Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity (original) (raw)
Abstract
Aire induces ectopic expression of peripheral tissue antigens (PTAs) in thymic medullary epithelial cells, which promotes immunological tolerance. Beginning with a broad screen of histone peptides, we demonstrate that the mechanism by which this single factor controls the transcription of thousands of genes involves recognition of the amino-terminal tail of histone H3, but not of other histones, by one of Aire's plant homeodomain (PHD) fingers. Certain posttranslational modifications of H3 tails, notably dimethylation or trimethylation at H3K4, abrogated binding by Aire, whereas others were tolerated. Similar PHD finger–H3 tail-binding properties were recently reported for BRAF-histone deacetylase complex 80 and DNA methyltransferase 3L; sequence alignment, molecular modeling, and biochemical analyses showed these factors and Aire to have structure–function relationships in common. In addition, certain PHD1 mutations underlying the polyendocrine disorder autoimmune polyendocrinopathy–candidiases–ectodermaldystrophy compromised Aire recognition of H3. In vitro binding assays demonstrated direct physical interaction between Aire and nucleosomes, which was in part buttressed by its affinity to DNA. In vivo Aire interactions with chromosomal regions depleted of H3K4me3 were dependent on its H3 tail-binding activity, and this binding was necessary but not sufficient for the up-regulation of genes encoding PTAs. Thus, Aire's activity as a histone-binding module mediates the thymic display of PTAs that promotes self-tolerance and prevents organ-specific autoimmunity.
Keywords: T cell tolerance, plant homeodomain finger, thymus, APS-1, APECED
Central mechanisms of tolerance induction are an important means of protecting an individual from autoimmunity (1). The breadth of central T cell tolerance reflects the spectrum of self-antigens presented to differentiating thymocytes, a spectrum now known to include thousands of peripheral tissue antigens (PTAs) representing essentially all parenchymal organs (2). Much of this broad repertoire is expressed by a small subset of thymic medullary epithelial cells (MECs), which somehow permit transcriptional access to otherwise tissue-specific genes, enabling these cells to purge tissue-reactive specificities from the T cell repertoire (3–5). Genetic analyses revealed the transcriptional regulator Aire to be the molecular determinant of autoimmune polyendocrinopathy–candidiases–ectodermal dystrophy (APECED) (6, 7); mechanistic studies on Aire-deficient mice, which also show polyendocrine autoimmunity (3, 8), demonstrated its control over a large fraction of PTAs representing a wide range of peripheral organs (3).
The molecular mechanism of Aire has been elusive, although studies on the particular genes it controls have provided some clues. Bioinformatic analyses revealed significant clustering of loci regulated by Aire in an interspersed pattern of Aire-independent, Aire-induced, and Aire-repressed genes (9, 10). This noncontiguous clustering may reflect shifts in looping and long-distance intrachromosomal and interchromosomal interactions (11). Aire-dependent dysregulation of the H19/Igf2 imprinted cluster (9) is consistent with this view, because the imprinting status is coordinated by higher-order chromatin configurations involving the action of CCCTC-binding factor (12). Additionally, the clustering of Aire-regulated genes may involve the recruitment of tissue-specific domains to euchromatic territories. Indeed, Aire is located adjacent to nuclear speckles in MECs (13), a structure enriched with RNA polymerase II (Pol II), transcriptional elongation factors, chromatin-remodeling complexes, and essentially all factors required for pre-mRNA splicing (14, 15).
The domain structure of Aire is also indicative of a chromatin-associated factor. The Sp100, Aire1, NucP41/75, DEAF1 (SAND) domain is homologous to regions in the Sp100 family of transcription factors that associate with the nuclear matrix (16). Interestingly, Aire interacts with matrix-associated proteins in transfection experiments, suggesting a potential mechanism of recruiting discrete chromosomal domains into active matrix-associated regions (17). A potential mechanism for Aire's interaction with chromatin has emerged from the recent characterization of plant homeodomain (PHD) zinc fingers as histone-binding modules that recognize specific posttranslational modifications (PTMs) on histone tails (18). Distinct patterns of PTM recognition facilitate the recruitment and/or stabilization of macromolecular machinery that effect changes in the dynamic and structural properties of the target loci. These properties largely determine the transcriptional programs important for the differentiation state of the cell (18). AIRE contains two PHD fingers that could potentially couple tissue-specific chromosomal domains featuring distinct PTMs with cognate effector machinery that can directly or indirectly modify the transcriptional state.
We exploited a broad in vitro screen to identify direct interactions between Aire and specific histone PTMs, confirmed and further defined these interactions through mutagenesis and biochemical experiments, and determined their in vivo contribution to ectopic up-regulation of PTAs. In brief, the results demonstrate a link between histone-binding modules and organ-specific tolerance mechanisms involved in human disease.
Results
Aire Recognizes the Unmodified N Terminus of Histone H3 Through Its PHD1 Domain.
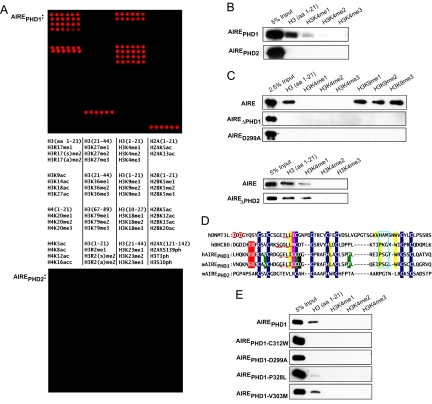
To investigate whether one or the other of the Aire PHD fingers can directly interact with histones, particularly histone PTMs, we probed microarrays containing ≈60 distinct “bare” and modified histone peptides with GST-tagged murine AirePHD1 (amino acids 294–350), AirePHD2 (amino acids 430–481), or AirePHD1+2 (amino acids 294–481). AirePHD1 and AirePHD1+2 recognized the unmodified N-terminal 21 residues of histone H3, but no other H3, H2A, H2B, H4, or H2AX fragments tested [Fig. 1A Top and supporting information (SI) Fig. S1]. In striking contrast, AirePHD2 did not detectably bind to any of the histone peptides (Fig. 1A Bottom). The binding of AirePHD1 to H31–21 was abrogated by methylation at Arg-2 (H3R2), phosphorylation at Thr-3 (H3T3), or di/trimethylation of Lys-4 (H3K4) (Fig. 1A). It also could not bind to peptides without the first nine residues of H3 but tolerated modifications at H3K9, Ser-10 (H3S10), H3K14, and H3R17, suggesting recognition of a motif spanning (at least) the first eight residues of H3. A peptide pulldown assay confirmed the interaction of AirePHD1 but not AirePHD2 with the unmodified H3 tail, as well as the inhibition of this binding by di/trimethylated H3K4 (Fig. 1B). Binding to H3K4me1 was inconsistent in these assays, likely reflecting a weak interaction sensitive to experimental variability. Pulldown experiments with full-length Aire also showed that PHD1 was required for H3 tail binding (Fig. 1C, upper), that PHD2 was dispensable (Fig. 1C, lower), and that Aire could interact with monomethylated, dimethylated, and trimethylated H3K9 (Fig. 1C Upper).
Fig. 1.
Aire recognizes the unmodified N terminus of H3 tail via PHD1. (A) Histone peptide microarray containing the indicated modifications was probed with GST-mAire294–350 or GST-mAire425–481. Red spots indicate GST-specific signals. H indicates histone; me, methylation; ac, acetylation; ph, phosphorylation; s, symmetric; a, asymmetric. (B) Anti-GST Western blots of histone peptide pulldowns with indicated biotinylated peptides and GST-mAirePHD1 or PHD2 fusion proteins. (C) Peptide pulldowns as in B except with full-length WT, D299A, or ΔPHD2 Aire-flag. (D) Comparisons of PHD fingers from BHC80, hAIRE, mAire, and DNMT3L. Blue indicates Zn2+-chelating residues. Red indicates H3K4me0-binding residues. Red underscore indicates antiparallel β-strand-engaging H3 peptide. Pink indicates residues that insert between H3R2me0 and H3K4me0 and interact with H3 side chains. Yellow indicates hydrophobic pocket specific to H3A1. Turquoise box indicates hydrogen bond cage recognizing N terminus of H3. Black indicates predicted H3R2me0-interacting residue. Green indicates APECED mutations: V301M (identified in patient with autoimmune Addison's disease, ref. 46), C311Y, and P326L/Q. (E) Peptide pulldowns as in B except with critical binding and APECED mutations.
Aire Has Structure–Function Properties in Common with BRAF-Histone Deacetylase Complex 80 (BHC80) and DNA Methyltransferase 3L (DNMT3L).
Aire's recognition of the unmodified N terminus of H3, in particular its sensitivity to H3K4 methylation, echoed the recently reported recognition properties of histone-binding modules of BHC80 and DNMT3L (19, 20). Therefore, we aligned the PHD fingers of murine Aire (mAire), human AIRE (hAIRE), human BHC80, and the Cys-rich PHD-like domain of human DNMT3L to determine whether the defined H3-recognition motifs of the latter proteins could reveal parallel determinants for Aire. H3 peptides show an induced-fit, antiparallel, β-strand conformation along the exposed edge of the β-sheet in BHC80 and DNMT3L (19, 20). Human AIRE and mAire share conserved residues that form this β-sheet scaffold (Fig. 1D, red underscore). L501/L106 of BHC80/DNMT3L, respectively, interact with downstream residues (BHC80: L512, W527, and P523; DNMT3L: V134 and W140) to form the hydrophobic pocket into which the side chain of H3A1 inserts. Human AIRE and mAire also contain these key downstream residues (L320/L322, P331/P333, and W335/W337), which potentially interact with I309/I311 to form the hydrophobic pocket (Fig. 1D, yellow highlights). Additionally, residues in hAIRE and mAire (P331–G333 and P333–G335) are consistent with the residues in BHC80 (P523–G525) that form the hydrogen bond cage via backbone carbonyls that recognize the N terminus of H3 (Fig. 1D, turquoise box). Furthermore, key glutamate and aspartate residues that form hydrogen bonds with the unmodified ε-amino group of H3K4 are conserved between hAIRE/mAire (E296, D297/E298, and D299) and BHC80 (E488 and D489) (19), whereas D88 and D90 of DNMT3L form similar bonds (20) (Fig. 1D, red highlight). These residues are the hallmark of the recognition specificity that mediates steric exclusion of methyl groups on H3K4. Moreover, the aspartate side chain is interposed between H3K4 and H3T6/H3R8, whereas a hydrophobic residue (M502 of BHC80, I107 of DNMT3L) inserts between H3R2 and H3K4 to form an interdigitation of H3 peptide and PHD finger side chains (19, 20). hAIRE/mAire's C310/C312 together with D297/D299 can form compatible bonds to feature a similar interdigitation, likely from H3R2 to H3T6 (Fig. 1D, pink highlight). Taken together, these observations predict that the AirePHD1–H31–21 interaction adopts folds highly similar to those of the BHC80PHD–H31–10 and DNMT3LPHD-like–H31–21 crystal structures.
To test this hypothesis, we created the mAirePHD1 point mutations D299A and C312W, which are predicted to abolish interactions with H3K4me0 and disrupt the antiparallel β-strand that engages the H3 tail. Using the histone peptide pulldown assay, we confirmed these predictions in the contexts of the PHD1 finger (Fig. 1E) and the full-length protein (Fig. 1C Upper). Isothermal titration calorimetry (ITC) of AirePHD1 and AireSAND+PHD1+2 (amino acids 185–481) revealed dissociation constants (_K_d) of ≈10 μM for unmodified H31–21 (Fig. 2 and Fig. S2). These values are comparable with the _K_d values determined from ITC experiments with BHC80PHD–H31–10 (≈30 μM) (19), as well as the _K_d values from fluorescence polarization experiments with DNMT3Lfull-length–H31–21 (≈2 μM) (20). The _K_d values for D299A and C312W were ≈150 μM or not detectable, respectively (Fig. 2). The D299A/C312W mutations parallel D489A/M502W in BHC80 and D90A/I107W in DNMT3L (Fig. 1D), which were all described to disrupt binding to H3K4me0 (19, 20). Thus, it appears that Aire adopts a structure similar to previously described protein modules that bind to unmodified H3.
Fig. 2.
Dissociation constants of mAire's recognition of the N terminus of the H3 tail as measured by ITC.
APECED AirePHD1 Mutations.
Four APECED-causing point mutations map to AirePHD1: V301M, C311Y, P326L, and P326Q, none of which is a residue modeled to be critical for AirePHD1's recognition of the H3 tail (Fig. 1D). C311Y impairs Zn2+ coordination and thereby destroys the fold of the entire domain, whereas P326L and P326Q result in partial disruption of the tertiary PHD fold (21). In contrast, V301M has no impact on the structure or stability of the PHD finger (21). We asked whether the murine homologs of certain of these mutations, namely, V303M, C313Y, and P328L (Fig. 1D), influence interactions between AirePHD1 and the H3 tail. As expected, C313Y disrupted the binding completely and P328L did partially, according to both peptide pulldown and ITC experiments (Figs. 1E and 2 and Fig. S2). In contrast, V303M binding was similar to that of the WT fragment. This finding may not be so surprising: according to the NMR structure of AirePHD1, this residue is partially solvent-exposed and has the potential to contribute to its interaction with partner proteins that form the transcriptional machinery driving ectopic expression of genes encoding PTAs. In the end, these studies confirm the importance of PHD1 and its relevance to the autoimmune disease, but they do not speak to the details of the molecular model for AirePHD1's recognition of the H3 tail.
Aire Directly Interacts with Nucleosomes.
Although Aire directly recognizes H3 tails, we also asked whether this interaction is relevant in the context of the nucleosome by testing full-length Aire's affinity for reconstituted mononucleosomes, using an EMSA. Aire bound robustly to nucleosomes as well as to the free DNA used to reconstitute them (Fig. S3_A Upper_). However, PHD1 deletions did not affect the affinity of the binding (Fig. S3_B_). Furthermore, tailless or H3K4me3 nucleosomes (reconstituted with trypsinized histones or methyl-lysine analogues, respectively) did not affect the binding of Aire vis à vis its interaction with unmodified nucleosomes (data not shown). These results are consistent with previous studies of polycomb and heterochromatin protein 1, which showed weak contributions by histone tail recognition to nucleosome-binding energy, despite the proteins' strong binding to histone tail peptides (22–24). Aire's affinity to DNA had a greater contribution to nucleosome-binding energy in the EMSA, consistent with PHD–nucleosome interactions elsewhere (25), because poly(dI-dC) effectively competed out Aire–nucleosome interactions (Fig. S3_A Lower_). This effect of poly(dI-dC), and the absence of previously reported Aire-specific motifs (26, 27) in the DNA used in our EMSAs (28), suggest a substantial non-sequence-dependent component to DNA binding, and raise questions about the importance of sequence specificity to the overall binding. Although we describe here the direct interaction between Aire and nucleosomes, we turned to more sensitive assays to probe Aire–chromatin interactions in vivo, because EMSA analyses cannot resolve small micromolar differences potentially contributed by H3 tail binding.
Aire Recognition of the H3 Tail Contributes to Its Interaction with Chromatin in Vivo.
Having established direct interactions between Aire and H3 tails in vitro, we tested their functional relevance in vivo. Aire-positive MECs are postmitotic and number fewer than 105 per thymus (29), rendering them refractory to ex vivo biochemical studies. Therefore, we turned to 4D6 human thymic epithelial cells, a nontransformed cloned line derived from postnatal human thymi (30). Although 4D6 does not express Aire, it has been previously used in transient transfection studies to investigate Aire control of human PTAs (31). We further validated the Aire-transfected 4D6 model through gene expression profiling, revealing thousands of up-regulated PTAs, a subset of which was also induced in WT vs. Aireo/o MECs (data not shown). Thus, we surmised that Aire operates by the same general mechanism in this cell line, but with cell type-to-cell type variability in its precise targets. We similarly characterized an Aire-transfected 293T model, which has been widely used to demonstrate Aire dimerization, subnuclear localization, transactivation potential, and induction of PTAs (32), with the same conclusion (data not shown).
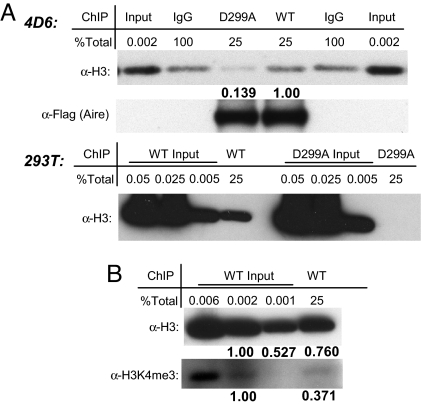
To assess the contribution of H3 tail binding to Aire's interaction with chromatin in vivo, we performed a cross-linked ChIP assay. Aire-containing chromatin from WT/D299A-transfected 4D6 or 293T cells was enriched by using anti-flag antibodies. After washing, we eluted Aire-interacting proteins and performed Western blot analysis using anti-H3 antibodies. A greater than 7-fold enrichment of H3 was associated with Aire in 4D6 transfected with plasmids expressing AireWT compared with AireD299A (Fig. 3A Upper). In 293T cells, H3 associated with AireD299A was not detectable vis à vis the robust signal for associated WT Aire (Fig. 3A Lower). Moreover, Aire-associated H3 was depleted for trimethylation at K4 (Fig. 3B). Thus, Aire preferentially interacts with chromatin regions depleted of H3K4me3, and its recognition of H3 tails contributes importantly to the stability of the interaction.
Fig. 3.
Recognition of H3 tail contributes to Aire's in vivo interaction with chromatin. (A) Aire ChIP. The 4D6 (Upper) or 293T (Lower) cells transfected with tagged WT or D299A were Flag-ChIPed, followed by Western blotting with the indicated antibodies (α-). Percentages of total ChIPed or input were loaded as indicated. Relative band intensities are in bold below respective bands. (B) Histones associated with Aire in vivo are H3K4me3-depleted. The 293T ChIP is as in A.
Aire Recognition of the H3 Tail Is Essential for Ectopic Expression of Genes Encoding PTAs.
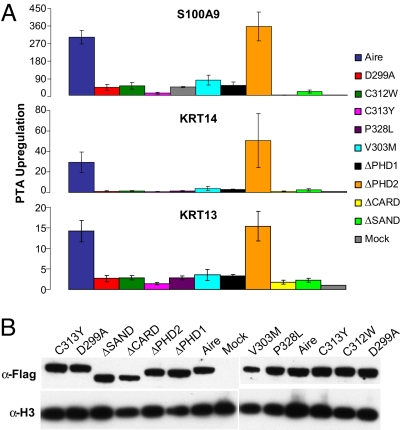
Having established the specificity of Aire's interaction with chromatin in vivo, we next tested whether H3 tail binding is important for Aire's regulation of endogenous genes encoding PTAs. Using transfected 4D6 cells, we evaluated the effects of the D299A/C312W mutations, domain deletions, and APECED-causing PHD1 mutations on the ectopic up-regulation of three genes previously described to be Aire-dependent (ref. 3; J.A., C.B., and D.M., unpublished results). All mutations except ΔPHD2 showed a dramatic inhibition of transcriptional up-regulation at all three Aire-dependent loci (Fig. 4A). Not surprisingly, the magnitude of inhibition was dependent on Aire expression levels (Fig. S4), although at any one transfected plasmid concentration, all mutants expressed comparable protein levels after transfection (Fig. 4B). These findings were also true of the 293T system (Fig. S4). The inhibitions seen with the C313Y, P328L, V303M, and ΔPHD1 mutations were comparable with those observed for D299A and C312W (Fig. 4A). The inhibitory effect of V303M suggests a role for AirePHD1 beyond H3 tail interactions, because this mutation did not affect binding to the H3 peptide (Figs. 1E and 2). In contrast, it is likely that the effects of C313Y and P328L are due to the disruption of the PHD finger, which may compromise binding to the H3 tail and, potentially, other interactions. The effects of the caspase recruitment domain (CARD) deletion, consistent with its described role for subnuclear localization (33), and the SAND domain deletion demonstrate that Aire's H3 tail binding activity is necessary but not sufficient for Aire regulation of genes encoding PTAs.
Fig. 4.
Recognition of H3 tail is crucial for Aire's ectopic expression of PTA in vivo. (A) Quantitative RT-PCR of total RNA from 4D6 cells transfected with WT or mutant Aire representative of four independent experiments. Enrichment of endogenous PTA mRNA represents normalized levels to internal HPRT and mock transfections. (B) Expression levels from transfection. Western blots of 4D6 whole-cell lysates with indicated antibodies.
Discussion
Aire exerts its influence on induction of immunological tolerance at the central level by regulating expression of PTAs in thymic MECs, and it is critical for keeping autoimmune disease at bay. This report offers insights into the molecular mechanism of Aire by exploring its activity as a histone-binding module that specifically recognizes the unmodified N terminus of H3. We demonstrate that this recognition is critical for Aire's interaction with chromatin in vivo and for its control of PTA expression in MECs.
Our study extends a previous report of Aire–H3 interactions (34) by: its broad screening of histone sequences and modifications, including the four core histones, the H2AX variant, and abrogating (H3R2me1/me2 and H3T3ph) and tolerated (H3K9ac, H3S10ph, H3K14ac, and H3R17me1/me2) H3 PTMs; its linkage of PHD1–H3 interactions to autoimmune disease through exploration of APECED-causing PHD1 mutations in in vitro binding and in vivo functional experiments; its data on Aire–nucleosome and Aire–DNA binding, raising questions about the importance of Aire specifically recognizing a particular DNA motif; and its establishment of the importance of Aire's H3 tail recognition for in vivo chromatin interactions and regulation of PTA expression in the context of a functionally validated thymic epithelial cell line. Findings from the two reports implicate a chromatin-associated mechanism mediating central induction of organ-specific tolerance.
Recognition of the H3 Tail: Aire and BHC80.
The recognition of unmodified H3 by a PHD finger has been described in other contexts: BHC80 interaction with the unmodified ε-amino group of H3K4, forming bonds with the aspartate N-terminal to the first cysteine of the PHD finger that sterically excludes dimethyl and trimethyl groups (19). Our data indicate that AirePHD1 contains the conserved residues needed to form all of the features of this recognition motif, and thus suggest that AirePHD1 and BHC80PHD have similar structures.
Yet, subtle differences do exist: both Aire and BHC80 employ an antiparallel β-sheet to engage the H3 tail; however, the β-strand of BHC80 extends to interactions with the backbone of H3R8 (S497), whereas Aire's β-strand is likely limited to H3T6, because G307 (parallel to BHC80's S497) cannot engage the backbone H3R8 (Fig. 1D, red underscore). A model of the hAIREPHD1–H3 peptide complex built from the bromodomain and PHD transcription factor (BPTF)–H3 crystal structure (35) confirms these features. The model also predicts that D312 of hAIRE forms a salt bridge with H3R2 (34) (Fig. 1D, black highlight). This interaction was validated through ITC and fluorescence spectroscopy experiments as well as by functional data showing that hAIRE D312A abolishes all recognition of the H3 tail (34). BHC80 does not interact with H3R2 (19). It is possible that substitution of the Met502BHC80 → Cys310AIRE and S497BHC80 → G307AIRE collectively shifts the orientation of the antiparallel β-sheet to accommodate the interaction of D312 of AIRE with H3R2. These features require structural confirmation; however, the crystal structure of Aire remains elusive.
Aire: Reader of Histone Modifications.
Eight types of histone PTMs have been reported, entailing more than 70 different sites ranging from N-terminal tails to the internal nucleosome core (36). Evidence that histone PTMs demarcate distinct chromosomal domains that translate into specific transcriptional outcomes has been provided by recent global genome studies (37, 38). PHD fingers have emerged as important “readers” of histone PTMs, recruiting transcriptional or chromatin-remodeling machinery to distinct chromosomal domains (18). This report highlights Aire as a member of the growing PHD family that recognizes histone PTMs. The _K_d of binding is ≈10 μM, a relatively weak interaction that is consistent with those of other histone-binding modules (18, 23, 24), a situation that favors dynamic control of the modules' residence times at target loci, encouraging competition between chromatin-associated macromolecular assemblages (18). Another consideration is that in vivo, histone PTMs tend to occur in domains spread across many kilobases (37, 38), and thus an increased avidity to a specific PTM state might be amplified by the close proximity of large numbers of nucleosomes with that PTM. Beyond tail binding, we also established direct physical interaction between Aire and nucleosomes as well as Aire and DNA. Aire's apparently strong affinity for nonspecific DNA sequences calls into question whether the sequence-specific motifs reported elsewhere (26, 27) are important for Aire specificity.
The family of PHD fingers recognizing the unmodified N terminus of H3 includes BHC80 and DNMT3L, and potentially extends to Sp110 and Sp140, which share key recognition residues (34). Histone PTM readers interacting with the unmodified amino termini of H3 and H4 tails have been associated with hypoacetylation and transcriptional silencing (39), which is consistent with BHC80's role as a subunit of the lysine demethylase-1 corepressor complex associated with transcriptional repression of neuron-specific genes (40). Likewise, DNMT3L controls de novo DNA methylation that results in gene silencing and heterochromatin formation (20). However, the module-associated machinery that Aire recruits to genes encoding PTAs effect transcriptional activation. Here, this activation was dependent on Aire's recognition of the H3 tail (Fig. 4) and, importantly, homologues of the APECED-causing AirePHD1 mutations C311Y and P326L showed no or partial binding to the H3 tail (Figs. 1 and 2). In contrast, the disease-causing V301M mutation is more likely to be involved in interactions with partner proteins important in assembling the transcription-activating machinery.
Linking Aire Histone-Binding Activity and Tissue-Specific Transcriptional Activation.
Aire displays punctate subnuclear localization adjacent to nuclear speckles in MECs (13), structures often close to highly active transcription sites and enriched in phosphorylated Pol II, transcription elongation factors (e.g., P-TEFb), and pre-mRNA splicing factors (14, 15). Concordant with this localization, a study on trichostatin A-induced cell lines suggested that Aire physically interacts with and recruits P-TEFb to promoters of tissue-specific genes (41). Moreover, native Aire coimmunoprecipitation assays reveal direct interactions with RNA splicing/processing factors that are required for the Aire-dependent up-regulation of PTAs in 293T cells (J.A., C.B., and D.M., unpublished results). These observations favor an Aire-dependent mechanism that mediates interactions between nuclear speckles and genes encoding PTAs. Aire's activity as a histone-binding module may provide the targeting specificity for such a mechanism, whereas Aire-associated complexes may modify the speckle proteins themselves (e.g., phosphorylation) to affect their release and recruitment to proximal sites (14). Any proposed mechanism, however, will also need to account for the observation that Aire mediates the expression of only a small subset of PTAs per cell (42, 47). Whether thymic MECs sequester PTA genes in chromosomal domains by binding to H3 needs to be directly demonstrated, along with the genome-wide localization of Aire, to establish a direct link between Aire targeting and Aire effector functions. Above all, a validation of the contribution of H3 tail recognition in the whole-mouse setting is paramount.
Materials and Methods
Peptide Microarray.
Peptide microarray experiments were performed as described previously (43). Briefly, biotinylated histone peptides were printed in six replicates onto a streptavidin-coated slide before being incubated with GST-Aire. Details are given in SI Methods.
Biotinylated Peptide-Binding Assays.
Biotinylated Peptide pulldown assays were performed as described previously (44). Briefly, biotinylated peptides were incubated with GST-Aire overnight, then subjected to streptavidin beads for 1 h before washing and Western blotting. Details are given in SI Methods.
Isothermal Titration Calorimetry.
ITC was conducted as previously reported (19). Measurements were carried out from 40 to 80 μM His-tagged Aire and 1 mM H3 peptide at 24°C, as detailed in SI Methods.
Chromatin Immunoprecipitation.
ChIP was done as described previously (45), except eluates were precipitated with trichloroacetic acid and cross-links were reversed by 30 min at 99°C. See SI Methods.
Quantitative RT-PCR.
Quantitative RT-PCR was performed as previously described (3) and is detailed in SI Methods.
Supplementary Material
Supporting Information
Acknowledgments.
We thank Dr. A. G. W. Matthews for critical suggestions, and the Aire group and Kingston lab for inspiration and helpful discussions. This work was supported by National Institutes of Health (NIH) Grant P30 DK36836 to the Joslin Diabetes Center's Diabetes and Endocrinology Research Center core facilities; NIH Grant R01 DK60027 and Young Chair funds to D.M. and C.B.; NIH Grant R01 GM48405 to R.E.K.; and NIH Grant R01 GM079641 and a Searle Scholar Award to O.G. A.S.K. was supported by NIH Grant T32 DK07260, A.J.K. by a predoctoral fellowship from Genentech, S.Y.P. by National Research Service Award F32 DK774852, and J.A. by a Juvenile Diabetes Research Foundation Advanced Postdoctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
References
- 1.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: Learning self-control in the thymus. Nat Rev Immunol. 2005;5:772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 2.Kyewski B, Klein L. A central role for central tolerance. Annu Rev Immunol. 2006;24:571–606. doi: 10.1146/annurev.immunol.23.021704.115601. [DOI] [PubMed] [Google Scholar]
- 3.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the Aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 4.Liston A, et al. Aire regulates negative selection of organ-specific T cells. Nat Immunol. 2003;4:350–354. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- 5.Anderson MS, et al. The cellular mechanism of Aire control of T cell tolerance. Immunity. 2005;23:227–239. doi: 10.1016/j.immuni.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 6.The Finnish-German APECED Consortium. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet. 1997;17:399–403. doi: 10.1038/ng1297-399. [DOI] [PubMed] [Google Scholar]
- 7.Nagamine K, et al. Positional cloning of the APECED gene. Nat Genet. 1997;17:393–398. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- 8.Ramsey C, et al. Aire deficient mice develop multiple features of APECED phenotype and show altered immune response. Hum Mol Genet. 2002;11:397–409. doi: 10.1093/hmg/11.4.397. [DOI] [PubMed] [Google Scholar]
- 9.Derbinski J, et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnnidis JB, et al. Chromosomal clustering of genes controlled by the aire transcription factor. Proc Natl Acad Sci USA. 2005;102:7233–7238. doi: 10.1073/pnas.0502670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurst LD, Pal C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- 12.Gaszner M, Felsenfeld G. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 13.Su MA, et al. Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire. J Clin Invest. 2008;118:1712–1726. doi: 10.1172/JCI34523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamond AI, Spector DL. Nuclear speckles: A model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 15.Saitoh N, et al. Proteomic analysis of interchromatin granule clusters. Mol Biol Cell. 2004;15:3876–3890. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akiyoshi H, et al. Subcellular expression of autoimmune regulator is organized in a spatiotemporal manner. J Biol Chem. 2004;279:33984–33991. doi: 10.1074/jbc.M400702200. [DOI] [PubMed] [Google Scholar]
- 17.Tao Y, et al. AIRE recruits multiple transcriptional components to specific genomic regions through tethering to nuclear matrix. Mol Immunol. 2006;43:335–345. doi: 10.1016/j.molimm.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 18.Taverna SD, et al. How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan F, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bottomley MJ, et al. NMR structure of the first PHD finger of autoimmune regulator protein (AIRE1). Insights into autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) disease. J Biol Chem. 2005;280:11505–11512. doi: 10.1074/jbc.M413959200. [DOI] [PubMed] [Google Scholar]
- 22.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 23.Ringrose L, Ehret H, Paro R. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol Cell. 2004;16:641–653. doi: 10.1016/j.molcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 24.Eskeland R, Eberharter A, Imhof A. HP1 binding to chromatin methylated at H3K9 is enhanced by auxiliary factors. Mol Cell Biol. 2007;27:453–465. doi: 10.1128/MCB.01576-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B, et al. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 26.Kumar PG, et al. The autoimmune regulator (AIRE) is a DNA-binding protein. J Biol Chem. 2001;276:41357–41364. doi: 10.1074/jbc.M104898200. [DOI] [PubMed] [Google Scholar]
- 27.Ruan QG, et al. The autoimmune regulator directly controls the expression of genes critical for thymic epithelial function. J Immunol. 2007;178:7173–7180. doi: 10.4049/jimmunol.178.11.7173. [DOI] [PubMed] [Google Scholar]
- 28.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 29.Gray DH, Abramson J, Benoist C, Mathis D. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J Exp Med. 2007;204:2521–2528. doi: 10.1084/jem.20070795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez E, et al. Establishment and characterization of cloned human thymic epithelial cell lines. Analysis of adhesion molecule expression and cytokine production. Blood. 1994;83:3245–3254. [PubMed] [Google Scholar]
- 31.Giraud M, et al. An IRF8-binding promoter variant and AIRE control CHRNA1 promiscuous expression in thymus. Nature. 2007;448:934–937. doi: 10.1038/nature06066. [DOI] [PubMed] [Google Scholar]
- 32.Villasenor J, Benoist C, Mathis D. AIRE and APECED: Molecular insights into an autoimmune disease. Immunol Rev. 2005;204:156–164. doi: 10.1111/j.0105-2896.2005.00246.x. [DOI] [PubMed] [Google Scholar]
- 33.Ferguson BJ, et al. AIRE's CARD revealed, a new structure for central tolerance provokes transcriptional plasticity. J Biol Chem. 2008;283:1723–1731. doi: 10.1074/jbc.M707211200. [DOI] [PubMed] [Google Scholar]
- 34.Org T, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 2008;9:370–376. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Pokholok DK, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 38.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Edmondson DG, Smith MM, Roth SY. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 40.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 41.Oven I, et al. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol Cell Biol. 2007;27:8815–8823. doi: 10.1128/MCB.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Derbinski J, et al. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc Natl Acad Sci USA. 2008;105:657–662. doi: 10.1073/pnas.0707486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi X, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthews AG, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soderbergh A, et al. Autoantibodies against aromatic L-amino acid decarboxylase identifies a subgroup of patients with Addison's disease. J Clin Endocrinol Metab. 2000;85:460–463. doi: 10.1210/jcem.85.1.6266. [DOI] [PubMed] [Google Scholar]
- 47.Villasenor J, Benoist C, Mathis D. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: Probabilistic, monoallelic, misinitiated. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0808069105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information