Specific Molecular Chaperone Interactions and an ATP-dependent Conformational Change Are Required during Posttranslational Protein Translocation into the Yeast ER (original) (raw)
Abstract
The posttranslational translocation of proteins across the endoplasmic reticulum (ER) membrane in yeast requires ATP hydrolysis and the action of hsc70s (DnaK homologues) and DnaJ homologues in both the cytosol and ER lumen. Although the cytosolic hsc70 (Ssa1p) and the ER lumenal hsc70 (BiP) are homologous, they cannot substitute for one another, possibly because they interact with specific DnaJ homologues on each side of the ER membrane. To investigate this possibility, we purified Ssa1p, BiP, Ydj1p (a cytosolic DnaJ homologue), and a GST–63Jp fusion protein containing the lumenal DnaJ region of Sec63p. We observed that BiP, but not Ssa1p, is able to associate with GST–63Jp and that Ydj1p stimulates the ATPase activity of Ssa1p up to 10-fold but increases the ATPase activity of BiP by <2-fold. In addition, Ydj1p and ATP trigger the release of an unfolded polypeptide from Ssa1p but not from BiP. To understand further how BiP drives protein translocation, we purified four dominant lethal mutants of BiP. We discovered that each mutant is defective for ATP hydrolysis, fails to undergo an ATP-dependent conformational change, and cannot interact with GST–63Jp. Measurements of protein translocation into reconstituted proteoliposomes indicate that the mutants inhibit translocation even in the presence of wild-type BiP. We conclude that a conformation- and ATP-dependent interaction of BiP with the J domain of Sec63p is essential for protein translocation and that the specificity of hsc70 action is dictated by their DnaJ partners.
INTRODUCTION
The first step in secretory protein biogenesis, the translocation of newly synthesized polypeptides across the endoplasmic reticulum (ER) membrane, is a highly intricate, multistep process (for reviews, see Corsi and Schekman, 1996; Rapoport et al., 1996; Johnson, 1997). Protein translocation may occur in one of two ways, either cotranslationally or posttranslationally. In cotranslational translocation, nascent secretory polypeptides are targeted to the ER membrane before translation is complete. Posttranslational translocation proceeds after the secretory polypeptide has been fully synthesized and released from the ribosome. In either case, various proteins in the cytosol, in the ER membrane, and in the lumen of the ER are required for protein translocation. Among these are members of the 70-kDa class of heat shock cognate proteins (Hsc70s).
Hsc70s consist of a highly conserved N-terminal ATPase domain and a variable C-terminal region that mediates peptide binding and can act as molecular chaperones by disallowing the aggregation of exposed hydrophobic regions and preventing nonproductive folding pathways (for review, see Gething et al., 1995). Homologues of the bacterial DnaJ molecular chaperone activate the ATPase activity of hsc70s, can present polypeptide substrates to hsc70s, and have also been shown to play a role in protein translocation into both mitochondria and the ER (for reviews, see Hartl, 1996; Cheetham and Caplan, 1998).
The ER lumenal hsc70 BiP was originally identified in the ER of mammalian cells as a protein bound noncovalently to immunoglobulin heavy chains that had been synthesized in the absence of light chain (Haas and Wabl, 1983). In the yeast Saccharomyces cerevisiae, BiP has been shown to be essential for protein translocation. Cytosolic preproteins (secretory precursors) accumulate when BiP is depleted from the cell, and strains containing a temperature-sensitive mutation in the KAR2 gene, which encodes BiP (Normington et al., 1989; Rose et al., 1989), accumulate untranslocated preproteins when incubated at the nonpermissive temperature (Vogel et al., 1990; Nguyen et al., 1991). We and others have shown that BiP is required in vitro for both co- and posttranslational protein translocation (Sanders et al., 1992; Brodsky et al., 1995). A role for BiP early in the translocation process was demonstrated by Sanders et al. (1992) because mutations in KAR2 prevent the association of an ER-targeted preprotein with the translocation channel Sec61p. Additionally, BiP and ATP have been shown to be required for the release of a preprotein from an initial recognition complex of Sec proteins on the cytosolic face of the ER (Lyman and Schekman, 1997). BiP also acts later in the process of protein translocation, when BiP is in contact with a translocation-arrested polypeptide (Sanders et al., 1992), and certain mutant alleles of KAR2 arrest protein translocation after the polypeptide has entered the translocation channel (Sanders et al., 1992; Lyman and Schekman, 1995). Matlack et al. (1997) have observed that BiP is required for the completion of preprotein passage through the translocation channel in a solubilized system that recapitulates the translocation reaction.
BiP requires a DnaJ homologue to facilitate protein translocation into the ER in yeast. Sec63p, a multispanning integral membrane protein of the ER, harbors a lumenal region that is 42% identical to the N terminus of DnaJ (Sadler et al., 1989; Feldheim et al., 1992). Cells containing a temperature-sensitive mutation in SEC63 accumulate untranslocated preproteins in the cytosol (Rothblatt et al., 1989), and ER microsomes derived from the sec63-1 strain are defective for both post- and cotranslational translocation in vitro (Rothblatt et al., 1989; Brodsky et al., 1995). Genetic (Scidmore et al., 1993) as well as biochemical interactions between BiP and Sec63p are well established (Brodsky and Schekman, 1993; Lyman and Schekman, 1995, 1997; Corsi and Schekman, 1997; Matlack et al., 1997), and Sec63p is required, along with BiP and ATP, to promote preprotein release from the recognition complex at the ER (Lyman and Schekman, 1997). The complexity of BiP action is underscored further by the recent observation that BiP may gate Sec61p in the mammalian ER during protein translocation (Hamman et al., 1998).
Another hsc70 required during posttranslational protein translocation is the cytosolic protein Ssa1p. Ssa1p depletion in vivo resulted in the accumulation of untranslocated prepro-α factor (ppαf), a yeast mating pheromone precursor that is translocated posttranslationally (Deshaies et al., 1988). Also, Ssa1p was identified as one of the active components of a cytosolic fraction that stimulated the posttranslational translocation of wheat germ–synthesized ppαf into yeast microsomes (Chirico et al., 1988). Bush and Meyer (1996) have suggested that Ssa1p recognizes preproteins that have prematurely folded in the cytosol and restores them to a translocation-competent conformation.
Whereas BiP is functionally coupled with the DnaJ homologue Sec63p, Ssa1p may also be paired with a DnaJ homologue that modulates its function. The most likely candidate for this DnaJ homologue is Ydj1p. A role for Ydj1p in protein translocation is supported by the observation that cells containing a temperature-sensitive allele of YDJ1 accumulate untranslocated ppαf at the nonpermissive temperature (Caplan et al., 1992). Ydj1p has been shown to stimulate the ATPase activity of Ssa1p and also to catalyze the release of bound protein substrate from Ssa1p in the presence of ATP (Cyr et al., 1992; Srinivasan et al., 1997). Additionally, Becker et al. (1996) have shown that SSA1 and YDJ1 may interact.
Despite the fact that BiP and Ssa1p are ∼63% identical and because hsc70s may be expected to perform similar functions, they are unable to substitute for one another during posttranslational translocation (Brodsky et al., 1993). In this manuscript we demonstrate for the first time that this is caused by specific interactions with unique DnaJ homologues. To understand further how BiP uniquely engineers events required for protein translocation, we have biochemically characterized a collection of four dominant lethal BiP mutants (G274D, G246D, G247D, and K117Q). Two of these mutants are analogous to previously characterized mammalian BiP mutants, G226D and G227D, that were found to be defective for ATP binding and release of bound peptide substrates (Wei et al., 1995); these results suggest that ATP binding and the accompanying conformational change are important for modulating substrate protein binding and release. We have found that all four mutant BiP proteins are defective for ATP hydrolysis, are unable to undergo a nucleotide-dependent conformational change, and fail to interact in a stable, ATP-dependent manner with the DnaJ domain of Sec63p. Because these mutants cannot support protein translocation in vitro, we suggest that an ATP-dependent conformational change in BiP is required both to bind Sec63p and to facilitate the translocation reaction.
MATERIALS AND METHODS
Purification of Ssa1p
Ssa1p was purified as previously described (Brodsky et al., 1993), with minor modifications. Yeast strain RSY169 (MW141 [Werner-Washburne et al., 1987]) was grown at 30°C in YP (1% Bacto-yeast extract and 2% Bacto-peptone; Difco, Detroit, MI) containing 2% galactose to an OD600 of 0.5-2. Cells were harvested, washed once with water, and resuspended to 20 ml per 2 l of original culture in buffer B (40 mM HEPES, pH 6.8, 5 mM MgOAc, 75 mM KCl, and 1 mM DTT) containing 1 mM PMSF and 0.5 μg/ml pepstatin-A. One-half vol of glass beads was added, and the cells were agitated on a Vortex mixer six times for 60 s with a 2 min incubation on ice between each disruption. The supernatant was centrifuged to remove unbroken cells and then centrifuged at 22,000 × g for 10 min at 4°C (Sorvall SS34 rotor; Newtown, CT). The resulting supernatant was centrifuged at 100,000 × g for 45 min in a Beckman SW28 rotor (Fullerton, CA) at 4°C. The high-speed supernatant was applied to a 1:2 ATP-agarose (Sigma Chemical, St. Louis, MO):Sephadex G-50 (Pharmacia, Piscataway, NJ) column equilibrated in buffer C (20 mM HEPES, pH 6.8, 2 mM MgOAc, 25 mM KCl, and 1 mM DTT). The ATP-agarose was diluted so that it contained ∼1-2 μmol of ATP per ml of resin. The column was washed sequentially with 20 ml of buffer C, 20 ml of buffer C containing 1 M KCl, and then 10 ml of buffer C. Ssa1p was eluted with 20 ml of buffer C containing 5 mM ATP (Sigma Chemical), and 1 ml fractions were collected. Peak fractions, determined by SDS-PAGE and immunoblot analysis, were pooled and loaded onto a 5 ml DEAE Sepharose (Pharmacia) column equilibrated in buffer C. The column was washed with 10 vol of buffer C, and Ssa1p was eluted with a gradient of 15 ml of buffer C to 15 ml of buffer C containing 750 mM KCl. Ssa1p-containing fractions were pooled and dialyzed against 500 vol of dialysis buffer (50 mM Tris, pH 7.4, 50 mM NaCl, 0.8 mM DTT, 2 mM MgOAc, and 5% glycerol) for 12 h. Dialyzed protein was snap-frozen in liquid nitrogen and stored at −70°C. When thawed, aliquots containing Ssa1p were used immediately.
Purification of Ydj1p
Ydj1p was purified as previously described (Caplan et al., 1992), with minor modifications. BL21 (DE3) cells expressing Ydj1p were grown at 26°C in Luria-Bertani medium (1% Bacto-tryptone, 0.5% Bacto-yeast extract, and 1% NaCl, pH 7) containing 25 μg/ml kanamycin to an OD600 of ∼0.5. Ydj1p expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM, and the cells were grown for another 2 h. Cells were harvested, washed once with water, and recentrifuged, and the cell pellet was resuspended in buffer A (20 mM MOPS, pH 7.5, 0.5 mM EDTA, 10 mM DTT, and 0.5 mM PMSF). The cells were sonicated (Fisher Scientific Sonic Dismembrator, Pittsburgh, PA) three times for 30 s with a 2 min incubation on ice between sonications. Sonicated cells were centrifuged at 100,000 × g for 30 min in a Beckman SW28 rotor at 4°C. The resulting supernatant was applied to a 5 ml DE52 column (Whatman, Maidstone, United Kingdom) equilibrated in buffer A. The column was washed with 25 ml of buffer A and eluted with a gradient of 15 ml of buffer A to 15 ml of buffer A containing 300 mM NaCl, and 1 ml fractions were collected. Peak fractions, determined by SDS-PAGE and Coomassie brilliant blue staining, were pooled, diluted 1:5 in buffer B (5 mM KPi, pH 7, and 10 mM DTT), and loaded onto a 5 ml hydroxylapatite column (Bio-Rad, Richmond, CA) equilibrated in buffer B. The column was washed with 25 ml of buffer B, and Ydj1p was eluted with a 10 ml of buffer B to 10 ml of buffer B containing 400 mM KPi gradient, and 1 ml fractions were collected. Peak fractions, again determined by SDS-PAGE and Coomassie brilliant blue staining, were pooled and dialyzed against 500 vol of buffer C (10 mM HEPES, pH 7, 50 mM NaCl, 10 mM DTT, and 10% glycerol) for 12 h. Dialyzed Ydj1p was snap-frozen in liquid nitrogen and stored at −70°C. When thawed, aliquots containing Ydj1p were used immediately.
Construction of BiP Mutants
Dominant mutations in the KAR2 gene were generated and isolated as described by Vogel (1993). In brief, the KAR2 gene was placed on a _LEU2_-containing centromeric plasmid under the control of a galactose-regulated promoter. This plasmid, pMR2245, was mutagenized with hydroxylamine as previously described (Rose and Fink, 1987). Mutagenized plasmids were transformed into MS37 (leu2-3,-112 ura3-52 ade2-11 MAT [Vogel, 1993]), and synthetic medium lacking leucine was used to select cells containing the plasmid. Potential dominant lethal KAR2 alleles were isolated by identifying colonies that grew on glucose but that failed to grow on galactose.
Construction of Hexahistidine-tagged BiP
Constructs used to purify the dominant mutant forms of BiP were created as described by Vogel (1993). In brief, a restriction site was introduced immediately downstream of the region encoding the signal sequence cleavage site so that, upon subcloning, the signal sequence coding region was absent. Ultimately, wild-type and mutagenized KAR2 were cloned into a plasmid containing the hexahistidine tag portion of vector pQE9 (Qiagen, Hilden, Germany) and under the control of the lacIQ promoter that was subcloned from pGEX-2T (Pharmacia). The fusion resulted in the addition of 12 amino acids, MRGSHHHHHHGS, before amino acid 43 (alanine) in mature BiP.
Sequencing of Dominant Lethal BiP Mutants
The locations of the amino acid substitutions in the four dominant lethal KAR2 alleles were determined by dideoxy sequencing using the T7 Sequenase version 2.0 Sequencing Kit (Amersham, Arlington Heights, IL) according to the provided protocol. The upstream primer, corresponding to bases 280-299 of the KAR2 gene as numbered by Normington et al. (1989) (also see Rose et al., 1989), had the sequence GAA AGA TTG ATT GGT GAT GC. The downstream primer, of sequence CCA TGC TTC TTG AAA GC, corresponded to bases 865-885 of KAR2.
Purification of Wild-Type or Dominant Mutant Hexahistidine-tagged BiP
Bacterial strain RR1 expressing pMR2623 (wild-type BiP), pMR2618 (G274D), pMR2619 (G246D), pMR2620 (G247D), or pMR2622 (K117Q) (see Table 2) was grown in 50 ml of Luria-Bertani medium containing 50 μg/ml ampicillin for ∼16 h and then added to 1 l of the same medium and grown for 2 h at 26°C. IPTG was added to a final concentration of 0.5 mM, and the cells were grown for an additional 3-4 h at 26°C. Cells were harvested, washed once with water, respun, and resuspended in ∼20 ml of sonication buffer (50 mM HEPES, pH 7.4, 300 mM NaCl, 10 mM imidazole, and 5 mM β-mercaptoethanol) containing 1 mM PMSF and 0.5 μg/l pepstatin-A. The cells were sonicated three times for 60 s with a 2 min incubation on ice between sonications. Sonicated cells were centrifuged at 16,000 × g for 10 min, and the supernatant was loaded onto a 5 ml nickel nitriloacetic acid (Ni2+-NTA)-agarose column (Qiagen) equilibrated in sonication buffer. The column was washed sequentially with 30 ml of 1) sonication buffer, 2) sonication buffer containing 1% Triton X-100 and 5% glycerol, 3) sonication buffer containing 1 M NaCl and 5% glycerol, 4) sonication buffer containing 5 mM ATP and 5% glycerol, 5) sonication buffer containing 0.5 M Tris, pH 7.4, and 5% glycerol, and 6) sonication buffer containing 25 mM imidazole and 5% glycerol. BiP was eluted with 30 ml of sonication buffer containing 250 mM imidazole and 5% glycerol, and 1 ml fractions were collected. Fractions were analyzed by SDS-PAGE followed by Coomassie brilliant blue staining and immunoblot analysis using anti-Kar2p antibody (Brodsky and Schekman, 1993) and were found to lack contaminating DnaK chaperone (our unpublished observations). Peak fractions were pooled, diluted 1:4 in buffer 88 (20 mM HEPES, pH 6.8, 150 mM KOAc, 250 mM sorbitol, and 5 mM MgOAc), and loaded onto a 10 ml Q-Sepharose column (Pharmacia) equilibrated in buffer 88. The column was washed with 30 ml of buffer 88 before elution with a 15 ml by 15 ml gradient of buffer 88 to buffer 88 containing 2 M KOAc. Peak fractions, determined by SDS-PAGE and Coomassie brilliant blue staining, were pooled and dialyzed against 500 vol of dialysis buffer for 12 h. Dialyzed BiP was snap-frozen in liquid nitrogen and stored at −70°C. When thawed, aliquots containing BiP were used immediately.
Table 2.
Amino acid changes in BiP mutants
| Dominant lethal BiP mutants | BiP amino acid substitution | Corresponding bovine brain hsc70 residue | Analogous mutations (hamster BiP) | References |
|---|---|---|---|---|
| D3 | G274D | Gly 229 | ||
| D4 | G246D | Gly 201 | G226D | Wei et al., 1995 |
| D21 | G247D | Gly 202 | G227D | Wei et al., 1995 |
| D29 | K117Q | Lys 71 |
Purification of GST–63J and the GST–63J–Binding Assay
GST–63J was purified essentially as described by Corsi and Schekman (1997), except that the protease inhibitors used were PMSF (to a final concentration of 1 mM) and pepstatin-A (to a final concentration of 0.5 μg/ml).
The GST–63J–binding assay was performed essentially as described by Corsi and Schekman (1997). A total of 3 μg of GST–63J was incubated with 10 μl of glutathione–cross-linked agarose (Sigma Chemical) in GST-binding buffer (20 mM Tris, pH 8.0, 100 mM KCl, 5 mM MgCl2, 0.1% NP40, 2% glycerol, 1 mM DTT, 1 mM EDTA, and 1 mM PMSF) and rotated at 4°C for 1 h in a total volume of 50 μl. Reactions were centrifuged at 16,000 × g for 2 min, the supernatant was removed, and the pellet was washed with 50 μl of GST-binding buffer. This step was repeated three times. GST-binding buffer and 2 μg of wild-type BiP, Ssa1p, or dominant mutant BiP isolates were added to the final pellet along with 1 mM ATP or ADP to a total volume of 50 μl. The reactions were rotated for 2 h at 4°C and then centrifuged at 16,000 × g for 2 min. The supernatant was collected, and the pellet was washed three times as described above. SDS-PAGE sample buffer was added to the supernatant and pellet fractions that were then analyzed by SDS-PAGE using 15% polyacrylamide gels followed by Coomassie brilliant blue staining.
ATPase Assay
ATPase assays were performed essentially as described (Cyr et al., 1992; Srinivasan et al., 1997). Briefly, 1 μg of the indicated hsc70 was incubated with 1 nmol of cold ATP and 0.01 μCi of [α-32P]ATP (Amersham) in ATPase buffer (50 mM HEPES, pH 7.4, 50 mM NaCl, 10 mM DTT, and 2 mM MgCl2) in a total volume of 20 μl at 30°C. Where indicated, Ydj1p or GST–63Jp was also included in this incubation. After 1 h, during which time ATPase activity was linear (our unpublished observations), 1 μl of each reaction was spotted in triplicate on polyethyleneimine cellulose TLC plates (Selecto Scientific, Norcross, GA), and the plates were developed in 1 M formic acid and 0.5 M LiCl (Shlomai and Kornberg, 1980). Plates were examined for conversion of [α-32P]ATP to [α-32P]ADP by phosphorimage analysis, and results were quantified using MacBas software (Fuji Medical Systems USA, Stamford, CT). In calculations of specific activity (nmoles of ATP converted to ADP per minute per milligram of hsc70), the percent of ATP converted to ADP in control reactions containing no protein was subtracted from that in hsc70-containing reactions, and control reactions containing Ydj1p or GST–63Jp alone were subtracted from hsc70 and Ydj1p or GST–63Jp-containing reactions. Lineweaver–Burk analyses based on the results of ATPase assays in which the amount of nonradioactive ATP added to reactions was titrated from 0.0375 to 100 μM were performed. Linear regression analyses were performed by the use of the Kaleidagraph software package (version 3.0.4; Abelbeck Software, Reading, PA).
125I-Carboxymethylated α-Lactalbumin–Binding Assay
Carboxymethylated α-lactalbumin (CMLA; Sigma Chemical) was labeled with Na125I as previously described (Cyr et al., 1992). Reactions containing 4 μg of Ssa1p or BiP were preincubated with ∼0.4 μCi of 125I-CMLA in hsc70-binding buffer (50 mM HEPES, pH 6.8, 50 mM NaCl, 1 mM DTT, and 0.1 mM EDTA) for 20 min at 30°C in a total volume of 10 μl. To these mixtures, 8 μg of Ydj1p (where indicated), 1 mM MgCl2, an ATP-regenerating system (Brodsky et al., 1993), and hsc70-binding buffer in a total volume of 20 μl were added, and the reactions were incubated for an additional 20 min at 30°C. Reactions were quenched by the addition of 10 μl of 4× ice-cold sample buffer lacking SDS (65 mM Tris, pH 6.8, 0.05 mg/ml bromophenol blue, and 10% glycerol) and were subjected to nondenaturing PAGE on a 5-15% gradient gel containing 1 mM ATP that was run for 16 h (9 mA) at 4°C. Dried gels were analyzed by phosphorimage analysis as described above.
Protease Protection Assay
The ability of wild-type or mutant BiP to undergo a nucleotide-dependent conformational change was assessed using a procedure modified from that of Kassenbrock and Kelly (1989). A total of 5 μg of BiP and ATP or ADP at a final concentration of 1 mM were incubated in assay buffer (20 mM HEPES, pH 7.2, 25 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, and 0.5 mM DTT) for 1 h at 20°C in a final volume of 64 μl. Proteinase K was then added to a final concentration of 0.3 μg/l, and the reactions were incubated for 5 min at 20°C before being quenched with ice-cold trichloroacetic acid (TCA) to a final concentration of 20%. TCA-precipitated proteins were resuspended in SDS sample buffer and subjected to SDS-PAGE analysis on 10% polyacrylamide gels followed by Coomassie brilliant blue staining.
Reactions were also performed in buffer containing 25 mM NaCl instead of 25 mM KCl, because a previous study found that Ssa1p required potassium to undergo a complete conformational change in the presence of nucleotide (Fung et al., 1996). However, we observed no such difference with BiP whether our reactions were performed in potassium- or sodium-containing buffers (our unpublished observations).
Microsome Preparation, Reconstitution of Proteoliposomes, and Translocation Assay
Yeast microsomes were prepared and proteoliposomes were reconstituted as previously described (Brodsky et al., 1993; Brodsky and Schekman, 1993). The amount of BiP present in reconstituted proteoliposomes was determined by floating reconstituted proteoliposomes through a sucrose step gradient to separate unincorporated protein from protein contained in proteoliposomes (Brodsky et al., 1993; Brodsky, 1997). The floated proteoliposomes were TCA-precipitated, and the proteins were resuspended in SDS sample buffer and assayed by SDS-PAGE followed by immunoblot analysis using anti-Kar2p as the primary antibody and 125I-protein A as the secondary antibody. The amount of BiP present in reconstituted proteoliposomes either lacking or supplemented with BiP was quantified by phosphorimage analysis. We found that ∼1.6-fold more BiP was present in reconstituted proteoliposomes when 17 μg of exogenous BiP was added to the reactions. Translocation reactions in which the transport of ppαf into vesicles was assayed were performed as described (McCracken and Brodsky, 1996).
RESULTS
Specificity of Hsc70–DnaJ Cognate Interactions
We have shown previously that the cytosolic hsc70 Ssa1p and the ER lumenal hsc70 BiP are unable to substitute for one another in two distinct in vitro assays that measure posttranslational protein translocation (Brodsky et al., 1993). To identify the basis of this compartmental specificity, we purified Ssa1p, BiP, the cytosolic DnaJ homologue Ydj1p, and a GST fusion protein containing the DnaJ domain of Sec63p, GST–63Jp (Caplan et al., 1992; Brodsky et al., 1993; Corsi and Schekman, 1997).
ATP hydrolysis is necessary for hsc70 function, and the modulation of hsc70 ATPase activity by DnaJ partner proteins can mediate hsc70 action (for review, see Hartl, 1996). Previous studies have shown that Ydj1p substantially stimulates ATP hydrolysis by Ssa1p (Cyr et al., 1992; Ziegelhoffer et al., 1995; Srinivasan et al., 1997). Although stable complexes between Ydj1p and Ssa1p have not been observed, it is generally assumed that the stimulation of the ATPase activity of an hsc70 by its DnaJ cognate arises from transient interactions.
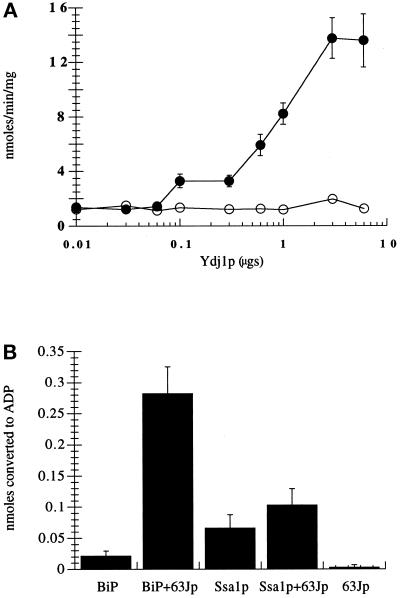
We first chose to examine whether Ydj1p could similarly stimulate ATP hydrolysis by BiP. Titrating the amount of Ydj1p included in steady-state ATPase reactions demonstrated that Ydj1p specifically stimulated the rate of ATP hydrolysis by Ssa1p up to 10-fold but increased the activity of BiP <2-fold (Figure 1A). At 6 μg of added Ydj1p, the molar ratio of Ydj1p to Ssa1p is ∼10:1; thus, half-maximal stimulation of Ssa1p was achieved at a Ydj1p:Ssa1p ratio of ∼1.4:1. We next examined whether BiP, but not Ssa1p, was specifically activated by BiP’s DnaJ partner Sec63p (Brodsky and Schekman, 1993; Scidmore et al., 1993). An ∼3-fold molar excess of GST–63Jp (Corsi and Schekman, 1997) resulted in a 13-fold stimulation of BiP ATPase activity. However, the ATPase activity of Ssa1p was enhanced only 1.6-fold by GST–63Jp (Figure 1B). Taken together, these results indicate that the ATPase activities of BiP and Ssa1p are significantly enhanced only by the DnaJ proteins that are localized to the compartment in which the hsc70s reside; these data further suggest that the specificity of hsc70 action during protein translocation (Brodsky et al., 1993) may be attributable to the requirement for unique hsc70–DnaJ pairs.
Figure 1.
Ydj1p specifically activates the ATPase activity of Ssa1p. (A) Ssa1p (filled circles) or BiP (open circles) was incubated with increasing amounts of Ydj1p for 1 h at 30°C. ATP hydrolysis was assessed by determining the fraction of [α-32P]ATP converted to [α-32P]ADP as described in MATERIALS AND METHODS. Data represent the means of at least six independent assays. The mean specific activities (nanomoles of ATP hydrolyzed per minute per milligram of hsc70) of Ssa1p and BiP in these experiments were 1.38 and 1.22 nmol·min−1·mg−1, respectively. (B) Ssa1p or BiP was incubated with or without GST–63Jp for 1 h at 30°C, and ATP hydrolysis was assessed as described above. Data represent the means of at least four independent assays (± SD). The mean specific activities (nmoles of ATP hydrolyzed per minute per milligram of hsc70) of Ssa1p and BiP in these experiments were 1.1 and 0.36 nmol·min−1·mg−1, respectively.
In examining the effects of salt conditions on ATPase activity, we made three observations. First, the inherent ATPase activities of Ssa1p and BiP were elevated six- and threefold, respectively, when 20 mM KCl was included in the reaction buffer (our unpublished observations), as anticipated (O’Brien and McKay, 1995; Ziegelhoffer et al., 1995). Second, we found that the level of activation of Ssa1p and BiP by their respective DnaJ partner proteins decreased ∼50% in the presence of potassium (Table 1). Third, although the fold stimulation of BiP ATPase activity by Ydj1p was potassium independent, the inclusion of potassium resulted in similar levels of Ssa1p ATPase activity stimulation by both Ydj1p and GST–63Jp. Interestingly, Levy et al. (1995) demonstrated that Ydj1p and Escherichia coli DnaJ stimulated the ATPase activity of Ssa1p to nearly the same extent in the presence of potassium. These results suggest that Ssa1p may be more amenable to transient interactions with multiple DnaJ partner proteins than is BiP.
Table 1.
Fold stimulation of hsc70 ATPase activities by DnaJ homologues
| Ydjlp | 63Jp | |
|---|---|---|
| Ssalp | 6.1 (3)a | 1.5 (3.1) |
| BiP | 1.4 (1.7) | 13.1 (6.1) |
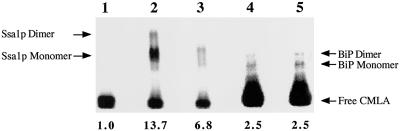
Hsc70s bind to solvent-exposed hydrophobic regions of partially unfolded proteins, and this interaction is regulated by cycles of ATP binding and hydrolysis and by DnaJ homologues (for review, see Hartl, 1996). Ydj1p, in the presence of Mg2+ and ATP, catalyzes the release of the unfolded polypeptide CMLA from Ssa1p (Cyr et al., 1992). Thus, we examined the specificity of Ydj1p-catalyzed release of CMLA from Ssa1p and BiP in the presence of ATP. Both monomeric and dimeric forms of Ssa1p and BiP were found to bind 125I-CMLA (Figure 2). We observed, however, that Ydj1p released CMLA only from Ssa1p (Figure 2, lane 3) but not from BiP (Figure 2, lane 5) in the presence of ATP and Mg2+, suggesting further that BiP and Ydj1p do not form an active chaperone pair. Although relatively efficient binding between mammalian BiP and CMLA has been observed (Fourie et al., 1994), the basis for the inefficient binding of CMLA by yeast BiP is unknown (see DISCUSSION).
Figure 2.
Ydj1p specifically catalyzes protein substrate release by Ssa1p. Ssa1p (lanes 2 and 3) or BiP (lanes 4 and 5) was preincubated with 125I-CMLA and incubated either in the absence (lanes 2 and 4) or presence (lanes 3 and 5) of Ydj1p and an ATP-regenerating system. Lane 1 contains only 125I-CMLA. Values below the figure indicate the relative amounts of the CMLA–Hsc70 complex above background as determined by phosphorimager analysis. To better detect the chaperone–CMLA complexes, we cut the gel so that only a portion of the free CMLA signal was evident.
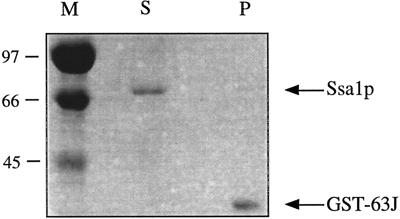
The existence of a stable, ATP-dependent interaction between BiP and the lumenal DnaJ domain of the integral membrane protein Sec63p has been shown to be necessary for posttranslational protein translocation (Brodsky and Schekman, 1993; Scidmore et al., 1993; Lyman and Schekman, 1995, 1997; Corsi and Schekman, 1997; Matlack et al., 1997). Therefore, the inability of Ssa1p to substitute for BiP in protein translocation may derive from the inability of Ssa1p to interact stably with Sec63p. To test this hypothesis, we again used GST–63Jp. BiP has been shown to interact with this fusion protein, in the presence of ATP, when GST–63Jp is immobilized on glutathione-conjugated agarose beads (Corsi and Schekman, 1997) (also see Figure 7 below). Figure 3 shows the results of such an experiment performed with Ssa1p. In the presence of 1 mM ATP, Ssa1p remains in the supernatant and fails to associate with glutathione-bound GST–63Jp. This result was confirmed with multiple preparations of Ssa1p and was unaltered by the use of ADP or ATPγS in place of ATP (our unpublished observations). Although some preparations of Ssa1p exhibited limited binding, the association between Ssa1p and GST–63Jp in these experiments was nucleotide independent, indicating that the interaction was nonspecific (our unpublished observations). These data suggest that Ssa1p may not be able to replace BiP during protein translocation because of the inability of Ssa1p to stably interact with the DnaJ domain of Sec63p.
Figure 7.
The dominant lethal BiP mutants are unable to bind to the Sec63p DnaJ domain. GST–63Jp was prebound to glutathione–cross-linked agarose, and BiP and either ATP or ADP were added. Proteins remaining in the supernatant (S) or associated with the pellet (P) were resolved by SDS-PAGE and visualized by Coomassie brilliant blue staining. Signals corresponding to BiP and GST–63Jp are indicated, as are molecular weight markers (M; × 10−3).
Figure 3.
Ssa1p cannot stably interact with the lumenal domain of Sec63p. GST–63Jp was prebound to glutathione–cross-linked agarose before Ssa1p and ATP were added, as described in MATERIALS AND METHODS. Proteins remaining in the supernatant (S) and bound to the pellet (P) were resolved by SDS-PAGE and visualized by Coomassie brilliant blue staining. Molecular weight markers are shown (×10−3) to the left (in kilodaltons).
Experiments examining whether Ssa1p and BiP form stable complexes with Ydj1p using native polyacrylamide gels were unsuccessful (our unpublished observations). To date, stable complexes between unique hsc70s and their cognate DnaJ homologues have only been observed for BiP and Sec63p (Brodsky and Schekman, 1993; Corsi and Schekman, 1997; this manuscript), hsc70 and auxilin (Jiang et al., 1997), and hsc70/hsp70 and the cysteine-string protein (Chamberlain and Burgoyne, 1997).
Dominant Lethal Mutations Map to the ATPase Domain of BiP
To better characterize the functions of BiP that are necessary for protein translocation, we generated four dominant lethal BiP mutants (see MATERIALS AND METHODS) that contain unique amino acid substitutions in the ATP-binding domain of BiP. Expression of the dominant mutants in yeast using a regulatable promoter resulted in lethality and defects in protein translocation (Vogel, 1993). The positions of the amino acid residues substituted in the four mutants were identified by DNA sequence analysis and are depicted in Figure 4. Highlighted are the bovine brain hsc70 residues that correspond to the mutated residues in yeast BiP. The altered amino acids, which are conserved among all hsc70s, and the specific amino acid changes in the BiP mutants are designated in Table 2. To study the defects of these mutant proteins in vitro, we developed a means to purify them. In brief, the N-terminal signal sequence was removed and replaced with a hexahistidine tag, and the proteins were expressed from an IPTG-inducible promoter in bacteria. Proteins were subsequently purified from whole-cell extracts by Ni2+-NTA and Q-Sepharose chromatography (see MATERIALS AND METHODS) to >90% purity as determined by SDS-PAGE analysis and Coomassie blue staining.
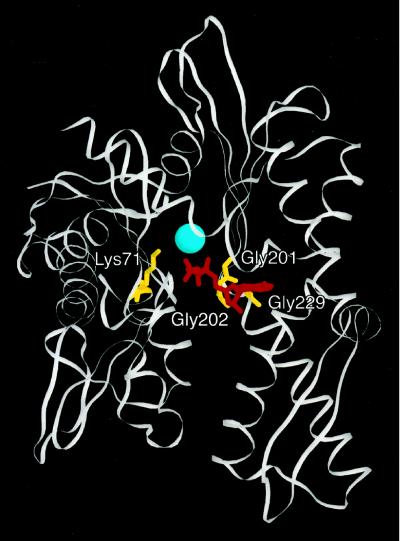
Figure 4.
Dominant lethal mutations map to the ATPase domain of BiP. Mutated residues (depicted in yellow) were mapped onto the crystal structure of the ATPase domain of bovine brain hsc70 (Flaherty et al., 1990). Adenylyl-imidodiphosphate is shown in red, and the associated magnesium ion is displayed in blue. Numbers correspond to amino acid positions in bovine brain hsc70 (see Table 2).
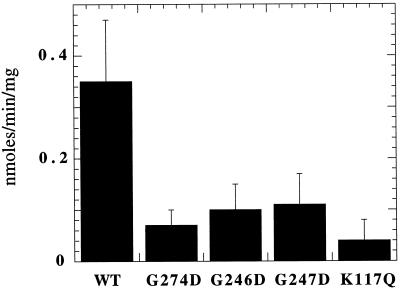
Because ATP hydrolysis is critical for posttranslational protein translocation (Hansen et al., 1986; Rothblatt and Meyer, 1986; Waters and Blobel, 1986) and because BiP is the only ATPase in the yeast translocation complex (for review, see Brodsky, 1996), we examined whether the dominant lethal BiP mutants were competent for ATP hydrolysis. Steady-state ATPase assays were performed in which the concentration of ATP was titrated and the data were analyzed by Lineweaver–Burk plots to estimate the _K_m and Vmax for wild-type BiP and each of the mutant BiP proteins. As shown in Table 3, we found that the wild-type and mutant BiP proteins exhibit a _K_m for ATP between 0.6 and 8 μM, values that correspond well to that for wild-type mammalian BiP (Wei and Hendershot, 1995). In addition, wild-type BiP exhibited a _K_d for ATP of 1 μM as determined by Scatchard analysis (our unpublished observations). Because cellular ATP levels in yeast are well above the range of _K_m values we observed (>2 mM [Brindle et al., 1990]), the major defect of these mutant proteins could, in theory, arise from apparent differences in Vmax. In fact, the mutant proteins exhibited Vmax values between 16.5- and 50-fold lower than that observed for wild-type BiP (Table 3). We next conducted steady-state ATPase assays using ATP at a final concentration of 50 μM, which is well in excess of the _K_m (Table 3), and found that the steady-state ATPase activity of wild-type BiP was consistently at least threefold greater than that of any of the mutants (Figure 5). These results are also in accordance with those obtained by Wei et al. (1995) who noted that there was an approximately threefold decrease in the Vmax of the mammalian G226D and G227D BiP mutants, proteins that correspond to the yeast G246D and G247D mutants, respectively (see Table 2).
Table 3.
Results of Lineweaver-Burk analyses of WT and dominant mutant BiP proteins
| Vmax | _K_m | R2 value | |
|---|---|---|---|
| WT | 1.65 | 8 | 0.999 |
| G274D | 0.055 | 0.72 | 0.981 |
| G246D | 0.058 | 0.91 | 0.977 |
| G247D | 0.1 | 2 | 0.997 |
| K117Q | 0.03 | 0.65 | 0.981 |
Figure 5.
The BiP mutants are defective for ATP hydrolysis. Wild- type or the dominant mutant BiP isolates were incubated for 1 h at 30°C, and ATP hydrolysis was assessed as described in MATERIALS AND METHODS. Data represent the means of at least four independent assays (± SD).
Dominant Mutant Forms of BiP Are Unable to Exhibit an ATP-dependent Conformational Change
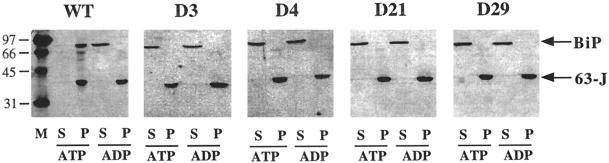
Protease digestion of ATP- or ADP-bound hsp70/hsc70 results in characteristic nucleotide-dependent patterns of protease protection (Chappell et al., 1987; Kassenbrock and Kelly, 1989; Kamath-Loeb et al., 1995; Wei and Hendershot, 1995; Fung et al., 1996). These patterns indicate that hsc70s undergo a conformational change upon binding ATP. This conformational change may be necessary to modulate hsc70-specific functions such as binding to unfolded proteins or, in the case of yeast BiP, interacting with Sec63p. We found that an ∼45-kDa fragment of wild-type yeast BiP was protected in the presence of ATP after limited digestion with proteinase K (Figure 6, open arrow). By determining that the ∼45-kDa fragment bound to Ni2+-NTA and thus retained the N-terminal hexahistidine tag (our unpublished observations), we concluded that this fragment represented the N-terminal ATPase domain. In the presence of ADP, an ∼60-kDa fragment encompassing the ATPase domain and a portion of the peptide-binding domain was protected from proteinase K (Figure 6, closed arrow). These results suggest that the C-terminal peptide-binding domain is more accessible to protease when BiP is bound to ATP. Similar results have been obtained with related hsc70s (Kamath-Loeb et al., 1995; Wei and Hendershot, 1995).
Figure 6.
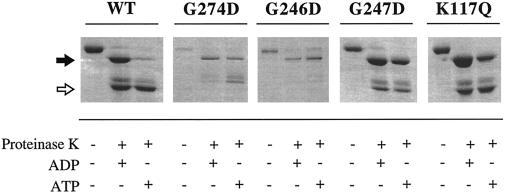
Dominant mutant forms of BiP are unable to undergo an ATP-dependent conformational change. Either wild-type or dominant mutant BiP (as indicated) was preincubated with either ATP or ADP before the addition of proteinase K, as described in MATERIALS AND METHODS. The resulting cleavage products were resolved by SDS-PAGE and visualized by Coomassie brilliant blue staining. The filled arrow indicates the position of the ∼60-kDa fragment (corresponding to the ATPase domain and a portion of the C-terminal peptide-binding domain), and the open arrow indicates the position of the ∼45-kDa fragment (comprised primarily of the ATPase domain).
When the dominant lethal BiP mutant proteins were treated with proteinase K in the presence of either ATP or ADP, the proteolytic patterns obtained resembled the ADP-bound conformation of wild-type BiP. Although the ∼60-kDa fragment in the K117Q mutant was more susceptible to protease in the presence of ATP than were the other three mutant proteins, there was an approximately fourfold increase in degradation of the ∼60-kDa band from wild-type BiP but less than a twofold increase for the K117Q mutant in this experiment. We conclude that the BiP mutants are deficient to different extents in their ability to undergo a conformational change in the presence of ATP.
The Dominant Lethal Mutants Are Unable to Bind to GST–63Jp and Fail to Support Protein Translocation into Reconstituted Proteoliposomes
The ability of BiP to interact with the lumenal domain of Sec63p in an ATP-dependent manner is essential for posttranslational protein translocation as well as for cell viability at elevated temperatures (Feldheim et al., 1992; Brodsky and Schekman, 1993; Nelson et al., 1993; Scidmore et al., 1993; Lyman and Schekman, 1995, 1997; Corsi and Schekman, 1997; Matlack et al., 1997). Thus, we examined whether the dominant lethal proteins associated with GST–63Jp. We initially confirmed the results of Corsi and Schekman (1997) by showing that wild-type BiP associates with GST–63Jp bound to glutathione-agarose in the presence of 1 mM ATP but not in the presence of 1 mM ADP (Figure 7). However, each of the dominant mutants was unable to stably associate with GST–63Jp in the presence of either nucleotide (Figure 7), suggesting that the conformational change accompanying ATP binding, and not simply the ability to bind ATP, is imperative for a stable interaction with Sec63p.
Protein translocation can be reconstituted in vitro either by inserting detergent-solubilized microsomal proteins and BiP into phospholipid vesicles (Brodsky et al., 1993; Brodsky and Schekman, 1993) or by directly reconstituting the purified translocation complex into vesicles (Panzner et al., 1995). In both cases, the resulting reconstituted proteoliposomes support the ATP-dependent posttranslational translocation of a radiolabeled yeast mating pheromone, ppαf. The amount of ppαf successfully translocated is determined by SDS-PAGE to detect protease-protected and signal sequence–cleaved ppαf.
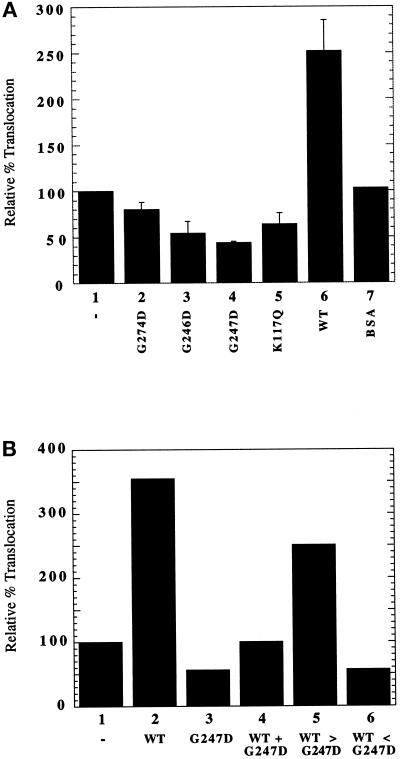
To verify that the dominant lethal phenotype of the BiP mutants in vivo was directly related to their ability to compromise protein translocation, we inserted wild-type BiP and the mutant proteins into reconstituted proteoliposomes as previously described (Brodsky et al., 1993; Brodsky and Schekman, 1993). The translocation of ppαf into proteoliposomes lacking added protein was normalized to represent 100% translocation (Figure 8A). As shown previously, the addition of wild-type BiP to 5% of total protein stimulated ppαf translocation approximately threefold (Brodsky and Schekman, 1993). In contrast, if mutant BiP protein was added, ppαf translocation was inhibited to levels lower than that seen in reconstituted vesicles lacking wild-type BiP (Figure 8A). As a negative control, we showed that ppαf translocation was unaffected by BSA.
Figure 8.
The dominant lethal BiP mutants compromise protein translocation in reconstituted proteoliposomes. (A) The translocation of ppαf into proteoliposomes reconstituted from solubilized wild-type microsomes was examined either in the absence or presence of wild-type or mutant BiP protein or in the presence of BSA, as described in MATERIALS AND METHODS. Bar 1, no added protein; bar 2, G274D; bar 3, G246D; bar 4, G247D; bar 5, K117Q; bar 6, wild-type BiP; bar 7, BSA. The amount of in vitro–translated 35S-ppαf that was translocated into reconstituted proteoliposomes to which no exogenous BiP was added (bar 1) was normalized to 100% and corresponded to a translocation efficiency of ∼10%. Data represent the means of three independent determinations (± SD). (B) The translocation of ppαf into reconstituted proteoliposomes containing the following proteins was examined: bar 1, no added protein; bar 2, 17 μg of wild-type BiP; bar 3, 17 μg of G247D; bar 4, 17 μg each of wild-type BiP and G247D; bar 5, 25 μg of wild-type BiP and 8 μg of G247D; bar 6, 8 μg of wild-type BiP and 25 μg of G247D. Relative percent translocation was determined as described in A.
To define further the mechanism of this inhibition, we titrated the amounts of wild-type BiP and of the G247D mutant relative to one another in the reconstituted vesicles and assayed for ppαf translocation. We found that the addition of equimolar amounts of wild-type BiP and G247D resulted in 3.5-fold less ppαf translocation than the level that was achieved when only additional wild-type protein was added (Figure 8B, compare bars 2 and 4). Also, the addition of a greater than threefold excess of wild-type BiP over G247D was unable to restore translocation to wild-type levels (Figure 8B, compare bars 2 and 5). On the basis of these results, we conclude that the dominant lethal proteins are acting such that they directly interfere with the action of wild-type BiP. Models for this interference are presented in the DISCUSSION.
DISCUSSION
These studies were initiated to better understand the function of yeast BiP during protein translocation. To this end, two unique approaches were taken. First, purified proteins and established in vitro analyses were used to elucidate the basis for molecular chaperone specificity during protein translocation, and second, the characterization of dominant BiP mutants indicated that an ATP-dependent conformational change may be coupled to Sec63p binding.
We have found that four dominant lethal BiP mutants, each of which has a single amino acid change in the ATPase domain (Figure 4 and Table 2), are defective for ATP hydrolysis (Figure 5 and Table 3) and cannot perform an ATP-dependent conformational change (Figure 6). Because wild-type BiP and related hsc70s exhibit different ATP- and ADP-bound conformations (Chappell et al., 1987; Kassenbrock and Kelly, 1989; Kamath-Loeb et al., 1995; Wei and Hendershot, 1995; Fung et al., 1996), some aspect of hsc70 function may rely on these conformational changes. Liberek et al. (1991) have proposed that such a conformational change triggers the release of substrates from DnaK. In the case of yeast BiP, we suggest that this conformational change is required for association with its DnaJ partner protein Sec63p, because each of the dominant mutants examined here cannot interact with the Sec63p DnaJ domain (Figure 7). Because the ability of BiP to interact with Sec63p in an ATP-dependent manner is essential for posttranslational translocation (Brodsky and Schekman, 1993; Scidmore et al., 1993; Lyman and Schekman, 1995, 1997; Corsi and Schekman, 1997; Matlack et al., 1997), we had anticipated that the BiP mutants would be defective for this process in vitro. Indeed, we found that each dominant lethal BiP mutant inhibited the translocation of ppαf into reconstituted proteoliposomes (Figure 8A) and that the addition of equimolar amounts of wild-type BiP and G247D to reconstituted proteoliposomes resulted in basal levels of ppαf translocation, even though adding the same amount of wild-type BiP in the absence of G247D increased ppαf translocation ∼3.5-fold (Figure 8B, compare lanes 1, 2, and 4). We conclude that the dominant BiP mutant proteins prevent posttranslational protein translocation by directly interfering with the function of wild-type BiP.
The question remains as to why these mutant proteins are dominant. Our results suggest that the proteins do not interfere with wild-type BiP by competing for Sec63p binding because they are unable to interact with the lumenal, DnaJ domain of Sec63p (Figure 7). The dominant mutant proteins are also unable to interfere directly with ATP hydrolysis by wild-type BiP because experiments in which equimolar amounts of wild-type and mutant protein were included in an ATPase reaction demonstrated no inhibition of wild-type activity (our unpublished observations). Our data instead suggest three possible scenarios. First, the mutant BiP proteins may lock onto translocating polypeptides and halt further import. In support of this hypothesis, it has been shown that the mammalian BiP mutant G227D, analogous to our G247D mutant (see Table 2), binds irreversibly to murine immunoglobulin lambda light chains in the ER and prevents their secretion (Hendershot et al., 1996). In examining whether this is the case for the yeast mutant proteins, we and others have observed polypeptide binding to both the wild-type and mutant BiP proteins but were unable to achieve substrate release in the presence of GST–63Jp and ATP (Argon, personal communication; our unpublished observations). This raises the intriguing possibility that another ER lumenal DnaJ homologue, perhaps Scj1p or Jem1p (Schlenstedt et al., 1995; Nishikawa and Endo, 1997), may interact with BiP to modulate its interaction with polypeptide substrates. Second, the dominant mutants may compromise the function of wild-type BiP by forming mixed dysfunctional dimers. In accordance with this hypothesis, we observed that both monomeric and dimeric BiPs bind to an unfolded polypeptide substrate (Figure 2), and we have identified monomers, dimers, and higher order oligomers of wild-type and each of the dominant mutant BiP isolates when the purified proteins were subjected to nondenaturing PAGE (our unpublished observations). Although previous studies with mammalian BiP and the mitochondrial hsp70 found that the addition of synthetic peptides or ATP converted dimers to monomers (Blond-Elguindi et al., 1993; Wei et al., 1995; Azem et al., 1997), our attempts to catalyze such a conversion, and to address whether the dominant mutants prevent this conversion, have been unsuccessful. Third, we cannot eliminate the possibility that the dominant mutants bind irreversibly to another component of the translocation complex, such as the translocation channel itself (Sec61p; Brodsky, 1996), an effect that would result in translocation arrest and cell death. Recent data from Hamman et al. (1998) demonstrating a nucleotide-dependent, stoichiometric interaction of BiP with the translocation pore in mammalian microsomes support this hypothesis. Future experiments will seek to define which of these three scenarios is most likely to be correct.
We are confident that the results we have obtained with the dominant mutants are not an artifact of the use of bacterially expressed hexahistidine-tagged forms of BiP. First, the ATPase activity of wild-type BiP purified from yeast is ∼0.52 nmol·min−1·mg−1 at 30°C (Goeckeler and Brodsky, unpublished observations; also see Brodsky et al., 1995), and the ATPase activities we report in this study for bacterially expressed hexahistidine-tagged BiP range from 0.36 to 1.22 nmol·min−1·mg−1 at 30°C. Second, BiP purified from yeast and bacterially expressed recombinant BiP behaved identically in their abilities to rescue ppαf translocation in in vitro translocation reactions (Brodsky et al., 1993) (Figure 8). Third, the biochemical activities of bacterially expressed hexahistidine-tagged mammalian BiP from which the hexahistidine tag had or had not been cleaved were nearly identical when analyzed for ATPase activity, oligomerization status, and the ability to undergo an ATP-dependent conformational change (Wei and Hendershot, 1995).
We have also shown in this report that specific DnaK–DnaJ homologue interactions exist on opposing sides of the ER membrane, thus providing an explanation for the observation that Ssa1p and BiP are unable to substitute for one another during protein translocation (Brodsky et al., 1993). This result further defines the necessity of specific molecular chaperone interactions during preprotein import. We found that the ATPase activity of the cytosolic hsc70 Ssa1p is specifically activated by the cytosolic protein Ydj1p but that Ydj1p is unable to similarly enhance the ATPase activity of BiP (Figure 1A). Also, we showed that Ydj1p effected polypeptide release from Ssa1p but not from BiP (Figure 2) and that Ssa1p is unable to stably interact with the lumenal domain of Sec63p in an ATP-dependent manner (Figure 3).
The notion that specific functions exist for eukaryotic hsc70s and that the specificity of these functions can be regulated by unique DnaJ-like partner proteins is well established (Brodsky et al., 1993; Wiech et al., 1993; Cyr and Douglas, 1994; Cyr et al., 1994; Cyr, 1995; King et al., 1995; Levy et al., 1995). Interestingly, the DnaJ domain of another ER lumenal DnaJ homologue, Scj1p, was able to functionally replace the DnaJ domain of Sec63p (Schlenstedt et al., 1995). These results suggest that the specificity of DnaK–DnaJ interactions may also depend on the context in which the conserved DnaJ domain lies. The identification of new ER-associated chaperones in yeast (Baxter et al., 1996; Hamilton and Flynn, 1996; Nishikawa and Endo, 1997; Saris et al., 1997) dictates that continued biochemical analyses are warranted to define the specificity of molecular chaperone action.
ACKNOWLEDGMENTS
We thank Ann Corsi and Randy Schekman for generously providing the strain expressing GST–63Jp, Lois Greene and Evan Eisenberg for advice on kinetic analysis, Tom Harper for assistance with computer-generated images, and Susan Lyman for comments on the manuscript. This work was supported by grant MCB-9506002 from the National Science Foundation to J.L.B. and grant GM-37739 from the National Institutes of Health to M.D.R. A.J.M. acknowledges the support of a Department of Defense predoctoral training grant, J.B.E. received support from a Research Experience for Undergraduates award from the National Science Foundation, and J.L.B. acknowledges a Junior Faculty Research Award grant from the American Cancer Society.
REFERENCES
- Azem A, Oppliger W, Lustig A, Jeno P, Feifel B, Schatz G, Horst M. The mitochondrial hsp70 chaperone system. J Biol Chem. 1997;272:20901–20906. doi: 10.1074/jbc.272.33.20901. [DOI] [PubMed] [Google Scholar]
- Baxter BK, James P, Evans T, Craig EA. SSI1 encodes a novel hsp70 of the Saccharomyces cerevisiae endoplasmic reticulum. Mol Cell Biol. 1996;16:6444–6456. doi: 10.1128/mcb.16.11.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Walter W, Yan W, Craig EA. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blond-Elguindi S, Fourie AM, Sambrook JF, Gething M-J. Peptide-dependent stimulation of the ATPase activity of the molecular chaperone BiP is the result of conversion of oligomers to active monomers. J Biol Chem. 1993;268:12730–12735. [PubMed] [Google Scholar]
- Brindle K, Braddock P, Fulton S. 31P NMR measurements of the ADP concentration in yeast cells genetically modified to express creatine kinase. Biochemistry. 1990;29:3295–3302. doi: 10.1021/bi00465a021. [DOI] [PubMed] [Google Scholar]
- Brodsky JL. Post-translational protein translocation: not all hsc70s are created equal. Trends Biochem Sci. 1996;21:122–126. [PubMed] [Google Scholar]
- Brodsky JL. Quantitation of membrane proteins in reconstituted vesicles prepared from yeast. Anal Biochem. 1997;246:262–263. doi: 10.1006/abio.1997.2028. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Goeckeler J, Schekman R. Sec63p and BiP are required for both co- and post-translational protein translocation into yeast microsomes. Proc Natl Acad Sci USA. 1995;92:9643–9646. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Hamamoto S, Feldheim D, Schekman R. Reconstitution of protein translocation from solubilized yeast membranes reveals topologically distinct roles for BiP and cytosolic hsc70. J Cell Biol. 1993;120:95–102. doi: 10.1083/jcb.120.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Schekman R. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush GL, Meyer DI. The refolding activity of the yeast heat shock proteins Ssa1 and Ssa2 defines their role in protein translocation. J Cell Biol. 1996;135:1229–1237. doi: 10.1083/jcb.135.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AJ, Cyr DM, Douglas MG. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- Chamberlain LH, Burgoyne RD. Activation of the ATPase activity of heat-shock proteins Hsc70/Hsp70 by cysteine-string protein. Biochem J. 1997;322:853–858. doi: 10.1042/bj3220853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell TG, Konforti BB, Schmidt SL, Rothman JE. The ATPase core of a clathrin uncoating protein. J Biol Chem. 1987;262:746–751. [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure, function, and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress & Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirico WJ, Waters MG, Blobel G. 70K heat shock related proteins stimulate protein translocation into microsomes. Nature. 1988;332:805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- Corsi AK, Schekman R. Mechanism of polypeptide translocation into the endoplasmic reticulum. J Biol Chem. 1996;271:30299–30302. doi: 10.1074/jbc.271.48.30299. [DOI] [PubMed] [Google Scholar]
- Corsi AK, Schekman R. The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. J Cell Biol. 1997;137:1483–1493. doi: 10.1083/jcb.137.7.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr DM. Cooperation of the molecular chaperone Ydj1 with specific Hsp70 homologs to suppress protein aggregation. FEBS Lett. 1995;359:129–132. doi: 10.1016/0014-5793(95)00024-4. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Douglas MG. Differential regulation of hsp70 subfamilies by the eukaryotic DnaJ homologue YDJ1. J Biol Chem. 1994;269:9798–9804. [PubMed] [Google Scholar]
- Cyr DM, Langer T, Douglas MG. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Lu X, Douglas MG. Regulation of hsp70 function by a eukaryotic DnaJ homolog. J Biol Chem. 1992;267:20927–20931. [PubMed] [Google Scholar]
- Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor proteins. Nature. 1988;332:800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- Feldheim D, Rothblatt J, Schekman R. Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol Cell Biol. 1992;12:3288–3296. doi: 10.1128/mcb.12.7.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- Fourie AM, Sambrook JF, Gething M-J. Common and divergent peptide binding specificities of hsp70 molecular chaperones. J Biol Chem. 1994;269:30470–30478. [PubMed] [Google Scholar]
- Fung KL, Hilgenberg L, Wang NM, Chirico WJ. Conformations of the nucleotide and polypeptide binding domains of a cytosolic hsp70 molecular chaperone are coupled. J Biol Chem. 1996;271:21559–21565. doi: 10.1074/jbc.271.35.21559. [DOI] [PubMed] [Google Scholar]
- Gething M-J, Blond-Elguindi S, Buchner J, Fourie A, Knarr G, Modrow S, Nanu L, Segal M, Sambrook J. Protein Kinesis: The Dynamics of Protein Trafficking and Stability, Cold Spring Harbor Symposia on Quantitative Biology. LX. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1995. Binding sites in hsp70 molecular chaperones in natural proteins; pp. 417–428. [DOI] [PubMed] [Google Scholar]
- Haas IG, Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983;306:387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- Hamilton TG, Flynn GC. Cer1p, a novel hsp70-related protein required for posttranslational endoplasmic reticulum translocation in yeast. J Biol Chem. 1996;271:30610–30613. doi: 10.1074/jbc.271.48.30610. [DOI] [PubMed] [Google Scholar]
- Hamman BD, Hendershot LM, Johnson AE. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 1998;92:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- Hansen W, Garcia PD, Walter P. In vitro protein translocation across the yeast endoplasmic reticulum: ATP-dependent posttranslational translocation of the prepro–factor. Cell. 1986;45:397–406. doi: 10.1016/0092-8674(86)90325-9. [DOI] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hendershot L, Wei J-Y, Gaut J, Melnick J, Aviel S, Argon Y. Inhibition of immunoglobulin folding and secretion by dominant negative BiP ATPase mutants. Proc Natl Acad Sci USA. 1996;93:5269–5274. doi: 10.1073/pnas.93.11.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R-F, Greener T, Barouch W, Greene L, Eisenberg E. Interaction of auxilin with the molecular chaperone, Hsc70. J Biol Chem. 1997;272:6141–6145. doi: 10.1074/jbc.272.10.6141. [DOI] [PubMed] [Google Scholar]
- Johnson AE. Protein translocation at the ER membrane: a complex process becomes more so. Trends Cell Biol. 1997;7:90–95. doi: 10.1016/S0962-8924(97)01029-5. [DOI] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Lu CZ, Suh W-C, Lonetto MA, Gross CA. Analysis of three DnaK mutant proteins suggests that progression through the ATPase cycle requires conformational changes. J Biol Chem. 1995;270:30051–30059. doi: 10.1074/jbc.270.50.30051. [DOI] [PubMed] [Google Scholar]
- Kassenbrock CK, Kelly RB. Interaction of heavy chain binding protein (BiP/GRP78) with adenine nucleotides. EMBO J. 1989;8:1461–1467. doi: 10.1002/j.1460-2075.1989.tb03529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C, Eisenberg E, Greene L. Polymerization of 70-kDa heat shock protein by yeast DnaJ in ATP. J Biol Chem. 1995;270:22535–22540. doi: 10.1074/jbc.270.38.22535. [DOI] [PubMed] [Google Scholar]
- Levy EJ, McCarty J, Bukau B, Chirico WJ. Conserved ATPase and luciferase refolding activities between bacteria and yeast hsp70 chaperones and modulators. FEBS Lett. 1995;368:435–440. doi: 10.1016/0014-5793(95)00704-d. [DOI] [PubMed] [Google Scholar]
- Liberek K, Skowyra D, Zylicz M, Johnson C, Georgopoulos C. The Escherichia coli DnaK chaperone, the 70-kDa heat shock protein eukaryotic equivalent, changes conformation upon ATP hydrolysis, thus triggering its dissociation from a bound target protein. J Biol Chem. 1991;266:14491–14496. [PubMed] [Google Scholar]
- Lyman SK, Schekman R. Interaction between BiP and Sec63p is required for the completion of protein translocation into the ER of Saccharomyces cerevisiae. J Cell Biol. 1995;131:1163–1171. doi: 10.1083/jcb.131.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman SK, Schekman R. Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell. 1997;88:85–96. doi: 10.1016/s0092-8674(00)81861-9. [DOI] [PubMed] [Google Scholar]
- Matlack KES, Plath K, Misselwitz B, Rapoport TA. Protein transport by purified yeast Sec complex and Kar2p without membranes. Science. 1997;277:938–941. doi: 10.1126/science.277.5328.938. [DOI] [PubMed] [Google Scholar]
- McCracken AA, Brodsky JL. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MK, Kurihara T, Silver PA. Extragenic suppressors of mutations in the cytoplasmic C terminus of SEC63 define five genes in Saccharomyces cerevisiae. Genetics. 1993;134:159–173. doi: 10.1093/genetics/134.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TH, Law DTS, Williams DB. Binding protein BiP is required for translocation of secretory proteins into the endoplasmic reticulum in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:1565–1569. doi: 10.1073/pnas.88.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S, Endo T. The yeast JEM1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. J Biol Chem. 1997;272:12889–12892. doi: 10.1074/jbc.272.20.12889. [DOI] [PubMed] [Google Scholar]
- Normington K, Kohno K, Kozutsumi Y, Gething M-J, Sambrook J. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell. 1989;57:1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- O’Brien MC, McKay DB. How potassium affects the activity of the molecular chaperone Hsc70. J Biol Chem. 1995;270:2247–2250. doi: 10.1074/jbc.270.5.2247. [DOI] [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 1995;81:561–570. doi: 10.1016/0092-8674(95)90077-2. [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membrane. Annu Rev Biochem. 1996;65:271–304. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- Rose MD, Fink GR. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987;48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Rothblatt JA, Deshaies RJ, Sanders S, Daum G, Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol. 1989;109:2641–2652. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothblatt JA, Meyer DI. Secretion in yeast: translocation and glycosylation of prepro–factor in vitro can occur via an ATP-dependent post-translational mechanism. EMBO J. 1986;5:1031–1036. doi: 10.1002/j.1460-2075.1986.tb04318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler I, Chiang A, Kurihara T, Rothblatt J, Way J, Silver P. A yeast gene important for protein assembly into the endoplasmic reticulum and the nucleus has homology to DnaJ, an Escherichia coli heat shock protein. J Cell Biol. 1989;109:2665–2675. doi: 10.1083/jcb.109.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Whitfield KM, Vogel JP, Rose MD, Schekman R. Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell. 1992;69:353–365. doi: 10.1016/0092-8674(92)90415-9. [DOI] [PubMed] [Google Scholar]
- Saris N, Holkeri H, Craven RA, Stirling CJ, Makarow M. The hsp70 homologue Lhs1p is involved in a novel function of the yeast endoplasmic reticulum, refolding and stabilization of heat-denatured protein aggregates. J Cell Biol. 1997;137:813–824. doi: 10.1083/jcb.137.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Harris S, Risse B, Lill R, Silver PA. A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/Kar2p via a conserved domain that specifies interactions with hsp70s. J Cell Biol. 1995;129:979–988. doi: 10.1083/jcb.129.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scidmore M, Okamura HH, Rose MD. Genetic interactions between KAR2 and SEC63, encoding eukaryotic homologues of DnaK and DnaJ in the endoplasmic reticulum. Mol Biol Cell. 1993;4:1145–1159. doi: 10.1091/mbc.4.11.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai J, Kornberg A. A prepriming DNA replication enzyme of Escherichia coli. II. Purification of protein n′: a sequence specific, DNA-dependent ATPase. J Biol Chem. 1980;255:6789–6793. [PubMed] [Google Scholar]
- Srinivasan A, McClellan AJ, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky JL, Pipas JM. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol. 1997;17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP. Kar2, the Yeast Homologue of Mammalian BiP/GRP78. Ph.D. Thesis. Princeton, NJ: Princeton University; 1993. [Google Scholar]
- Vogel JP, Misra LM, Rose MD. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J Cell Biol. 1990;110:1885–1895. doi: 10.1083/jcb.110.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MG, Blobel G. Secretory protein translocation in a yeast cell-free system can occur posttranslationally and requires ATP hydrolysis. J Cell Biol. 1986;102:1543–1550. doi: 10.1083/jcb.102.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J-Y, Gaut JR, Hendershot LM. In vitro dissociation of BiP-peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J Biol Chem. 1995;270:26677–26682. doi: 10.1074/jbc.270.44.26677. [DOI] [PubMed] [Google Scholar]
- Wei J-Y, Hendershot LM. Characterization of the nucleotide binding properties and ATPase activity of recombinant hamster BiP purified from bacteria. J Biol Chem. 1995;270:26670–26676. doi: 10.1074/jbc.270.44.26670. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M, Stone DE, Craig EA. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiech H, Buchner J, Zimmerman M, Zimmerman R, Jakob U. Hsc70, immunoglobulin heavy chain binding protein, and hsp90 differ in their ability to stimulate transport of precursor proteins into mammalian microsomes. J Biol Chem. 1993;268:7414–7421. [PubMed] [Google Scholar]
- Ziegelhoffer T, Lopez-Buesa P, Craig EA. The dissociation of ATP from hsp70 of Saccharomyces cerevisiae is stimulated by both Ydj1p and peptide substrates. J Biol Chem. 1995;270:10412–10419. doi: 10.1074/jbc.270.18.10412. [DOI] [PubMed] [Google Scholar]