Phylogenomic analyses of lophophorates (brachiopods, phoronids and bryozoans) confirm the Lophotrochozoa concept (original) (raw)
Abstract
Based on embryological and morphological evidence, Lophophorata was long considered to be the sister or paraphyletic stem group of Deuterostomia. By contrast, molecular data have consistently indicated that the three lophophorate lineages, Ectoprocta, Brachiopoda and Phoronida, are more closely related to trochozoans (annelids, molluscs and related groups) than to deuterostomes. For this reason, the lophophorate groups and Trochozoa were united to Lophotrochozoa. However, the relationships of the lophophorate lineages within Lophotrochozoa are still largely unresolved. Maximum-likelihood and Bayesian analyses were performed based on a dataset comprising 11 445 amino acid positions derived from 79 ribosomal proteins of 39 metazoan taxa including new sequences obtained from a brachiopod and a phoronid. These analyses show that the three lophophorate lineages are affiliated with trochozoan rather than deuterostome phyla. All hypotheses claiming that they are more closely related to Deuterostomia than to Protostomia can be rejected by topology testing. Monophyly of lophophorates was not recovered but that of Bryozoa including Ectoprocta and Entoprocta and monophyly of Brachiozoa including Brachiopoda and Phoronida were strongly supported. Alternative hypotheses that are refuted include (i) Brachiozoa as the sister group of Mollusca, (ii) ectoprocts as sister to all other Lophotrochozoa including Platyzoa, and (iii) ectoprocts as sister or to all other protostomes except chaetognaths.
Keywords: Brachiopoda, Bryozoa, Lophophorata, Metazoa, Phoronida, phylogeny
1. Introduction
Phylogenetic analyses of molecular markers have substantially changed our view of animal evolution in the past two decades (Halanych 2004). The new subdivision of Protostomia into two main groups, Lophotrochozoa and Ecdysozoa, originally based on 18S rDNA sequences (Halanych et al. 1995; Aguinaldo et al. 1997), has been corroborated by sequences of single nuclear protein-encoding genes (e.g. Ruiz-Trillo et al. 2002; Anderson et al. 2004), datasets combining multiple nuclear protein-encoding sequences (Peterson et al. 2004; Helmkampf et al. 2008) and phylogenomic approaches (Philippe et al. 2005; Philippe & Telford 2006; Baurain et al. 2007; Hausdorf et al. 2007).
However, the relationships within Lophotrochozoa could not be resolved robustly so far, neither with a large dataset of combined small and large subunit rDNAs (Passamaneck & Halanych 2006), nor with a dataset including several nuclear protein-encoding sequences (Helmkampf et al. 2008). Phylogenomic data were able to resolve some disputed relationships within Lophotrochozoa (Hausdorf et al. 2007), but such data are still missing for some phylogenetically important phyla such as Brachiopoda and Phoronida.
The placement of the lophophorate taxa within Lophotrochozoa as indicated by molecular phylogenetic studies is particularly inconsistent with the morphological evidence (Lüter & Bartolomaeus 1997). As originally defined based on morphology, Lophophorata consists of Ectoprocta, Brachiopoda and Phoronida, taxa that share a ciliated, tentacular feeding apparatus around the mouth opening called lophophore. Based on embryological and morphological characters, Lophophorata was traditionally considered the sister or paraphyletic stem group of Deuterostomia (Hennig 1979; Schram 1991; Ax 1995; Lüter & Bartolomaeus 1997; Sørensen et al. 2000; Brusca & Brusca 2003). However, Nielsen (2001) argued that the lophophore of Bryozoa is not homologous to that of Brachiopoda plus Phoronida, and considered Lophophorata diphyletic. He suggested that ectoprocts are more closely related to entoprocts within Spiralia, whereas he still considered Brachiopoda+Phoronida as the sister group of Deuterostomia sensu stricto (his Neorenalia). By contrast, the molecular phylogenetic studies have shown Ectoprocta as well as Brachiopoda and Phoronida to be more closely related to Trochozoa, i.e. Annelida, Mollusca and related groups, than to Deuterostomia; these include analyses that used rDNA (Halanych et al. 1995; Mackey et al. 1996; Littlewood et al. 1998; Cohen 2000; Giribet et al. 2000; Peterson & Eernisse 2001; Mallatt & Winchell 2002; Passamaneck & Halanych 2006), Hox genes (de Rosa et al. 1999; Passamaneck & Halanych 2004), mitochondrial protein genes (Stechmann & Schlegel 1999; Helfenbein & Boore 2004; Waeschenbach et al. 2006), single nuclear protein genes (e.g. Ruiz-Trillo et al. 2002; Anderson et al. 2004) and sets of multiple nuclear protein genes (Helmkampf et al. 2008). For this reason, Halanych et al. (1995) united the lophophorate groups and Trochozoa into Lophotrochozoa.
Yet the morphological similarities between Brachiozoa (Brachiopoda+Phoronida=Phoronozoa) and Deuterostomia seem so strong that they affect the topology of the trees even in some analyses considering both 18S rDNA sequences and morphological characters. In the total-evidence analysis of Zrzavý et al. (1998), Brachiozoa clustered with Deuterostomia, while in the analysis of Eernisse & Peterson (2004) deuterostomes were the sister group of Lophotrochozoa. There, the brachiozoans were sister to the remaining lophotrochozoan groups. However, this was not the case in some other total-evidence analyses (Giribet et al. 2000; Peterson & Eernisse 2001) in which Brachiopoda and Phoronida were part of Lophotrochozoa (or Trochozoa), and Deuterostomia did not appear as the sister group of Lophotrochozoa. As a caveat to these findings, the above-mentioned studies did not include many genes at all.
To provide a more robust resolution of the relationships of Brachiopoda, Phoronida and Bryozoa, we supplemented a previously compiled dataset of 79 sequences encoding ribosomal proteins with new expressed sequence tag (EST) sequences of a brachiopod and a phoronid.
2. Material and methods
(a) EST generation and processing
Specimens of the brachiopod Novocrania anomala (Müller 1776) and the phoronid Phoronis muelleri Selys-Longchamps 1903 were collected in the Gullmarsfjord near Kristineberg, Sweden. To minimize potential contamination sources, care was taken to remove epibionts growing on the shells and tubes, respectively. Total RNA was isolated from pools of 20 living adult individuals each with the TRIzol Plus purification system (Invitrogen, Karlsruhe, Germany). The mRNA of Novocrania was purified by the Dynabeads mRNA Purification Kit (Invitrogen) before it was transcribed by primer extension. The products were size fractioned and cloned directionally using CloneMiner technology (Invitrogen) to construct a cDNA library. In Phoronis, the PolyATtract mRNA Isolation System III (Promega, Mannheim, Germany) was used, followed by transcription and long-distance PCR amplification, size fractioning and directional cloning employing the Creator SMART cDNA Library Construction Kit (Clontech, Heidelberg, Germany).
From these libraries, ESTs were generated by sequencing 2247 (Novocrania) and 2315 (Phoronis) clones from the 5′ end on the automated capillary sequencer system ABI 3730XL (Applied Biosystems, Darmstadt, Germany) using BigDye chemistry (Applied Biosystems). EST processing was accomplished as described previously (Hausdorf et al. 2007), with the addition of a second clustering step after quality clipping to improve contig assembly. The final number of contigs acquired from each organism amounted to 1699 (Novocrania) and 1467 (Phoronis).
(b) Extraction and alignment of ribosomal protein sequences
Ribosomal protein sequences were retrieved from the new datasets using 79 human ribosomal protein sequences as local BLAST search queries. A total of 42 and 54 (at least partial) ribosomal protein sequences were identified in Novocrania and Phoronis, respectively. These sequences, available in GenBank under the accession nos. EU558289–EU558330 (Novocrania) and EU558331–EU558384 (Phoronis), were individually aligned to orthologous riboprotein sequences of 36 additional taxa compiled previously (Hausdorf et al. 2007) and of a nemertean (Struck & Fisse 2008) using the ClustalW algorithm (Thompson et al. 1994). The resulting single-gene alignments were inspected and adjusted manually, and concatenated into a single multiple sequence alignment. Ambiguously aligned positions were automatically removed by Gblocks (Castresana 2000) applying low stringency parameters. The resulting alignment included 5458 amino acids of Novocrania (47.8% of the total alignment length minus gap positions) and 7922 amino acids of Phoronis (69.3%). More extensive information about the number of genes and amino acids present per taxon is reported in the electronic supplementary material. The final alignment has been deposited at TreeBase (http://www.treebase.org, study accession no. S2050).
(c) Phylogenetic analyses
Maximum-likelihood analyses were conducted with Treefinder (Jobb et al. 2004; Jobb 2007). The rtRev+G+F model of protein evolution was used for the maximum-likelihood analyses because its fit to the present dataset was superior to other models according to the Akaike information criterion with a correction term for small sample size. Confidence values for the edges of the maximum-likelihood tree were computed by bootstrapping (100 replications; Felsenstein 1985).
To test predefined phylogenetic hypotheses, we used constrained trees and the ‘resolve multifurcations’ option of Treefinder to obtain the maximum-likelihood tree for a specified hypothesis. Then we investigated whether the maximum-likelihood trees for these hypotheses are part of the confidence set of trees applying the approximately unbiased test (Shimodaira 2002) and the expected likelihood weights method (Strimmer & Rambaut 2002).
Bayesian analyses were performed using PhyloBayes v. 2.3 (Blanquart & Lartillot 2006) based on the site-heterogeneous CAT model (Lartillot & Philippe 2004). Four independent Markov chains, starting from random points of the parameter space, were run simultaneously for 20 000 cycles each. Chain stationarity was evaluated by monitoring key parameters for long-term trends (e.g. log likelihood, alpha parameter). The first 2000 points were consequently discarded as burn-in. Both runs reached convergence, indicated by the maximal and mean difference of split frequencies amounting to 0.21 and 0.007, respectively. Subsampling every tenth tree from each chain, a 50% majority rule consensus tree was finally computed. We accept Bayesian posterior probabilities larger than 95% and bootstrap values larger than 70% as significant.
3. Results and Discussion
(a) Deuterostome versus lophotrochozoan relationships of lophophorates
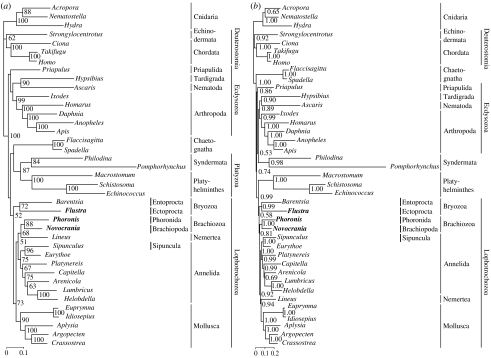
The results of our maximum-likelihood (figure 1a), as well as Bayesian analyses (figure 1b), based on concatenated sequences of 79 ribosomal proteins encompassing 11 445 amino acid positions from 39 taxa, demonstrate that the three lophophorate lineages, Ectoprocta, Brachiopoda and Phoronida, are more closely related to trochozoan phyla than to deuterostomes. They do not form a monophyletic group. Thus, our analyses confirm the results of previous molecular phylogenetic studies (see §1).
Figure 1.
Phylogenetic analyses of lophophorate relationships based on 11 445 amino acid positions derived from 79 concatenated ribosomal proteins. Lophophorate lineages appear in bold. (a) Maximum-likelihood tree. Bootstrap support values larger than 50% are shown to the right of the nodes. (b) Bayesian inference reconstruction. Bayesian posterior probabilities are shown to the right of the nodes.
Characters that were traditionally used to unite Lophophorata with deuterostomes include the following: a body organization with three distinct coelomic cavities, namely protocoel, mesocoel and metacoel (archimery); a mesosomal tentacular apparatus; entomesoderm derived from the archenteron by enterocoely, larvae with upstream-collecting ciliary bands; and heterogeneously assembled metanephridia (Hennig 1979; Schram 1991; Ax 1995; Lüter & Bartolomaeus 1997; Sørensen et al. 2000; Brusca & Brusca 2003). However, the hypothesis that all lophophorate lineages are more closely allied to Deuterostomia than Protostomia can be rejected by topology tests based on our ribosomal protein data (table 1, hypothesis 1). Nielsen (2001) argued that ectoprocts show no trace of archimery and that only Brachiopoda and Phoronida form a monophyletic group with Deuterostomia sensu stricto (his Neorenalia). Lüter (2000) suggested that the origin of the coelomic anlage from differentiated archenteral epithelium, which he defined as enterocoely, is a synapomorphy of Brachiopoda and Deuterostomia; he therefore considered these two taxa as sister groups. Consequently, we tested the hypotheses that Brachiozoa or Brachiopoda alone are the sister groups of Deuterostomia. Both possibilities were rejected (table 1, hypotheses 2–3).
Table 1.
Topology test results.
| no.a | phylogenetic hypothesis | references claiming the hypothesis | likelihood | Δlikelihoodb | AUc | ELWd |
|---|---|---|---|---|---|---|
| ML tree (figure 1a) | −273 512 | 0 | 0.6421 | 0.5010 | ||
| 1 | Lophophorata+Deuterostomia | Hennig (1979), Schram (1991), Ax (1995), Lüter & Bartolomaeus (1997), Sørensen et al. (2000) and Brusca & Brusca (2003) | −273 957 | 445 | 0.0000* | 0.0000* |
| 2 | Brachiozoa+Deuterostomia | Nielsen (2001) | −273 977 | 465 | 0.0000* | 0.0000* |
| 3 | Brachiopoda+Deuterostomia | Lüter (2000) | −273 805 | 293 | 0.0000* | 0.0000* |
| 4 | Ectoprocta sister to all other Lophotrochozoa inclusive Platyzoa | Halanych et al. (1995), Halanych (2004) and Passamaneck & Halanych (2006) | −273 599 | 87 | 0.0000* | 0.0052* |
| 5 | Ectoprocta sister to all other protostomes except chaetognaths | Giribet et al. (2000) | −273 624 | 112 | 0.0000* | 0.0002* |
| 6 | Trochozoa (Entoprocta+Eutrochozoa) | Zrzavý et al. (1998), Giribet et al. (2000) and Peterson & Eernisse (2001) | −273 592 | 80 | 0.0634 | 0.0107* |
| 7 | Eutrochozoa (Neotrochozoa+Nemertea) | Zrzavý et al. (1998), Giribet et al. (2000) and Peterson & Eernisse (2001) | −273 535 | 23 | 0.4114 | 0.1817 |
| 8 | Neotrochozoa (Annelida+Mollusca) | Zrzavý et al. (1998), Giribet et al. (2000) and Peterson & Eernisse (2001) | −273 520 | 8 | 0.5929 | 0.2777 |
| 9 | Conchozoa (Brachiozoa+Mollusca) | Cavalier-Smith (1998) and Halanych (2004) | −273 571 | 59 | 0.0000* | 0.0015* |
| 10 | Lophophorata monophyly | Emig (1984) | −273 571 | 59 | 0.1111 | 0.0219* |
The conflicting results concerning the phylogenetic relationships of the lophophorates is a major incongruity between morphological and molecular phylogenetic approaches. However, in the last decade, the morphological evidence for a close relationship between the lophophorate groups and the deuterostomes has become weaker by careful re-examinations of the characters. It has been shown that neither brachiopods nor phoronids possess three coelomic cavities, because a protocoel is lacking in all lophophorate groups (Lüter 2000; Bartolomaeus 2001). Thus, the archicoelomate concept (Siewing 1980) uniting Lophophorata and Deuterostomia, founded on the similarities of three distinct coelomic cavities, lost its basis. Additionally, the finding that Pterobranchia may nest within the enteropneusts (Cameron et al. 2000; Peterson & Eernisse 2001; Winchell et al. 2002) suggests that the ancestral deuterostome more closely resembled a mobile worm-like enteropneust than a sessile colonial pterobranch. This means that the similar tentacular feeding apparatuses of lophophorates and pterobranchs are not a synapomorphy of lophophorates and deuterostomes as supposed previously (Hennig 1979; Schram 1991; Ax 1995; Lüter & Bartolomaeus 1997), but evolved independently as convergent adaptations to the sessile lifestyle (Halanych 1996). Moreover, Lüter (2000) argued that the mesoderm does not originate by enterocoely in Ectoprocta and Phoronida, but that this is the case in Brachiopoda and Deuterostomia only. What is more, whether the mesoderm of brachiopods originates by enterocoely is also in dispute. Jenner (2004) tentatively concluded that reports of true enterocoely, i.e. mesoderm origin by epithelial folding, in brachiopods appear unsupported and that no fundamental difference in the source of mesoderm and mode of coelomogenesis exists between brachiopods and various protostomes. To conclude, there are fewer morphological characters arguing against protostome affiliations of brachiopods and phoronids than traditionally assumed.
(b) Relationships of lophophorates within Lophotrochozoa
The phylogenetic analyses of our ribosomal protein dataset (figure 1) strongly indicate that Brachiopoda and Phoronida constitute a monophyletic group, Brachiozoa (=Phoronozoa; bootstrap support 88%, Bayesian posterior probability 1.00). This corroborates previous results based on rDNA (Mackey et al. 1996; Cohen et al. 1998; Littlewood et al. 1998; Cohen 2000; Mallatt & Winchell 2002; Halanych 2004; Cohen & Weydmann 2005; but see Passamaneck & Halanych 2006), sodium–potassium ATPase α-subunit (Anderson et al. 2004), morphology (Nielsen 2001) and a combination of morphological and 18S rDNA datasets (Zrzavý et al. 1998; Giribet et al. 2000; Peterson & Eernisse 2001).
Our previous study (Hausdorf et al. 2007) recovered Ectoprocta as the sister group of Entoprocta, but this finding was considered tentative until a phoronid and a brachiopod could be added to the analysis, which was done here (figure 1). Indeed Ectoprocta and Entoprocta remain strongly united (bootstrap support 72%, Bayesian posterior probability 0.99). This agrees with the hypothesis that Bryozoa sensu lato is monophyletic (Nielsen 1971, 1985, 2001; Cavalier-Smith 1998). Alternative hypotheses concerning the phylogenetic position of ectoprocts, namely that they are sister to all other Lophotrochozoa including Platyzoa, i.e. Platyhelminthes, Syndermata and related groups (Halanych et al. 1995; Littlewood et al. 1998; Halanych 2004; Passamaneck & Halanych 2006), or that they are sister to all other protostomes except chaetognaths (Giribet et al. 2000) could be rejected (table 1, hypotheses 4–5).
Peterson & Eernisse (2001) defined several nested clades within Lophotrochozoa, namely (i) Neotrochozoa, which unites Mollusca and Annelida (with the annelids including Echiura and Sipuncula; see Hausdorf et al. (2007) and Struck et al. (2007)), (ii) Eutrochozoa, which includes Neotrochozoa and Nemertea, and (iii) Trochozoa, which comprises Eutrochozoa and Entoprocta. This last hypothesis, which we could not rule out with the previous dataset (Hausdorf et al. 2007), is now rejected by the expected likelihood weights method relying on the enlarged dataset (table 1, hypothesis 6). Although the more conservative approximately unbiased test is still marginally insignificant, this strengthens the evidence for the monophyly of Bryozoa sensu lato. On the other hand, neither the Neotrochozoa hypothesis nor the Eutrochozoa hypothesis is rejected by either test method (table 1, hypotheses 7–8).
Brachiopods plus phoronids appear as the sister group of nemerteans in the maximum-likelihood tree (figure 1a). By contrast, the Bayesian inference analysis shows a sister-group relationship of Brachiozoa and Eutrochozoa (figure 1b). The relationships of Brachiozoa within Lophotrochozoa thus remain uncertain. However, we can dismiss the Conchozoa hypothesis (Cavalier-Smith 1998; Mallatt & Winchell 2002), according to which Brachiozoa is the sister group of Mollusca (table 1, hypothesis 9).
As mentioned earlier, the three traditional lophophorate lineages, Ectoprocta, Phoronida and Brachiopoda, did not join into a monophyletic clade in our trees (figure 1). The monophyly of Lophophorata was rejected with the expected likelihood weights method, but not with the approximately unbiased test (table 1, hypothesis 10). If we constrain the monophyly of Lophophorata, it becomes the sister group of Eutrochozoa in the resulting maximum-likelihood tree (not shown). In this tree, Entoprocta is the sister group of Lophophorata plus Eutrochozoa. Even if this topology should prove correct, the radial cleavage of Lophophorata would be a secondary modification derived from spiral cleavage, given that the spiral cleavage of Entoprocta is homologous to that of Annelida and Mollusca.
When we constrain the monophyly of Eutrochozoa (table 1, hypothesis 7), then Brachiozoa and Bryozoa (including Ectoprocta and Entoprocta) form a monophyletic group in the resulting maximum-likelihood tree. The same maximum-likelihood tree results if we constrain the monophyly of Brachiozoa and Bryozoa. Thus, the test results (table 1, hypothesis 7) apply to this hypothesis as well. This extended version of ‘Lophophorata’ including Entoprocta is therefore part of the confidence set of trees, given our ribosomal protein dataset, a possibility that is especially interesting, because it is in better agreement with morphological data than topologies that suggest independent origins of Ectoprocta and Brachiozoa within Lophotrochozoa. Potential synapomorphies of Brachiozoa and Bryozoa are the transition to a sessile lifestyle accompanied by the evolution of a horseshoe-shaped, tentacular feeding apparatus and a hydrostatic skeleton consisting of a lophophore coelom and a trunk coelom. In this view, both coelomic cavities were connected in the common ancestor of the two bryozoan subgroups and then were lost in Entoprocta. Most potential synapomorphies of Brachiozoa and Bryozoa are characters that were once thought to support a sister-group relationship between Lophophorata and Deuterostomia, but in light of the present evidence that these two groups are unrelated, must have originated by convergence (see above). Hypotheses that suppose that Ectoprocta and Brachiozoa originated independently of each other from different lophotrochozoan ancestors would require additional convergences of these characters.
Despite the progress presented herein, the resolution achieved in our analyses is still insufficient to fully reconstruct the evolutionary history of Lophotrochozoa. This lack of resolution could neither be avoided by the inclusion of many riboprotein genes and all major lophotrochozoan taxa, nor by the use of the CAT model, which has been shown often to overcome long-branch attraction artefacts when other models fail (Baurain et al. 2007; Lartillot et al. 2007). Actually, the grouping of taxa with the longest branches in the maximum-likelihood tree (figure 1a), namely Syndermata and Platyhelminthes, is dissolved in the Bayesian inference reconstruction calculated with the CAT model (figure 1b). Further systematic errors unaccounted for by the present tree reconstruction methods, aggravated by the presumably rapid radiation of the lophotrochozoan taxa in the Late Precambrian and the limited taxon sampling within many phyla, might be responsible for the lack of resolution within Lophotrochozoa, which has been observed both here and in other studies (Halanych et al. 1995; Giribet et al. 2000; Peterson & Eernisse 2001; Mallatt & Winchell 2002; Ruiz-Trillo et al. 2002; Anderson et al. 2004; Passamaneck & Halanych 2006; Helmkampf et al. 2008). Improved models of molecular evolution and further taxonomic sampling within lophophorates and other lophotrochozoans will hopefully solve these issues in the future.
Added in preparation. While our manuscript was submitted, Dunn et al. (2008) published an important phylogenomic analysis of a huge number of new metazoan EST data. Regarding the relationships of brachiopods and phoronids, our maximum-likelihood tree (figure 1a) corresponds closely with the results presented by Dunn et al. (2008). In both analyses, brachiopods and phoronids form a clade with nemerteans (clade A in Dunn et al. 2008) that is the sister group of annelids (including sipunculans). These groups together (clade B in Dunn et al. 2008) are sister to the molluscs (together called clade C in Dunn et al. 2008). However, the results of our analyses differ from those of Dunn et al. (2008) with regard to the relationships of ectoprocts and entoprocts. Whereas these two groups form a well-supported clade in our analyses, their position is unstable in the analyses of Dunn et al. (2008). In the 77-taxon analysis of Dunn et al. (2008; figure 1), ectoprocts are sister to Platyzoa and entoprocts are sister to clade C.
Acknowledgments
We thank M. Obst (Kristineberg Marine Research Station) for providing logistical support in collecting specimens, P. Grobe (Freie Universität Berlin) for his help in identifying specimens and T. Struck (University of Osnabrück) for contributing ribosomal protein sequences prior to public release. We are also grateful to M. Kube and R. Reinhardt (Max Planck Institute for Molecular Genetics, Berlin) for the construction and sequencing of cDNA libraries, and to I. Ebersberger, S. Strauss and A. von Haeseler (Max F. Perutz Laboratories, Center for Integrative Bioinformatics, Vienna) for processing of our ESTs. An anonymous referee and the editor provided a wealth of helpful comments on the manuscript. This study was funded by the priority program ‘Deep Metazoan Phylogeny’ of the Deutsche Forschungsgemeinschaft (HA 2763/5-1).
Supplementary Material
Table S1. Taxa versus gene matrix
Taxa versus gene matrix. Data coverage is indicated as follows: 75% or more (++), more than 50 but less than 75% (+), more than 25 but less than 50% (≈), or less than 25% (–) of the amino acid positions of the gene represented in the Gblocks-edited alignment is present. The number of taxa in which more than 50% of the gene is present (# taxa), the length of the gene represented in the alignment (length), the number of genes which are covered by more than 50% per taxon (# genes) and the number (# Aa) and percentage of amino acids present in each taxon (% Aa) are also given. The last value refers to the total length of the alignment minus the number of gap positions in this taxon
References
- Aguinaldo A.M.A, Turbeville J.M, Linford L.S, Rivera M.C, Garey J.R, Raff R.A, Lake J.A. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–493. doi: 10.1038/387489a0. doi:10.1038/387489a0 [DOI] [PubMed] [Google Scholar]
- Anderson F.E, Cordoba A.J, Thollesson M. Bilaterian phylogeny based on analyses of a region of the sodium–potassium ATPase α-subunit gene. J. Mol. Evol. 2004;58:252–268. doi: 10.1007/s00239-003-2548-9. doi:10.1007/s00239-003-2548-9 [DOI] [PubMed] [Google Scholar]
- Ax P. Gustav Fischer Verlag; Stuttgart, Germany: 1995. Das System der Metazoa I. [Google Scholar]
- Bartolomaeus T. Ultrastructure and formation of the body cavity lining in Phoronis muelleri (Phoronida, Lophophorata) Zoomorphology. 2001;120:135–148. doi:10.1007/s004350000030 [Google Scholar]
- Baurain D, Brinkmann H, Philippe H. Lack of resolution in the animal phylogeny: closely spaced cladogeneses or undetected systematic errors? Mol. Biol. Evol. 2007;24:6–9. doi: 10.1093/molbev/msl137. doi:10.1093/molbev/msl137 [DOI] [PubMed] [Google Scholar]
- Blanquart S, Lartillot N. A Bayesian compound stochastic process for modeling nonstationary and nonhomogeneous sequence evolution. Mol. Biol. Evol. 2006;23:2058–2071. doi: 10.1093/molbev/msl091. doi:10.1093/molbev/msl091 [DOI] [PubMed] [Google Scholar]
- Brusca R.C, Brusca G.J. 2nd edn. Sinauer Associates; Sunderland, MA: 2003. Invertebrates. [Google Scholar]
- Cameron C.B, Garey J.R, Swalla B.J. Evolution of the chordate body plan: new insights from phylogenetic analyses of deuterostome phyla. Proc. Natl Acad. Sci. USA. 2000;97:4469–4474. doi: 10.1073/pnas.97.9.4469. doi:10.1073/pnas.97.9.4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. A revised six-kingdom system of life. Biol. Rev. 1998;73:203–266. doi: 10.1017/s0006323198005167. doi:10.1017/S0006323198005167 [DOI] [PubMed] [Google Scholar]
- Cohen B.L. Monophyly of brachiopods and phoronids: reconciliation of molecular evidence with Linnaean classification (the subphylum Phoroniformea nov.) Proc. R. Soc. B. 2000;267:225–231. doi: 10.1098/rspb.2000.0991. doi:10.1098/rspb.2000.0991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B.L, Weydmann A. Molecular evidence that phoronids are a subtaxon of brachiopods (Brachiopoda: Phoronata) and that genetic divergence of metazoan phyla began long before the Early Cambrian. Organ. Diver. Evol. 2005;5:253–273. doi:10.1016/j.ode.2004.12.002 [Google Scholar]
- Cohen B.L, Gawthrop A, Cavalier-Smith T. Molecular phylogeny of brachiopods and phoronids based on nuclear-encoded small subunit ribosomal RNA gene sequences. Phil. Trans. R. Soc. B. 1998;353:2039–2061. doi:10.1098/rstb.1998.0351 [Google Scholar]
- de Rosa R, Grenier J.K, Andreeva T, Cook C.E, Adoutte A, Akam M, Carroll S.B, Balavoine G. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399:772–776. doi: 10.1038/21631. doi:10.1038/21631 [DOI] [PubMed] [Google Scholar]
- Dunn C.W, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. doi:10.1038/nature06614 [DOI] [PubMed] [Google Scholar]
- Eernisse D.J, Peterson K.J. The history of animals. In: Cracraft J, Donoghue M.J, editors. Assembling the tree of life. Oxford University Press; New York, NY: 2004. pp. 197–208. [Google Scholar]
- Emig C. On the origin of the Lophophorata. Z. Zool. Syst. Evolutionsforsch. 1984;22:91–94. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. doi:10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Giribet G, Distel D.L, Polz M, Sterrer W, Wheeler W.C. Triploblastic relationships with emphasis on the acoelomates and the position of Gnathostomulida, Cycliophora, Plathelminthes, and Chaetognatha: a combined approach of 18S rDNA sequences and morphology. Syst. Biol. 2000;49:539–562. doi: 10.1080/10635159950127385. doi:10.1080/10635159950127385 [DOI] [PubMed] [Google Scholar]
- Halanych K.M. Convergence in the feeding apparatuses of lophophorates and pterobranch hemichordates revealed by 18S rDNA: an interpretation. Biol. Bull. 1996;190:1–5. doi: 10.2307/1542669. doi:10.2307/1542669 [DOI] [PubMed] [Google Scholar]
- Halanych K.M. The new view of animal phylogeny. Annu. Rev. Ecol. Evol. Syst. 2004;35:229–256. doi:10.1146/annurev.ecolsys.35.112202.130124 [Google Scholar]
- Halanych K.M, Bacheller J.D, Aguinaldo A.A, Liva S.M, Hillis D.M, Lake J.A. Evidence from 18S ribosomal DNA that the Lophophorates are protostome animals. Science. 1995;267:1641–1642. doi: 10.1126/science.7886451. doi:10.1126/science.7886451 [DOI] [PubMed] [Google Scholar]
- Hausdorf B, Helmkampf M, Meyer A, Witek A, Herlyn H, Bruchhaus I, Hankeln T, Struck T.H, Lieb B. Spiralian phylogenomics supports the resurrection of Bryozoa comprising Ectoprocta and Entoprocta. Mol. Biol. Evol. 2007;24:2723–2729. doi: 10.1093/molbev/msm214. doi:10.1093/molbev/msm214 [DOI] [PubMed] [Google Scholar]
- Helfenbein K.G, Boore J.L. The mitochondrial genome of Phoronis architecta—comparisons demonstrate that phoronids are lophotrochozoan protostomes. Mol. Biol. Evol. 2004;21:153–157. doi: 10.1093/molbev/msh011. doi:10.1093/molbev/msh011 [DOI] [PubMed] [Google Scholar]
- Helmkampf M, Bruchhaus I, Hausdorf B. Multigene analysis of lophophorate and chaetognath phylogenetic relationships. Mol. Phylogenet. Evol. 2008;46:206–214. doi: 10.1016/j.ympev.2007.09.004. doi:10.1016/j.ympev.2007.09.004 [DOI] [PubMed] [Google Scholar]
- Hennig W. 4th edn. Fischer; Jena, Germany: 1979. Wirbellose I (ausgenommen Gliedertiere). Taschenbuch der Speziellen Zoologie. [Google Scholar]
- Jenner R.A. Towards a phylogeny of the Metazoa: evaluating alternative phylogenetic positions of Platyhelminthes, Nemertea, and Gnathostomulida, with a critical reappraisal of cladistic characters. Contrib. Zool. 2004;73:3–163. [Google Scholar]
- Jobb, G. 2007 Treefinder, version of June 2007, Munich, Germany. Distributed by the author. See www.treefinder.de
- Jobb G, von Haeseler A, Strimmer K. Treefinder: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol. Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. doi:10.1186/1471-2148-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. doi:10.1093/molbev/msh112 [DOI] [PubMed] [Google Scholar]
- Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol. Biol. 2007;7:S4. doi: 10.1186/1471-2148-7-S1-S4. doi:10.1186/1471-2148-7-S1-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood D.T.J, Telford M.J, Clough K.A, Rohde K. Gnathostomulida—an enigmatic metazoan phylum from both morphological and molecular perspective. Mol. Phylogenet. Evol. 1998;9:72–79. doi: 10.1006/mpev.1997.0448. doi:10.1006/mpev.1997.0448 [DOI] [PubMed] [Google Scholar]
- Lüter C. The origin of the coelom in Brachiopoda and its phylogenetic significance. Zoomorphology. 2000;120:15–28. doi:10.1007/s004359900019 [Google Scholar]
- Lüter C, Bartolomaeus T. The phylogenetic position of Brachiopoda—a comparison of morphological and molecular data. Zool. Scr. 1997;26:245–254. doi:10.1111/j.1463-6409.1997.tb00414.x [Google Scholar]
- Mackey L.Y, Winnepenninckx B, De Wachter R, Backeljau T, Emschermann P, Garey J.R. 18S rRNA suggests that Entoprocta are protostomes, unrelated to Ectoprocta. J. Mol. Evol. 1996;42:552–559. doi: 10.1007/BF02352285. doi:10.1007/BF02352285 [DOI] [PubMed] [Google Scholar]
- Mallatt J, Winchell C.J. Testing the new animal phylogeny: first use of combined large-subunit and small-subunit rRNA gene sequences to classify the protostomes. Mol. Biol. Evol. 2002;19:289–301. doi: 10.1093/oxfordjournals.molbev.a004082. [DOI] [PubMed] [Google Scholar]
- Nielsen C. Entoproct life-cycles and the entoproct/ectoproct relationship. Ophelia. 1971;9:209–341. [Google Scholar]
- Nielsen C. Animal phylogeny in the light of the trochaea theory. Biol. J. Linn. Soc. 1985;25:243–299. doi:10.1111/j.1095-8312.1985.tb00396.x [Google Scholar]
- Nielsen C. 2nd edn. Oxford University Press; Oxford, UK: 2001. Animal evolution. Interrelationships of the living phyla. [Google Scholar]
- Passamaneck Y.J, Halanych K.M. Evidence from Hox genes that bryozoans are lophotrochozoans. Evol. Dev. 2004;6:275–281. doi: 10.1111/j.1525-142X.2004.04032.x. doi:10.1111/j.1525-142X.2004.04032.x [DOI] [PubMed] [Google Scholar]
- Passamaneck Y.J, Halanych K.M. Lophotrochozoan phylogeny assessed with LSU and SSU data: evidence of lophophorate polyphyly. Mol. Phylogenet. Evol. 2006;40:20–28. doi: 10.1016/j.ympev.2006.02.001. doi:10.1016/j.ympev.2006.02.001 [DOI] [PubMed] [Google Scholar]
- Peterson K.J, Eernisse D.J. Animal phylogeny and the ancestry of bilaterians: inferences from morphology and 18S rDNA gene sequences. Evol. Dev. 2001;3:170–205. doi: 10.1046/j.1525-142x.2001.003003170.x. doi:10.1046/j.1525-142x.2001.003003170.x [DOI] [PubMed] [Google Scholar]
- Peterson K.J, Lyons J.B, Nowak K.S, Takacs C.M, Wargo M.J, McPeek M.A. Estimating metazoan divergence times with a molecular clock. Proc. Natl Acad. Sci. USA. 2004;101:6536–6541. doi: 10.1073/pnas.0401670101. doi:10.1073/pnas.0401670101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Telford M.J. Large-scale sequencing and the new animal phylogeny. Trends Ecol. Evol. 2006;21:614–620. doi: 10.1016/j.tree.2006.08.004. doi:10.1016/j.tree.2006.08.004 [DOI] [PubMed] [Google Scholar]
- Philippe H, Lartillot N, Brinkmann H. Multigene analyses of bilaterian animals corroborate the monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia. Mol. Biol. Evol. 2005;22:1246–1253. doi: 10.1093/molbev/msi111. doi:10.1093/molbev/msi111 [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Paps J, Loukota M, Ribera C, Jondelius U, Baguñà J, Riutort M. A phylogenetic analysis of myosin heavy chain type II sequences corroborates that Acoela and Nemertodermatida are basal bilaterians. Proc. Natl Acad. Sci. USA. 2002;99:11 246–11 251. doi: 10.1073/pnas.172390199. doi:10.1073/pnas.172390199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schram F.R. Cladistic analysis of metazoan phyla and the placement of fossil problematica. In: Simonetta A.M, Conway-Morris S, editors. The early evolution of Metazoa and the significance of problematic taxa. Cambridge University Press; Cambridge, UK: 1991. pp. 35–46. [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. doi:10.1080/10635150290069913 [DOI] [PubMed] [Google Scholar]
- Siewing R. Das Archicoelomatenkonzept. Zool. Jahrb. Anat. 1980;103:439–482. [Google Scholar]
- Sørensen M.V, Funch P, Willerslev E, Hansen A.J, Olesen J. On the phylogeny of the Metazoa in the light of Cycliophora and Micrognathozoa. Zool. Anz. 2000;239:297–318. [Google Scholar]
- Stechmann A, Schlegel M. Analysis of the complete mitochondrial DNA sequence of the brachiopod Terebratulina retusa places Brachiopoda within the protostomes. Proc. R. Soc. B. 1999;266:2043–2052. doi: 10.1098/rspb.1999.0885. doi:10.1098/rspb.1999.0885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K, Rambaut A. Inferring confidence sets of possibly misspecified gene trees. Proc. R. Soc. B. 2002;269:137–142. doi: 10.1098/rspb.2001.1862. doi:10.1098/rspb.2001.1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck T.H, Fisse F. Phylogenetic position of Nemertea derived from phylogenomic data. Mol. Biol. Evol. 2008;25:728–736. doi: 10.1093/molbev/msn019. doi:10.1093/molbev/msn019 [DOI] [PubMed] [Google Scholar]
- Struck T.H, Schult N, Kusen T, Hickman E, Bleidorn C, McHugh D, Halanych K.M. Annelid phylogeny and the status of Sipuncula and Echiura. BMC Evol. Biol. 2007;7:57. doi: 10.1186/1471-2148-7-57. doi:10.1186/1471-2148-7-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D, Higgins D.G, Gibson T.J. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. doi:10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waeschenbach A, Telford M.J, Porter J.S, Littlewood D.T.J. The complete mitochondrial genome of Flustrellidra hispida and the phylogenetic position of Bryozoa among the Metazoa. Mol. Phylogenet. Evol. 2006;40:195–207. doi: 10.1016/j.ympev.2006.03.007. doi:10.1016/j.ympev.2006.03.007 [DOI] [PubMed] [Google Scholar]
- Winchell C.J, Sullivan J, Cameron C.B, Swalla B.J, Mallatt J. Evaluating hypotheses of deuterostomes phylogeny and chordate evolution with new LSU and SSU ribosomal DNA data. Mol. Biol. Evol. 2002;19:762–776. doi: 10.1093/oxfordjournals.molbev.a004134. [DOI] [PubMed] [Google Scholar]
- Zrzavý J, Mihulka S, Kepka P, Bezděk A, Tietz D. Phylogeny of the Metazoa based on morphological and 18S ribosomal DNA evidence. Cladistics. 1998;14:249–285. doi: 10.1111/j.1096-0031.1998.tb00338.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Taxa versus gene matrix
Taxa versus gene matrix. Data coverage is indicated as follows: 75% or more (++), more than 50 but less than 75% (+), more than 25 but less than 50% (≈), or less than 25% (–) of the amino acid positions of the gene represented in the Gblocks-edited alignment is present. The number of taxa in which more than 50% of the gene is present (# taxa), the length of the gene represented in the alignment (length), the number of genes which are covered by more than 50% per taxon (# genes) and the number (# Aa) and percentage of amino acids present in each taxon (% Aa) are also given. The last value refers to the total length of the alignment minus the number of gap positions in this taxon