Hoxd11 specifies a program of metanephric kidney development within the intermediate mesoderm of the mouse embryo (original) (raw)
. Author manuscript; available in PMC: 2009 Jul 15.
Abstract
The mammalian kidney consists of an array of tubules connected to a ductal system that collectively function to control water/salt balance and to remove waste from the organisms’ circulatory system. During mammalian embryogenesis, three kidney structures form within the intermediate mesoderm. The two most anterior structures, the pronephros and the mesonephros, are transitory and largely non-functional, while the most posterior, the metanephros, persists as the adult kidney. We have explored the mechanisms underlying regional specific differentiation of the kidney forming mesoderm. Previous studies have shown a requirement for Hox11 paralogs (Hoxa11, Hoxc11 and Hoxd11) in metanephric development. Mice lacking all Hox11 activity fail to form metanephric kidney structures. We demonstrate that the Hox11 paralog expression is restricted in the intermediate mesoderm to the posterior, metanephric level. When Hoxd11 is ectopically activated in the anterior mesonephros, we observe a partial transformation to a metanephric program of development. Anterior Hoxd11 + cells activate Six2, a transcription factor required for the maintenance of metanephric tubule progenitors. Additionally, Hoxd11 + mesonephric tubules exhibit an altered morphology and activate several metanephric specific markers normally confined to distal portions of the functional nephron. Collectively, our data support a model where Hox11 paralogs specify a metanephric developmental program in responsive intermediate mesoderm. This program maintains tubule forming progenitors and instructs a metanephric specific pattern of nephron differentiation.
Keywords: kidney, mesonephros, metanephros, Hoxd11, Hox genes
Introduction
The vertebrate kidney functions to maintain the organism’s homeostasis by controlling water/salt balance, eliminating nitrogenous waste and regulating blood pressure and blood composition. The kidney serves as a well-established model system to study multiple aspects of organogenesis such as reciprocal interactions between mesenchymal and epithelial populations (Grobstein, 1953; Vega et al., 1996), mesenchymal to epithelial transitions (MET) (Barasch et al., 1999; Carroll et al., 2005) and branching morphogenesis (Basson et al., 2006; Majumdar et al., 2003).
Around embryonic day 8.0 (E8.0), the murine intermediate mesoderm (IM) gives rise to two lateral paired ducts, the nephric ducts (ND), that extend caudally along the anterior-posterior (A-P) axis of the embryo (Grote et al., 2006; Pedersen et al., 2005). Three different kidney structures, the pronephros, the mesonephros and the metanephros, form in association with, and are dependent on, the ductal epithelium (Saxén, 1987). The pronephros forms around E8.5 in the most anterior portion of the IM, but quickly degenerates (Bouchard et al., 2002; Saxén, 1987). The mesonephros arises just caudal to the pronephros beginning around E9.5 and consists of arrays of cranial and caudal tubules that lie adjacent to the ND (Sainio etal.,1997; Sainio and Raatikainen-Ahokas, 1999; Saxén, 1987). Cranial tubules form connections to the ND, and in the mouse, caudal tubules remain disconnected. Sainio et al., (1997) have suggested that cranial tubules are derived from outgrowths of the mesonephric ND whereas caudal tubules form due to a MET within the adjacent IM-derived mesonephric mesenchyme. However, direct evidence of a ND contribution to cranial tubules has not been demonstrated. The mesonephric kidney of the mouse is not thought to play a role in embryonic physiology. The eventual fate of the mesonephros depends on the sex of the animal. Cranial tubules are remodeled to form epididymal ducts around E12.5 in the male mouse embryo. In contrast, the female mesonephric duct and mesonephric tubules undergo apoptosis (Sainio and Raatikainen-Ahokas, 1999; Saxén, 1987; Tilmann and Capel, 2002; Vize et al., 2002).
The metanephric kidney, the functional kidney of the fetus and permanent kidney in amniotes, develops from the posterior IM at the level of the hindlimb around E10.5 of mouse development. Several lines of evidence support a model whereby Gdnf expression is restricted to the metanephric mesenchyme (Grieshammer et al., 2004; Kume et al., 2000; Sanchez et al., 1996). GDNF then induces the metanephric (posterior) portion of the ND to invade (Basson et al., 2005; Michos et al., 2007; Pepicelli et al., 1997; Vega et al., 1996) and thereafter, branch repetitively within the mesenchyme, forming the network of the collecting duct system (Basson et al., 2006; Majumdar et al., 2003). An inductive interaction at the tips of the branching ureteric epithelium induces a sub-population of adjacent metanephric mesenchyme capping each tip to undergo a MET establishing the renal vesicle, the epithelial precursor of the main body of the nephron (Barasch et al., 1999; Carroll et al., 2005; Grieshammer et al., 2005; Kobayashi et al., 2005b; Park et al., 2007; Stark et al., 1994). Importantly, maintenance of a nephron progenitor population requires the activity of a transcriptional regulator, Six2 (Self et al., 2006). Upon MET, a portion of the cap mesenchyme downregulates Six2 (Self et al., 2006), while the remainder maintains Six2 expression, ensuring that each future ureteric tip will be associated with a tubule precursor population.
The ND is critical for the induction of both mesonephric and metanephric tubules (Carroll et al., 2005; Grobstein, 1953; Gruenweld, 1952; Saxén, 1987). Recent studies indicate that ND-derived Wnt9b is a key factor required for induction of both tubule types (Carroll et al., 2005). Wnt9b acts via the canonical Wnt signaling pathway (Park et al., 2007) to induce a common, tubule promoting developmental program involving Wnt4 (Stark et al., 1994; Vainio et al., 1999) and FGF8 (Crossley et al., 1996; Perantoni et al., 2005). While both the mesonephric and metanephric kidneys share a common regulatory pathway of tubulogenesis, the nephrons that form are distinct in organization, scale and differentiation as mesonephric tubules lack both a juxtaglomerular apparatus and loop-of-Henle (Vize et al., 2002). What determines the differences in response to a common inductive input is unclear.
One model supposes that the IM mesenchyme is differentially patterned along its A-P axis and that this pre-pattern instructs either a mesonephric or metanephric response to inductive signals. Amongst candidate pattern regulators, Hox genes are of particular interest given that Hox gene function has been widely demonstrated to regulate differential patterning along the body axis during metazoan development (Deschamps and van Nes, 2005; Krumlauf, 1994; Wellik, 2007). Evidence in favor of a potential role for a Hox mediated pre-patterning of the IM mesenchyme comes from analysis of Hox11 paralog mutants (Patterson et al., 2001; Wellik et al., 2002). When activity of all three Hox11 paralogs (Hoxa11, Hoxc11 and Hoxd11) is removed in the mouse, there is a complete failure of both Gndf and Six2 expression in the position where metanephric mesenchyme normally forms, resulting in renal agenesis (Wellik et al., 2002). In contrast, mesonephric development is largely unaffected. Though mutations in Pax2 (Bouchard et al., 2002; Grote et al., 2006; Narlis et al., 2007; Torres et al., 1995), Osr1 (James et al., 2006; Wang et al., 2005), Sall1 (Nishinakamura et al., 2001; Ott et al., 2001), and Six1 (Kobayashi et al., 2007; Xu et al., 2003) result in similar metanephric phenotypes, mesonephric development is also perturbed. Sajithlal et al. (2005) hypothesize that _Eya1_specifies the metanephros, however they also demonstrate Eya1 expression in mesonephric mesenchyme, suggesting a more general role for Eya1 in IM development. Thus, Hox11 paralogs are strong candidates for the specific regulation of a metanephric response in the IM.
We have investigated the differential specification of mesonephric and metanephric kidneys. We demonstrate that mesonephric tubules are formed by MET of the mesonephric mesenchyme and that these tubules do not express markers of distal metanephric segments. Unlike other transcription factors required for metanephric development, Hox11 paralogs are not expressed in the mesonephros, only in the metanephros. We demonstrate that ectopic Hoxd11 activity in the mesonephric mesenchyme is sufficient to activate cell-autonomous ectopic expression of Six2. Furthermore, Hoxd11+ mesonephric tubules adopt a morphology and genetic profile reminiscent of metanephric tubules. Taken together, our data support a model where the posterior metanephric mesenchyme is specified as such by Hox11 paralogs. These paralogs simultaneously activate distinct molecular programs, the first being required for the maintenance of the cap mesenchyme while the second instructs metanephric-specific tubule development in response to inductive signals.
Materials and Methods
Ontolology
All urogenital ontological terms are used as defined in Little et al. (2007).
Animals and genotyping
Animal care and research protocols were performed in accordance with Harvard University’s institutional guidelines, following approval by Harvard University’s institutional committee on animal use. For staging of embryos, the morning of vaginal plug was designated as embryonic day 0.5 (E0.5). Swiss Webster mice (Taconic) were used for all wild type whole mount in situ hybridization. Tg(Hoxb7-cre)12Amc (Yu et al., 2002), Tg(Rarb-cre)1Bhr (Kobayashi et al., 2005a), Gt(ROSA)26 tm1(EYFP)Cos (Srinivas et al., 2001) and Gt(ROSA)26 tm1Sor (Soriano, 1999) (Jackson Laboratories) were genotyped as previously described. The Osr1eGFPCreERt2/+ line was generated by knocking an eGFPCreERt2 construct into the endogenous Osr1 locus and will be described in more detail in a future publication. Osr1eGFPCreERt2/+ mice and embryos were genotyped with the primers Osr1ER3’GenoFw: ACCCGTGATATTGCTGAAGAGCTTG and Osr1ER3’GenoRv: TGAAGAGCGCTGAAACCATAC. The eGFPCreERt2 protein driven by the Osr1 promoter was activated by intraperitoneal injection of Tamoxifen (Sigma) dissolved in corn oil (Sigma) into dams at a dose of 3mg Tamoxifen per 40g mouse body weight at day E7.75.
The R26Hoxd11 line was created by inserting a cassette containing mouse Hoxd11::IRES2::nuclear LacZ into the NheI and XhoI sites of pBigT (Srinivas et al., 2001), releasing the construct with AscI and PacI and subcloning it into pRosa26PAS (Mao et al., 2005) cut with AscI and PacI. The construct was linearized with SwaI and electroporated into YFP 3.1 embryonic stem cells (Mao et al., 2005). Two neomycin resistant colonies demonstrated ubiquitous YFP expression and no β-gal activity. Nuclear β-gal activity was detected in these two clones only upon addition of 4OH-Tamoxifen to the ES cell media. The clones were expanded and confirmed on the 5’ end by PCR using the primers Rosa26–5armFlanking: CCTAAAGAAGAGGCTGTGCTTTGG and Rosa26-SA: CATCAAGGAAACCCTGGACTACTG. The presence of the targeted Hoxd11 transcript was also detected only with addition of 4OH-Tamoxifen with the following primers: cHD11Fw: AAAAGCGCTGTCCCTACACCAAGTAC and cHD11Rv: TCAACAGACCTTGCATTCCTTTGGC. One clone was injected into host (C57BL/6J) blastocysts by the Genome Manipulation Facility, MCB, Harvard University. The line was maintained on a C57BL/6J background (Jackson Laboratories). Mice and embryos were genotyped by PCR with the cHD11Fw and cHD11Rv primers.
Generation of a Hoxd11 polyclonal antibody
Rabbits were immunized with a KHL-conjugated peptide PEGAADKGDPKPG corresponding to amino acids 202–214 of mouse Hoxd11 and anti-serum was affinity purified against immobilized Hoxd11 peptide (Covance Research Products). The purified anti-serum was tested by transiently transfecting (Lipofectamine 2000, Invitrogen) COS7 cells with a mouse Hoxd11 expression plasmid followed by immunostaining. Immunostaining was also performed on E15.5 metanephric cryosections. Specificity was tested by pre-incubation of purified anti-serum with Hoxd11 peptide followed by immunostaining on E15.5 metanephric cryosections.
Whole mount analysis
For in situ hybridization, embryos were collected in PBS, fixed in 4% paraformaldehyde at 4°C overnight, washed thoroughly in PBS, dehydrated through a methanol series into 100% methanol and stored at −20°C. Prior to in situ hybridization, head and trunk regions above the forelimb were removed from E9.5 or E10.5 embryos and embryos were split in half along the dorsal-ventral axis. E11.5 to E15.5 mesonephros, gonads and metanephros were dissected from the rest of the embryo prior to in situ hybridization. Whole mount in situ hybridization was performed as previously described (Park et al., 2007). For whole mount X-gal staining, embryos were dissected in PBS and fixed (1% formaldehyde, 0.2% gluteraldehyde, 2mM MgCl2, 5mM EGTA, 0.02% NP-40) for 30 minutes and washed thoroughly in PBS + 0.02% NP-40. Prior to staining, embryos were dissected as described above. Staining was carried out in staining solution (5mM K3Fe(CN)6, 5mM K4Fe(CN)6, 2mM MgCl2, 0.01% NaDeoxycholate, 0.02% NP-40, 1mg/ml X-gal) at 37°C for 6–8 hours. Staining was stopped by thorough washing in PBS followed by fixation in 4% paraformaldehyde + 0.02% gluteraldehyde at 4°C overnight. Whole mount samples were cleared in 80% glycerol and imaged on a Nikon SMZ1500 Stereoscope with a Nikon DXM1200C camera (Nikon Instruments).
Histological analysis
For immunofluorescence, embryos were dissected in PBS, fixed for one hour on ice in cold 4% PFA, washed thoroughly in PBS and cryopreserved overnight at 4°C in 30% sucrose. Tissue was frozen in OCT (Tissue-Tek) on a dry ice/ethanol bath. 20µm cryosections were collected on Superfrost coated slides (VWR), dried and used immediately or frozen at −20°C. Slide mounted sections were incubated in PBS for 5 minutes, blocked for 30 minutes in 3%BSA, 1% serum in PBS + 0.25% TritonX100 and incubated with anti-Hoxd11 (1:500), anti-Six2 (1:1000, J.W. Mugford and A.P. McMahon, unpublished), anti-Pax2 (1:500, Covance), anti-GFP (1:500, AvesLabs), anti-cytokeratin (1:500, Sigma) or anti-β-galactosidase (1:500, AbCam) in block solution overnight at 4°C. Sections were washed 3 times in PBS + 0.25% TritonX100 (PBTX) and then incubated at room temperature with appropriate Cy2 (1:500, Jackson Immuno), Alexa488, Alexa568 or Alexa647 (1:500, Invitrogen) conjugated secondary antibodies in block solution for 1.5 hours. Sections were washed 3 times in PBTX, rinsed once in PBS, stained with 1µg/ml Hoechst 33342 (Invitrogen) for 5 minutes and rinsed once in PBS. Sections were coverslipped in Vectashield (Vector Labs) and imaged on a Zeiss LSM 510 META confocal microscope (Zeiss). After the whole mount in situ hybridization or X-gal staining procedure, 50 µm vibrotome sections were cut, mounted in 80% glycerol and imaged on a Nikon Eclipse 90i compound microscope with a Nikon DXM1200C camera (Nikon Instruments).
Results
Cranial mesonephric tubules are primarily derived from mesonephric mesenchyme
As a first step towards understanding the differential specification of the IM towards a mesonephric or metanephric tubulogenic program, we first sought to clarify the origin of mesonephric tubules. Sainio et al. (1997) proposed a ND origin for cranial mesonephric tubules, in contrast to more posterior mesonephric tubules that are physically detached from the ND. We addressed the fate of cranial mesonephric tubules using Tg(Hoxb7-cre)12Amc (Yu et al., 2002) and Tg(Rarb-cre)1Bhr (Kobayashi et al., 2005a) Cre driver lines, referred to here as Hoxb7-Cre and Rarb-Cre, respectively. Hoxb7-Cre expresses Cre recombinase throughout the entire ND prior to either mesonephric or metanephric tubule formation whereas Rarb-Cre expresses Cre recombinase in the mesonephric (from E9.5) and also the metanephric (from E10.5) mesenchyme. To fate map the mesonephric tubules, we crossed Hoxb7-Cre and Rarb-Cre to the Gt(ROSA)26tm1Sor (Soriano, 1999) reporter line, referred to here as R26R, and examined the location of X-gal + cells in the mesonephros of both Hoxb7-Cre;R26R and Rarb-Cre;R26R embryos.
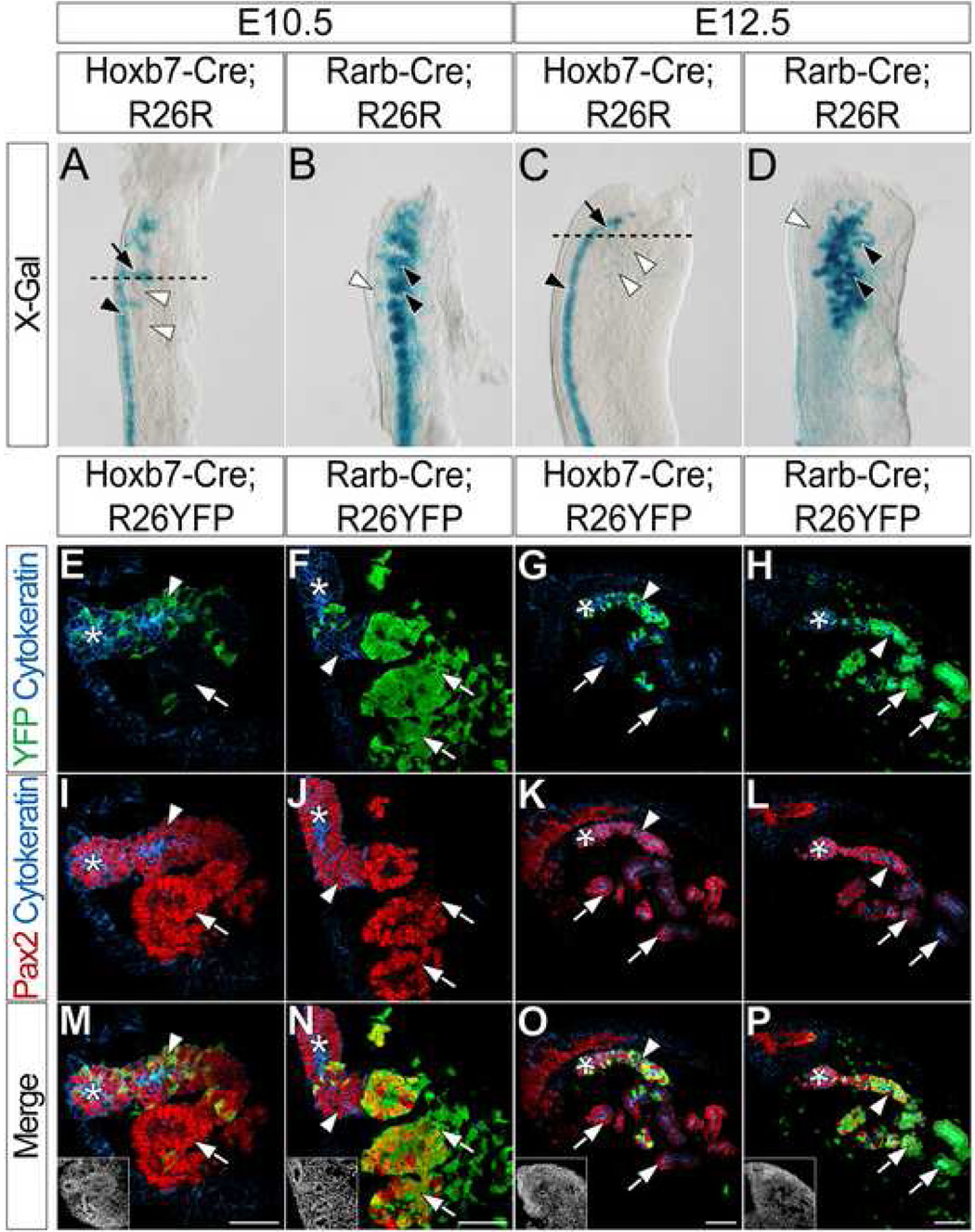
In the E10.5 mesonephros of Hoxb7-Cre;R26R embryos, X-gal + cells are located mainly in the ND (Fig. 1A black arrowhead). Additionally, X-gal+ ND outgrowths project into the mesonephric mesenchyme (Fig. 1A arrow) toward developing X-gal − cranial mesonephric tubules (Fig. 1A white arrowheads). Conversely, developing mesonephric tubules of Rarb-Cre;R26R embryos are X-gal+ (Fig. 1B black arrowheads). In addition, a small number of X-gal+ cells are found in or around the ND (Fig. 1B white arrowhead). At E12.5, cranial and caudal mesonephric tubules of Hoxb7-Cre;R26R embryos are largely X-gal− (Fig. 1C white arrowheads); X-gal+ cells are located in the ND (Fig. 1C black arrowhead). Conversely, X-gal staining in E12.5 Rarb-Cre;R26R embryos is complimentary to that of Hoxb7-Cre;R26R embryos; all mesonephric tubules are X-gal+ (Fig. 1D black arrowheads), while the ND remains X-gal− (Fig. 1D white arrowhead).
Figure 1. Cranial mesonephric tubules are primarily derived from anterior intermediate mesoderm mesenchyme.
(A–D) X-gal staining in the mesonephros of E10.5 (A) and E12.5 (C) Hoxb7-Cre;R26R or E10.5 (B) and E12.5 (D) Rarb-Cre;R26R embryos. Black arrowheads (A, C) indicate X-gal staining in the ND. Black arrows (A, C) indicate X-gal staining in ND outgrowths. Black arrowheads (B, D) indicate X-gal staining in the mesonephric tubules. White arrowheads (B, D) indicate lack of X-gal staining in the ND. Dashed lines (A, C) indicate approximate planes of section (E–T). (E–T) Immunofluorescent confocal microscopy of transverse sections of anterior mesonephros in E10.5 (E, I, M) and E12.5 (G, K, O) Hoxb7-Cre;R26YFP or E10.5 (F, J, N) and E12.5 (H, L, P) Rarb-Cre;R26YFP embryos stained for YFP, Cytokeratin and Pax2. Insets (M, N, O, P) indicate nuclei. Asterisks indicate the ND. Arrowheads indicate ND outgrowths (E, F, I, J, M, N) or mesonephric tubule connecting segments (G, H, K, L, O, P). Arrows (E–P) indicate developing mesonephric tubules. Scale bars in M, N, O and P = 50µm.
To examine cell fates with single cell resolution, we next crossed mice carrying Hoxb7-Cre and Rarb-Cre alleles to the Gt(ROSA)26 tm1(EYFP)Cos (Srinivas et al., 2001) reporter line, referred to here as R26YFP, and performed immunofluorescence for Cytokeratin, Pax2 and YFP on transverse sections through cranial mesonephros. At E10.5, the ND and ND outgrowths are both Cytokeratin+ and Pax2+ (Fig. 1I, J, M, N asterisks and arrowheads, respectively), whereas the developing mesonephric tubules produce only Pax2 (Fig. 1I, J, M, N arrows). In the E10.5 mesonephros of Hoxb7-Cre;R26YFP embryos, YFP is detected in Cytokeratin+ and Pax2+ cells (Fig. 1E, M arrowheads) and is rarely observed in cells that only produce Pax2 (Fig. 1E, M arrows). Conversely, YFP is largely restricted in E10.5 Rarb-Cre;R26YFP embryos to cells that are only Pax2+ (Fig. 1F, N arrows), in addition to a few cells of the surrounding mesonephric mesenchyme. Minimal YFP is observed in cells that produce both Cytokeratin and Pax2 (Fig. 1F, N arrowheads).
At E12.5, the ND and all mesonephric tubules are both Cytokeratin+ and Pax2+ (Fig. 1K, L). The ND is located lateral to the mesonephric tubules (Fig. 1G, H, K, L, O, P asterisks). YFP is detected in the connections between anterior cranial tubules and the ND in the E12.5 cranial mesonephros of Hoxb7-Cre;R26YFP embryos (Fig. 1G, O arrowheads), whereas the majority of the remaining mesonephric tubules are YFP− (Fig. 1G, O arrows). Interestingly, YFP is also detected in the connections between the cranial mesonephric tubules and the ND in the E12.5 mesonephros of Rarb-Cre;R26YFP embryos (Fig. 1H, P arrowheads). Additionally, YFP is present throughout the remainder of both cranial and caudal E12.5 mesonephric tubules of Rarb-Cre;R26R embryos (Fig. 1H, P arrows and data not shown). In summary, these results demonstrate that the majority of cranial mesonephric tubules are derived from the mesonephric mesenchyme. The ND contributes to a connecting segment attaching the ND to a mesonephric tubule, creating a structure that is likely a mosaic of ND and mesonephric mesenchyme-derived cells.
Mesonephric tubules do not express markers of metanephric distal segments
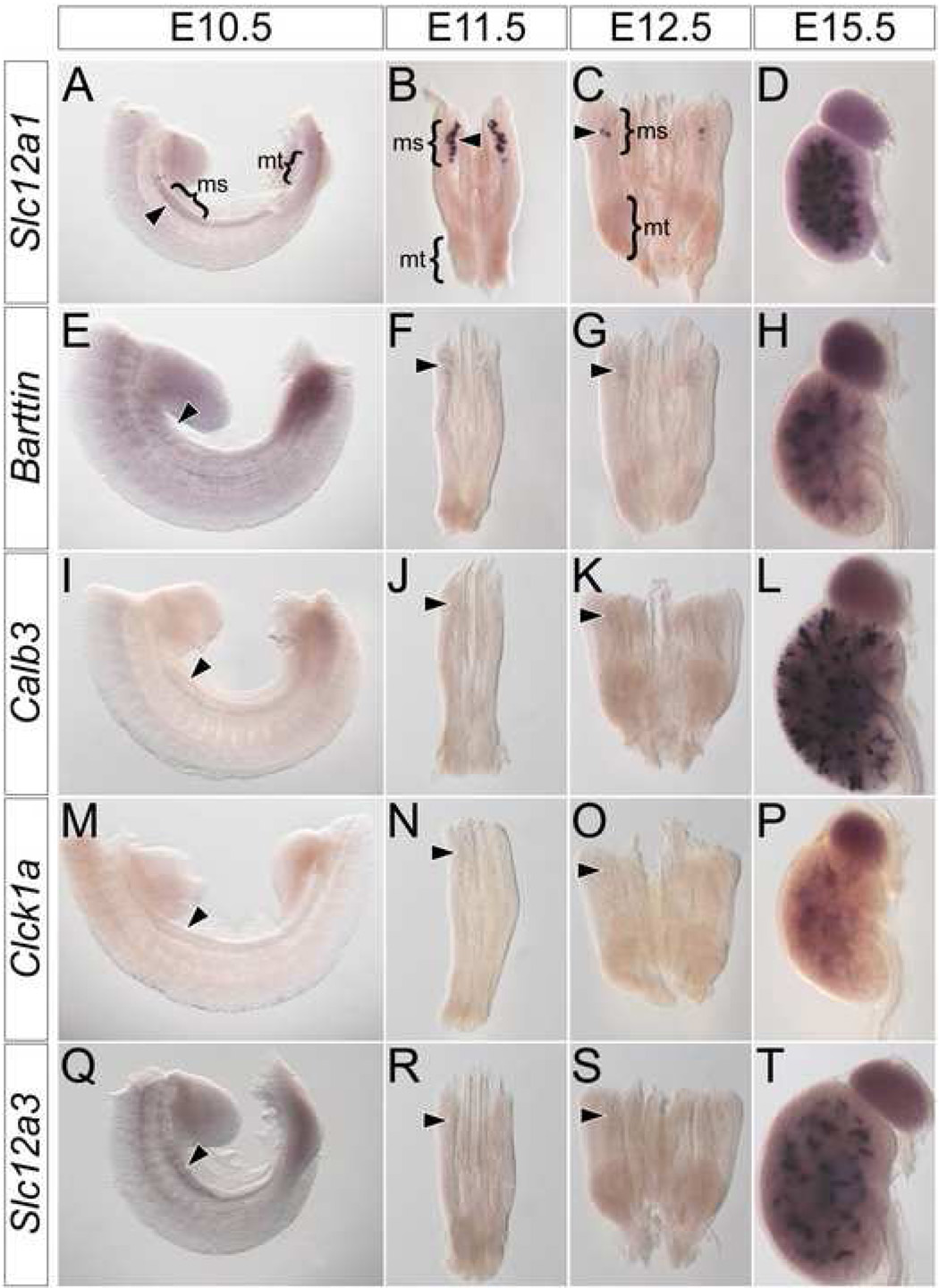
Next, we screened for molecular markers that may potentially distinguish between a mesonephric and metanephric program of renal tubule development. Initially, we examined mesonephric tubules for genes with known segment-specific expression patterns along the proximal-distal (glomerular-collecting duct) axis of the metanephric nephron. Of these, six genes are expressed in both mesonephric and metanephric tubules, forming two classes according to their relative expression levels. Both classes initiate expression in developing mesonephric tubules at E11.5 (Fig. 2A, B, E, F arrowheads). Class one genes are expressed at high levels and include Slc12a1 (Fig. 2A–D), a marker of the thick ascending loop-of-Henle (Nakai et al., 2003), Brn1 (data not shown), a marker of the thin ascending loop-of-Henle (Nakai et al., 2003) and Slc34a1 (data not shown), a marker of proximal convoluted tubules (Murer et al., 2003). Class two genes are expressed at lower levels than class one genes and include Barttin (Fig. 2E–H), a maker of the ascending loop-of-Henle (Wolf et al., 2003), Ihh (data not shown), a marker of proximal straight tubules (Valentini et al., 1997) and Gsh1 (data not shown), a marker of early podocytes (McMahon et al., 2006). Neither class one nor class two genes are expressed in the mesonephros after mesonephric tubule remodeling (data not shown).
Figure 2. Mesonephric tubules do not express markers of metanephric distal segments.
Whole mount in situ hybridization for Slc12a1 (A–D), Barttin (E–H), Calb3 (I–L), Clck1a (M–P) and Slc12a3 (Q–T) in wild type E10.5 (A, E, I, M, Q) trunks, E11.5 (B, F, J, N, R) and E12.5 (C, G, K, O, S) IM and E15.5 (D, H, L, P, T) metanephros. Arrowheads indicate location of mesonephric tubules. ms – mesonephros, mt – metanephros.
Three genes were expressed exclusively in metanephric nephrons. This group includes Calb3 (Fig. 2I–L), a marker of connecting tubules (Chen et al., 2006), Clck1a (Fig. 2 M–P), a marker of the thin ascending loop-of-Henle (Wolf et al., 2003) and Slc12a3 (Fig. 2 Q–T), a marker of distal convoluted tubules (Nakai et al., 2003). Expression of these genes was never observed in developing mesonephric tubules (Fig.2 I–K, M–O, Q–S arrowheads) but each was expressed with a distinct temporal program in metanephric tubules (Fig. 2L, P, T); Calb3 expression initiates at E13.5 (Fig. S1A, B arrowhead), and Slc12a3 (Fig. S1C, D arrowhead) and Clck1a (Fig. S1E, F arrowhead) at E14.5. In general, the genetic profile of mesonephric and metanephric tubules are remarkably similar. The principle difference reflects genes associated with distal metanephric distal segments that arise late in metanephric nephron patterning.
Hox11 paralog expression implicates their role in metanephric specification
In order to clearly define Hox11 paralog expression in conjunction with kidney development, we compared the expression of Hoxa11, Hoxc11 and Hoxd11 in the IM with that of known regulators of metanephric development such as Osr1 (James et al., 2006; Wang et al., 2005), Pax2 (Narlis et al., 2007; Torres et al., 1995), Eya1 (Sajithlal et al., 2005; Xu et al., 1999), Six1 (Kobayashi et al., 2007; Xu et al., 2003), WT1 (Donovan et al., 1999; Sainio et al., 1997), Six2 (Self et al., 2006) and Sall1 (Nishinakamura et al., 2001; Nishinakamura and Osafune, 2006).
Our results demonstrate that the transcription factors examined can be grouped into three classes. The first class includes Osr1 (Fig. S2A–E), Pax2 (Fig. S2F–J), Sall1 and WT1 (data not shown). As exemplified by Osr1 and Pax2, class one genes are expressed from E9.5 through E11.5 in both the developing mesonephros (Fig. S2A–J, arrowheads) and metanephros (Fig. S2C, H, E, Jarrows). Differences exist among the mesonephric expression patterns of the different transcription factors. For instance, Osr1 is expressed in mesenchymal populations (Fig. S2B, D arrowheads), whereas Pax2 is expressed in both epithelial and mesenchymal populations (Fig. S1G, H, I arrowheads).
The second class includes Eya1 (Fig. S2K–O), Six1 and Six2 (data not shown). As exemplified by Eya1, class two genes are also expressed in both the mesonephros (Fig. S2K, L, N arrowheads) and metanephros (Fig. S2M, O arrows). Unlike class one genes, their expression is down-regulated in the mesonephric mesenchyme at E10.5 (Fig. S2N arrowhead) and absent by E11.5 (Fig. S2O arrowhead). Eya1 and Six2 are robustly expressed in the metanephric mesenchyme from E10.5 onwards (Fig. S2O arrow), while Six1 is completely down-regulated by E11.5 (data not shown).
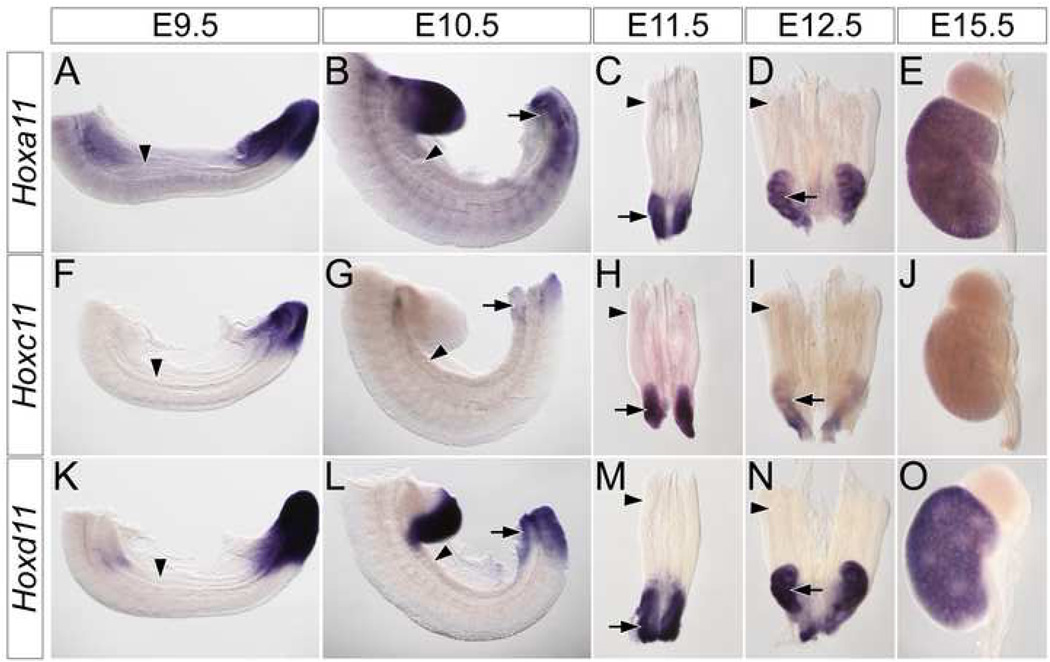
The third class is restricted to members of the Hox11 paralog group (Fig. S2P–T and Fig.3). Within the IM, these three genes are expressed exclusively in the metanephric mesenchyme (arrows in Fig. S2R, T and Fig. 3B–E, G–I, L–O). Interestingly, as the metanephros develops, individual Hox11 paralog members have slightly differing expression patterns. Hoxa11 (Fig. 3A–E) and Hoxd11 (Fig. 3K–O) are consistently expressed at higher levels than Hoxc11 (FIG. 3F–J). Hoxc11 is undetectable by E15.5 (Fig. 3J and data not shown). In addition, Hoxd11 (Fig. 3N arrow and O) is expressed more robustly Hoxa11 (Fig. 3D arrow and E) in both the cortical stroma and cap mesenchyme. In summary, our results demonstrate that the expression patterns of the Hox11 paralogs are unique among transcription factors required for metanephric development and suggestive of a class of molecules that may differentially regulate mesonephric and metanephric responses within the IM mesenchyme to inductive signals.
Figure 3. Hox11 paralogs are exclusively expressed in the metanephros.
Whole mount in situ hybridization for Hoxa11 (A–E), Hoxc11 (F–J) and Hoxd11 (K–O) in wild type E9.5 (A, F, K) and E10.5 (B, G, L) trunks, E11.5 (C, H, M) and E12.5 (D, I, N) IM and E15.5 (E, J, O) metanephros. Arrowheads indicate location of the mesonephros. Arrows indicate the location of the metanephros.
Six2, but not Gdnf, is activated in mesonephric mesenchyme expressing Hoxd11
To directly address the function of Hox11 genes, we developed two different mouse lines to enable ectopic activation of Hoxd11 in anterior IM prior to mesonephric tubule induction. Hoxd11 was selected amongst the three Hox11 paralogs based on the relative contribution of Hoxd11 to the metanephric kidney program in compound mutants (Wellik et al., 2002) and the observation that Hoxd11 shows strong, broad early expression that is maintained throughout subsequent metanephric development (Fig. 3L–O). An R26Hoxd11 line was generated by introducing a full-length mouse Hoxd11 cDNA and a nuclear LacZ reporter into the ROSA26 locus in a configuration that enables Cre dependent regulation in any cell type (Fig. S3). A second line, Osr1 eGFPCreERt2/+, referred to here as Osr1-GCE (J.W. Mugford, P. Sipilä;, J. McMahon and A.P. McMahon, unpublished), contains an eGFPCreERt2 construct knocked into the Osr1 locus. When Osr1-GCE;R26R compound heterozygotes were induced with Tamoxifen at E7.75 through intraperitoneal injection of the dam, X-gal+ cells contribute to the IM from E8.5, strongly labeling the mesonephros, gonad and metanephros at E12.5 (Fig. S4A and data not shown).
R26Hoxd11 mice were intercrossed with mice carrying Osr1-GCE and Rarb-Cre alleles. In Rarb-Cre;R26Hoxd11 embryos, Hoxd11 is activated in mesonephric tubules and their derivatives beginning at E9.5 (Kobayashi et al., 2005a). In contrast Osr1-GCE;R26Hoxd11 embryos injected with Tamoxifen at E7.75 activate Hoxd11 throughout the IM as early as E8.5. Immunofluorescence using anti-Hoxd11 (Fig. S4C), anti-Cytokeratin and anti-β-gal on sections of E13.5 mesonephros from R26Hoxd11, Rarb-Cre;R26Hoxd11 and Osr1-GCE;HoxD11 embryos demonstrates that both Hoxd11+ and β-gal+ cells co-localize with Cytokeratin+ mesonephric tubules only when Cre alleles are present (Fig. S4B).
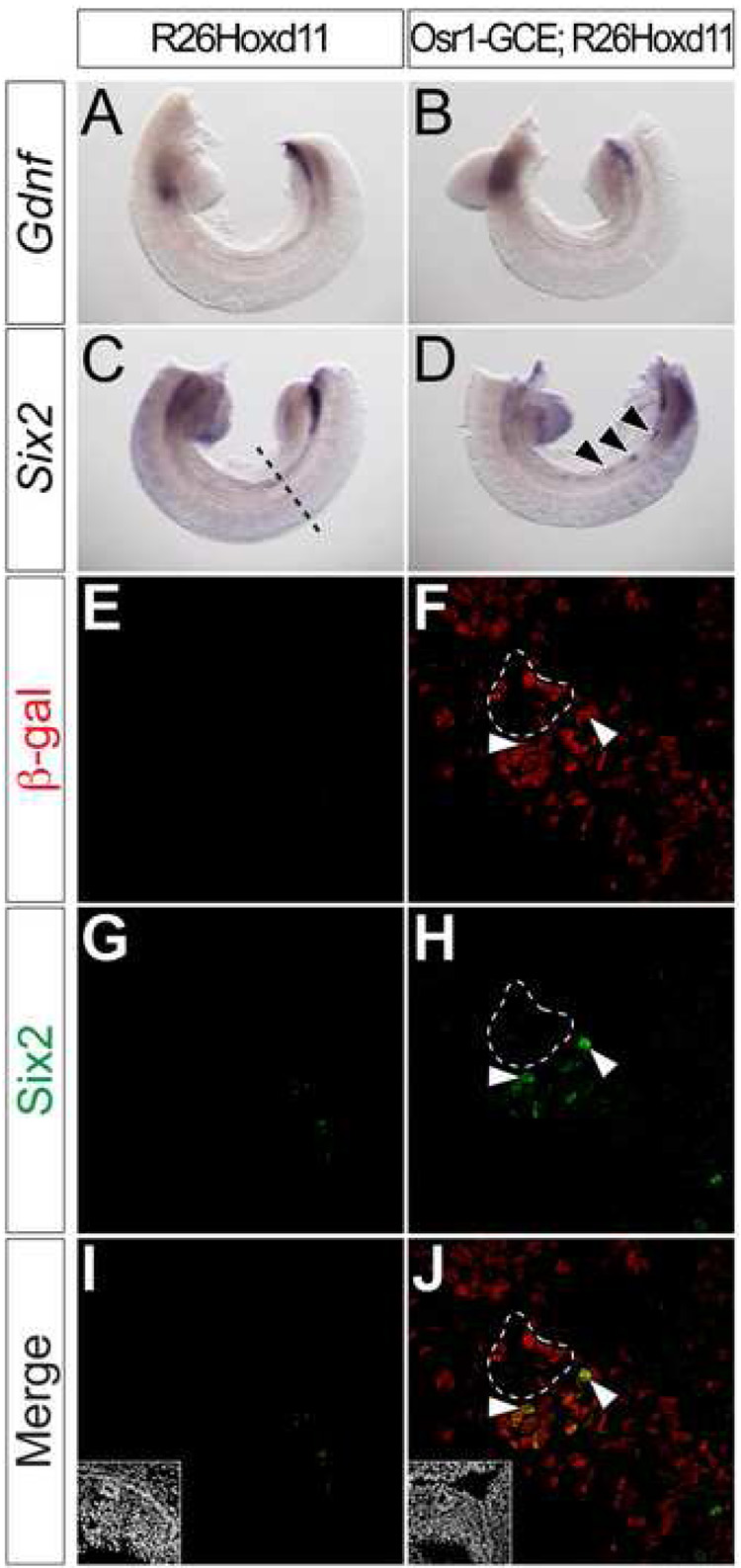
Both Six2 and Gdnf expression are absent from the posterior IM of Hox11 mutants (Wellik et al., 2002). We examined their expression in the E10.5 mesonephros of Rarb-Cre;R26Hoxd11 and Osr1-GCE;R26Hoxd11 embryos. Neither was activated in the E10.5 mesonephros of Rarb-Cre;R26Hoxd11 embryos (data not shown). In contrast, Six2 was cell-autonomously activated in small clusters of cells following Osr1-GCE mediated activation of R26Hoxd11 at E7.75 (Fig. 4D) arrows. _ Six2 expression is normally absent in the mesonephric region at E10.5 (Fig. 4C, E, G, I) and all Six2+ cells are β-gal+ (Fig. 4 F, H, J arrowheads). As expected, β-gal was not detected in the absence of Cre activity (Fig. 4E, G, I). Ectopic Six2 was restricted to mesenchymal mesonephric cells; no Six2 was observed in either the ND or developing mesonephric tubules (Fig.4 F, H, J dashed line and data not shown). These results suggest that the maintenance of Six2 expression, a distinct feature of the metanephric mesenchyme, is regulated by Hoxd11 and likely other Hox11 paralog members (Wellik et al., 2002), consistent with Hoxd11 regulation of a metanephric mesenchymal program. The differential results between the two IM drivers likely reflect the early activation from the Osr1-GCE driver allele. In contrast to the Six2 findings, we did not observe activation of Gdnf (Fig. 4A, B) using the Osr1-GCE driver allele. In an attempt to increase the levels of Hoxd11, we intercrossed Osr1-GCE;R26Hoxd11 males with R26Hoxd11 homozygous females and examined E10.5 embryos harboring the Osr1-GCE allele and two R26Hoxd11 alleles, but found no evidence of ectopic branching of the ND (data not shown). Thus, Hoxd11 may not be sufficient for full programming of a metanephric pathway (see Discussion).
Figure 4. Hoxd11 activates cell-autonomous activation of Six2, but not Gdnf, in mesonephric mesenchyme.
(A–D) Whole mount in situ hybridization for Gdnf (A, B) and Six2 (C, D) in E10.5 control (A, C) or Osr1-GCE;R26Hoxd11 (B, D) embryos. Arrowheads (D) indicate sites of ectopic Six2 expression. Dashed line (C) indicates approximate plane of section (E–L). (E–L) Immunofluorescent confocal microscopy of transverse sections of the mesonephros in E10.5 control (E, G, I) or Osr1-GCE;R26HoxD11 (F, H, J) embryos stained for β-gal and Six2. Arrowheads (F, H, J) indicate examples of β-gal and Six2 co-localization. White dashed line (F, H, J) indicates ND epithelia positive for β-gal, but not Six2. Insets (I, J) indicate nuclei.
Ectopic Hoxd11 activity alters the morphology and differentiation of mesonephric tubules
Hoxd11 expression is normally down-regulated as metanephric tubules develop (Patterson and Potter, 2004). In order to ensure that metanephric tubule development occurs normally in the presence of constitutive Hoxd11 expression from the ROSA26 locus, we examined the expression of Slc12a1, Barttin, Calb3, Clck1a and Slc12a3 in the E15.5 metanephros following activation of the R26Hoxd11 allele. All tubule markers were detected (data not shown), thus deregulated Hoxd11 expression does not alter, in an obvious way, induction or subsequent development of the metanephric tubules.
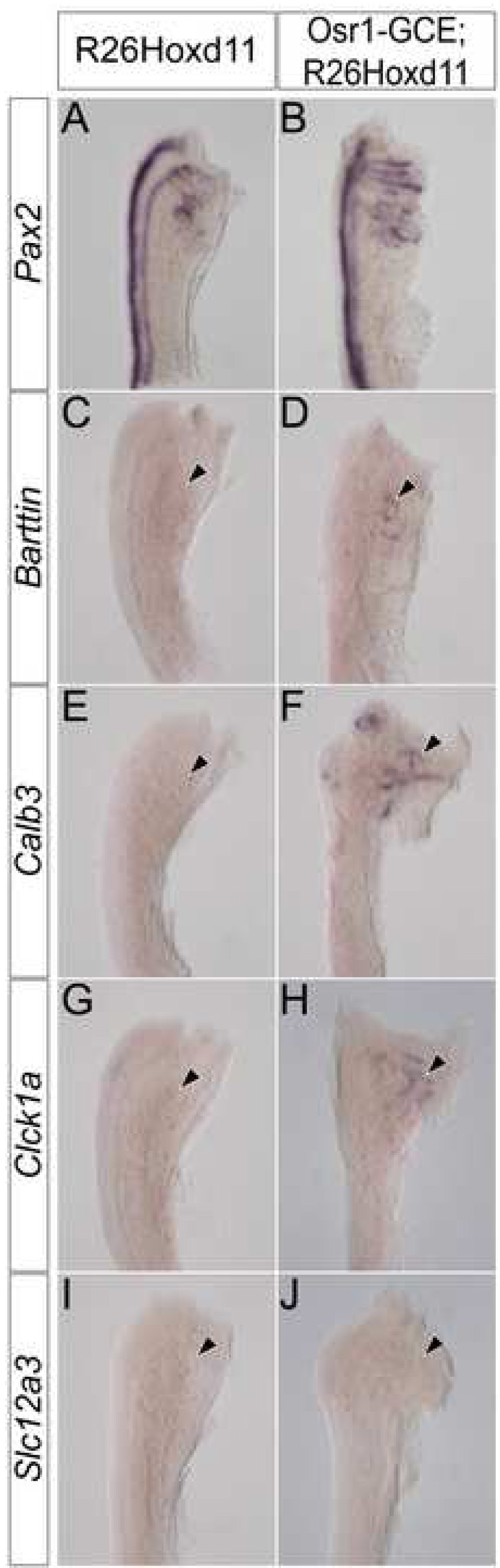
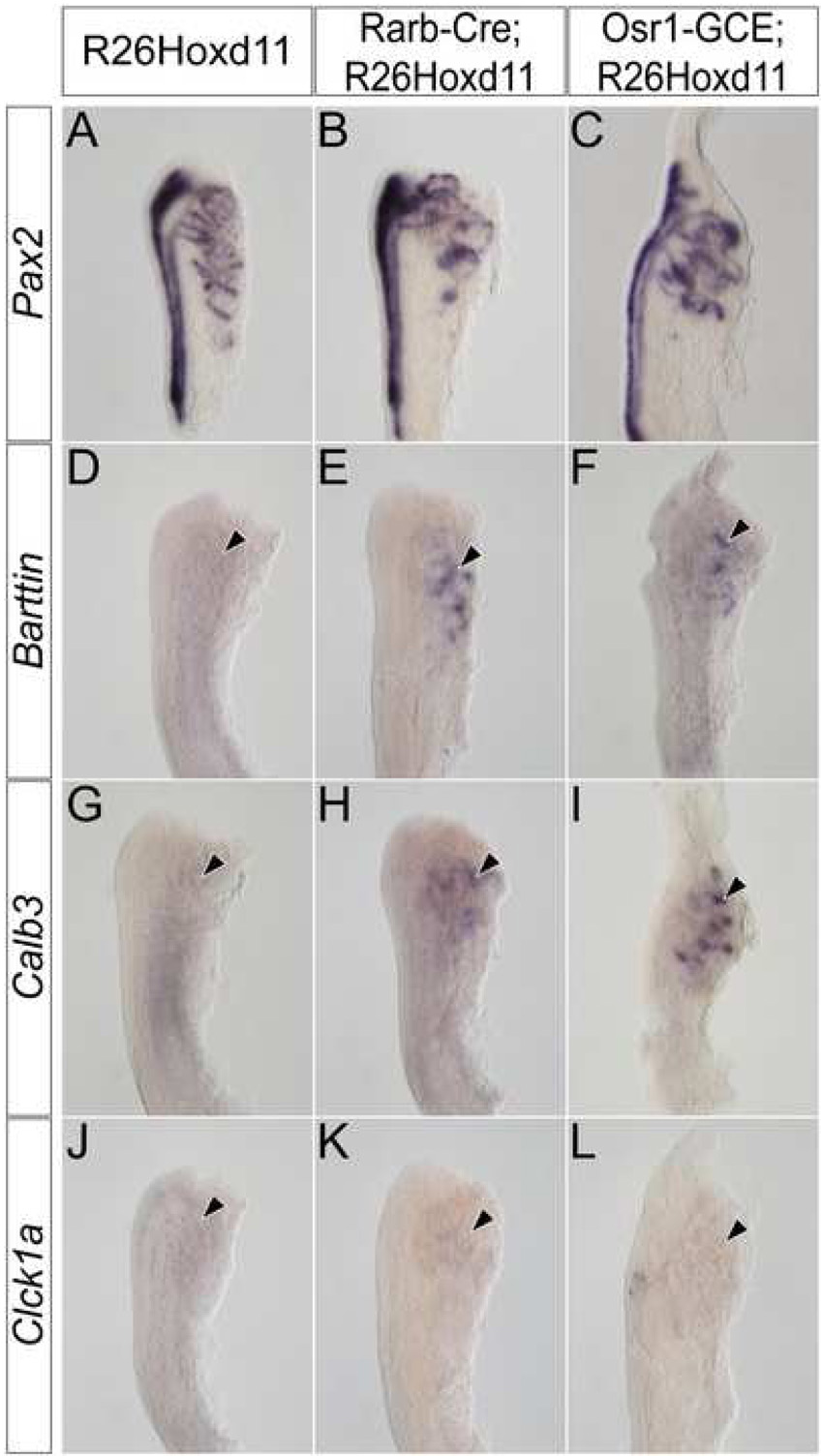
To examine the affect of ectopic Hoxd11 activity in the mesonephros, we used Pax2 expression to assess the morphology of E13.5 mesonephric tubules in Osr1-GCE;R26Hoxd11 embryos. While the overall number and spatial arrangement of mesonephric tubules of both males and females is relatively normal, mesonephric tubules of Osr1-GCE;R26Hoxd11 embryos exhibit an altered, more elaborate morphology as compared to control littermates at E13.5 (Fig. 5A, B). Further, Barttin, whose expression is absent in control embryos at E13.5, was maintained in transgenic tubules (Fig. 5C, D arrowhead). Additionally, Calb3 (Fig. 5E,F arrowhead) and Clck1a (Fig. 5G, H arrowhead), which are never expressed in mesonephric structures, were activated upon Hoxd11 expression. No Slc12a3 expression was observed (Fig. 5I, J arrowhead). Pax2 expression in the E12.5 mesonephros of Rarb-Cre;R26Hoxd11 and Osr1-GCE;R26Hoxd11 reveals a similarly altered morphology (Fig. 6A–C). Embryos of both genotypes express Barttin (Fig. 6D–F arrowheads) and Calb3 (Fig. 6G–I arrowheads), but not Clck1a at this time (Fig. 6J–L arrowheads). Identical results were obtained in the mesonephros of E12.5 embryos harboring the Osr1-GCE allele and two R26Hoxd11 alleles (data not shown). Thus, mesonephric tubules expressing Hoxd11 activate a transcriptional program reflective of a partial metanephric conversion.
Figure 5. Mesonephric tubules expressing Hoxd11 have a morphology and gene expression reminiscent of metanephric tubules.
Whole mount in situ hybridization for Pax2 (A, B), Barttin (C, D), Calb3 (E, F), Clck1a (G, H) and Slc12a3 (I, J) in the E13.5 mesonephros of control (A, C, E, G, I) or Osr1-GCE;R26HoxD11 (B, D, F, H, J) embryos. Arrowheads indicate location of mesonephric tubules.
Figure 6. Hoxd11 activity in the mesonephros alters the morphology and gene expression of E12.5 mesonephric tubules.
Whole mount in situ hybridization for Pax2 (A–C), Barttin (D–F), Calb3 (G–I), and Clck1a (J–L) in the E12.5 mesonephros of control (A, D, G, J), Rarb-Cre;R26HoxD11 (B, E, H, K) or Osr1-GCE;R26HoxD11 (C, F, I, L) embryos. Arrowheads indicate location of mesonephric tubules.
Discussion
Mesonephric and metanephric tubules utilize identical inductive and early developmental programs
The cellular origin of mesonephric and metanephric tubules has been a matter of debate. Sainio et al., (1997) have suggested that cranial mesonephric tubules are derivatives of the ND, whereas the caudal mesonephros forms due to a MET of the mesonephric mesenchyme, implying a distinct, regional specific origin of tubular structures. Our fate mapping analysis solves this issue. The ND only contributes to a connecting segment of a cranial mesonephric tubule, whereas the remainder of the tubule is derived from the mesonephric mesenchyme. The connecting segment is a mosaic of ND and mesonephric mesoderm-derived cell types, whereas the remainder of the mesonephric tubule is entirely derived from mesenchymal progenitors. Thus, as in metanephric nephrons, all mesonephric tubules derive from a MET within mesenchyme adjacent to the ND.
Previous studies have demonstrated that Wnt9b induces the metanephric mesenchyme to form tubules via the canonical Wnt pathway (Carroll et al., 2005; Park et al., 2007) and mesonephric tubules are absent from Wnt9b mutants (Carroll et al., 2005). Thus, it is likely that the mesonephric mesenchyme is also induced to undergo MET by a similar inductive events. That Wnt4 and FGF8 are also downstream of Wnt9b in both mesonephric and metanephric tubulogenesis lends additional evidence for a common program of tubule formation within distinct regions of the IM (Grieshammer et al., 2005; Perantoni et al., 2005; Stark et al., 1994; Vainio et al., 1999). Further, these findings support the hypothesis that the differences observed between the mesonephros and metanephros are due to a pre-pattern of the IM mesenchyme rather than regionally distinct inductive signals that govern distinct tubule differentiation programs.
Interestingly, mutants in WT1 (Sainio et al., 1997) and Osr1 (James et al., 2006) are reported to develop cranial, but not caudal mesonephric tubules. However, these studies fail to distinguish between the connecting segment and the more distal regions of the cranial tubule. In both mutants, the morphology of the cranial tubules is grossly abnormal. Thus, the remaining cranial mesonephric tubules observed in these mutants may actually represent arrested ND outgrowths that lack any mesenchymal contribution. Cell fate analysis in the context of each mutant would shed light on this issue.
Our analysis demonstrates that mesonephric and metanephric tubules express many of the same genes. Interestingly, though mesonephric tubules are reported to lack metanephric structures, notably the loop-of-Henle (Vize et al., 2002), mesonephric tubules do express markers such as Barttin and Brn1, that are present in the loop-of-Henle (Nakai et al., 2003; Wolf et al., 2003). In contrast, mesonephric tubules do not express Calb3, Clck1a and Slc12a3, markers of connecting tubules, ascending thin loop-of-Henle, and distal convoluted tubules, respectively (Chen et al., 2006; Nakai et al., 2003; Wolf et al., 2003). Thus, mesonephric and metanephric tubules differ largely with respect to distal segment markers.
The Hox11 paralogs specify the metanephric mesenchyme
Remarkably, the Hox11 paralogs are unique in displaying metanephric mesenchyme specific expression prior to metanephric tubulogenesis. Consistent with roles in the development of both kidney types, Osr1, Pax2, WT1, Sall1, Eya1 and Six1 are all expressed, at least briefly, in both the mesonephros and later, with the exception of Six1, in the metanephric mesenchyme throughout metanephric kidney development. Indeed, mutants in Osr1, Pax2, WT1, Sall1, and Six1 all display mesonephric and metanephric phenotypes (Grote et al., 2006; James et al., 2006; Kobayashi et al., 2007; Narlis et al., 2007; Nishinakamura et al., 2001; Sainio et al., 1997; Torres et al., 1995; Wang et al., 2005; Xu et al., 2003). The role of Eya1 is less clear. A recent report suggests that Eya1 specifies the metanephros (Sajithlal et al., 2005) as Eya1 mutants lack metanephric kidneys (Xu et al., 1999), but mesonephric tubules are present at E10.5. However Hoxd11 is expressed in Eya1 mutants (Gong et al., 2007), thus regional specification of the IM is intact in Eya1 mutants. Interestingly, Eya1 expression remains in Hox11 mutants, suggesting that Eya1 has a more general role in IM nephric development that is at least partially independent of Hox11 paralog activity.
Consistent with Hox11 specification of the metanephros, our results demonstrate that ectopic Hoxd11 activity in the mesonephros is sufficient to drive ectopic expression of a key regulator, Six2, in mesonephric IM mesenchyme. Several additional lines of evidence support a likely role for Hox11 regulation of Six2. Six2 expression is absent in Hox11 mutants (Wellik et al., 2002). Further, a recent report demonstrates that Pax2, Eya1, and Hoxa11 can synergistically activate a reporter under the control of a Six2 enhancer element in MDCK cells (Gong et al., 2007). Our results demonstrate that Hoxd11 activity in the E10.5 mesonephric mesenchyme activates Six2 in a cell-autonomous manner. Furthermore, Six2 expression in the mesonephros was only observed in mesenchymal cells and not in β-gal+ epithelial cells of the ND or in mesonephric tubules. Thus, some other mesenchymally contributed factors must ensure an appropriate cell type specific response in conjunction with Hoxd11.
In the metanephros, Six2 is down-regulated as the cap mesenchyme epithelializes to form nephron precursors, renal vesicles (Self et al., 2006). We have demonstrated that mesonephric tubules are derivatives of the mesonephric mesenchyme. The presence of β-gal+, Six2− cells within mesonephric tubules indicates that these cells also down-regulate Six2 expression upon epithelialization. This likely explains why Rarb-Cre;R26Hoxd11 embryos do not activate Six2 as Rarb-Cre is not active until mesonephric mesenchyme induction is underway. Taken together, these results demonstrate that Hoxd11 activates Six2, potentially directly, and that the addition of Hoxd11 activity in cells of the mesonephric mesenchyme causes these cells to behave in a similar manner to metanephric cap mesenchyme maintaining Six2 activity. Six2, in turn, ensures the continued expression of a metanephric nephron progenitor population (Self et al., 2006).
Importantly, the morphology and onset of metanephric marker expression in mesonephric tubules expressing Hoxd11 mimics that of metanephric tubules. During metanephric development, Calb3 and Barttin expression precede that of Slc12a3 and Clck1a by one day. The same timing is observed in developing Hoxd11+ mesonephric nephrons. Though Slc12a3 was not detected in mesonephric tubules expressing Hoxd11, it is possible that these tubules are still receiving sex-specific molecular cues from the adjacent gonads. Gonad derived signals could compete with the metanephric program activated by Hoxd11 and not allow for the completion of a bona fide metanephric tubule program. Furthermore, the E12.5 mesonephric tubules of Osr1-GCE;R26Hoxd11 and Rarb-Cre;R26Hoxd11 embryos express identical metanephric markers. This suggests that the metanephric tubule program activated by Hoxd11 is independent of Six2 function since Six2 is not ectopically activated in Rarb-Cre;R26Hoxd11 embryos. This indicates that the Hox11 paralogs simultaneously activate two separate and independent molecular programs in the metanephric mesenchyme. The activation of Six2 allows for the maintenance of the cap mesenchyme, while an independent program confers a metanephric-specific response downstream of common inductive events.
While ectopic Hoxd11 expression can result in several features of a metanephric program, we do not observe a whole scale conversion of the mesonephric region into a metanephric kidney. There may be many underlying reasons for this result. For example, the kinetics and mosaicism of Hoxd11 activation by Osr1-GCE may activate Hoxd11 too late and in too few cells for a global change. Alternatively, the levels of Hoxd11 may be insufficient; however when Hoxd11 dosage was increased by activating two copies of R26Hoxd11, we did not observe an enhancement of the mesonephric to metanephric, thus suggesting that this explanation is unlikely. Further, the roles of the Hox11 paralogs may not be simply additive. One critical factor in the failure of a more extensive metanephric conversion is likely to be the failure of Gdnf activation in Hoxd11+ mesonephric mesenchyme.
Gdnf expression is absent in the metanephric mesenchyme of Hox11 mutants, suggesting that Gdnf is a likely target of Hox11 action (Wellik et al., 2002). Additionally, Pax2, Eya1 and Hoxa11 can synergistically activate a reporter gene under the control of a Gdnf promoter element in MDCK cells (Gong et al., 2007). At the initiation of metanephric development, Gdnf expression must be tightly controlled in order to restrict ND invasion to a specific location in the metanephric mesenchyme. Gdnf activation in the metanephros requires Osr1, Pax2/Pax8, Eya1, Six1 and Hox11 paralog function (James et al., 2006; Kobayashi et al., 2007; Narlis et al., 2007; Sajithlal et al., 2005; Wellik et al., 2002; Xu et al., 2003). However, in addition to these activating factors, FoxC1 activity (Kume et al., 2000) and Robo2/Slit2 signaling (Grieshammer et al., 2004) are also required to restrict Gdnf to the metanephric mesenchyme. Hoxd11 activity in mesonephric mesenchyme may be insufficient to override the negative regulation of FoxC1 or Robo2/Slit2 signaling in vivo, whereas these negative signals may not be present in MDCK cell culture.
It is tempting to speculate that the development of the metanephric kidney requires the convergence of parallel molecular pathways upon the posterior IM. In this model, the early IM is specified by genes such as Osr1, Pax2, WT1 and Eya1. Hox11 paralog expression in the posterior IM mesenchyme functions with Pax2 and Eya1 to activate Gdnf and Six2, allowing for ND branching and self-renewal of nephron progenitors, respectively. Gdnf expression is restricted to a single site by FoxC1 and Robo2/Slit2 signaling, providing the exact location for ND invasion into the metanephric mesenchyme. Simultaneously, the Hox11 paralogs regulate the metanephric-specific tubule response of the induced cap mesenchyme, resulting in the formation of an appropriately organized nephron structure.
Supplementary Material
01. Supplemental Figure 1 – Calb3 expression precedes expression of Clck1a and Slc12a3 in developing metanephric tubules.
Whole mount in situ hybridization for Calb3 (A, B), Clck1a (C, D) and Slc12a3 (E, F) in E13.5 (A, C, E) and E14.5 (B, D, F) metanephric kidneys. Arrowheads indicate expression (A, B, D, F) or lack of expression (C, E).
02. Supplemental Figure 2 – The Hox11 IM expression pattern differs from that of other transcription factors required for metanephric development.
Whole mount in situ hybridization for Osr1 (A–E), Pax2 (F–J), Eya1 (K–O) and Hoxd11 (P–T) in E9.5 (A, B, F, G, K, L, P, Q) and E10.5 (C, H, M, R) trunks, E10.5 (D, I, N, S) mesonephros and E11.5 (E, J, O, T) IM of wild type embryos. Arrowheads indicate location of the mesonephros. Arrows indicate the location of the metanephros. Dashed line (A) indicates approximate plane of section of embryos (B, G, L, Q).
03. Supplemental Figure 3 – Creation of the R26Hoxd11 line.
(A) Schematic of the inducible Hoxd11 IRES2 nuclear LacZ construct in the R26 locus before and after Cre mediated recombination. (B) RT-PCR detection of the Hoxd11 construct transcript from targeted YFP 3.1 ES cells untreated or treated with 4OH-Tamoxifen using primers cHD11Fw and cHD11Rv. (C–F) Images of an expanded Hoxd11 YFP 3.1 ES cell clone. Bright field (C), YFP expression (D) and X-gal staining without (E) and with addition (F) of 4OH-Tamoxifen.
04. Supplemental Figure 4 – Osr1-GCE is active in the early IM and the R26Hoxd11 allele behaves as expected in vivo.
(A) X-gal staining in whole mount and vibrotome sections of Osr1-GCE;R26R E8.5, E9.5 and E12.5 embryos induced with Tamoxifen at E7.75. Arrowheads indicate expression in intermediate mesoderm. Dashed lines indicate approximate planes of section. (B) Immunofluorescent confocal microscopy of the mesonephros of E13.5 control, Rarb-Cre;R26HD11 and Osr1-GCE;R26Hoxd11 embryos stained for Hoxd11, Cytokeratin, β-gal and Hoechst 33342. (C) Section in situ analysis for Hoxd11 mRNA and immunofluorescent confocal microscopy of Hoxd11 and Cytokeratin protein in wild-type E15.5 metanephric kidneys.
Acknowledgements
We thank Denis Duboule for the gift of full-length mouse Hoxd11 cDNA. P.S. was funded by grants from the Finnish Academy (#107827), Helsingin Sanomain 100-vuotis and Alfred Kordelin Foundations. R.R.B was funded by NIH grant HD30284. Work in A.P.M.’s laboratory is supported by NIH grant DK054364.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barasch J, Yang J, Ware CB, Taga T, Yoshida K, Erdjument-Bromage H, Tempst P, Parravicini E, Malach S, Aranoff T, Oliver JA. Mesenchymal to epithelial conversion in rat metanephros is induced by LIF. Cell. 1999;99:377–386. doi: 10.1016/s0092-8674(00)81524-x. [DOI] [PubMed] [Google Scholar]
- Basson MA, Akbulut S, Watson-Johnson J, Simon R, Carroll TJ, Shakya R, Gross I, Martin GR, Lufkin T, McMahon AP, Wilson PD, Costantini FD, Mason IJ, Licht JD. Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev Cell. 2005;8:229–239. doi: 10.1016/j.devcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Basson MA, Watson-Johnson J, Shakya R, Akbulut S, Hyink D, Costantini FD, Wilson PD, Mason IJ, Licht JD. Branching morphogenesis of the ureteric epithelium during kidney development is coordinated by the opposing functions of GDNF and Sprouty1. Dev Biol. 2006;299:466–477. doi: 10.1016/j.ydbio.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Bouchard M, Souabni A, Mandler M, Neubuser A, Busslinger M. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 2002;16:2958–2970. doi: 10.1101/gad.240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9:283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Chen Z, Chen J, Weng T, Jin N, Liu L. Identification of rat lung--prominent genes by a parallel DNA microarray hybridization. BMC Genomics. 2006;7:47. doi: 10.1186/1471-2164-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley PH, Minowada G, MacArthur CA, Martin GR. Roles for FGF8 in the induction, initiation, and maintenance of chick limb development. Cell. 1996;84:127–136. doi: 10.1016/s0092-8674(00)80999-x. [DOI] [PubMed] [Google Scholar]
- Deschamps J, van Nes J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development. 2005;132:2931–2942. doi: 10.1242/dev.01897. [DOI] [PubMed] [Google Scholar]
- Donovan MJ, Natoli TA, Sainio K, Amstutz A, Jaenisch R, Sariola H, Kreidberg JA. Initial differentiation of the metanephric mesenchyme is independent of WT1 and the ureteric bud. Dev Genet. 1999;24:252–262. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<252::AID-DVG8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Duboule D. The rise and fall of Hox gene clusters. Development. 2007;134:2549–2560. doi: 10.1242/dev.001065. [DOI] [PubMed] [Google Scholar]
- Ferrier DE, Minguillon C. Evolution of the Hox/ParaHox gene clusters. Int J Dev Biol. 2003;47:605–611. [PubMed] [Google Scholar]
- Gong KQ, Yallowitz AR, Sun H, Dressler GR, Wellik DM. A hox-eya-pax complex regulates early kidney developmental gene expression. Mol Cell Biol. 2007;27:7661–7668. doi: 10.1128/MCB.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieshammer U, Cebrian C, Ilagan R, Meyers E, Herzlinger D, Martin GR. FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development. 2005;132:3847–3857. doi: 10.1242/dev.01944. [DOI] [PubMed] [Google Scholar]
- Grieshammer U, Le M, Plump AS, Wang F, Tessier-Lavigne M, Martin GR. SLIT2-mediated ROBO2 signaling restricts kidney induction to a single site. Dev Cell. 2004;6:709–717. doi: 10.1016/s1534-5807(04)00108-x. [DOI] [PubMed] [Google Scholar]
- Grobstein C. Inductive Epithelio-mesenchymal Interaction in Cultured Organ Rudiments of the Mouse. Science. 1953;118:52–55. doi: 10.1126/science.118.3054.52. [DOI] [PubMed] [Google Scholar]
- Grote D, Souabni A, Busslinger M, Bouchard M. Pax 2/8-regulated Gata 3 expression is necessary for morphogenesis and guidance of the nephric duct in the developing kidney. Development. 2006;133:53–61. doi: 10.1242/dev.02184. [DOI] [PubMed] [Google Scholar]
- Gruenweld P. Development of the Excretory System. Annals of the New York Academy of Sciences. 1952;55:142–146. doi: 10.1111/j.1749-6632.1952.tb26529.x. [DOI] [PubMed] [Google Scholar]
- James RG, Kamei CN, Wang Q, Jiang R, Schultheiss TM. Odd-skipped related 1 is required for development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Development. 2006;133:2995–3004. doi: 10.1242/dev.02442. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kwan KM, Carroll TJ, McMahon AP, Mendelsohn CL, Behringer RR. Distinct and sequential tissue-specific activities of the LIM-class homeobox gene Lim1 for tubular morphogenesis during kidney development. Development. 2005a;132:2809–2823. doi: 10.1242/dev.01858. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Shawlot W, Kania A, Behringer RR. Requirement of Lim1 for female reproductive tract development. Development. 2004;131:539–549. doi: 10.1242/dev.00951. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Kawakami K, Asashima M, Nishinakamura R. Six1 and Six4 are essential for Gdnf expression in the metanephric mesenchyme and ureteric bud formation, while Six1 deficiency alone causes mesonephric-tubule defects. Mech Dev. 2007;124:290–303. doi: 10.1016/j.mod.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Tanaka H, Kuwana H, Inoshita S, Teraoka H, Sasaki S, Terada Y. Wnt4-transformed mouse embryonic stem cells differentiate into renal tubular cells. Biochem Biophys Res Commun. 2005b;336:585–595. doi: 10.1016/j.bbrc.2005.08.136. [DOI] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Kume T, Deng K, Hogan BL. Murine forkhead/winged helix genes Foxc1 (Mf1) and Foxc2 (Mfh1) are required for the early organogenesis of the kidney and urinary tract. Development. 2000;127:1387–1395. doi: 10.1242/dev.127.7.1387. [DOI] [PubMed] [Google Scholar]
- Lemons D, McGinnis W. Genomic evolution of Hox gene clusters. Science. 2006;313:1918–1922. doi: 10.1126/science.1132040. [DOI] [PubMed] [Google Scholar]
- Little MH, Brennan J, Georgas K, Davies JA, Davidson DR, Baldock RA, Beverdam A, Bertram JF, Capel B, Chiu HS, Clements D, Cullen-McEwen L, Fleming J, Gilbert T, Herzlinger D, Houghton D, Kaufman MH, Kleymenova E, Koopman PA, Lewis AG, McMahon AP, Mendelsohn CL, Mitchell EK, Rumballe BA, Sweeney DE, Valerius MT, Yamada G, Yang Y, Yu J. A high-resolution anatomical ontology of the developing murine genitourinary tract. Gene Expr Patterns. 2007;7:680–699. doi: 10.1016/j.modgep.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A, Vainio S, Kispert A, McMahon J, McMahon AP. Wnt11 and Ret/Gdnf pathways cooperate in regulating ureteric branching during metanephric kidney development. Development. 2003;130:3175–3185. doi: 10.1242/dev.00520. [DOI] [PubMed] [Google Scholar]
- Mao J, Barrow J, McMahon J, Vaughan J, McMahon AP. An ES cell system for rapid, spatial and temporal analysis of gene function in vitro and in vivo. Nucleic Acids Res. 2005;33:e155. doi: 10.1093/nar/gni146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AP, Davidson D, Davies J, Gaido K, Lessard J, Little M, Grimmond S, Potter SS, Zhang P. GUDMAP Home Page. 2006 www.gudmap.org.
- Michos O, Goncalves A, Lopez-Rios J, Tiecke E, Naillat F, Beier K, Galli A, Vainio S, Zeller R. Reduction of BMP4 activity by gremlin 1 enables ureteric bud outgrowth and GDNF/WNT11 feedback signalling during kidney branching morphogenesis. Development. 2007;134:2397–2405. doi: 10.1242/dev.02861. [DOI] [PubMed] [Google Scholar]
- Murer H, Hernando N, Forster I, Biber J. Regulation of Na/Pi transporter in the proximal tubule. Annu Rev Physiol. 2003;65:531–542. doi: 10.1146/annurev.physiol.65.042902.092424. [DOI] [PubMed] [Google Scholar]
- Nakai S, Sugitani Y, Sato H, Ito S, Miura Y, Ogawa M, Nishi M, Jishage K, Minowa O, Noda T. Crucial roles of Brn1 in distal tubule formation and function in mouse kidney. Development. 2003;130:4751–4759. doi: 10.1242/dev.00666. [DOI] [PubMed] [Google Scholar]
- Narlis M, Grote D, Gaitan Y, Boualia SK, Bouchard M. Pax2 and pax8 regulate branching morphogenesis and nephron differentiation in the developing kidney. J Am Soc Nephrol. 2007;18:1121–1129. doi: 10.1681/ASN.2006070739. [DOI] [PubMed] [Google Scholar]
- Nishinakamura R, Matsumoto Y, Nakao K, Nakamura K, Sato A, Copeland NG, Gilbert DJ, Jenkins NA, Scully S, Lacey DL, Katsuki M, Asashima M, Yokota T. Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development. 2001;128:3105–3115. doi: 10.1242/dev.128.16.3105. [DOI] [PubMed] [Google Scholar]
- Nishinakamura R, Osafune K. Essential roles of Sall family genes in kidney development. J Physiol Sci. 2006;56:131–136. doi: 10.2170/physiolsci.M95. [DOI] [PubMed] [Google Scholar]
- Ott T, Parrish M, Bond K, Schwaeger-Nickolenko A, Monaghan AP. A new member of the spalt like zinc finger protein family, Msal-3, is expressed in the CNS and sites of epithelial/mesenchymal interaction. Mech Dev. 2001;101:203–207. doi: 10.1016/s0925-4773(00)00552-9. [DOI] [PubMed] [Google Scholar]
- Park JS, Valerius MT, McMahon AP. Wnt/beta-catenin signaling regulates nephron induction during mouse kidney development. Development. 2007;134:2533–2539. doi: 10.1242/dev.006155. [DOI] [PubMed] [Google Scholar]
- Patterson LT, Pembaur M, Potter SS. Hoxa11 and Hoxd11 regulate branching morphogenesis of the ureteric bud in the developing kidney. Development. 2001;128:2153–2161. doi: 10.1242/dev.128.11.2153. [DOI] [PubMed] [Google Scholar]
- Patterson LT, Potter SS. Atlas of Hox gene expression in the developing kidney. Dev Dyn. 2004;229:771–779. doi: 10.1002/dvdy.10474. [DOI] [PubMed] [Google Scholar]
- Pedersen A, Skjong C, Shawlot W. Lim 1 is required for nephric duct extension and ureteric bud morphogenesis. Dev Biol. 2005;288:571–581. doi: 10.1016/j.ydbio.2005.09.027. [DOI] [PubMed] [Google Scholar]
- Pepicelli CV, Kispert A, Rowitch DH, McMahon AP. GDNF induces branching and increased cell proliferation in the ureter of the mouse. Dev Biol. 1997;192:193–198. doi: 10.1006/dbio.1997.8745. [DOI] [PubMed] [Google Scholar]
- Perantoni AO, Timofeeva O, Naillat F, Richman C, Pajni-Underwood S, Wilson C, Vainio S, Dove LF, Lewandoski M. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development. 2005;132:3859–3871. doi: 10.1242/dev.01945. [DOI] [PubMed] [Google Scholar]
- Sainio K, Hellstedt P, Kreidberg JA, Saxen L, Sariola H. Differential regulation of two sets of mesonephric tubules by WT-1. Development. 1997;124:1293–1299. doi: 10.1242/dev.124.7.1293. [DOI] [PubMed] [Google Scholar]
- Sainio K, Raatikainen-Ahokas A. Mesonephric kidney--a stem cell factory? Int J Dev Biol. 1999;43:435–439. [PubMed] [Google Scholar]
- Sajithlal G, Zou D, Silvius D, Xu PX. Eya 1 acts as a critical regulator for specifying the metanephric mesenchyme. Dev Biol. 2005;284:323–336. doi: 10.1016/j.ydbio.2005.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MP, Silos-Santiago I, Frisen J, He B, Lira SA, Barbacid M. Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature. 1996;382:70–73. doi: 10.1038/382070a0. [DOI] [PubMed] [Google Scholar]
- Saxén L. Organogenesis of the kidney. New York: Cambridge University Press, Cambridge [Cambridgeshire]; 1987. [Google Scholar]
- Self M, Lagutin OV, Bowling B, Hendrix J, Cai Y, Dressler GR, Oliver G. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. Embo J. 2006;25:5214–5228. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark K, Vainio S, Vassileva G, McMahon AP. Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature. 1994;372:679–683. doi: 10.1038/372679a0. [DOI] [PubMed] [Google Scholar]
- Tilmann C, Capel B. Cellular and molecular pathways regulating mammalian sex determination. Recent Prog Horm Res. 2002;57:1–18. doi: 10.1210/rp.57.1.1. [DOI] [PubMed] [Google Scholar]
- Torres M, Gomez-Pardo E, Dressler GR, Gruss P. Pax-2 controls multiple steps of urogenital development. Development. 1995;121:4057–4065. doi: 10.1242/dev.121.12.4057. [DOI] [PubMed] [Google Scholar]
- Vainio S, Heikkila M, Kispert A, Chin N, McMahon AP. Female development in mammals is regulated by Wnt-4 signalling. Nature. 1999;397:405–409. doi: 10.1038/17068. [DOI] [PubMed] [Google Scholar]
- Valentini RP, Brookhiser WT, Park J, Yang T, Briggs J, Dressler G, Holzman LB. Post-translational processing and renal expression of mouse Indian hedgehog. J Biol Chem. 1997;272:8466–8473. doi: 10.1074/jbc.272.13.8466. [DOI] [PubMed] [Google Scholar]
- Vega QC, Worby CA, Lechner MS, Dixon JE, Dressler GR. Glial cell line-derived neurotrophic factor activates the receptor tyrosine kinase RET and promotes kidney morphogenesis. Proc Natl Acad Sci U S A. 1996;93:10657–10661. doi: 10.1073/pnas.93.20.10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vize PD, Woolf AS, Bard JBL. The kidney: from normal development to congenital diseases. Boston: Academic Press, Amsterdam; 2002. [Google Scholar]
- Wang Q, Lan Y, Cho ES, Maltby KM, Jiang R. Odd-skipped related 1 (Odd 1) is an essential regulator of heart and urogenital development. Dev Biol. 2005;288:582–594. doi: 10.1016/j.ydbio.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellik DM, Hawkes PJ, Capecchi MR. Hox11 paralogous genes are essential for metanephric kidney induction. Genes Dev. 2002;16:1423–1432. doi: 10.1101/gad.993302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellik DM. Hox patterning of the vertebrate axial skeleton. Dev Dyn. 2007;236:2454–2463. doi: 10.1002/dvdy.21286. [DOI] [PubMed] [Google Scholar]
- Wolf K, Meier-Meitinger M, Bergler T, Castrop H, Vitzthum H, Riegger GA, Kurtz A, Kramer BK. Parallel down-regulation of chloride channel CLC-K1 and barttin mRNA in the thin ascending limb of the rat nephron by furosemide. Pflugers Arch. 2003;446:665–671. doi: 10.1007/s00424-003-1098-8. [DOI] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- Xu PX, Zheng W, Huang L, Maire P, Laclef C, Silvius D. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130:3085–3094. doi: 10.1242/dev.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Carroll TJ, McMahon AP. Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development. 2002;129:5301–5312. doi: 10.1242/dev.129.22.5301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01. Supplemental Figure 1 – Calb3 expression precedes expression of Clck1a and Slc12a3 in developing metanephric tubules.
Whole mount in situ hybridization for Calb3 (A, B), Clck1a (C, D) and Slc12a3 (E, F) in E13.5 (A, C, E) and E14.5 (B, D, F) metanephric kidneys. Arrowheads indicate expression (A, B, D, F) or lack of expression (C, E).
02. Supplemental Figure 2 – The Hox11 IM expression pattern differs from that of other transcription factors required for metanephric development.
Whole mount in situ hybridization for Osr1 (A–E), Pax2 (F–J), Eya1 (K–O) and Hoxd11 (P–T) in E9.5 (A, B, F, G, K, L, P, Q) and E10.5 (C, H, M, R) trunks, E10.5 (D, I, N, S) mesonephros and E11.5 (E, J, O, T) IM of wild type embryos. Arrowheads indicate location of the mesonephros. Arrows indicate the location of the metanephros. Dashed line (A) indicates approximate plane of section of embryos (B, G, L, Q).
03. Supplemental Figure 3 – Creation of the R26Hoxd11 line.
(A) Schematic of the inducible Hoxd11 IRES2 nuclear LacZ construct in the R26 locus before and after Cre mediated recombination. (B) RT-PCR detection of the Hoxd11 construct transcript from targeted YFP 3.1 ES cells untreated or treated with 4OH-Tamoxifen using primers cHD11Fw and cHD11Rv. (C–F) Images of an expanded Hoxd11 YFP 3.1 ES cell clone. Bright field (C), YFP expression (D) and X-gal staining without (E) and with addition (F) of 4OH-Tamoxifen.
04. Supplemental Figure 4 – Osr1-GCE is active in the early IM and the R26Hoxd11 allele behaves as expected in vivo.
(A) X-gal staining in whole mount and vibrotome sections of Osr1-GCE;R26R E8.5, E9.5 and E12.5 embryos induced with Tamoxifen at E7.75. Arrowheads indicate expression in intermediate mesoderm. Dashed lines indicate approximate planes of section. (B) Immunofluorescent confocal microscopy of the mesonephros of E13.5 control, Rarb-Cre;R26HD11 and Osr1-GCE;R26Hoxd11 embryos stained for Hoxd11, Cytokeratin, β-gal and Hoechst 33342. (C) Section in situ analysis for Hoxd11 mRNA and immunofluorescent confocal microscopy of Hoxd11 and Cytokeratin protein in wild-type E15.5 metanephric kidneys.