O-Linked β-N-Acetylglucosaminyltransferase Substrate Specificity Is Regulated by Myosin Phosphatase Targeting and Other Interacting Proteins (original) (raw)
Abstract
O_-GlcNAc-transferase (OGT) substrate specificity is regulated by transiently interacting proteins. To further examine the regulation of OGT, we have identified 27 putative OGT-interacting proteins through a yeast two-hybrid screen. Two of these proteins, Trak1 (OIP106) and_O_-GlcNAcase, have been shown previously to interact with and regulate OGT. We demonstrate here that MYPT1 and CARM1 also interact with and target OGT. MYPT1 and CARM1 are substrates of OGT in vitro and in vivo. MYPT1 and CARM1 also function to alter OGT substrate specificity_in vitro. Furthermore depletion of MYPT1 in Neuro-2a neuroblastoma cells alters GlcNAcylation of several proteins under basal conditions, suggesting that MYPT1 regulates OGT substrate specificity in vivo.
In metazoans, nuclear and cytoplasmic proteins are post-translationally modified on serines and threonines with_O_-GlcNAc3 in response to different extracellular signals, altering their biochemical and functional properties (for a review, see Ref.1). The addition of the monosaccharide moiety is catalyzed by _O_-GlcNAc-transferase (OGT) using UDP-GlcNAc as the nucleotide sugar donor (2), whereas the hydrolysis of the _O_-GlcNAc residue is catalyzed by _O_-GlcNAcase (3).
In many respects, protein GlcNAcylation is analogous to Ser/Thr phosphorylation. However, a primary difference is that, unlike phosphorylation, which is catalyzed by almost 400 Ser/Thr kinases encoded by the human genome (4), GlcNAcylation is catalyzed by the products of a single human gene (5,6). Although kinase substrate selectivity is partially genetically encoded, the mechanism of OGT substrate specificity is not clear. More than 600 GlcNAcylated proteins have now been identified (1), yet the question of how OGT is able to distinguish its substrates from other proteins is not well understood.
Using synthetic peptide substrates, OGT does seem to show sequence specificity in vitro. In addition, OGT is responsive to a wide range of UDP-GlcNAc concentrations, and the affinity of OGT for peptide substrates is altered with different UDP-GlcNAc concentrations (7). As a result, it has been suggested that the substrate specificity of OGT in vivo may be regulated by UDP-GlcNAc concentrations, which have been shown to be responsive to the nutritional state of the cell (8–10). Although this hypothesis does partially explain hyperglycemia-mediated changes in protein GlcNAcylation, it cannot account for changes in GlcNAcylation of specific proteins that are independent of alterations in UDP-GlcNAc concentration.
The crystal structure of an OGT homolog has been solved recently along with a partial structure of OGT, providing insight into its regulation (11,12). The tetratricopeptide repeat domain of OGT forms a large superhelical structure very similar to that of importin α (11,12), suggesting that OGT may interact with a diverse group of proteins through its N terminus (13,14).
In support of this hypothesis, it has been shown that Trak1 (previously known as OIP106) interacts with the tetratricopeptide repeat domain of OGT, recruiting OGT to RNA polymerase II (15). Other studies have since revealed OGT to be regulated by its interacting proteins (16–18). Recently we have demonstrated that glucose deprivation in Neuro-2a cells increases the OGT-mediated GlcNAcylation of neurofilament H via targeting mediated by activated p38 (19). However, in this case, instead of interacting with the tetratricopeptide repeat domain of OGT, p38 interacts with the C terminus of OGT. Interestingly this same region of OGT has also been shown to interact with phosphatidylinositol 3,4,5-trisphosphate, leading to recruitment of OGT to the plasma membrane during insulin signaling (20).
Thus, it is becoming apparent that recruitment of OGT through its interaction domains may be a key mode of regulating OGT activity and substrate specificity. To further explore this hypothesis, we sought to identify additional OGT-interacting proteins using a conventional yeast two-hybrid screen. Here not only do we identify candidate OGT regulators, but also we demonstrate that MYPT1 and CARM1 can both function to alter OGT substrate specificity.
EXPERIMENTAL PROCEDURES
_Cell Culture_—Neuro-2a murine neuroblastoma cells (ATCC) were grown in Dulbecco's modified Eagle's medium (5 mm glucose; Mediatech) containing 10% (v/v) fetal bovine serum (Gemini Bio-Products) and penicillin/streptomycin (Mediatech) in a humidified incubator at 37 °C with 5% CO2.
Plasmids, siRNAs, and Transfections_—pJL59-OGT, the yeast expression plasmid encoding full-length rat OGT fused to GAL4 DNA binding domain, was created by standard PCR methods and transformed into the_Saccharomyces cerevisiae AH109 (_MAT_a) strain using the polyethylene glycol/lithium acetate procedure.
pGEX-MYPT1, the prokaryotic expression vector encoding full-length chicken MYPT1, was provided as a kind gift from Dr. Anne A. Wooldridge and Dr. Timothy A. J. Haystead (Duke University Medical Center, Durham, NC) (21). The mammalian expression vector pEF-HA-MYPT1 was created by subcloning from pGEX-MYPT1.
pSG5.HA-CARM1 was provided as a kind gift from Dr. Michael R. Stallcup (University of Southern California, Los Angeles, CA) (22). pGEX-CARM1 was created by subcloning from pSG5.HA-CARM1. pEF-HA-CARM1 was created by standard PCR cloning methods.
The prokaryotic expression vector encoding full-length human OGT was a kind gift of Dr. Suzanne Walker (Harvard Medical School, Boston, MA) (23).
Predesigned control non-targeting siRNAs and siRNAs specific for MYPT1 were obtained from Dharmacon. Neuro-2a cells were transfected with plasmid and siRNAs using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. siRNAs were transfected at a final concentration of 50 nm for a total of 48 h.
_Recombinant Protein Expression and Purification_—ncOGT was expressed and purified as described previously (23). GST was expressed using the pGEX-5x1 plasmid (GE Healthcare) and purified over glutathione-Sepharose (GE Healthcare) according to the manufacturer's instructions.
GST-MYPT1 was expressed and purified as described previously with minor modifications (21). Briefly bacteria were lysed in 25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 75 μg/ml hen egg lysozyme with protease inhibitors. The lysate was purified over glutathione-Sepharose, and the eluant was fractionated over a Mono S high pressure liquid chromatography column developed with a linear gradient of NaCl up to 600 mm. The fractions containing GST-MYPT1 were pooled and desalted into 25 mm Tris-HCl, pH 7.5, 150 mm NaCl.
GST-CARM1 was expressed and purified over glutathione-Sepharose according to the manufacturer's instructions with minor modifications. Briefly the bacterial cell pellet was lysed with 10 mm Tris-HCl, pH 8.0, 200 mm NaCl, 1% Triton X-100 with protease inhibitors. The eluant was dialyzed against 10 mm Tris-HCl, pH 8.0, 200 mm NaCl.
_Protein Analysis and Antibodies_—Neuro-2a cells were washed with ice-cold phosphate-buffered saline, collected using cell scrapers, frozen on dry ice, and stored at –80 °C until lysis. Neuro-2a cell pellets were lysed, immunoprecipitated, and immunoblotted as described previously (19).
For immunoprecipitation experiments, antibodies against HA (12CA5; Roche Applied Science), OGT (AL-25), MYPT1 (Covance), or normal rabbit or mouse IgG were used. For immunoblotting, antibodies against OGT (DM-17; Sigma), HA (HA.11; Covance), PP1β (also known as PP1δ) (Upstate), MYPT1 (Covance), _O_-GlcNAc (CTD110.6), _O_-GlcNAcase (a kind gift of Dr. Stewart Whiteheart, University of Kentucky, Lexington, KY) (24), actin (Sigma), and Thr(P) (Cell Signaling Technology Inc.) were used. For immunoprecipitation experiments in yeast, clones were grown in 100 ml of yeast-extract peptone complete medium; lysed in 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Nonidet P-40 using mechanical disruption by glass beads; and immunoprecipitated for HA using HA.11 (Covance).
_Yeast Two-hybrid Screen_—The human fetal brain MATCHMAKER cDNA library fused to the GAL4 activation domain in pACT2 was obtained pretransformed into the S. cerevisiae Y189 (MATα) strain (BD Biosciences). The yeast two-hybrid screen was performed according to the manufacturer's instructions.
The AH109 strain containing pJL59-OGT was mated with the Y189 strain pretransformed with the pACT2 cDNA library. Mating efficiency was determined to be within the acceptable limits according to the manufacturer's protocol. Next diploid yeast were assayed for β-galactosidase reporter gene expression by filter assay. Yeast colonies positive for β-galactosidase expression were selected for growth on SD/–His/–Leu/–Trp minimal medium (for low stringency selection). Yeast colonies were replica plated onto SD/–Ade/–His/–Leu/–Trp/5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-α-gal) minimal medium to select for medium (white) and high (blue) stringency clones. Positive clones were retested according to the manufacturer's instructions. Plasmid DNA purified from the clones surviving the high stringency selection that were confirmed by retesting were sequenced. A list of these clones is found in Table 1.
TABLE 1.
List of 27 putative OGT-interacting proteins identified by yeast two-hybrid screen from a human fetal brain cDNA library categorized by function
| GenBank™ accession no. | Symbol | Alias/description |
|---|---|---|
| Cytoskeletal | ||
| NM_023019 | DCTN1 | Dynactin 1 |
| NM_021069 | SORBS2 | Sorbin and SH3 domain-containing protein 2 |
| Development | ||
| NM_022467 | CHST8 | Carbohydrate (_N_-acetylgalactosamine 4-O)-sulfotransferase 8 |
| NM_178568 | NGRH2 | Nogo-66 receptor homolog 2 |
| NM_016948 | PARD6A | Partitioning-defective protein 6 |
| Ion transport | ||
| NM_001681 | ATP2A2 | ATPase, Ca2+-transporting, cardiac muscle, slow twitch 2 |
| NM_020659 | TTYH1 | Tweety homolog 1 |
| Metabolism | ||
| NM_001823 | CKB | Creatine kinase, brain |
| NM_002611 | PDK2 | Pyruvate dehydrogenase kinase, isoenzyme 2 |
| Scaffolding | ||
| NM_198889 | ANKRD17 | Ankyrin repeat domain 17 |
| NM_014764 | DAZAP2 | DAZ-associated protein 2 |
| NM_005493 | RANBP9 | RAN-binding protein 9 |
| Signal transduction | ||
| NM_004639 | BAT3 | HLA-B-associated transcript 3 |
| NM_198589 | BSG | Basigin |
| NM_016408 | CDK5RAP1 | CDK5 regulatory subunit-associated protein 1 |
| NM_001893 | CSNK1D | Casein kinase 1, δ |
| NM_198269 | HIPK1 | Homeodomain-interacting protein kinase 1 |
| NM_002480 | MYPT1 | Myosin phosphatase targeting subunit 1 |
| NM_012215 | _O_-GlcNAcase | _O_-GlcNAc-specific glucosaminidase (25) |
| NM_002972 | SBF1/MTMR5 | SET binding factor 1/myotubularin-related protein 5 |
| Trafficking | ||
| NM_004710 | SYNGR2 | Synaptogyrin 2 |
| Transcription | ||
| NM_199141 | CARM1 | Coactivator-associated arginine methyltransferase 1 |
| NM_022728 | NEUROD6 | Neurogenic differentiation 6 |
| NM_024545 | SAP130 | Sin3A-associated protein 130 |
| NM_013260 | SAP30BP | Sin3A-associated protein 30-binding protein |
| NM_014965 | TRAK1 | Trafficking protein and kinesin-binding protein 1 (15) |
| Unknown | ||
| NM_018835 | RC3H2 | Ring finger and CCCH-type zinc finger domains 2 |
_OGT and O-GlcNAcase Assays_—OGT assays were performed as described previously with minor modifications (19). 3 μg of ncOGT was assayed toward 1 μg of GST, GST-MYPT1, or GST-CARM1 in buffer containing 50 mm Tris-HCl, pH 7.5, 1 μg of purified bovine serum albumin, 1 unit of calf intestinal alkaline phosphatase, 0.5 μCi of UDP-[3H]GlcNAc. Reactions were performed at room temperature for 1 h, stopped with Laemmli buffer, and separated by SDS-PAGE. The gels were then fixed, stained for protein using Coomassie Brilliant Blue G-250, treated with En3Hance autofluorography solution (PerkinElmer Life Sciences), dried, and exposed to film.
For OGT assays toward lysate, rat brain lysate was enriched over Sepharose covalently coupled to MYPT1 or CARM1. Proteins were eluted with 8 m urea, concentrated, and buffer-exchanged into 20 mm Tris-HCl, pH 7.5. 20 μg of the enriched lysate was assayed in the above conditions with or without 1 μg GST, GST-MYPT1, or GST-CARM1.
OGT activity measurements from whole cell lysate were performed as described previously with minor modifications (19). Cell lysate was concentrated and buffer-exchanged into 50 mm Tris-HCl, pH 7.5, using 30-kDa molecular mass cutoff centrifugal filter devices (Millipore). 140 μg of concentrated lysate was assayed for [3H]GlcNAc incorporation from UDP-[3H]GlcNAc into a substrate peptide derived from casein kinase II (1 mm) for 30 min at room temperature. The reactions were purified over C18 MacroSpin columns (The Nest Group) and counted by scintillation counting. _O_-GlcNAcase activity was assayed from whole cell lysates as described previously (19).
_Statistical Measurements_—All experiments were performed on at least three separate occasions (n ≥ 3). Error bars represent S.E.
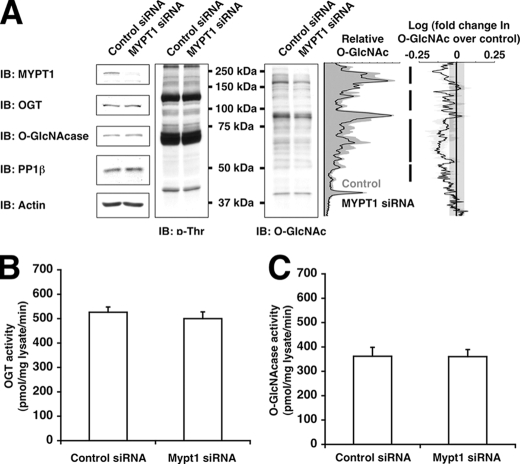
For densitometric lane profiles, the plot profile function in NIH Image v1.63 was used. The log(-fold change in _O_-GlcNAc over control) was calculated as follows. First, lane profiles were normalized against actin, and then the normalized lane profile for the experimental sample was divided by its paired control sample, resulting in the -fold change over control. The logarithm of the -fold change over the lane profile was taken. Values greater than 0 indicate increases over control, whereas values less than 0 indicate decreases compared with control. The plot inFig. 5 represents the average of three independent experiments. The normal experimental deviation across control samples was measured to be ±0.05 log(-fold change) units or ∼12% and indicated on the plot by a gray bar between –0.05 and +0.05.
FIGURE 5.
Knockdown of MYPT1 decreases GlcNAcylation of several proteins in vivo. A, lysates from Neuro-2a cells transfected with control siRNA or MYPT1 siRNA were immunoblotted (IB) for MYPT1, OGT,_O_-GlcNAcase, PP1β, actin, phosphothreonine (p-Thr), and_O_-GlcNAc. Densitometric lane profiles from the representative_O_-GlcNAc immunoblot (normalized to actin) are shown in the left plot. The right plot displays the logarithm of the -fold change in _O_-GlcNAc signal profiles of MYPT1 siRNA samples over control samples. The black line is the average of three independent experiments with S.E. bars in light gray, and the dark gray bar represents the normal deviation between control samples.B, lysates from Neuro-2a cells transfected with control siRNA or MYPT1 siRNA were assayed for OGT activity using 1 mm casein kinase II peptide as substrate. C, lysates from Neuro-2a cells transfected with control siRNA or MYPT1 siRNA were assayed for _O_-GlcNAcase activity using _p_-nitrophenyl-GlcNAc as substrate. Data shown are representative of n ≥ 3 experiments.
RESULTS
_Putative OGT-interacting Proteins Were Identified Using a Yeast Two-hybrid Screen_—To identify candidate OGT regulatory proteins, we performed a conventional yeast two-hybrid screen of a human fetal brain cDNA library using full-length OGT as bait. Using high stringency reporter selection, we were able to identify 27 putative binding partners for OGT (Table 1). In our screen, two previously known interacting partners for OGT were successfully identified, namely _O_-GlcNAcase (25) and Trak1 (previously known as OIP106) (15). It is interesting to note that the proteins identified belong to a wide range of functional classes from ion transporting integral membrane proteins to signaling and transcriptional regulators.
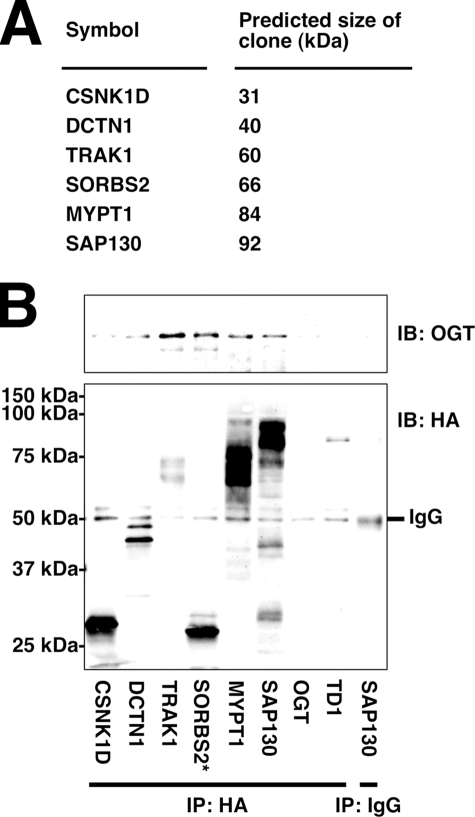
To confirm the results of the yeast two-hybrid screen, we performed co-immunoprecipitations from several of the yeast clones isolated (Fig. 1_A_). As expected, all of the clones tested were confirmed to interact with OGT, whereas the TD1 negative control protein did not interact with OGT (Fig. 1_B_). Using this assay, we were also able to detect SORBS2 as a false positive result of the screen because the size of the protein failed to match the predicted size based on the cDNA sequence of the clone.
FIGURE 1.
Several putative OGT binding partners interact with OGT when co-expressed in yeast. A, predicted size of selected HA-GAL4 activation domain fusion proteins based on cDNA sequence. B, lysates from yeast transformed with expression plasmids encoding OGT and the indicated proteins fused to HA-GAL4 activation domain were immunoprecipitated (IP) for HA and immunoblotted (IB) for OGT and HA. * indicates a false positive result due to incorrect size of fusion protein. Data shown are representative of n ≥ 3 experiments.
_MYPT1 and CARM1 Both Interact with OGT_—We then chose two putative OGT interactors from the yeast two-hybrid screen, MYPT1 and CARM1, for further analysis. MYPT1 is a known targeting regulatory subunit of PP1β (also known as PP1δ) (for a review, see Ref.26), whereas CARM1 is a known transcriptional coactivating arginine methyltransferase (for a review, see Ref. 27).
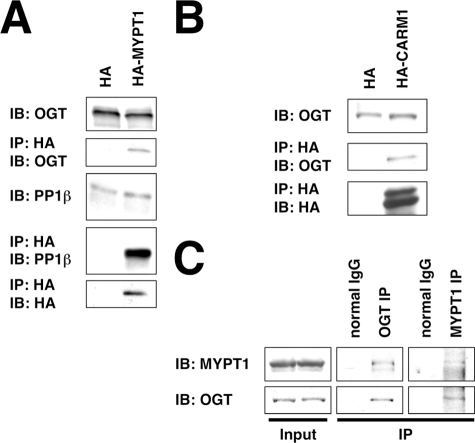
We transfected Neuro-2a cells with plasmids expressing HA, HA-tagged MYPT1, or HA-tagged CARM1 and assayed their ability to interact with endogenously expressed OGT. Indeed both MYPT1 and CARM1 were found to co-immunoprecipitate with OGT (Fig. 2, A and_B_).
FIGURE 2.
MYPT1 and CARM1 interact with OGT in Neuro-2a cells. A, lysates from Neuro-2a cells transfected with HA or HA-MYPT1 were immunoprecipitated (IP) for HA and immunoblotted (IB) for OGT, PP1β, and HA. B, lysates from Neuro-2a cells transfected with HA or HA-CARM1 were immunoprecipitated for HA and immunoblotted for OGT and HA. C, lysates from rat brain were immunoprecipitated for OGT, for MYPT1, or using normal rabbit IgG and immunoblotted for OGT and MYPT1. Data shown are representative of n ≥ 3 experiments.
Next we confirmed that MYPT1 interacts with OGT in vivo. Using OGT-specific and MYPT1-specific antibodies, we were able to co-immunoprecipitate endogenous MYPT1 with OGT from rat brain lysate (Fig. 2_C_).
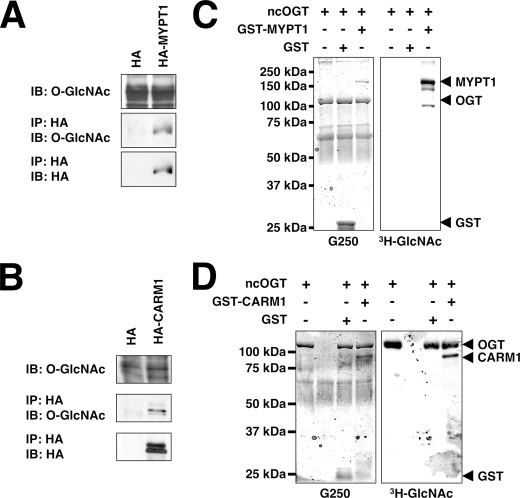
MYPT1 and CARM1 Are GlcNAcylated_—Because both MYPT1 and CARM1 interact with OGT, we decided to determine whether they were substrates of OGT. Toward this end, we transfected Neuro-2a cells with plasmids expressing HA, HA-MYPT1, or HA-CARM1 and assayed their GlcNAcylation using an_O_-GlcNAc-specific antibody. Both MYPT1 and CARM1 are GlcNAcylated_in vivo (Fig. 3, A and_B_).
FIGURE 3.
MYPT1 and CARM1 are substrates of OGT in vivo and in vitro. A, lysates from Neuro-2a cells transfected with HA or HA-MYPT1 were immunoprecipitated (IP) for HA and immunoblotted (IB) for _O_-GlcNAc and HA. B, lysates from Neuro-2a cells transfected with HA or HA-CARM1 were immunoprecipitated for HA and immunoblotted for _O_-GlcNAc and HA. C, recombinant GST and GST-MYPT1 (1 μg) were assayed as substrates for recombinant ncOGT (3 μg) in the presence of UDP-[3H]GlcNAc. The right panel shows an autofluorograph of the gel stained with Coomassie Brilliant Blue G-250 seen in the left panel. D, recombinant GST and GST-CARM1 (1 μg) were assayed as substrates for recombinant ncOGT (3 μg) in the presence of UDP-[3H]GlcNAc. The right panel shows an autofluorograph of the gel stained with Coomassie Brilliant Blue G-250 seen in the left panel. Data shown are representative of n ≥ 3 experiments.
To confirm that MYPT1 and CARM1 are OGT substrates, we performed in vitro OGT assays using purified recombinant GST-tagged proteins. Indeed both GST-tagged MYPT1 and GST-tagged CARM1, but not GST, are substrates for OGT in vitro (Fig. 3, C and D). MYPT1 appears to be a better substrate than CARM1 because there was greater [3H]GlcNAc incorporation into MYPT1 than CARM1 when both were compared with OGT.
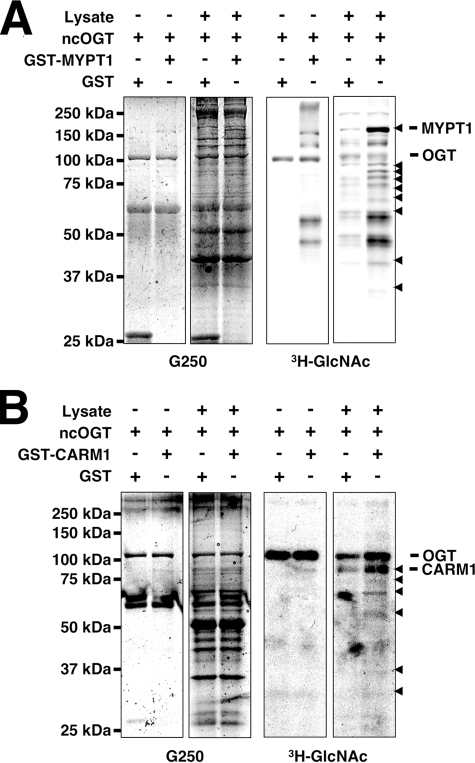
_MYPT1 and CARM1 Affect OGT Substrate Specificity in Vitro_—Recent experiments have suggested that OGT is targeted to its substrates by interacting or bridging proteins (14–19). Thus, we sought to determine whether MYPT1 or CARM1 would be able to affect the substrate selectivity of OGT. We first assayed OGT activity toward whole rat brain lysate in the presence of either GST or GST-MYPT1; however, differences were difficult to detect and quantify because of the very large number of OGT substrates (data not shown). Therefore, we undertook an approach to enrich the rat brain lysate for MYPT1-binding proteins by passing the lysate over a MYPT1 affinity column. The proteins bound to this column were eluted and subsequently tested as substrates for OGT in the presence of GST or GST-MYPT1.
Indeed there are several proteins that seem to be better OGT substrates in the presence of GST-MYPT1 (Fig. 4_A_, arrowheads), suggesting that MYPT1 can alter OGT substrate specificity in vitro. Interestingly GST-MYPT1 itself was a better substrate for OGT in the presence of MYPT1-binding proteins, implying that there are proteins that also increase the affinity of OGT for MYPT1. We also tested to see whether MYPT1 could increase OGT activity toward purified myosin in vitro. MYPT1 had no effect on the GlcNAcylation of myosin in vitro (data not shown).
FIGURE 4.
MYPT1 and CARM1 alter OGT substrate selectivity in vitro. A, rat brain lysate enriched over a MYPT1 column (20 μg of protein) was assayed as substrate for recombinant ncOGT (3 μg) in the presence of UDP-[3H]GlcNAc and GST or GST-MYPT1 (1 μg). The_right panel_ shows an autofluorograph of the gel stained with Coomassie Brilliant Blue G-250 seen in the left panel. B, rat brain lysate enriched over a CARM1 column (20 μg of protein) was assayed as substrate for recombinant ncOGT (3 μg) in the presence of UDP-[3H]GlcNAc and GST or GST-CARM1 (1 μg). The right panel shows an autofluorograph of the gel stained with Coomassie Brilliant Blue G-250 seen in the left panel. Data shown are representative of n ≥ 3 experiments.
Next we tested whether CARM1 would function similarly in an in vitro OGT assay. However, it appears that there are only a few CARM1-binding proteins that are better OGT substrates in the presence of GST-CARM1 (Fig. 4_B_, arrowheads), confirming that CARM1 can alter OGT substrate specificity in vitro. Interestingly it also appears that CARM1 increases GlcNAcylation of OGT itself.
MYPT1 Affects OGT Substrate Specificity in Vivo_—To determine whether MYPT1 alters OGT substrate selectivity in vivo, we decided to assay the effects of RNA interference-mediated knockdown of MYPT1 on protein GlcNAcylation. Although depletion of MYPT1 in Neuro-2a cells had little effect on OGT, O_-GlcNAcase, PP1β, or phosphorylated threonine levels, protein GlcNAcylation was altered significantly (Fig. 5_A_). In fact, it appears that depletion of MYPT1 decreased the in vivo GlcNAcylation of many proteins (Fig. 5_A_, black bars) without affecting the specific catalytic activity of OGT or _O_-GlcNAcase (Fig. 5, B and_C_), suggesting that MYPT1 is responsible for maintaining GlcNAcylation of many proteins under basal conditions. In contrast, knockdown of CARM1 had little to no detectable effect on protein GlcNAcylation under basal conditions (data not shown). However, association of OGT with CARM1 does seem to affect CARM1 activity toward histones.4
DISCUSSION
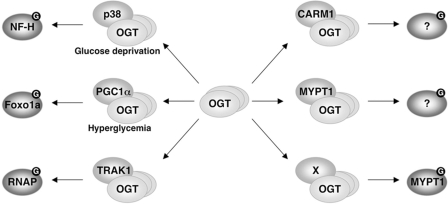
The ability to selectively GlcNAcylate proteins in response to different conditions is essential for the function of OGT as a signaling molecule. One mechanism for achieving context-dependent substrate specificity is by using a number of bridging or adaptor proteins to recruit OGT directly to its targets (Fig. 6). Here we describe the identification of putative OGT-interacting proteins and the characterization of MYPT1 and CARM1 as OGT recruitment factors. Although MYPT1 appears to be a key regulator of OGT under basal conditions, CARM1 is likely to regulate OGT under other conditions.
FIGURE 6.
OGT is recruited by its interacting proteins under different conditions to GlcNAcylate-specific substrates. NF-H, neurofilament H;RNAP, RNA polymerase II.
Current efforts are focused on determining the cellular state(s) under which CARM1 regulates OGT. CARM1 is an arginine methyltransferase whose substrates include many components of the transcription initiation complex (27). When nuclear hormone receptors are activated, coactivator proteins recruit CARM1 to the site of activation to methylate proteins leading to full transcriptional activation (27). It is possible that under these same conditions CARM1 may recruit OGT to GlcNAcylate components of the transcriptional machinery to affect activation. Although OGT has been shown to regulate the activity of many transcription factors, there appears to be a number of different mechanisms used, making it difficult to generalize a single role for GlcNAcylation in transcription. Nonetheless understanding the context of the recruitment of OGT by CARM1 may help to elucidate the role of OGT and protein GlcNAcylation in the regulation of transcription by certain nuclear hormone receptors at individual promoters.
We are also currently working to identify the GlcNAcylated proteins that are recruited to OGT by MYPT1 and CARM1. The identification of these substrates promises to clarify the functional roles of MYPT1 and CARM1 in regulating OGT. Although myosin subunits are GlcNAcylated (28,29), MYPT1 did not affect the activity of OGT toward myosin. Also although MYPT1 and OGT are known to interact with PP1β (30,31), MYPT1 does not seem to be required for the interaction of OGT with PP1β (data not shown). Although the function and sites of phosphorylation on OGT are not known (32), it is possible that MYPT1 may also serve to dephosphorylate OGT under certain conditions.
It has been shown that MYPT1 is essential for cellular survival. MYPT1-null mouse embryos die prior to 7.5 days postcoitum, and cells completely lacking MYPT1 have yet to be isolated (33). In fact, it has recently been shown that knockdown of MYPT1 results in slight defects in mitotic progression in HeLa and SW962 cells consistent with a role in regulating mitosis and cytokinesis by regulating Polo-like kinase 1 (34–36). We have recently shown that disruption of _O_-GlcNAc cycling by overexpression of _O_-GlcNAcase or OGT alters mitotic progression and cytokinesis also at least partially via Polo-like kinase 1 (24). Whether or not the mitotic defect seen during knockdown of MYPT1 is also related to altered protein GlcNAcylation is currently under investigation. Although we have shown here that MYPT1 regulates OGT during basal conditions, it is possible that MYPT1 may also regulate OGT during mitosis, recruiting OGT to a different subset of proteins.
The idea that a single protein may be capable of recruiting OGT to different sets of substrates depending on the cellular conditions is an intriguing one consistent with known mechanisms of regulating protein phosphatases. For example, MYPT1 recruits the dephosphorylation of myosin during cell migration (37), yet recruits the dephosphorylation of Polo-like kinase 1 during cell division (34). Identifying cell division machinery, which is targeted to OGT by MYPT1, may be a promising direction for future research.
Furthermore the disruption of the interaction of OGT with its individual binding partners may aid in studying GlcNAcylation of different proteins and signaling pathways, eventually leading to promising protein GlcNAcylation-targeted inhibitors and therapeutics. Here we present a number of candidate OGT-interacting proteins. It will be very interesting to see whether more of these proteins serve functional roles as OGT recruitment factors, helping to define substrate specificity.
Acknowledgments
We greatly appreciate the gift of the prokaryotic expression plasmid encoding MYPT1 from Dr. Anne A. Wooldridge and Dr. Timothy A. J. Haystead (Duke University Medical Center, Durham, NC), the prokaryotic expression plasmid encoding ncOGT from Dr. Suzanne Walker (Harvard Medical School, Boston, MA), and the plasmid containing CARM1 cDNA from Dr. Michael R. Stallcup (University of Southern California, Los Angeles, CA). We acknowledge Dr. Stewart Whiteheart (University of Kentucky, Lexington, KY) for providing the O-GlcNAcase antibodies and Dr. Sai Iyer for creating the yeast expression plasmid encoding OGT fused to GAL4 binding domain. We also thank laboratory members for critical reading of this manuscript.
*
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HD13563 and R01 CA42486 (to G. W. H.). This work was also supported by a National Science Foundation graduate research fellowship (to W. D. C.). Under a licensing agreement between The Johns Hopkins University, Covance Research Products, Sigma-Aldrich, and Santa Cruz Biotechnology, G. W. H. receives royalties from the sale of the CTD110.6 _O_-GlcNAc antibody. The terms of this agreement are managed by The Johns Hopkins University. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “_advertisement_” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
3
The abbreviations used are: _O_-GlcNAc, _O_-linked β-_N_-acetylglucosamine; _O_-GlcNAcase, _O_-linked β-_N_-acetylglucosaminidase (β-_N_-acetylhexosaminidase, EC 3.2.1.52); OGT, _O_-linked β-_N_-acetylglucosaminyltransferase (protein_N_-acetylglucosaminyltransferase, EC 2.4.1.94); MYPT1, myosin phosphatase targeting subunit 1; CARM1, coactivator-associated arginine methyltransferase 1; siRNA, small interfering RNA; GST, glutathione_S_-transferase; PP1β, protein phosphatase 1β; HA, hemagglutinin.
4
K. Sakabe and G. W. Hart, manuscript in preparation.
References
- 1.Copeland, R. J., Bullen, J. W., and Hart, G. W. (2008) Am. J. Physiol. 295 E17–E28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haltiwanger, R. S., Blomberg, M. A., and Hart, G. W. (1992) J. Biol. Chem. 267 9005–9013 [PubMed] [Google Scholar]
- 3.Dong, D. L., and Hart, G. W. (1994) J. Biol. Chem. 269 19321–19330 [PubMed] [Google Scholar]
- 4.Manning, G., Whyte, D. B., Martinez, R., Hunter, T., and Sudarsanam, S. (2002) Science 298 1912–1934 [DOI] [PubMed] [Google Scholar]
- 5.Nolte, D., and Muller, U. (2002) Mamm. Genome 13 62–64 [DOI] [PubMed] [Google Scholar]
- 6.Shafi, R., Iyer, S. P., Ellies, L. G., O'Donnell, N., Marek, K. W., Chui, D., Hart, G. W., and Marth, J. D. (2000) Proc. Natl. Acad. Sci. U. S. A 97 5735–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreppel, L. K., and Hart, G. W. (1999) J. Biol. Chem. 274 32015–32022 [DOI] [PubMed] [Google Scholar]
- 8.Wang, J., Liu, R., Hawkins, M., Barzilai, N., and Rossetti, L. (1998) Nature 393 684–688 [DOI] [PubMed] [Google Scholar]
- 9.Marshall, S., Nadeau, O., and Yamasaki, K. (2004) J. Biol. Chem. 279 35313–35319 [DOI] [PubMed] [Google Scholar]
- 10.McClain, D. A., and Crook, E. D. (1996) Diabetes 45 1003–1009 [DOI] [PubMed] [Google Scholar]
- 11.Jinek, M., Rehwinkel, J., Lazarus, B. D., Izaurralde, E., Hanover, J. A., and Conti, E. (2004) Nat. Struct. Mol. Biol. 11 1001–1007 [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Fleites, C., Macauley, M. S., He, Y., Shen, D. L., Vocadlo, D. J., and Davies, G. J. (2008) Nat. Struct. Mol. Biol. 15 764–765 [DOI] [PubMed] [Google Scholar]
- 13.Kreppel, L. K., Blomberg, M. A., and Hart, G. W. (1997) J. Biol. Chem. 272 9308–9315 [DOI] [PubMed] [Google Scholar]
- 14.Iyer, S. P., and Hart, G. W. (2003) J. Biol. Chem. 278 24608–24616 [DOI] [PubMed] [Google Scholar]
- 15.Iyer, S. P., Akimoto, Y., and Hart, G. W. (2003) J. Biol. Chem. 278 5399–5409 [DOI] [PubMed] [Google Scholar]
- 16.Yang, X., Zhang, F., and Kudlow, J. E. (2002) Cell 110 69–80 [DOI] [PubMed] [Google Scholar]
- 17.Marz, P., Stetefeld, J., Bendfeldt, K., Nitsch, C., Reinstein, J., Shoeman, R. L., Dimitriades-Schmutz, B., Schwager, M., Leiser, D., Ozcan, S., Otten, U., and Ozbek, S. (2006) J. Biol. Chem. 281 20263–20270 [DOI] [PubMed] [Google Scholar]
- 18.Riu, I. H., Shin, I. S., and Do, S. I. (2008) Biochem. Biophys. Res. Commun. 372 203–209 [DOI] [PubMed] [Google Scholar]
- 19.Cheung, W. D., and Hart, G. W. (2008) J. Biol. Chem. 283 13009–13020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang, X., Ongusaha, P. P., Miles, P. D., Havstad, J. C., Zhang, F., So, W. V., Kudlow, J. E., Michell, R. H., Olefsky, J. M., Field, S. J., and Evans, R. M. (2008) Nature 451 964–969 [DOI] [PubMed] [Google Scholar]
- 21.Wooldridge, A. A., MacDonald, J. A., Erdodi, F., Ma, C., Borman, M. A., Hartshorne, D. J., and Haystead, T. A. (2004) J. Biol. Chem. 279 34496–34504 [DOI] [PubMed] [Google Scholar]
- 22.Lee, Y. H., Koh, S. S., Zhang, X., Cheng, X., and Stallcup, M. R. (2002) Mol. Cell. Biol. 22 3621–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross, B. J., Kraybill, B. C., and Walker, S. (2005) J. Am. Chem. Soc. 127 14588–14589 [DOI] [PubMed] [Google Scholar]
- 24.Slawson, C., Zachara, N. E., Vosseller, K., Cheung, W. D., Lane, M. D., and Hart, G. W. (2005) J. Biol. Chem. 280 32944–32956 [DOI] [PubMed] [Google Scholar]
- 25.Whisenhunt, T. R., Yang, X., Bowe, D. B., Paterson, A. J., Van Tine, B. A., and Kudlow, J. E. (2006) Glycobiology 16 551–563 [DOI] [PubMed] [Google Scholar]
- 26.Matsumura, F., and Hartshorne, D. J. (2008) Biochem. Biophys. Res. Commun. 369 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stallcup, M. R., Kim, J. H., Teyssier, C., Lee, Y. H., Ma, H., and Chen, D. (2003) J. Steroid Biochem. Mol. Biol. 85 139–145 [DOI] [PubMed] [Google Scholar]
- 28.Cieniewski-Bernard, C., Bastide, B., Lefebvre, T., Lemoine, J., Mounier, Y., and Michalski, J. C. (2004) Mol. Cell. Proteomics 3 577–585 [DOI] [PubMed] [Google Scholar]
- 29.Hedou, J., Cieniewski-Bernard, C., Leroy, Y., Michalski, J. C., Mounier, Y., and Bastide, B. (2007) J. Biol. Chem. 282 10360–10369 [DOI] [PubMed] [Google Scholar]
- 30.Terrak, M., Kerff, F., Langsetmo, K., Tao, T., and Dominguez, R. (2004) Nature 429 780–784 [DOI] [PubMed] [Google Scholar]
- 31.Wells, L., Kreppel, L. K., Comer, F. I., Wadzinski, B. E., and Hart, G. W. (2004) J. Biol. Chem. 279 38466–38470 [DOI] [PubMed] [Google Scholar]
- 32.Song, M., Kim, H. S., Park, J. M., Kim, S. H., Kim, I. H., Ryu, S. H., and Suh, P. G. (2008) Cell. Signal. 20 94–104 [DOI] [PubMed] [Google Scholar]
- 33.Okamoto, R., Ito, M., Suzuki, N., Kongo, M., Moriki, N., Saito, H., Tsumura, H., Imanaka-Yoshida, K., Kimura, K., Mizoguchi, A., Hartshorne, D. J., and Nakano, T. (2005) Transgenic Res. 14 337–340 [DOI] [PubMed] [Google Scholar]
- 34.Yamashiro, S., Yamakita, Y., Totsukawa, G., Goto, H., Kaibuchi, K., Ito, M., Hartshorne, D. J., and Matsumura, F. (2008) Dev. Cell 14 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawano, Y., Fukata, Y., Oshiro, N., Amano, M., Nakamura, T., Ito, M., Matsumura, F., Inagaki, M., and Kaibuchi, K. (1999) J. Cell Biol. 147 1023–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Totsukawa, G., Yamakita, Y., Yamashiro, S., Hosoya, H., Hartshorne, D. J., and Matsumura, F. (1999) J. Cell Biol. 144 735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia, D., Stull, J. T., and Kamm, K. E. (2005) Exp. Cell Res. 304 506–517 [DOI] [PubMed] [Google Scholar]