context of a specific developmental process, Shh-mediated patterning of the limb. The methodology is sensitive requiring relatively low numbers of cells (∼2.3 × 106 cells per ChIP) and the general strategy of a genetically inducible epitope-tagged transcription factor is broadly applicable to other Hh-mediated regulatory events and other transcriptional networks. We detect ∼20,000 GBRs, focusing our analysis on a subset of ∼5000 GBRs that are greater than six standard deviations from the mean. The substantial enrichment of Gli motifs within this population gives credence to the view that these are most likely to include biologically significant regulatory regions. While it is likely that some sites are missed, for example, where Gli3 binding occurs within a subpopulation of limb mesenchyme below the threshold of detection, the numbers of binding sites are consistent with genome scale studies in mammalian cell culture (Carroll et al. 2006; Yang et al. 2006; Johnson et al. 2007).
Currently this represents the only large-scale data set for GBRs. The data incorporate a more limited set of 25 Gli1-binding regions identified in Shh patterning of neural tissue (Table 2), indicating that Gli activator and repressor forms have similar binding specificities in vivo as they exhibit in vitro (Kinzler and Vogelstein 1990; Hallikas et al. 2006; Vokes et al. 2007). These include neural specific target genes that are transcriptionally silent in the limb. Thus, their GBRs are accessible to Gli factors but Gli3 activity is not required for tissue-specific silencing. In light of this result, it is likely that a significant fraction of the GBRs may reflect genes regulated by Hh signals in other tissue contexts. Indeed, only 696 of the 5274 GBRs (12.4%) associate with Shh pathway-regulated genes in the limb. Whereas some of these may reflect genes regulated at earlier or later stages of limb development, a large number are probably “inert” binding sites as observed in whole-genome studies in Drosophila (Li et al. 2008).
Transgenic analyses of regulatory networks within mammalian systems have demonstrated that considerable distances often separate _cis_-regulatory regions from the TSSs they modulate and regulatory regions may lie 5′ or 3′ of a gene’s promoter. Existing promoter arrays contain _cis_-regulatory sequences that are relative close to the TSS (e.g., −7.5 kb upstream to +2.5 kb from the TSS). Because of this proximity, most studies have assumed that binding of a regulatory factor within this proximal domain associates with the regulation of that gene; they do not attempt to measure an explicit FDR. In whole-genome studies such as ours, however, the indeterminate distance between genes and their cognate regulatory regions makes assigning long-range binding sites a nontrivial problem. We estimated the FDR associated with varying distances and find that by including information about both the gene density of the region and clustering tendencies of binding regions, we are able to make and then validate predictions for direct Gli input into long-range _cis_-elements that are likely to regulate Gremlin (>100 kb from the 3′ end of the transcriptional unit), Blimp1 (>25 kb from the 3′ end of the transcriptional unit) and Hand2 (10 kb and 85 kb downstream from the transcriptional unit). Of the 656 GBRs associated with differentially expressed limb genes, 252 are ≥20 kb from a TSS with an FDR <50% (Supplemental Data Set 3). Thus, longer-range _cis_-regulatory interactions are relatively common in this Gli3 regulatory network and likely more generally in mammalian systems.
Gli regulatory circuitry and modes of Gli action
Our analysis of several Gli3-binding regions in mediating _cis_-regulatory control of endogenous target genes validate the approach and data set, providing interesting new insights into cross-regulatory mechanisms that contribute to limb patterning. In the mouse, Blimp1 expression precedes Shh (Vincent et al. 2005) and analysis of Blimp1 mutants indicates that Blimp1 regulates Shh levels (Robertson et al. 2007). Our results suggest that a Shh–Gli regulatory input plays a reciprocal role in maintaining Blimp1 expression, a feedback mechanism essential for forming the most posterior digits (Robertson et al. 2007). In the pectoral fin of the zebrafish, Shh is also required for maintenance, but not induction, of Blimp1. Further, Shh lies genetically upstream of _Blimp1_-mediated regulation of slow twitch muscle fibers (Baxendale et al. 2004; Lee and Roy 2006). Given the critical role of Blimp1 in several stem/progenitor cell compartments and Shh’s roles in maintenance of stem/progenitor cells, we speculate that this regulatory module may play a broader role in other tissues.
Two major signaling pathways are specifically enriched among target genes of Gli regulation; as expected, the Hh pathway itself where feedback systems play in important role in modulating activity and output, and the BMP pathway (Supplemental Table S5). BMP signaling plays a crucial role in regulating limb outgrowth through the AER (see above) and is postulated to play a role in patterning digit identity when signaling is modulated in the chick limb (Dahn and Fallon 2000; Drossopoulou et al. 2000). However, removal of BMP activity in the mouse mesenchyme gives a normal range of digits but supernumerary preaxial and post-axial digits (Selever et al. 2004; Bandyopadhyay et al. 2006). We identify both Bmp2 and Gremlin as direct targets and provide functional evidence for the Shh–Gremlin interaction. A GBR >100 kb downstream from the gene encoding this critical BMP antagonist mediates a direct Gli activator activity in the posterior half of the normal Gremlin expression domain. Interestingly, although our data clearly demonstrate a Gli activator role, derepression of Gli3 in _Shh_−/−; _Gli3_−/− (Litingtung et al. 2002; te Welscher et al. 2002b) or _Smo_−/−;_Gli3_−/− (Fig. 4E) compound mutants appears to be sufficient for distal expression of Gremlin in both anterior and posterior domains. Bmp2 is expressed in these mutants and Bmp2 can activate Gremlin in a _Shh_-independent fashion (Capdevila et al. 1999; Litingtung et al. 2002; Nissim et al. 2006). Moreover, BMP signaling is essential for all Gremlin expression (Capdevila et al. 1999). Together these data suggest a model where BMP-input, through a regulatory module that remains to be determined, provides a tonic level of Gremlin activity throughout the distal limb mesenchyme. In the posterior compartment of the limb, close to the Shh source, a distinct Gli activator-dependent enhancer independently regulates Gremlin. This accounts for the higher level of Gremlin expression evident in both the chick and mouse on this posterior limb domain and its persistence in _Smo_−/−;_Gli3_−/− mutants (Figs. 4H,I,5).
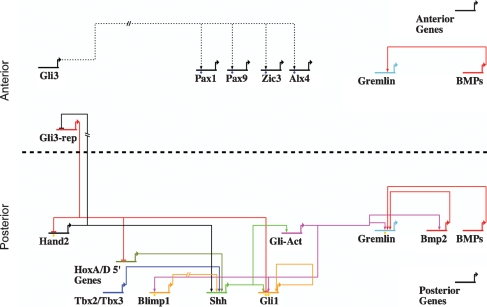
Figure 5.
Model depicting the _cis_-regulatory network underlying Gli-mediated limb patterning. Links with orange diamonds below signify regions tested in for expression in transgenic mice. Interactions between Gli3 and several anteriorly localized genes are depicted at the top of the diagram, with dashed lines and blue diamonds to indicate their speculative nature. Gremlin gene expression is regulated by both anterior and posterior inputs; this continuum of gene expression is represented as discrete enhancer circuits in the anterior and posterior compartments.
Our analysis suggests a model where Gli3 acts through a _cis_-acting silencer region to directly repress Hand2; Shh signaling attenuates Gli3 repressor production (Wang et al. 2000) enabling the maintenance of Hand2 expression in the posterior limb mesenchyme. The predicted silencer likely interacts with positive acting elements located outside of the GBR assayed. Chromosome capture conformation studies will likely shed important light on this and many other long-range interactions mediating Gli regulatory function. Further, new methodologies will likely improve silencer analysis, almost certainly an artificially underestimated component of regulatory networks.
Future studies
The present analysis provides a regulatory scaffold for the construction of Gli-dependent subcircuits in limb development (Fig. 5). In addition to an ongoing analysis of predicted targets and their roles in the Shh-dependent limb patterning process, a number of questions raised herein warrant further study. First, many putative Shh target genes have expression patterns that reflect multiple regulatory inputs. We know that at some regulatory level, the transcriptional processes must integrate additional signaling pathways including Wnt, FGF, BMP, and Notch. By analogy with other model systems, integration of these inputs at the level of _cis_-regulatory modules is an attractive proposition. However, we do not detect an enrichment for Smad (BMP pathway) or Lef/Tcf (Wing pathway)-binding motifs in GBRs. Second, nearly half of the binding regions do not contain a predicted Gli motif even though this subset is more conserved at the phylogenetic level than those containing direct Gli-binding sites. This may indicate Gli3 association without direct DNA binding. Gli proteins have been shown to associate with other transcriptional regulators such as Smads, β-catenin, and 5′HoxD proteins in various contexts (Liu et al. 1998; Chen et al. 2004; Ulloa et al. 2007). The enrichment of a composite bHLH/homeodomain bipartite motif may provide a clue to these interactions. Additional DNA/chromatin interaction and in vivo expression studies will be required to understand this interesting population of Gli targets.
Materials and methods
Generation of mouse strains and ChIP-on-chip
We generated a cDNA encoding a truncated form of mouse Gli3 containing the first 740 amino acids of mouse Gli3 (a gift from Dr. Ulrich Rüther) to make a Gli3 repressor (Gli3T) with a C-terminal 3XFlag epitope (Sigma). Gli3T was targeted to the Rosa26 locus (Supplemental Fig. S1A; Soriano 1999) in YFP3.1 embryonic stem (ES) cells (Supplemental Fig. S1B–C) (Mao et al. 2005) to generate a Cre-inducible Gli3 repressor line. Rosa Gli3TFlagc/c females mice were crossed with Prrx1Cre homozygous males and litters were assayed at E11.5. ChIP and LMPCR was performed as described previously (Vokes et al. 2007) with minor modifications (see the Supplemental Material) and DNA products were hybridzed to the Mouse Tiling 2.0R 7 array set or Mouse Promoter 1.0R array (single samples). The Histone H3K27Me3 antibody (Abcam ab6002) was incubated with anti-mouse IgG beads (Dynal #112.01) and the Pan Histone H3 antibody (Abcam ab1791) with anti-rabbit IgG beads (Dynal #112.03). ChIPs were processed as above using a single nonpooled ChIP from one litter of wild-type (Swiss-Webster) anterior or posterior limb fractions (two biological replicates). These were assayed on Affymetrix Mouse Promoter 1.0R arrays.
Probes in Affymetrix tiling arrays were remapped to the mm8 build 36 version of the mouse genome—all coordinates in this study are reported in mm8. Raw data were quantile-normalized and binding regions were determined using the new version of TileMap incorporated into CisGenome using a moving average (MA) (see the Supplemental Material; Ji and Wong 2005). All expression and tiling array data associated with this study have been deposited to the GEO database (GSE11062 and GSE11063).
Exon gene expression arrays
All gene expression experiments used E11.5 forelimbs. Samples were hybridized to Mouse Exon 1.0ST arrays (Affymetrix). The data were normalized and gene level expression indices were computed using GeneBASE software (Xing et al. 2006; Kapur et al. 2007). We then generated the following pairwise comparisons using PowerExpress, which implements the gene selection methods described in Paik et al. (2007): Shh < wild type; SmoM2 > wild type; Gli3 < wild type; Gli3 > wild type; Ant < Post; Ant > Post. Genes with an FDR ≤10% and a fold change ≥2 were selected. We did not generate pairwise comparisons for a certain combinations with SmoGli3 and Gli3 mutants because data from these arrays contained significant variability. To identify additional genes that were Shh-responsive, we performed the following multiple sample comparisons using an FDR ≤10% and a posterior probability cutoff of ≤25%: (1) Ant < Post and Shh < wild type < SmoM2, (2) Ant < Post and Shh < wild type < SmoM2 and Gli3 > SmoGli3, (3) Ant < Post and Shh < wild type < SmoM2 and wild type > SmoGli3. To define polarized gene sets representing both anterior and posterior compartments, we used the gene expression data on dissected anterior and posterior limb buds, Shh mutant limb buds, and SmoM2 limb buds (see Supplemental Table S6).
Assignment of Gli target genes
To calculate an initial intersection of binding regions with expression data, we identified 753 genes differentially expressed in at least one of the pairwise or multivariable comparisons in the exon array data. We counted the number of GBRs located within a given distance of their TSS, and we compared the observed number with random expectations (see the Supplemental Material; Supplemental Fig. S4A). To incorporate information about intervening transcripts (gene density), each of the 5274 GBRs was associated with a gene encoding the closest differentially expressed transcript. We counted the number of GBR–gene pairs separated by ≤Wbp and ≤K intervening promoters and compared the observed number with random expectations (see the Supplemental Material; Supplemental Fig. S4D). In order to explore binding regions located further away from genes, we collected the 689 GBRs in the 123 binding clusters reported in Figure 1F and repeated the same peak–gene association procedure for these 689 GBRs (Supplemental Table S4). To determine motif enrichment, GMS was run on various data sets (see the Supplemental Material). As a complementary motif analysis, we further mapped all TRANSFAC human and mouse motifs to GBRs and computed the relative enrichment r1 compared with matched genomic controls (see the Supplemental Material).
Transgenic experiments
To test for enhancer activity, we first visually scanned the annotated GBRs and adjusted fragment size in an attempt to recover an entire _cis_-regulatory domain based on visual inspection of conserved sequence using MultiZ alignments. These extended GBRs were PCR amplified inserted into pHSP68lacZ2XINS (Vokes et al. 2007) (coordinates are described in the Supplemental Material). The Hand2 constructs were tested for repressive activity in pSilencer, a modified version of pBSMhox—a precursor construct for Prx1Cre (Logan et al. 2002)—where a LacZ expression cassette was inserted downstream from the Prrx1 regulatory sequences that drive limb expression (Fig. 4P).
Acknowledgments
We thank Zhenjuan Wang and Manfred Baetscher (Harvard Genome Modification Facility) for pronuclear injections; Benjamin Allen, Jennifer Couget (FAS Center for Systems Biology), Joshua Mugford, Joo-Seop Park, and Courtney Yuen for advice; Yi Xing and Karen Kapur for providing exon array analysis software and expertise; Baolin Wang for providing the Gli3 antibody; and Renate Hellmiss for help with figures. Alex Schier and Cliff Tabin provided helpful advice either during the project or on the manuscript. We acknowledge support from the Helen Hay Whitney Foundation and Charles A. King Trust, Bank of America, Co-Trustee (Boston, MA) (to S.A.V.); the JHSPH Richard Gelb Faculty Innovation Award (to H.J.); and funding from the NIH, HG003903 (to A.P.M. and W.H.W.) and NS033642 (to A.P.M.).
Footnotes
References
- Bai C.B., Stephen D., Joyner A.L. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell. 2004;6:103–115. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A., Tsuji K., Cox K., Harfe B.D., Rosen V., Tabin C.J. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2:e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxendale S., Davison C., Muxworthy C., Wolff C., Ingham P.W., Roy S. The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat. Genet. 2004;36:88–93. doi: 10.1038/ng1280. [DOI] [PubMed] [Google Scholar]
- Capdevila J., Tsukui T., Rodriquez Esteban C., Zappavigna V., Izpisua Belmonte J.C. Control of vertebrate limb outgrowth by the proximal factor Meis2 and distal antagonism of BMPs by Gremlin. Mol. Cell. 1999;4:839–849. doi: 10.1016/s1097-2765(00)80393-7. [DOI] [PubMed] [Google Scholar]
- Carroll J.S., Meyer C.A., Song J., Li W., Geistlinger T.R., Eeckhoute J., Brodsky A.S., Keeton E.K., Fertuck K.C., Hall G.F., et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Charité J., McFadden D.G., Olson E.N. The bHLH transcription factor dHAND controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development. 2000;127:2461–2470. doi: 10.1242/dev.127.11.2461. [DOI] [PubMed] [Google Scholar]
- Chen Y., Knezevic V., Ervin V., Hutson R., Ward Y., Mackem S. Direct interaction with Hoxd proteins reverses Gli3-repressor function to promote digit formation downstream of Shh. Development. 2004;131:2339–2347. doi: 10.1242/dev.01115. [DOI] [PubMed] [Google Scholar]
- Dahn R.D., Fallon J.F. Interdigital regulation of digit identity and homeotic transformation by modulated BMP signaling. Science. 2000;289:438–441. doi: 10.1126/science.289.5478.438. [DOI] [PubMed] [Google Scholar]
- Dai P., Akimaru H., Tanaka Y., Maekawa T., Nakafuku M., Ishii S. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 1999;274:8143–8152. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- Drossopoulou G., Lewis K.E., Sanz-Ezquerro J.J., Nikbakht N., McMahon A.P., Hofmann C., Tickle C. A model for anteroposterior patterning of the vertebrate limb based on sequential long- and short-range Shh signalling and Bmp signalling. Development. 2000;127:1337–1348. doi: 10.1242/dev.127.7.1337. [DOI] [PubMed] [Google Scholar]
- Hallikas O., Palin K., Sinjushina N., Rautiainen R., Partanen J., Ukkonen E., Taipale J. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Harfe B.D., Scherz P.J., Nissim S., Tian H., McMahon A.P., Tabin C.J. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Horsley V., O’Carroll D., Tooze R., Ohinata Y., Saitou M., Obukhanych T., Nussenzweig M., Tarakhovsky A., Fuchs E. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell. 2006;126:597–609. doi: 10.1016/j.cell.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Mao J., Tenzen T., Kottmann A.H., McMahon A.P. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes & Dev. 2004;18:937–951. doi: 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Wong W.H. TileMap: Create chromosomal map of tiling array hybridizations. Bioinformatics. 2005;21:3629–3636. doi: 10.1093/bioinformatics/bti593. [DOI] [PubMed] [Google Scholar]
- Ji H., Vokes S.A., Wong W.H. A comparative analysis of genome-wide chromatin immunoprecipitation data for mammalian transcription factors. Nucleic Acids Res. 2006;34:e146. doi: 10.1093/nar/gkl803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein–DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kapur K., Xing Y., Ouyang Z., Wong W.H. Exon array assessment of gene expression. Genome Biol. 2007;8:R82. doi: 10.1186/gb-2007-8-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha M.K., Hsu D., Brunet L.J., Dionne M.S., Harland R.M. Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nat. Genet. 2003;34:303–307. doi: 10.1038/ng1178. [DOI] [PubMed] [Google Scholar]
- Kiefer S.M., Ohlemiller K.K., Yang J., McDill B.W., Kohlhase J., Rauchman M. Expression of a truncated Sall1 transcriptional repressor is responsible for Townes-Brocks syndrome birth defects. Hum. Mol. Genet. 2003;12:2221–2227. doi: 10.1093/hmg/ddg233. [DOI] [PubMed] [Google Scholar]
- Kinzler K.W., Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol. Cell. Biol. 1990;10:634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijper S., Feitsma H., Sheth R., Korving J., Reijnen M., Meijlink F. Function and regulation of Alx4 in limb development: Complex genetic interactions with Gli3 and Shh. Dev. Biol. 2005;285:533–544. doi: 10.1016/j.ydbio.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Laufer E., Nelson C.E., Johnson R.L., Morgan B.A., Tabin C. Sonic hedgehog and Fgf-4 act through a signaling cascade and feedback loop to integrate growth and patterning of the developing limb bud. Cell. 1994;79:993–1003. doi: 10.1016/0092-8674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- Lee B.C., Roy S. Blimp-1 is an essential component of the genetic program controlling development of the pectoral limb bud. Dev. Biol. 2006;300:623–634. doi: 10.1016/j.ydbio.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P.M., Dunn M.P., McMahon J.A., Logan M., Martin J.F., St-Jacques B., McMahon A.P. Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell. 2001;105:599–612. doi: 10.1016/s0092-8674(01)00369-5. [DOI] [PubMed] [Google Scholar]
- Li X.Y., Macarthur S., Bourgon R., Nix D., Pollard D.A., Iyer V.N., Hechmer A., Simirenko L., Stapleton M., Hendriks C.L., et al. Transcription factors bind thousands of active and inactive regions in the Drosophilablastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litingtung Y., Dahn R.D., Li Y., Fallon J.F., Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418:979–983. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- Liu F., Massague J., Ruiz i Altaba A. Carboxy-terminally truncated Gli3 proteins associate with Smads. Nat. Genet. 1998;20:325–326. doi: 10.1038/3793. [DOI] [PubMed] [Google Scholar]
- Logan M., Martin J.F., Nagy A., Lobe C., Olson E.N., Tabin C.J. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- Mao J., Barrow J., McMahon J., Vaughan J., McMahon A.P. An ES cell system for rapid, spatial and temporal analysis of gene function in vitro and in vivo. Nucleic Acids Res. 2005;33:e155. doi: 10.1093/nar/gni146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.F., Olson E.N. Identification of a prx1 limb enhancer. Genesis. 2000;26:225–229. [PubMed] [Google Scholar]
- McGlinn E., Tabin C.J. Mechanistic insight into how Shh patterns the vertebrate limb. Curr. Opin. Genet. Dev. 2006;16:426–432. doi: 10.1016/j.gde.2006.06.013. [DOI] [PubMed] [Google Scholar]
- McGlinn E., van Bueren K.L., Fiorenza S., Mo R., Poh A.M., Forrest A., Soares M.B., Bonaldo M.d.e.F., Grimmond S., Hui C.C., et al. Pax9 and Jagged1 act downstream of Gli3 in vertebrate limb development. Mech. Dev. 2005;122:1218–1233. doi: 10.1016/j.mod.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Naiche L.A., Papaioannou V.E. Tbx4 is not required for hindlimb identity or post-bud hindlimb outgrowth. Development. 2007;134:93–103. doi: 10.1242/dev.02712. [DOI] [PubMed] [Google Scholar]
- Nissim S., Hasso S.M., Fallon J.F., Tabin C.J. Regulation of Gremlin expression in the posterior limb bud. Dev. Biol. 2006;299:12–21. doi: 10.1016/j.ydbio.2006.05.026. [DOI] [PubMed] [Google Scholar]
- Nissim S., Allard P., Bandyopadhyay A., Harfe B.D., Tabin C.J. Characterization of a novel ectodermal signaling center regulating Tbx2 and Shh in the vertebrate limb. Dev. Biol. 2007;304:9–21. doi: 10.1016/j.ydbio.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswander L., Jeffrey S., Martin G.R., Tickle C. A positive feedback loop coordinates growth and patterning in the vertebrate limb. Nature. 1994;371:609–612. doi: 10.1038/371609a0. [DOI] [PubMed] [Google Scholar]
- Ohinata Y., Payer B., O’Carroll D., Ancelin K., Ono Y., Sano M., Barton S.C., Obukhanych T., Nussenzweig M., Tarakhovsky A., et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I., Loots G.G., Nobrega M.A., Hardison R.C., Miller W., Stubbs L. Evolution and functional classification of vertebrate gene deserts. Genome Res. 2005;15:137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik J.H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J.W., Carrasco D.R., et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S., Niswender K.D., Ji Q., van der Meer R., Keeney D., Magnuson M.A., Wisdom R. Polydactyly and ectopic ZPA formation in Alx-4 mutant mice. Development. 1997;124:3999–4008. doi: 10.1242/dev.124.20.3999. [DOI] [PubMed] [Google Scholar]
- Robertson E.J., Charatsi I., Joyner C.J., Koonce C.H., Morgan M., Islam A., Paterson C., Lejsek E., Arnold S.J., Kallies A., et al. Blimp1 regulates development of the posterior forelimb, caudal pharyngeal arches, heart and sensory vibrissae in mice. Development. 2007;134:4335–4345. doi: 10.1242/dev.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo R.F., Rajan L., Li X., Lufkin T. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes & Dev. 2002;16:1089–1101. doi: 10.1101/gad.988402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz P.J., Harfe B.D., McMahon A.P., Tabin C.J. The limb bud Shh–Fgf feedback loop is terminated by expansion of former ZPA cells. Science. 2004;305:396–399. doi: 10.1126/science.1096966. [DOI] [PubMed] [Google Scholar]
- Selever J., Liu W., Lu M.F., Behringer R.R., Martin J.F. Bmp4 in limb bud mesoderm regulates digit pattern by controlling AER development. Dev. Biol. 2004;276:268–279. doi: 10.1016/j.ydbio.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Tarchini B., Duboule D., Kmita M. Regulatory constraints in the evolution of the tetrapod limb anterior–posterior polarity. Nature. 2006;443:985–988. doi: 10.1038/nature05247. [DOI] [PubMed] [Google Scholar]
- te Welscher P., Fernandez-Teran M., Ros M.A., Zeller R. Mutual genetic antagonism involving GLI3 and dHAND prepatterns the vertebrate limb bud mesenchyme prior to SHH signaling. Genes & Dev. 2002a;16:421–426. doi: 10.1101/gad.219202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Welscher P., Zuniga A., Kuijper S., Drenth T., Goedemans H.J., Meijlink F., Zeller R. Progression of vertebrate limb development through SHH-mediated counteraction of GLI3. Science. 2002b;298:827–830. doi: 10.1126/science.1075620. [DOI] [PubMed] [Google Scholar]
- Towers M., Mahood R., Yin Y., Tickle C. Integration of growth and specification in chick wing digit-patterning. Nature. 2008;452:882–886. doi: 10.1038/nature06718. [DOI] [PubMed] [Google Scholar]
- Ulloa F., Itasaki N., Briscoe J. Inhibitory Gli3 activity negatively regulates Wnt/β-catenin signaling. Curr. Biol. 2007;17:545–550. doi: 10.1016/j.cub.2007.01.062. [DOI] [PubMed] [Google Scholar]
- Vincent S.D., Dunn N.R., Sciammas R., Shapiro-Shalef M., Davis M.M., Calame K., Bikoff E.K., Robertson E.J. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;32:1315–1325. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- Vokes S.A., Ji H., McCuine S., Tenzen T., Giles S., Zhong S., Longabaugh W.J., Davidson E.H., Wong W.H., McMahon A.P. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development. 2007;134:1977–1989. doi: 10.1242/dev.001966. [DOI] [PubMed] [Google Scholar]
- Wang B., Fallon J.F., Beachy P.A. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Xing Y., Kapur K., Wong W.H. Probe selection and expression index computation of affymetrix exon arrays. PLoS ONE. 2006;1:e88. doi: 10.1371/journal/pone.0000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Drossopoulou G., Chuang P.T., Duprez D., Marti E., Bumcrot D., Vargesson N., Clarke J., Niswander L., McMahon A., et al. Relationship between dose, distance and time in Sonic Hedgehog-mediated regulation of anteroposterior polarity in the chick limb. Development. 1997;124:4393–4404. doi: 10.1242/dev.124.21.4393. [DOI] [PubMed] [Google Scholar]
- Yang A., Zhu Z., Kapranov P., McKeon F., Church G.M., Gingeras T.R., Struhl K. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Yuen C.M., Rodda S.J., Vokes S.A., McMahon A.P., Liu D.R. Control of transcription factor activity and osteoblast differentiation in mammalian cells using an evolved small-molecule-dependent intein. J. Am. Chem. Soc. 2006;128:8939–8946. doi: 10.1021/ja062980e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakany J., Duboule D. Synpolydactyly in mice with a targeted deficiency in the HoxD complex. Nature. 1996;384:69–71. doi: 10.1038/384069a0. [DOI] [PubMed] [Google Scholar]
- Zhu J., Nakamura E., Nguyen M.T., Bao X., Akiyama H., Mackem S. Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev. Cell. 2008;14:624–632. doi: 10.1016/j.devcel.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga A., Haramis A.P., McMahon A.P., Zeller R. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature. 1999;401:598–602. doi: 10.1038/44157. [DOI] [PubMed] [Google Scholar]
- Zuniga A., Michos O., Spitz F., Haramis A.P., Panman L., Galli A., Vintersten K., Klasen C., Mansfield W., Kuc S., et al. Mouse limb deformity mutations disrupt a global control region within the large regulatory landscape required for Gremlin expression. Genes & Dev. 2004;18:1553–1564. doi: 10.1101/gad.299904. [DOI] [PMC free article] [PubMed] [Google Scholar]