Genome-wide association defines more than thirty distinct susceptibility loci for Crohn's disease (original) (raw)
. Author manuscript; available in PMC: 2009 May 1.
Published in final edited form as: Nat Genet. 2008 Jun 29;40(8):955–962. doi: 10.1038/NG.175
Abstract
Several new risk factors for Crohn's disease have been identified in recent genome-wide association studies. To advance gene discovery further we have combined the data from three studies (a total of 3,230 cases and 4,829 controls) and performed replication in 3,664 independent cases with a mixture of population-based and family-based controls. The results strongly confirm 11 previously reported loci and provide genome-wide significant evidence for 21 new loci, including the regions containing STAT3, JAK2, ICOSLG, CDKAL1, and ITLN1. The expanded molecular understanding of the basis of disease offers promise for informed therapeutic development.
The first genome-wide association studies (GWAS) have identified many common variants associated with complex diseases, and have rapidly expanded our knowledge of the genetic architecture of these traits. Progress in Crohn's disease (CD), a common idiopathic inflammatory bowel disease (IBD) with high heritability (λs ∼ 20-35), has been especially striking, with recent GWAS publications increasing the number of confirmed associated loci from two to more than ten 1. The results have identified new pathogenic mechanisms of IBD and promise to advance fundamentally our understanding of CD biology. These recent discoveries highlight, for instance, the key importance of autophagy and innate immunity2-5 as determinants of the dysregulated host-bacterial interactions implicated in disease pathogenesis. Furthermore, genetic associations have been shown to be shared between CD and other auto-inflammatory conditions – for example, IL23R variants 6 are also associated with psoriasis7 and ankylosing spondylitis8, and PTPN2 variants with type 1 diabetes3,5. As in other complex diseases, restricted sample sizes have resulted in early CD studies focusing on only the strongest effects, which turn out to explain only a fraction of the heritability of disease.
We recently published three separate GWA scans for CD in European-derived populations – the details of which are shown in Table 14,5,9. Motivated by the need for larger datasets to improve power to detect loci of modest effect, we carried out a genome-wide meta-analysis from our three CD scans. These analyses, together with a replication study in an equivalently sized, independent panel, have enabled us to identify at genome-wide levels of significance 21 novel Crohn's disease susceptibility genes and loci. This brings the total number of independent loci conclusively associated with Crohn's disease to more than 30 and provides unprecedented insight into both CD pathogenesis as well as the general genetic architecture of a multifactorial disease.
Table 1. Samples used (post QC) in this study.
| NIDDK | BEL/FR | UKIBDGC | Total | |
|---|---|---|---|---|
| Scan cases | 946 | 536 | 1,748 | 3,230 |
| Scan controls | 977 | 914 | 2,938 | 4,829 |
| Replication cases | 0 | 1,082 | 1,243 | 2,325 |
| Replication controls | 0 | 787 | 1,022 | 1,809 |
| Replication Trios | 720 | 619 | 0 | 1,339 |
| Nationality | USA/Canadian | Belgian/French | British | |
| Scan Platform | Illumina HumanHap300 | Illumina HumanHap300 | Affymetrix GeneChip 500K | |
| Replication Platform | Sequenom | Illumina GoldenGate | Sequenom |
Results
Meta-analysis of three genome-wide association scans
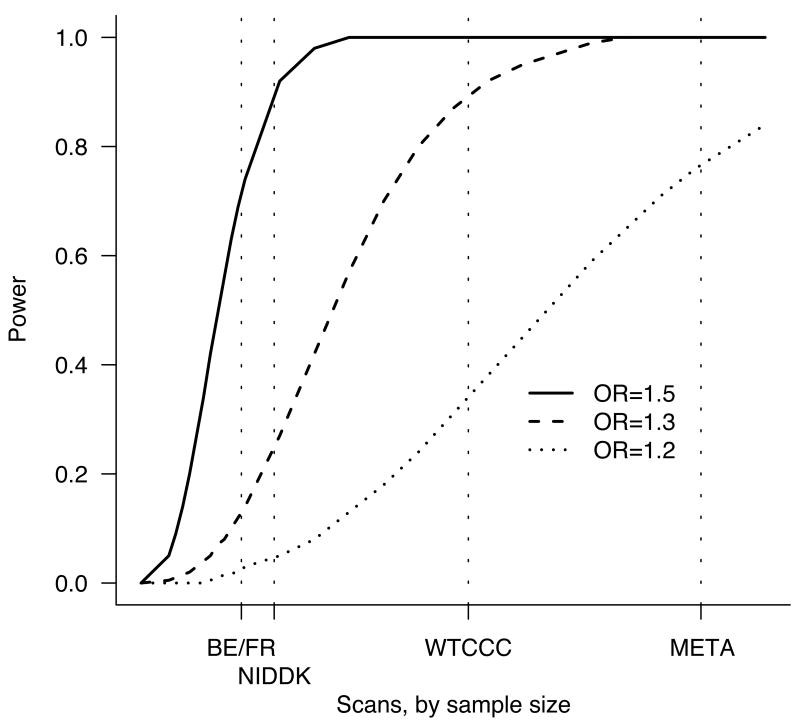
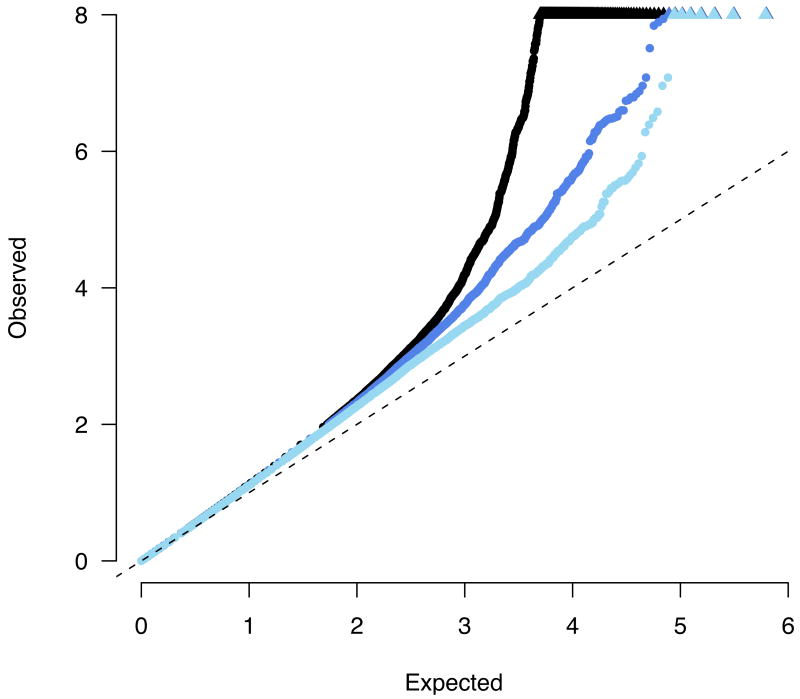
The combined GWAS study samples (Table 1) consisted of 3,230 cases and 4,829 controls, all of European descent. While the individual scans did identify new risk factors, they were only well-powered to discover common alleles with odds-ratios (ORs) above 1.3 (in the case of the WTCCC) or 1.5 (the smaller two scans, Figure 1). By contrast, the combined sample has 74% power at an OR of 1.2, allowing evaluation of the role of alleles with smaller effect sizes for the first time. As two different genotyping technologies were used in the constituent scans, we utilized recently developed imputation10,11 methods to assess association across all three studies at 635,547 SNPs contained on one or both platforms. A quantile-quantile (Q-Q) plot of the primary meta-statistic (single SNP Z-scores, Figure 2) shows a striking excess of significant associations, well beyond what would be attributable to the modest overall distributional inflation (genomic control λ < 1.16). Despite the large sample size, the overall inflation is modest because (1) each group had separately tested for evidence of population stratification, and the meta-analysis used a test that combined the results from each study (rather than mixing the raw data and compromising the case-control matching of each study), and (2) imputation was done on all samples ignoring case status and thus would not introduce artifactual differences between cases and controls12.
Figure 1.
Power to detect a genetic effect of various sizes (odds ratio 1.2, 1.3, 1.5) versus study sample size. Power is reported here as the probability (given a multiplicative model and risk allele frequency of 20%) of p < 5×10-5 in a scan – the value used to define regions for attempting replication in a larger sample set. Vertical dotted lines show the sample sizes for the three constituent scans and the meta-analysis. Relatively large effects are likely to be detected by any of these scans, whereas only the combined analysis is well powered to detect more modest effects.
Figure 2.
A quantile-quantile plot of observed -log10 p values versus the expectation under the null. Black points represent the complete meta-analysis, with a substantial departure from the null at the tail (values > 8 are represented along the top of the plot as triangles). Dark blue points show the distribution after removing 11 previously published loci, demonstrating a still notable excess. Light blue points show the distribution after removing all 40 loci which replicate at least nominally. In all the cases the overall distribution is marginally inflated (λGC < 1.16).
We focus our attention in this study specifically on the 526 SNPs from 74 distinct genomic loci which were associated with p < 5×10-5 – more than 7 times the number of SNPs expected by chance even after correction for the modest overall inflation detected. This threshold for follow-up is not meant to imply that there are no genuine associations among SNPs with less significant association in the meta-analysis, but rather reflects a practical desire to prioritize as many true positives as possible for immediate replication. Eleven associations previously replicated and established at genome-wide significance levels (Methods, Table 2), including both “historical” associations at NOD213,14 and 5q31 (IBD5) 15 as well as recent replicated findings from individual GWA scans such as IL23R, ATG16L1, IRGM, TNFSF15 and PTPN22-6,16, were among the 74 regions represented in this tail of the distribution of association statistics. Even after removing all SNPs in LD with these eleven loci, however, there continued to be a substantial excess of associated alleles beyond that which would be expected by chance (Figure 2).
Table 2. Convincingly (Bonferroni p < 0.05) replicated CD risk loci.
| p values | Odds ratios | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Critical region | Scan | Replication | Combined | Num. genes | Gene of interest | RAF | Risk allele | Case Ctrl | TDT |
| (a) Previously published loci | |||||||||||
| rs11465804 | 1p31 | 67.4* | 1.01×10-35 | 3.1×10-29 | 6.66×10-63 | NA | IL23R | 0.933 | T | 2.50 | 2.77 |
| rs3828309 | 2q37 | 230.9* | 1.13×10-20 | 7.67×10-14 | 2.36×10-32 | NA | ATG16L1 | 0.533 | G | 1.28 | 1.30 |
| rs3197999 | 3p21 | 48.73 - 49.87 | 2.16×10-7 | 5.64×10-7 | 1.15×10-12 | 35 | MST150 | 0.271 | A | 1.20 | 1.20 |
| rs4613763 | 5p13 | 40.32 - 40.48 | 4.52×10-22 | 2.79×10-8 | 6.82×10-27 | 0 | PTGER4** | 0.125 | C | 1.32 | 1.28 |
| rs2188962 | 5q31 | 131.44 - 131.90 | 4.58×10-9 | 3.52×10-11 | 2.32×10-18 | 7 | 0.425 | T | 1.25 | 1.26 | |
| rs11747270 | 5q33 | 150.15 - 150.32 | 6.36×10-11 | 2.57×10-7 | 3.40×10-16 | 3 | IRGM | 0.090 | G | 1.33 | 1.31 |
| rs4263839 | 9q32 | 114.61 - 114.78 | 3.92×10-7 | 6.58×10-5 | 2.60×10-10 | 2 | TNFSF15 | 0.677 | G | 1.22 | 1.07 |
| rs10995271 | 10q21 | 64.05 - 64.12 | 1.90×10-11 | 1.61×10-10 | 4.46×10-20 | 1 | ZNF365 | 0.387 | C | 1.25 | 1.53 |
| rs11190140 | 10q24 | 101.26 - 101.32 | 1.71×10-10 | 1.69×10-7 | 3.06×10-16 | 1 | NKX2-3 | 0.478 | T | 1.20 | 1.28 |
| rs2066847 | 16q12 | 49.3* | NA | 1.49×10-24 | 2.98×10-24 | NA | NOD2 | 0.018 | C | 3.99 | 2.57 |
| rs2542151 | 18p11 | 12.73 - 12.88 | 1.19×10-11 | 2.41×10-7 | 5.10×10-17 | 1 | P TPN2 | 0.152 | G | 1.35 | 1.14 |
| (b) Novel loci | |||||||||||
| rs2476601 | 1p13 | 113.79 - 114.17 | 1.81×10-5 | 0.000101 | 1.46×10-8 | 7 | P TPN22 | 0.899 | G | 1.31 | 1.17 |
| rs2274910 | 1q23 | 157.65 - 157.72 | 3.50×10-7 | 0.000481 | 1.46×10-9 | 2 | ITLN1 | 0.682 | C | 1.14 | 1.62 |
| rs9286879 | 1q24 | 169.54 - 169.67 | 4.02×10-7 | 0.000321 | 1.53×10-9 | 0 | 0.243 | G | 1.19 | 1.08 | |
| rs11584383 | 1q32 | 197.60 - 197.77 | 6.82×10-7 | 2.34×10-6 | 1.43×10-11 | 3 | 0.697 | T | 1.18 | 1.20 | |
| rs10045431 | 5q33 | 158.69 - 158.76 | 8.80×10-9 | 3.66×10-6 | 3.86×10-13 | 1 | IL12B | 0.708 | C | 1.11 | 1.36 |
| rs6908425 | 6p22 | 20.63 - 20.84 | 2.52×10-7 | 0.000278 | 8.96×10-10 | 1 | CDKAL1 | 0.780 | C | 1.21 | 1.09 |
| rs7746082 | 6q21 | 106.52 - 106.62 | 3.70×10-6 | 7.7×10-6 | 2.44×10-10 | 0 | 0.289 | C | 1.17 | 1.19 | |
| rs2301436 | 6q27 | 167.32 - 167.52 | 3.30×10-7 | 3.26×10-7 | 1.04×10-12 | 3 | CCR6 | 0.463 | T | 1.21 | 1.16 |
| rs1456893 | 7p12 | 50.03 - 50.11 | 4.92×10-5 | 1.1×10-5 | 4.60×10-9 | 0 | 0.678 | A | 1.20 | 1.14 | |
| rs1551398 | 8q24 | 126.60 - 126.62 | 4.90×10-6 | 0.000109 | 4.50×10-9 | 0 | 0.619 | A | 1.08 | 1.25 | |
| rs10758669 | 9p24 | 4.94 - 5.26 | 6.80×10-7 | 0.00043 | 3.46×10-9 | 3 | JAK2 | 0.348 | C | 1.12 | 1.21 |
| rs17582416 | 10p11 | 35.30 - 35.60 | 8.48×10-6 | 2.53×10-5 | 1.79×10-9 | 3 | 0.345 | G | 1.16 | 1.26 | |
| rs7927894 | 11q13 | 75.80 - 76.02 | 1.43×10-7 | 0.000732 | 1.32×10-9 | 1 | C11orf30 | 0.386 | T | 1.16 | 1.07 |
| rs11175593 | 12q12 | 38.61 - 39.31 | 1.33×10-7 | 0.000165 | 3.08×10-10 | 3 | LRRK2, MUC19 | 0.017 | T | 1.54 | 1.44 |
| rs3764147 | 13q14 | 43.13 - 43.54 | 1.61×10-7 | 1.33×10-7 | 2.08×10-13 | 3 | 0.221 | G | 1.25 | 1.19 | |
| rs2872507 | 17q21 | 34.63 - 35.34 | 2.12×10-6 | 0.000292 | 5.00×10-9 | 17 | ORMDL3 | 0.473 | A | 1.12 | 1.24 |
| rs744166 | 17q21 | 37.74 - 37.95 | 5.94×10-6 | 9.15×10-8 | 6.82×10-12 | 4 | STAT3 | 0.565 | A | 1.18 | 1.25 |
| rs1736135 | 21q21 | 15.73 - 15.76 | 2.06×10-5 | 4.58×10-5 | 7.40×10-9 | 0 | 0.565 | T | 1.18 | 1.10 | |
| rs762421 | 21q22 | 44.43 - 44.48 | 1.08×10-5 | 1.59×10-5 | 1.41×10-9 | 1 | ICOSLG | 0.389 | G | 1.13 | 1.21 |
Replication of 21 new loci
As these 74 regions included the 11 already reported as independently replicated and meeting genome-wide significance thresholds, this replication experiment effectively explored 63 putative associations in novel regions with 11 positive controls (Supplementary Table 1). To identify the true risk factors from these 63 regions, we undertook a replication study involving a total of 2,325 additional Crohn's disease cases and 1,809 controls alongside an independent family-based dataset of 1,339 parent-parent-affected offspring trios.
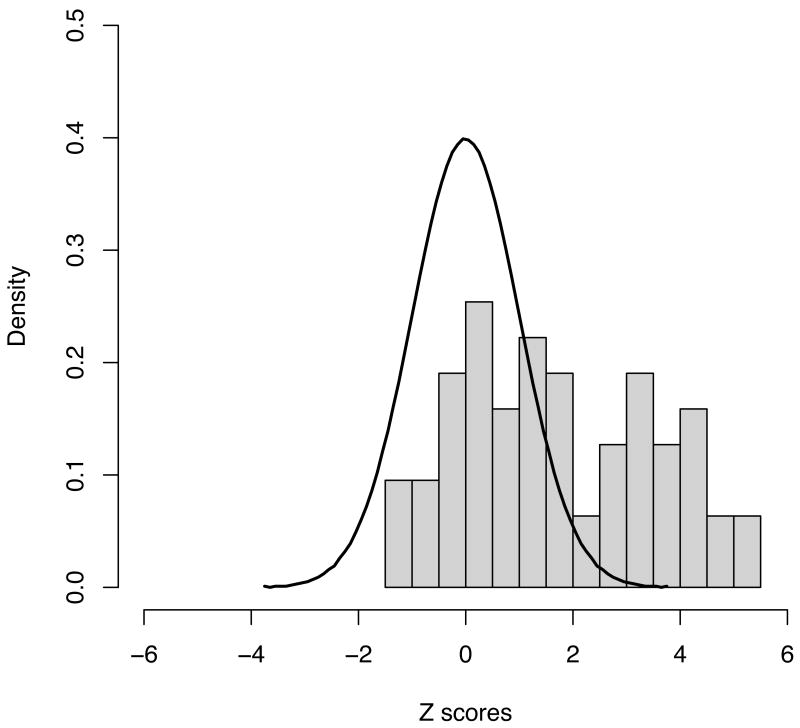
Results (significance levels and odds ratios) for strongly replicating loci, including all positive controls, are presented in Table 2. The distribution of Z-scores from the 63 putative regions shows a dramatic departure from the null distribution (Figure 3) with 19 novel regions showing significant replication (p < 0.0008 – a value of 0.05/63 representing a conservative threshold expected to be exceeded only once by chance in 20 such replication experiments). SNPs on chromosome 19p13 (replication p = 0.00347, combined p = 2.12×10-9) and in the MHC (replication p = 0.006, combined p = 5.2×10-9 - suspected but not previously conclusively established in Crohn's disease) did not reach this conservative threshold, but so convincingly satisfy proposed thresholds for genome-wide significance (p<5×10-8, Methods) that we propose these as the 20th and 21st additional Crohn's disease associated loci defined here. A further 8 of the 42 remaining loci showed nominal replication (Table 3).
Figure 3.
Distribution of observed Z scores from the 63 novel regions explored, along with the expected distribution under the null (a standard normal with mean 0 and variance 1). Even setting aside the 21 regions reaching genome-wide significance, the distribution is highly skewed – 4 more results exceed a Z of 2 (1 would be expected by chance under the null) whilst none showed a Z of less than -2 (same expectation under the null) suggesting that even more of the regions investigated here are likely to constitute true positive associations when additional data become available.
Table 3. Nominally (p < 0.05) replicated CD risk loci.
| p values | Odds ratios | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Critical region | Scan | Replication | Combined | Num. genes | Gene of interest | RAF | Risk allele | Case Ctrl | TDT |
| rs4807569 | 19p13 | 1.05 -1.15 | 1.16×10-8 | 0.00347 | 2.12×10-9 | 2 | 0.217 | C | 1.02 | 1.26 | |
| rs780094 | 2p23 | 27.30 - 27.77 | 3.82×10-6 | 0.00381 | 3.14×10-7 | 22 | GCKR | 0.397 | T | 1.08 | 1.13 |
| rs3763313 | 6p21 | 32.44-32.79 * | 1.45×10-8 | 0.00602 | 5.20×10-9 | 7 | BTNL2, DRA, DRB, DQA | 0.188 | C | 1.19 | 1.01 |
| rs13003464 | 2p16 | 61.09 - 61.14 | 3.44×10-5 | 0.00565 | 4.60×10-6 | 1 | CCDC139 | 0.376 | G | 1.16 | 1.08 |
| rs991804 | 17q12 | 29.57 - 29.70 | 4.02×10-6 | 0.0135 | 1.07×10-6 | 4 | CCL2, CCL7 | 0.726 | C | 1.1 | 1.08 |
| rs12529198 | 6p25 | 5.04 - 5.11 | 7.08×10-7 | 0.0192 | 6.96×10-7 | 1 | LYRM4 | 0.062 | G | 1.12 | 1.19 |
| rs17309827 | 6p25 | 3.36 - 3.42 | 2.08×10-6 | 0.0391 | 2.74×10-6 | 1 | SLC22A23 | 0.639 | T | 1.1 | 1.02 |
| rs7758080 | 6q25 | 149.54 - 149.65 | 7.28×10-6 | 0.044 | 8.78×10-6 | 0 | 0.274 | G | 1.12 | 0.99 | |
| rs8098673 | 18q11 | 17.74 - 17.93 | 3.18×10-5 | 0.0443 | 2.88×10-5 | 0 | 0.329 | C | 1.05 | 1.09 | |
| rs917997 | 2q11 | 102.31 - 102.64 | 2.16×10-5 | 0.0493 | 2.22×10-5 | 5 | IL18RAP | 0.222 | T | 1.05 | 1.11 |
It is possible that extreme population substructure in the replication sample could give rise to such a striking excess of hits. While unlikely, this was directly evaluated by the large family-based component of the replication study. Odds ratio estimates from the TDT analysis of the North American, French and Belgian families alone are consistent with those from the UK and Belgian case/control samples (Tables 2 & 3), with all 21 newly defined loci showing odds ratios in the same direction of association with the original scan in the family-based component (and nearly half showing greater OR than in the case-control arm). Importantly, none of the significantly or nominally replicating loci show significant evidence for heterogeneity (across studies or between family-based and population-based arms) when corrected for the number of tests performed. This independent family based evidence (Supplementary Table 6) confirms these alleles constitute true Crohn's disease loci.
For this newly expanded set of 32 unequivocally associated loci, we assessed whether there was evidence of significant pairwise interactions which could add further to the overall variance in liability explained by this set of loci. We performed a case-only analysis of the 3,664 cases in the replication study and observed no interactions that withstood a correction for the number of tests performed (Supplementary Table 2).
Deciphering the genetic architecture of CD
The contributions of the 32 loci to disease risk were computed using a standard liability threshold model and are displayed as a histogram of individual variances (Figure 4). The observations from this variance analysis that many loci were detected for which the current study had low power, and that only a minority of the variance in risk is explained by these 32 loci, suggest that many additional loci are yet to be identified. This is reinforced by the additional 8 nominal replications (Table 3) where only 2 or 3 would be expected by chance, and by the continued excess of small p values when these 40 total regions are removed (Figure 2).
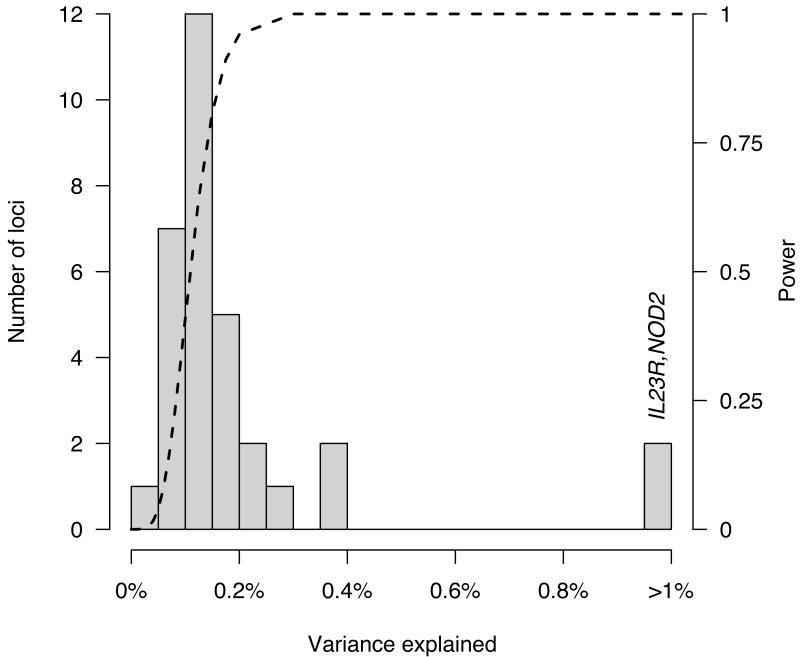
Figure 4.
Histogram of percent variance explained by each of the 32 established CD risk loci. The distribution resembles the long postulated exponential distribution of effect sizes. Dashed line shows the joint power for our meta-analysis to detect (p < 5×10-5), and for our replication sample to replicate (at Bonferroni corrected p values), a 20% variant explaining a given fraction of variance. Note how quickly this curve moves from nearly zero power to detect tiny effects (less than one tenth of one percent) to nearly full power to detect larger effects (presuming they are well covered by the current generation of GWAS chips). Complete power near the origin would likely reveal a more complete exponential distribution, with many very small effects. These are likely to increase somewhat once the causal variant or variants are identified in each locus. Indeed, NOD2 and IL23R are distant outliers, each explaining 1-2% of total variance, partially because multiple causal variants have already been discovered at these loci6,13.
While recognizing that fine-mapping is required to identify specific causal variants, we performed a series of analyses to gain some general insight into the CD associations. We first queried HapMap to discover any instances where a non-synonymous SNP (nsSNP) was correlated (r2 > 0.5) to the most associated variant discovered in this study. Accepting that HapMap is not a complete catalogue of nsSNPs, but including four loci where fine-mapping has identified coding variants, just 9 of the 32 genomewide significant associations were correlated with a known nsSNP (Supplementary Table 3). To explore whether any of the associations reflect a cis-acting regulatory effect on a nearby gene, we evaluated genotype-expression correlation using the panel of 400 lymphoblastoid cell lines described by Dixon et al.17. From all genes within 250 kb of the LD-based intervals defined in Table 2 and 3, five correlations between expression of a nearby gene and a CD-associated variant were identified (LOD > 2) (Supplementary Table 4). This was far in excess of chance (p∼0.001) (Supplementary Figure 1) and suggests that regulatory variation also contributes to the genetic architecture identified.
Discussion
Genome-wide association studies provide a systematic assessment of the contribution of common variation to disease pathogenesis. A limiting factor is often the size of the case-control dataset, and hence the power to detect any but the most strongly associated loci. Meta-analysis of existing data provides an obvious potential solution. As Figure 1 demonstrates, our expectation was that the additional power of the combined dataset would result in the identification of a substantially larger number of readily replicating associations than were derived from any of the smaller, constituent datasets. However, the paradigm of exploring common genetic variation with similar effects across studies (in this case all of European descent) needs testing before its results can be accepted as valid.
On the validity of the method our results are substantially reassuring. All 11 previously confirmed CD susceptibility loci were strongly replicated both in the meta-analysis and follow-up experiment. These include the two widely replicated findings from studies published in 200113-15 as well as all of the compelling findings from individual GWAS (Table 2 a). Significantly, we have also identified and replicated 21 new CD susceptibility loci. Using a conservative threshold for significance (only 1 such region would be expected by chance in 20 such experiments), the loci with clear evidence for association in the replication panel include a very high proportion of those showing strongest signals in the meta-analysis (Supplementary Table 1) – 9 of 9 previously unreported regions with p < 5×10-7 in the combined scan were replicated convincingly - emphasizing the validity of the meta-analysis results. Further emphasizing the robustness of these results, all 21 of these loci exceed a conservative genome-wide level of significance (p < 5×10-8) by a significant margin (all but two have p < 5×10-9) - and equivalent strength of association was observed in the family-based subset of our replication sample.
In keeping with other regions recently identified as associated with CD, the 21 new loci do not conform to any obvious pattern in terms of gene content. Thus, as shown in Table 2, some loci (defined by HapMap recombination hotspots flanking the set of correlated, associated variants) contain just a single gene, some contain many genes and others none. Clearly the first category provides the most immediate clues regarding pathogenic mechanisms. These genes are discussed briefly in Box 1, together with a number of genes which constitute striking candidates from regions with only a handful of transcripts. Included among these are compelling functional candidates such as STAT3, JAK2 and IL12B while others, such as CDKAL1 and PTPN22, highlight potentially intriguing contrasts between genetic susceptibility to Crohn's disease and some other complex disorders (Box 1). It is noteworthy – and consistent with previous findings from CD and other complex diseases – that we did not find any strong evidence of deviation from the model of multiplicative (random) effects when we tested for gene-gene interactions among the 32 confirmed associations. This is in spite of the fact that some of these genes seem to affect the same or overlapping pathways.
BOX 1. Noteworthy genes within loci newly implicated in Crohn's pathogenesis.
- Chemokine receptor 6 (CCR6): encoding a member of the G protein-coupled chemokine receptor family, this homing receptor is expressed by immature dendritic cells and memory T cells and is important for B-cell differentiation and tissue specific migration of dendritic and T cells during epithelial inflammatory and immunological responses 25. The ligand of this receptor is macrophage inflammatory protein 3 alpha (MIP-3 alpha); both genes are expressed in granulomas of pulmonary sarcoid 26. Recent studies have also demonstrated that CCR6, IL23R and RORγT are selectively expressed by IL-17 producing cells and IFNγ producing TH17/TH1 cells in CD27.
- Interleukin IL12B: encodes the p40 subunit which is a constituent of both heterodimeric interleukins IL-12 and IL-2328. Association with CD was previously reported5 but not confirmed, and it is also known to be associated with psoriasis7. The key role of the IL12/IL23 pathway in chronic intestinal inflammation is supported by the association between IL23R and CD3 and strong functional evidence from mouse models of colitis 29-32.
- Signal transducer and activator of transcription 3 (STAT3) and Janus kinase 2 (JAK2): the JAK-STAT pathway is a focal point in signal transmission downstream of cytokine and growth factor signals from cell surface receptors to the nucleus to modify transcription of various genes, notably in hematopoietic cells. The present findings are particularly significant, given the role of both genes in IL23R signaling 33, and the central role STAT3 in Th17 differentiation 34. However, JAK2 or STAT3 are also downstream of several other cytokines implicated in CD pathogenesis in addition to interleukin 23, highlighting the pathophysiologic complexity of these new associations. Further complexity is highlighted by the distinctly different roles of STAT3 in innate versus adaptive immunity in murine colitis models: activation of STAT3 in innate immune cells enhances mucosal barrier function whereas STAT3 activation in T-cells exacerbates colitis.
- Leucine-rich repeat kinase 2 (LRRK2). This gene encodes a multi-domain protein expressed mainly in the cytoplasm of neurons, myeloid cells and monocytes, and mutations in LRRK2 have been strongly associated with Parkinson's disease35. A recent study reported the induction of autophagy by mutant LRRK2,41 which is of interest given the strong associations between CD and the autophagy genes ATG16L1 and IRGM.2-5 The same locus also contains the gene MUC19, which encodes a large protein with multiple serine/threonine-rich repeats characteristic of the mucin gene family. The mucin proteins are core components of the mucus layer which protects the intestinal epithelia from injury, and mucin-deficiency potentiates intestinal inflammation in mouse models of colitis36.
- CDKAL1: the protein encoded by this gene is poorly characterized, but CDKAL1 is noteworthy for being recently confirmed as a type 2 diabetes susceptibility gene 24,37-39. In this study, we find that SNPs from the same intron of CDKAL1 that shows association with T2D are associated with CD, but the associated alleles for the two diseases are not correlated with each other.
- Inducible T-cell co-stimulator ligand (ICOSLG): this co-stimulatory molecule is expressed on intestinal (and other) epithelial cells and may play a role in their antigen presentation to and regulation of mucosal T lymphocytes40. Upon maturation, plasmacytoid dendritic cells express ICOSLG and drive the generation of IL-10 producing T regulatory cells41.
- Protein tyrosine phosphatase, non-receptor types 2 and 22 (PTPN2 and PTPN22). Both of these genes are associated with other autoimmune and inflammatory diseases and the effect described here for PTPN2 is similar to that previously described for type 1 diabetes (T1D) 42. However, the association of PTPN22 with CD, although mapping to the same coding variant (R602W) that is a risk factor for T1D and rheumatoid arthritis,43,44 is in the opposite direction, with the T1D and RA risk allele, 602W, offering protection from CD.
- Intelectin 1 (ITLN1) is known to be expressed in human small bowel and colon, and encodes a 120-kDa homotrimeric lectin recognizing galactofuranosyl residues found in cell walls of various microorganisms but not in mammals45. Human intelectin-1 is structurally identical to the lactoferrin receptor (LFR), expressed within the enterocyte brush border, and appears critical in membrane stabilization, preventing loss of digestive enzymes, and protecting the glycolipid microdomains from pathogens46. In addition, intelectin expression is reported in Paneth cells in both mouse and pig small intestine, further pointing to a role in innate immunity.
For loci containing multiple genes or no genes the picture is less well defined. The identified paucity of correlation between associated SNPs and coding variation suggests that these loci may, in particular, benefit from eQTL (expression quantitative trait locus) analysis. This seeks correlation between genotype and expression patterns – bearing in mind that such functional relationships need not respect the specific boundaries of LD around the association. One of our groups previously reported an eQTL effect incriminating PTGER4 at the 5p13 locus9. A striking outcome from our present analysis was at the established IBD5 locus 15, where CD-associated SNPs were associated with decreased SLC22A5 mRNA expression levels. While a SNP had previously been proposed as regulating SLC22A5 transcriptional activity18, these data suggest for the first time that the most disease-associated variants in the IBD5 region, including a coding variant in neighboring SLC22A4, are the same variants most associated with SLC22A5 expression. Equally striking, the most significant Crohn's disease associated eQTL reported here affects ORMDL3 (LOD = 20) on chromosome 17 and SNPs in precisely the same region were recently shown to be strongly associated with childhood asthma.19 This suggests that the same polymorphisms might underlie susceptibility to both CD and asthma, possibly by perturbing ORMDL3 expression.
The new loci that we have identified are of modest effect size, which is unsurprising given all loci with larger impact on disease risk were – as might be expected – discovered in the original scans. The small sizes of these effects explains the lack of overlap between linkage results in CD and these newly discovered loci (Supplementary Figure 2), with the possible exceptions of combined effects of multiple high ranking associations on chromosomes 5q and 6p. Indeed, the linkage evidence that led to the discovery of the IBD5 locus was very likely boosted by the nearby effects at IL12B and IRGM. As expected, the only gene conclusively discovered via linkage (NOD2) is one of two loci which stand well out from the remainder of the distribution of effect sizes (Figure 4). The other outlier, IL23R, illustrates an interesting characteristic of linkage – because (unlike NOD2) the most penetrant risk allele has very high frequency (93%), it is nearly invisible to linkage analysis despite the high OR; highly protective rare alleles are simply not present in multiplex affected families and thus do not influence allele sharing substantially.
Using a liability-threshold model, we estimate that the 32 loci identified to date explain about 10% of the overall variance in disease risk, which may be as much as a fifth of the genetic risk, given previous estimates of CD heritability of approximately 50%.20 This observation is consistent with the fact that these loci collectively contribute only a factor of two to sibling relative risk (λs), and even this figure is dominated by the substantial contribution of NOD2 variants. However, it should be emphasized that the full impact of the new loci cannot be determined until causal variants have been identified by directed sequencing and fine-mapping experiments. Until then the proportion of the variance in Crohn's disease risk explained must be measured from the confirmed SNPs, where association is due to LD with causal variants. Since multiple causal variants might exist at each locus (ranging in frequency from rare to common) our estimates of variance explained provide only a lower bound for the true contribution of each locus.
In conjunction with results from a very similar gene discovery effort in type 2 diabetes21, common lessons are beginning to emerge with respect to the genetic architecture of complex traits. In each example, substantial increase in sample size achieved through meta-analysis has led to dramatic success in gene discovery. In all cases, this progress has revealed an underlying architecture consistent with many individually modest effects which conventional genetic linkage analysis, and even the largest individual genome-wide association studies, are not well powered to detect. Common variants explaining more than 1% of the genetic variance are rare, whereas well-powered studies have found dozens of variants contributing 0.1% of overall variance in liability. Perhaps surprisingly, neither we nor others have yet to document a substantial role for epistasis among these loci and a number of associated loci are conclusively mapped to regions with no currently annotated protein coding genes. Despite the considerable concordant success, a distinct minority of the overall heritability has been explained by these documented associations.
Since our study is well-powered to identify loci that explain > 0.2% of the overall variance, but the sum of such loci explains a relatively small fraction of the total, it seems likely that many loci with even more modest effect sizes remain undiscovered. Of particular note is the continued excess of associations outside of the regions studied here, as well as the nominal replication of an additional 8 loci, notably greater than expected by chance. Overall, the distribution of Z scores in the replication experiment is clearly skewed towards replication – only 11 of the 63 Z-scores in this replication experiment generate Z<0. If only the 21 strongly confirmed loci were genuinely associated, half of the 42 remaining should end up with Z<0. Indeed, observing 8 of the 42 remaining tests with Z>1.5 is itself a highly significant observation (p < 0.0001). Although modest in terms of effect size, identification of such loci is likely to still provide important insights into pathogenic mechanisms, as biological importance need not be proportional to the statistical evidence for genetic association. Closer inspection of regions showing nominal association in the replication experiment reveals that a number of transcripts in these loci are of considerable interest, including CCL2/CCL722, IL18RAP23 and GCKR24.
It is important to note that the generation of GWAS arrays used in the scans here did not offer complete genome coverage of common variation (additional loci may reside in poorly covered intervals) and did not address either rare SNPs or copy number variation effectively. Thus in spite of the wealth of new susceptibility genes and loci identified by the current study, it seems implausible that there are not more to be found – albeit very large datasets are likely to be required to achieve robust statistical support for them. With respect to the present findings, there is much work to be done in resequencing and fine mapping to identify causal variants. While we do not yet have a complete understanding of the genetic architecture of Crohn's disease, dramatic progress has now been made towards this goal - and with it the prospect of directed functional exploration of the pathways identified, insight into how risk alleles interact with environmental modifiers, and the hope of new avenues for treatment.
Methods
Crohn's disease patients, controls, and GWAS
The meta-analysis was based on data from the 3 genome-wide scans of the NIDDK4, WTCCC5 and Belgian/French9 studies. Details of the numbers of cases and controls genotyped in the respective scans and of the genotyping platforms used are shown in Table 1, as are case/control and family cohorts genotyped in the replication study of the meta-analysis. Details of the ascertainment and characterization of these cohorts, as well as quality control procedures applied to the GWA datasets, were provided in the original scan and replication publications 3, 4, 5, 6, 9. Recruitment of study subjects was approved by local and national institutional review boards, and informed consent was obtained from all participants.
Imputation
Briefly, these methods rely on observed haplotype patterns in a set of reference data (the HapMap) and the actual genotype data from each project to make predictions (along with a measure of statistical certainty) at un-genotyped SNPs. We used the program MACH 10 with the NIDDK and Belgian/French data, and IMPUTE 11 with the WTCCC data. Comparisons between the two algorithms yielded very similar results (data not shown). We imputed the superset of polymorphic markers which passed QC in the original scans4,5,9. This set was comprised of SNPs on either the Affymetrix 500K only (n = 350,507), Illumina HumanHap300 version 1 only (n = 238,935), or both panels (n = 46,105) such that all association tests performed were at least partially based on observed genotype data.
Test for association, effect size estimation and interactions
Using the genotype probabilities (rather than best-guess genotypes) and empirical variances for imputed markers in the case and control tallies, we summarized the standard 1 d.f. allele-based test of association as a Z-score within each scan and combined scores across studies to produce a single meta-statistic for each SNP across all three datasets. Odds ratios were estimated separately in TDT samples and each case/control replication collection, and then combined and tested for heterogeneity. 47 Interaction tests were performed using the case-only epistasis test implemented in PLINK48.
Critical regions
Given that most associations contain many correlated SNPs showing signal, we demarcated independent loci by first defining the set of HapMap SNPs with r2 > 0.5 to the most significantly associated SNP. We then bounded the “critical region” by the flanking HapMap recombination hotspots which contained this set. These windows very likely contain the causal polymorphisms explaining the associations.
Replication
We defined loci to have been previously confirmed if an earlier study had both detected and replicated the association in independent samples and the association achieved p < 5 × 10-8 (recently proposed as an appropriate genome-wide significance level for GWAS49). For replication genotyping, we selected the most significantly associated SNP from each region along with a second, correlated SNP with p<0.0001 or a second assay on the opposite strand in order to have a technical backup should the first fail genotyping (Supplementary Table 1). Replication genotyping for the putatively associated loci was performed using primer extension chemistry and mass spectrometric analysis (iPLEX, Sequenom) using Sequenom Genetics Services (N. American panel) and Genome Research Limited, Wellcome Trust Sanger Institute (UK panel), and using a custom-made Golden Gate assay on a Beadstation500 (Illumina), following the manufacturer's recommendations (Belgian/French panel). The more completely genotyped SNP of the two from each region was chosen to represent that regional association in analysis (if both were completely typed, the SNP that was more strongly associated in the scan was used). Samples with >10% missing data (n = 267 for Belgian/French data, 111 for the UK data and 8 for the N. American data; these samples are not included in the tallies for Table 1), as well as SNPs with >10% missing data or Hardy-Weinberg p value < 0.001 were excluded from this analysis.
Regional Annotation: eQTL analysis
Effects of SNPs in Tables 2 & 3 on expression levels of neighbouring genes was studied using transcriptome data from the ∼400 lymphoblastoid cell lines described by Dixon et al.17. SNPs that were not genotyped on this panel (n=14) were replaced with a proxy with r2 > 0.95 when possible (n=12). LOD scores > 2 for genes (probe average) located within 250 Kb of the corresponding LD windows were retrieved from http://www.sph.umich.edu/csg/liang/asthma/. To evaluate the significance of the findings with the CD associated SNPs, we compared the observed (i) number of genes yielding LOD scores > 2, and (ii) sum of these LOD scores, with the corresponding frequency distributions for 1,000 randomly selected sets of 31SNPs, matched for allele frequency (± 0.02) and gene context. Window sizes determined for associated SNPs were used for the matched simulated SNPs.
URL
Meta-analysis test statistics and allele frequencies for all SNPs are available at: http://www.broad.mit.edu/∼jcbarret/ibd-meta/
Supplementary Material
Suplemental Material
Suplemental Figures
Acknowledgments
We acknowledge use of DNA from the 1958 British Birth Cohort collection (R.Jones, S. Ring, W. McArdle and M. Pembrey), funded by the Medical Research Council (grant G0000934) and The Wellcome Trust (grant 068545/Z/02) and the UK Blood Services Collection of Common Controls (W. Ouwehand) funded by the Wellcome Trust. We also acknowledge the National Association for Colitis and Crohn's disease and the Wellcome Trust for supporting the case DNA collections, and support from UCB Pharma (unrestricted educational grant) and the NIHR Cambridge Biomedical Research Centre. The National Institute of Diabetes and Digestive and Kidney Disease (NIDDK) IBD Genetics Consortium is funded by the following grants: DK62431 (S.R.B.), DK62422 (J.H.C.), DK62420 (R.H.D.), DK62432 and DK064869 (J.D.R.), DK62423 (M.S.S.), DK62413 (K.D.T.), NIH-AI06277 (R.J.X.) and DK62429 (J.H.C.). Additional support was provided by the Burroughs Wellcome Foundation (J.H.C.), the Crohn's and Colitis Foundation of America (S.R.B., J.H.C.). We thank Peter Gregersen and Annette Lee (Feinstein Medical Research Institute) for their efforts and the use of control samples. This work was supported by grants from (i) the DGTRE from the Walloon Region (n°315422 and CIBLES), (ii) from the Communauté Française de Belgique (Biomod ARC), and (iii) the Belgian Science Policy organisation (SSTC Genefunc and Biomagnet PAI). Edouard Louis, Sarah Hansoul, Denis Franchimont and Severine Vermeire are fellows of the Belgian FNRS and NFWO. Cynthia Sandor is a fellow of the FRIA. We are grateful to all the clinicians, consultants and nursing staff who recruited patients, including: Jean-Marc Maisin*, Vinciane Muls*, Jean Van Cauter*, Marc Van Gossum*, Philippe Closset*, Pierre Hayard* and Jean Michel Ghilain*; Paul Mainguet°, Faddy Mokaddem°, Fernand Fontaine°, Jacques Deflandre°, and Hubert Demolin°; Jean-Frédéric Colombel#, Marc Lemann#, Sven Almer#, Curt Tysk#, Yigael Finkel#, Miquel Gassul#, Colm O'Morain#, Vibeke Binder# and Jean-Pierre Cézard# (*Erasme-BBIH-IBD; ° Ulg Collaborators; #INSERM collaborators). Sincere thanks to L. Liang for his assistance in accessing the eQTL database, and to Françoise Merlin for expert technical assistance. Finally, we thank all subjects who contributed samples.
Footnotes
References
- 1.Mathew CG. New links to the pathogenesis of Crohn disease provided by genome-wide association scans. Nat Rev Genet. 2008;9:9–14. doi: 10.1038/nrg2203. [DOI] [PubMed] [Google Scholar]
- 2.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 3.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cargill M, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–90. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton PR, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–37. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Libioulle C, et al. A novel susceptibility locus for Crohn's disease identified by whole genome association maps to a gene desert on chromosome 5p13.1 and modulates the level of expression of the prostaglandin receptor EP4. Plos Genetics. 2007 doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Abecasis GRS. Mach 1.0: Rapid Haplotype Reconstruction and Missing Genotype Inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- 11.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 12.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37:1243–6. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 13.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 14.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 15.Rioux JD, et al. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet. 2001;29:223–8. doi: 10.1038/ng1001-223. [DOI] [PubMed] [Google Scholar]
- 16.Yamazaki K, et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn's disease. Hum Mol Genet. 2005;14:3499–506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
- 17.Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 18.Peltekova VD, et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36:471–5. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 19.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 20.Tysk C, Lindberg E, Jarnerot G, Floderus-Myrhed B. Ulcerative colitis and Crohn's disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut. 1988;29:990–6. doi: 10.1136/gut.29.7.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–45. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wedemeyer J, et al. Enhanced production of monocyte chemotactic protein 3 in inflammatory bowel disease mucosa. Gut. 1999;44:629–35. doi: 10.1136/gut.44.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinarello CA. Interleukin-18 and the pathogenesis of inflammatory diseases. Semin Nephrol. 2007;27:98–114. doi: 10.1016/j.semnephrol.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 25.Salazar-Gonzalez RM, et al. CCR6-mediated dendritic cell activation of pathogen-specific T cells in Peyer's patches. Immunity. 2006;24:623–32. doi: 10.1016/j.immuni.2006.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Facco M, et al. Expression and role of CCR6/CCL20 chemokine axis in pulmonary sarcoidosis. J Leukoc Biol. 2007;82:946–55. doi: 10.1189/jlb.0307133. [DOI] [PubMed] [Google Scholar]
- 27.Annunziato F, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204:1849–61. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oppmann B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–25. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 29.Hue S, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006;203:2473–83. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kullberg MC, et al. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J Exp Med. 2006;203:2485–94. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhlig HH, et al. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 2006;25:309–18. doi: 10.1016/j.immuni.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 32.Yen D, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–6. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parham C, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol. 2002;168:5699–708. doi: 10.4049/jimmunol.168.11.5699. [DOI] [PubMed] [Google Scholar]
- 34.Mathur AN, et al. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J Immunol. 2007;178:4901–7. doi: 10.4049/jimmunol.178.8.4901. [DOI] [PubMed] [Google Scholar]
- 35.Plowey ED, Cherra SJ, 3rd, Liu YJ, Chu CT. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008 doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van der Sluis M, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–29. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 37.Steinthorsdottir V, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–5. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 38.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–5. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeggini E, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–41. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakazawa A, et al. The expression and function of costimulatory molecules B7H and B7-H1 on colonic epithelial cells. Gastroenterology. 2004;126:1347–57. doi: 10.1053/j.gastro.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 41.Ito T, et al. Plasmacytoid dendritic cells prime IL-10-producing T regulatory cells by inducible costimulator ligand. J Exp Med. 2007;204:105–15. doi: 10.1084/jem.20061660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bottini N, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337–8. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 43.Criswell LA, et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet. 2005;76:561–71. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rieck M, et al. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J Immunol. 2007;179:4704–10. doi: 10.4049/jimmunol.179.7.4704. [DOI] [PubMed] [Google Scholar]
- 45.Tsuji S, et al. Human intelectin is a novel soluble lectin that recognizes galactofuranose in carbohydrate chains of bacterial cell wall. J Biol Chem. 2001;276:23456–63. doi: 10.1074/jbc.M103162200. [DOI] [PubMed] [Google Scholar]
- 46.Wrackmeyer U, Hansen GH, Seya T, Danielsen EM. Intelectin: a novel lipid raft-associated protein in the enterocyte brush border. Biochemistry. 2006;45:9188–97. doi: 10.1021/bi060570x. [DOI] [PubMed] [Google Scholar]
- 47.Kazeem GR, Farrall M. Integrating case-control and TDT studies. Ann Hum Genet. 2005;69:329–35. doi: 10.1046/j.1529-8817.2005.00156.x. [DOI] [PubMed] [Google Scholar]
- 48.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008 doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 50.Goyette P, et al. Gene-centric association mapping of chromosome 3p implicates MST1 in IBD pathogenesis. Mucosal Immunology. 2008;1:131–38. doi: 10.1038/mi.2007.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suplemental Material
Suplemental Figures