Acetylation of Histone H3 Lysine 56 Regulates Replication-Coupled Nucleosome Assembly (original) (raw)
. Author manuscript; available in PMC: 2008 Dec 8.
SUMMARY
Chromatin assembly factor 1 (CAF-1) and Rtt106 participate in the deposition of newly synthesized histones onto replicating DNA to form nucleosomes. This process is critical for the maintenance of genome stability and inheritance of functionally specialized chromatin structures in proliferating cells. However, the molecular functions of the acetylation of newly synthesized histones in this DNA replication-coupled nucleosome assembly pathway remain enigmatic. Here we show that histone H3 acetylated at lysine 56 (H3K56Ac) is incorporated onto replicating DNA and, by increasing the binding affinity of CAF-1 and Rtt106 for histone H3, H3K56Ac enhances the ability of these histone chaperones to assemble DNA into nucleosomes. Genetic analysis indicates that H3K56Ac acts in a nonredundant manner with the acetylation of the N-terminal residues of H3 and H4 in nucleosome assembly. These results reveal a mechanism by which H3K56Ac regulates replication-coupled nucleosome assembly mediated by CAF-1 and Rtt106.
INTRODUCTION
In eukaryotic cells, genomic DNA is packaged into chromatin, a highly organized complex of DNA and proteins that encodes epigenetic information governing gene expression, cell identity, and genome integrity (Luger, 2006). The basic unit of chromatin is the nucleosome core particle that consists of 147 base pairs of DNA wrapped around a histone octamer containing two molecules of each of four core histones, H2A, H2B, H3, and H4. During DNA replication, eukaryotic cells must temporarily disrupt nucleosomes to facilitate progression of the DNA replication machinery through chromatin (Falbo and Shen, 2006). Concomitant with the passage of the replication fork, newly synthesized histones as well as parental histones are deposited onto the nascent sister chromatids to ensure propagation of epigenetic information to daughter cells as well as maintenance of genome integrity (Groth et al., 2007b). While significant advances have been made to understand the duplication of genetic information, much less is known regarding inheritance of epigenetic information during S phase of the cell cycle (Goldberg et al., 2007).
The DNA replication-coupled nucleosome assembly pathway is important for inheritance of epigenetic information during DNA replication and repair (Groth et al., 2007b; Henikoff et al., 2004). Chromatin assembly factor 1 (CAF-1), a conserved three subunit protein complex, deposits newly synthesized H3 and H4 onto replicating DNA to form nucleosomes (Groth et al., 2007b; Stillman, 1986). CAF-1-mediated histone deposition onto DNA is assisted by Asf1 and Rtt106, two other H3-H4 histone chaperones that bind to CAF-1 (Huang et al., 2005; Tyler et al., 2001). However, it is not clear how the actions of these three histone chaperones are coordinated to achieve rapid deposition of histones onto nascent DNA during S phase of the cell cycle.
Newly synthesized histones H3 and H4 are acetylated before they are assembled into nucleosomes. For instance, new H4 molecules are acetylated at lysine residues 5 and 12, and this acetylation pattern is conserved from yeast to humans (Sobel et al., 1995). It has been hypothesized that acetylation of these two lysine residues may facilitate their assembly into nucleosomes. Supporting this idea, mutation of lysine residues 5, 8, and 12 of H4 in combination with a deletion of the N-terminal tail of H3 compromises cell viability and nucleosome assembly in the yeast S. cerevisiae (Ma et al., 1998). However, in vitro human CAF-1 can assemble H3-H4 lacking their N termini onto replicating DNA as efficiently as intact H3-H4 (Shibahara et al., 2000). These results argue that acetylation of lysine residues at the N termini of H3 and H4 is not essential for CAF-1 activity and may therefore have other functions in nucleosome assembly.
Acetylation of H3 lysine 56 (H3K56Ac) was recently described as a mark of newly synthesized H3 molecules (Masumoto et al., 2005). While some studies have indicated that H3K56Ac is involved in transcription (Rufiange et al., 2007; Xu et al., 2005), several laboratories have shown that H3K56Ac is also involved in the response to DNA damage during replication (Collins et al., 2007; Driscoll et al., 2007; Han et al., 2007a, 2007b; Masumoto et al., 2005; Tsubota et al., 2007). First, H3K56Ac increases during S phase progression and largely disappears during G2/M phase of the cell cycle (Masumoto et al., 2005; Zhou et al., 2006). Second, in addition to genotoxic agent sensitivity, cells lacking the H3K56 acetyltransferase Rtt109 (KAT11) (Allis et al., 2007) or cells expressing H3 with lysine 56 mutated to arginine (H3K56R) exhibit an increased frequency of spontaneous chromosome breaks (Driscoll et al., 2007; Han et al., 2007a). Third, mutations in RTT109 exhibit synthetic lethal/slow growth phenotypes with mutations in genes encoding proteins involved in DNA replication and double-strand break repair (Collins et al., 2007; Han et al., 2007a). Together, these results support the notion that H3K56Ac functions during DNA replication to maintain genome stability. However, the exact process in which H3K56Ac is involved during passage through S phase is not known. Here we show that H3K56 acetylation increases the binding affinity of H3 toward CAF-1 and Rtt106 both in vivo and in vitro and promotes nucleosome assembly during S phase of the cell cycle. Moreover, we demonstrate that H3K56Ac binds to a region of Rtt106 that is homologous to the unusual pleckstrin homology (PH) domain of Pob3, a subunit of the FACT chromatin assembly and disassembly complex (Reinberg and Sims, 2006; VanDemark et al., 2006). These results identify another acetyl-lysine binding motif and establish a molecular mechanism of action for H3K56Ac.
RESULTS
H3K56Ac and the Acetylation of Lysine Residues in the N Termini of H3 and H4 Function Nonredundantly in Nucleosome Assembly
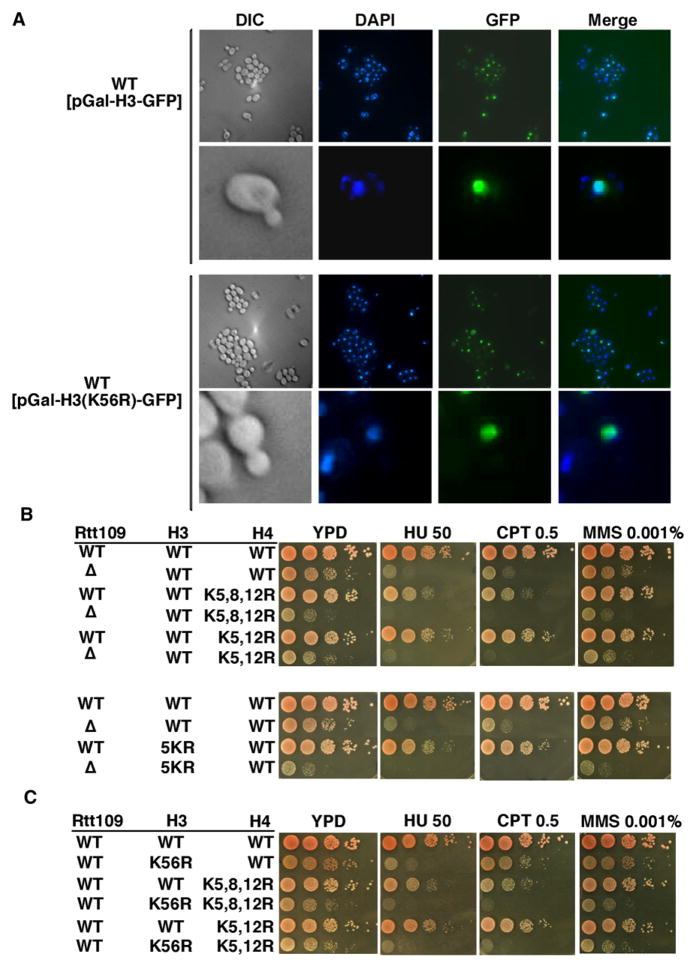
Recent studies have shown that acetylation of the N-terminal lysine residues of newly synthesized H3 and H4 regulates efficient nuclear import of H3 and H4 (Blackwell et al., 2007). Since H3K56Ac occurs in new H3 molecules (Masumoto et al., 2005), we first asked whether it modulates the nuclear localization of new H3 molecules. Mutations that abolish H3K56Ac (H3K56R or rtt109Δ) had no apparent effect on the nuclear localization of H3-GFP expressed from a galactose-inducible promoter (Figures 1A and S1 available online), suggesting that H3K56Ac is not essential for nuclear import of H3. We then compared the phenotypes of cells lacking H3K56Ac to those lacking acetylation at lysine residues at the N termini of H3 and H4. Mutations that eliminate H3K56Ac confer more severe sensitivity to genotoxic agents than mutations that abolish acetylation sites of either the H3 or H4 N terminus (Figures 1B and 1C). Moreover, combining mutations that eliminate H3K56Ac with either lysine-to-arginine mutations in the N termini of H3 or of H4 or a truncation of the N terminus of H3 (H3 Δ3-29) resulted in slow growth and DNA damage sensitivity far more pronounced than those observed in either rtt109Δ or H3K56R single mutants (Figures 1B, 1C, and S2). These results imply that H3K56Ac acts through a molecular mechanism that is genetically distinct from acetylation of the N termini of H3 and H4.
Figure 1. H3K56Ac Plays a Unique Role in Nucleosome Assembly.
(A) H3K56Ac is not required for nuclear localization of H3-GFP. H3-GFP or H3K56R-GFP under the control of the GAL1 promoter were expressed for 1 hr by the addition of 2% galactose and visualized by fluorescence microscopy.
(B and C) The rtt109Δ (B) and H3K56R (C) mutations exhibit synergistic defects in growth and sensitivity to DNA-damaging agents when combined with lysine-to-arginine mutations in the N terminus of H3 or H4. 5KR: lysine residues 9, 14, 18, 23, and 27 of H3 were mutated to arginine. Ten-fold serial dilutions of yeast cells of the indicated genotypes were plated onto nonselective YPD medium or medium containing DNA-damaging agents: hydroxyurea (HU, 50 mM), camptothecin (CPT, 0.5 μg/ml), and methyl methane sulfonate (MMS, 0.001%).
Histone Chaperones Associate with H3K56Ac during S Phase Progression
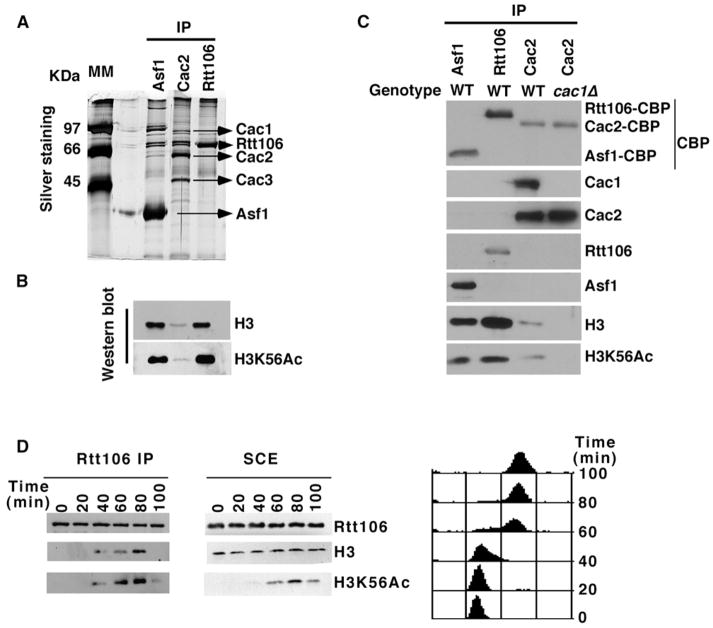
In yeast cells, H3 molecules associated with CAF-1 are acetylated at lysine 56 (Masumoto et al., 2005; Zhou et al., 2006). Therefore, we tested whether H3K56Ac was present in H3 molecules that copurified with Asf1 and Rtt106, two other histone chaperones that bind CAF-1 and are involved in nucleosome assembly (Huang et al., 2005; Tyler et al., 1999). Cac2- (a subunit of CAF-1), Rtt106-, and Asf1-containing complexes were purified from yeast cells by tandem affinity purification (TAP). Copurified proteins were detected by silver staining (Figure 2A) and by western blotting with antibodies against unmodified H3 or H3K56Ac (Figure 2B). While distinct proteins copurified with Cac2, Asf1, and Rtt106 (Figure 2A), all three histone chaperones were associated with H3 molecules acetylated at K56 (Figures 2B and 2C). Lower amounts of H3 were copurified with Cac2-TAP than with Asf1 or Rtt106 (Figure 2B). This was due to the lower amounts of CAF-1 than Asf1 or Rtt106 purified from the same amounts of yeast cells (Figure 2C). Moreover, little Asf1 and Rtt106 were copurified with Cac2 under these conditions (Figure 2C). Thus, all three histone chaperones bind H3K56Ac.
Figure 2. K56Ac Is Present in H3 Molecules Associated with Three Histone Chaperones: CAF-1, Rtt106, and Asf1.
(A and B) CAF-1, Rtt106, and Asf1 form distinct protein complexes in yeast cells. Cac2- (a subunit of CAF-1), Rtt106-, and Asf1-TAP were purified from equal amounts of yeast cells. Copurified proteins were detected by silver staining (A) or by western blotting using antibodies against unmodified H3 or H3K56Ac (B).
(C) Lower amounts of Cac2 than of Rtt106 or Asf1 were purified from the same number of yeast cells. The experiments were performed as in (B), and calmodulin-binding peptide (CBP) is present on all three proteins after purification.
(D) The association of Rtt106 with H3 increases during passage through S phase. Cells synchronized in G1 with α factor were released into the cell cycle. Every 20 min after release, cells were collected for affinity purification of Rtt106-TAP (left panel) or for analysis of DNA content using flow cytometry (right panel). Histones that copurified with Rtt106-TAP (left panel), as well as those in soluble cell extracts (SCE, middle panel), were detected by western blot.
In yeast cells, CAF-1 binding to H3 as well as H3K56Ac increase during passage through S phase (Masumoto et al., 2005; Zhou et al., 2006). We asked whether the association of Rtt106 with H3 was similarly regulated during cell-cycle progression. As previously reported, H3K56Ac progressively increased during S phase, reaching a maximum in either late S phase or G2 phase of the cell cycle (Figure 2D, middle panel) (Masumoto et al., 2005; Zhou et al., 2006). Importantly, although the levels of Rtt106-TAP were constant from G1 through G2, the association of Rtt106 with H3 increased with the same kinetics as the accumulation of H3K56Ac (Figure 2D). In G1 or G2, Rtt106 was associated with much lower levels of H3. Thus, the amount of H3 bound to Rtt106 increases as cells progress through S phase.
H3K56Ac Promotes the Association of H3 with CAF-1 and Rtt106 in Yeast Cells
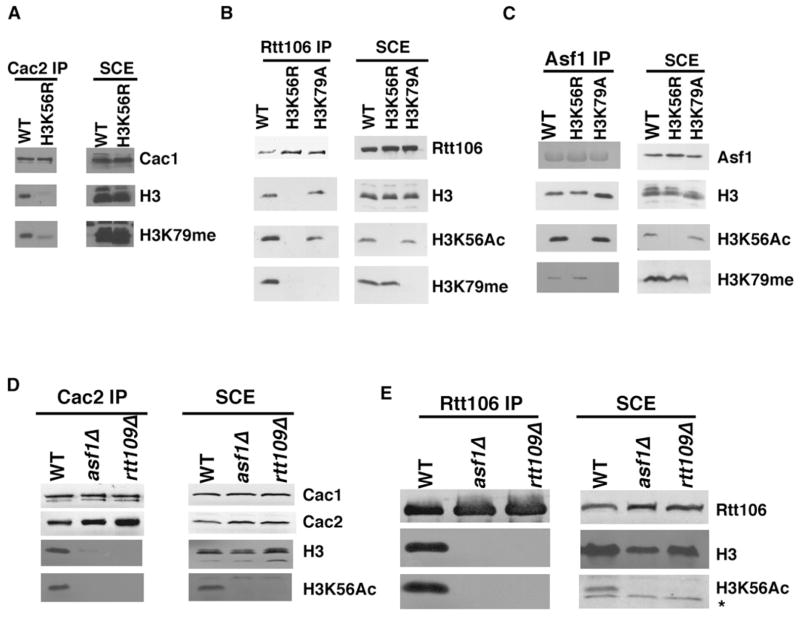
Because both CAF-1 and Rtt106 bind H3 predominantly during S phase when H3K56Ac is abundant, we determined whether H3K56Ac regulates the association of CAF-1, Rtt106, and Asf1 with histones. For this purpose, Cac2-, Rtt106-, and Asf1-TAP were purified from wild-type or H3K56R mutant cells and H3 associated with each chaperone were detected by western blotting. The amounts of H3 bound to CAF-1 and Rtt106 were significantly reduced in H3K56R mutant cells compared with wild-type cells (Figures 3A and 3B). In contrast, mutation of lysine 79 in H3, another lysine residue in the globular domain of H3, did not affect the binding of CAF-1 or Rtt106-TAP to H3 to a detectable degree (Figures 3B, S3A, and S3B). Interestingly, the H3K56R mutation had no detectable effect on the ability of Asf1-TAP to bind H3 in yeast cells (Figure 3C). This result is consistent with the fact that Asf1 binds H3 through its C-terminal region that does not contain K56 (English et al., 2006) and implies that the binding of CAF-1 and Rtt106 to histones is enhanced by H3K56Ac, whereas Asf1 binding to histones is independent of H3K56Ac.
Figure 3. H3K56Ac Enhances the Association of H3 with CAF-1 and Rtt106 but Not Asf1 In Vivo.
(A) Mutation of lysine 56 to arginine reduces the amounts of H3 bound to CAF-1 in yeast cells. Cac2-TAP was purified from wild-type or H3K56R mutant cells, and copurified proteins (left panel) as well as those in the soluble cell extracts (SCE, right panel) were analyzed by western blotting using antibodies against indicated proteins.
(B and C) Mutation of lysine 56 to arginine significantly reduces the binding of H3 to Rtt106 (B) but not Asf1 (C) in yeast cells. Experiments were performed as described in (A), except that Rtt106- and Asf1-TAP were purified.
(D and E) The association of CAF-1 and Rtt106 with H3 is significantly reduced in rtt109Δ and asf1Δ mutant cells that lack H3K56Ac. * marks a nonspecific band recognized by H3K56Ac antibodies in Figure 3E.
In S. cerevisiae, H3K56Ac is catalyzed by Rtt109 and dependent upon Asf1 (Celic et al., 2006; Collins et al., 2007; Driscoll et al., 2007; Han et al., 2007a; Recht et al., 2006). If H3K56Ac promotes the association of CAF-1 and Rtt106 with histones in yeast cells, this association should be compromised in rtt109Δ and asf1Δ mutant cells that lack H3K56Ac. Indeed, the amounts of H3 copurified with CAF-1 and Rtt106 were dramatically reduced in asf1Δ and rtt109Δ mutant cells when compared with wild-type cells (Figures 3D and 3E). These results strongly support the conclusion that H3K56Ac enhances the association of CAF-1 and Rtt106 with H3 in vivo.
H3K56Ac Increases the Binding Affinity of CAF-1 and Rtt106 for H3 In Vitro
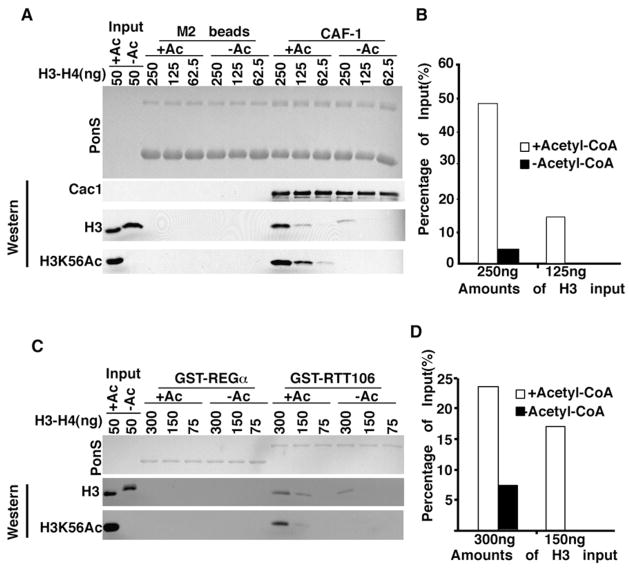
H3K56Ac increases the transcription of histone genes (Xu et al., 2005) and cells lacking Asf1 or Rtt109 grow slower than wild-type cells. It was therefore possible that the reduced binding of H3 to CAF-1 and Rtt106 in asf1Δ or rtt109Δ mutant cells was due either to reduced histone levels and/or altered cell-cycle progression. However, we could not detect any significant change in the total levels of H3 in cells lacking H3K56Ac compared with wild-type cells (Figure 3). Moreover, the interaction between CAF-1 and H3K56Ac was not decreased in cells lacking SPT10 (Figure S3C), which grow much more slowly than the _rtt109_D or _asf1_D mutant cells (data not shown). Spt10 is a transcriptional activator of histone genes and has been implicated in acetylation of H3 lysine 56 at histone gene promoters (Xu et al., 2005). These results argue that the reduced amounts of H3 bound to CAF-1 and Rtt106 in cells lacking H3K56Ac are not an indirect consequence of defects in histone gene expression or cell-cycle progression. Instead, H3K56Ac may directly promote the interactions of CAF-1 and Rtt106 with H3. To address this idea, we purified recombinant CAF-1 and GST-Rtt106 and tested how these proteins bound to H3-H4 acetylated in vitro by the Rtt109-Vps75 complex. As shown in Figures 4A and 4B, CAF-1 bound more H3 acetylated by Rtt109-Vps75 than unacetylated H3. In addition, Cac1, the large subunit of CAF-1, but not Cac2 or Cac3 bound to H3 and H3K56Ac in vitro under the same conditions tested (Figure S4). This result suggests that Cac1 mediates the interaction of CAF-1 with H3K56Ac. We also detected that GST-Rtt106 bound better to H3 acetylated at lysine 56 than to unmodified H3 (Figures 4C and 4D). These results indicate that H3K56Ac increases the binding affinity of CAF-1 and Rtt106 for H3 in vitro.
Figure 4. H3K56Ac Increases the Binding of H3 to CAF-1 and Rtt106 In Vitro.
(A and B) H3K56Ac enhances the binding of H3 to CAF-1. Flag-tagged recombinant CAF-1 bound to Flag antibody beads was used to bind different amounts of H3-H4 in reaction mixtures treated with Rtt109-Vps75 in the presence of (+Ac) or absence of acetyl-CoA (−Ac). Equal amounts of beads without CAF-1 were used as a negative control. Bound proteins were detected either by Ponceau S staining (A, upper panel) or by western blotting using antibodies against Cac1, unmodified H3, or H3K56Ac. The percentage of coprecipitated H3 from two different inputs was calculated (B).
(C and D) H3K56Ac increases the binding affinity of Rtt106 for H3. The experiments were performed as described above, except that GST-Rtt106 was used for binding assays and GST-REGα as negative controls. Representative results from three independent experiments are shown.
A Region of Rtt106 Homologous to the PH Domain of Pob3 Mediates the Interaction of Rtt106 with H3K56Ac
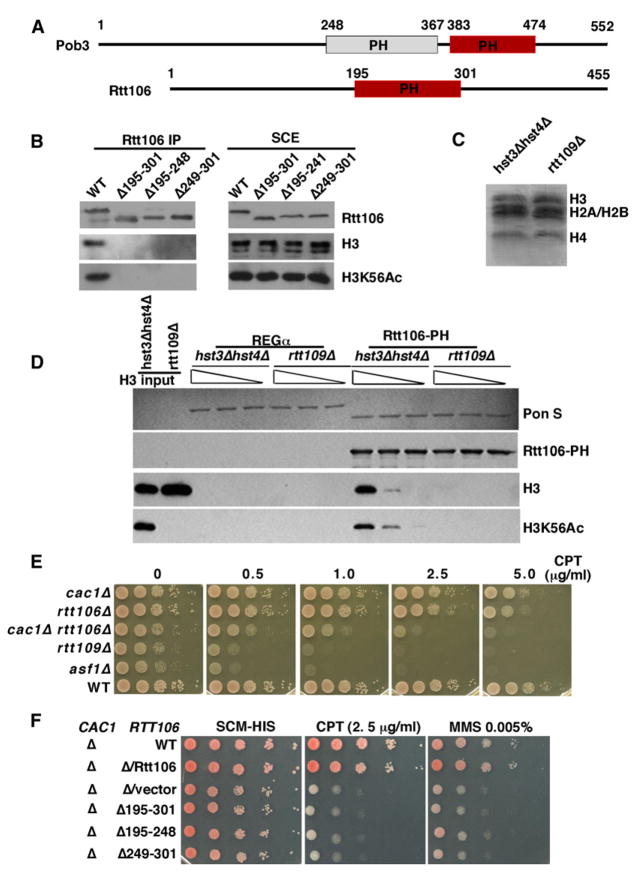
Some protein domains can “read” specific modifications of histones (Kouzarides, 2007). The bromodomain is the only domain thus far characterized that recognizes acetylated lysine residues. CAF-1 and Rtt106 do not contain classical bromodomains. We noticed, however, that Rtt106 contains a region (residues 195–301) that is related to the middle domain of Pob3 (Pob3-M) (Figure 5A). Structure determination of Pob3-M has identified two domains structurally related to a unique class of PH domains (VanDemark et al., 2006). Hence we will refer to this region of Rtt106 as a PH-like domain or simply Rtt106-PH. Because Pob3 is a subunit of FACT, a complex that promotes both nucleosome assembly and disassembly during transcription and DNA replication (Reinberg and Sims, 2006; VanDemark et al., 2006), we tested whether Rtt106-PH mediates interactions between Rtt106 and histones. Deletion of amino acids 195–301 of Rtt106 had no major effect on protein stability but largely abolished the interaction between Rtt106 and H3 in yeast cells (Figure 5B). Thus, Rtt106-PH is required for Rtt106 to bind H3 in vivo. Next, we tested whether Rtt106-PH is sufficient to bind H3K56Ac in vitro using histone octamers purified from yeast cells containing mutations in Hst3 and Hst4 or mutations in Rtt109 (Figure 5C). Hst3 and Hst4 are two H3K56Ac deacetylases, and in their absence, over 90% of H3 is K56 acetylated (Celic et al., 2006). Rtt106-PH bound more H3 purified from _hst3Δ hst4Δ_mutant cells than H3 isolated from the _rtt109Δ_mutant cells (Figure 5D). These results indicate that the Rtt106-PH domain is both necessary and sufficient to bind H3K56Ac.
Figure 5. The PH-like Domain of Rtt106 Is a Motif that Binds to H3K56Ac.
(A) Schematic representation of Rtt106 and Pob3. A region in Rtt106 is homologous to the second PH domain of Pob3 (red). The positions of each domain are marked by amino acid numbers.
(B) Deletion of the Rtt106-PH domain abolishes binding of Rtt106 to H3 in yeast cells. TAP-tagged Rtt106 or mutants with the indicated deletions were purified from equal amounts of yeast cells, and copurified proteins as well as proteins in soluble cell extracts (SCE) were analyzed by western blotting.
(C) Native yeast core histones were purified from hst3Δhst4Δ and rtt109Δ cells and detected by Coomassie blue staining.
(D) H3K56Ac increases binding of the Rtt106-PH domain to core histones in vitro. GST-Rtt106-PH or GST-REGα (negative control) were used to bind core histones from hst3Δhst4Δ cells or rtt109Δ cells (shown in C) and bound proteins were detected either by Ponceau S staining (Pon S) or by western blotting.
(E) cac1Δ rtt106Δ double mutants exhibit similar growth and DNA-damage sensitivity defects to the rtt109Δ mutant. Ten-fold serial dilutions of yeast cells of the indicated genotypes were spotted onto nonselective YPD medium or medium containing different concentrations of CPT. (F) Deletion of the PH domain of Rtt106 enhances the DNA-damage sensitivity of a cac1Δ mutant. The experiments were performed as described in (E), except that minimal media lacking histidine (-HIS) were used.
Mutations in the Rtt106-PH Domain Result in Increased Sensitivity to DNA-Damaging Agents
Cells lacking H3K56Ac are sensitive to genotoxic agents (Han et al., 2007a; Masumoto et al., 2005) (Figures 1B and 1C). To assess the role of histone chaperones in DNA damage resistance, we compared the sensitivity to genotoxic agents of cells lacking Rtt109 to those lacking Rtt106 or Cac1, the large subunit of CAF-1. The cac1Δ or rtt106Δ single mutant cells are generally much less sensitive to DNA-damaging agents (CPT, HU, and MMS) than asf1Δ or rtt109Δ mutant lacking H3K56Ac (Figures 5E and S5). However, the cac1Δ rtt106Δ double mutant cells were considerably more sensitive to these DNA-damaging agents than either cac1Δ or rt106Δ single mutants (Figures 5E and S5). Moreover, the sensitivity of rtt109Δ mutant cells to DNA-damaging agents (CPT, MMS, and HU) was not dramatically greater than that of cac1Δ rtt106Δ cells (Figures 5E and S5). These results support the notion that CAF-1 and Rtt106 are two important effectors that bind H3K56Ac.
We also tested the genotoxic agent sensitivity of cells expressing three _rtt106_-PH domain mutants. In the absence of Cac1, these three rtt106 mutants conferred a very similar degree of DNA-damage sensitivity to that observed in _cac1_Δ rtt106Δ cells (Figure 5F). Thus, in the absence of CAF-1, the association of Rtt106 with H3 acetylated at lysine 56 is important for cellular resistance to DNA-damaging agents.
H3K56Ac Facilitates Nucleosome Assembly In Vitro
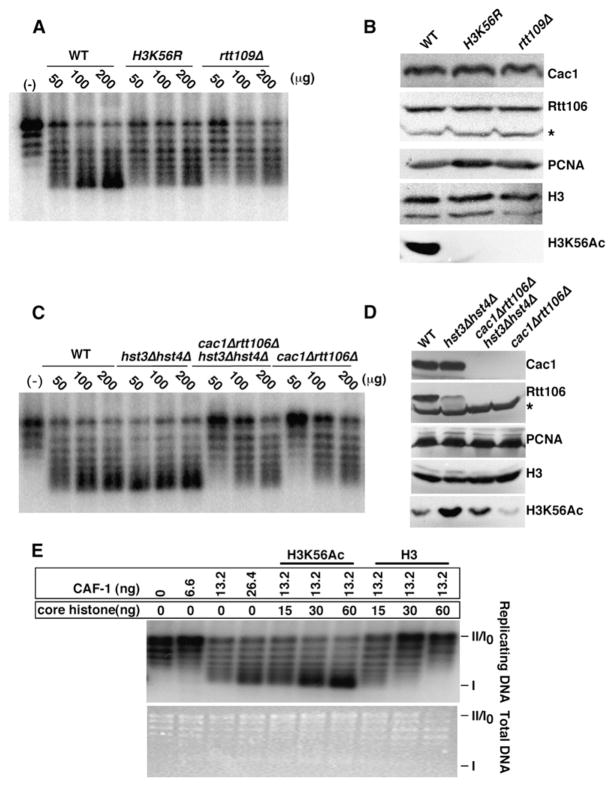
CAF-1 and Rtt106 mediate the deposition of histones onto DNA to form nucleosomes (Huang et al., 2005; Stillman, 1986). Since H3K56Ac is needed for the association of these two histone chaperones with H3 in vivo and in vitro, we tested whether H3K56Ac enhances nucleosome assembly using a DNA synthesis-independent plasmid supercoiling assay (Ma et al., 1998). In this assay, the wrapping of DNA around histones introduces one negative superhelical turn per nucleosome core particle. Radio-labeled plasmid DNA relaxed by topoisomerase I was incubated with cell extracts prepared from wild-type, H3K56R, or rtt109Δ mutant cells. Upon addition of increasing amounts of cell extracts prepared from wild-type cells, supercoiled DNA generated by nucleosome assembly progressively appeared (Figure 6A). Interestingly, extracts prepared from H3K56R or rtt109Δ cells assembled nucleosomes much less efficiently than extracts from wild-type cells (Figure 6A). This deficiency was not due to reduced amounts of histone chaperones (CAF-1 and Rtt106) or histone H3 in rtt109Δ or H3K56R mutant extracts (Figure 6B). Moreover, the nucleosome assembly defect observed in rtt109Δ extracts was suppressed more effectively by adding back H3-H4 acetylated in vitro by Rtt109-Vps75 than unacetylated H3-H4 (Figure S6). These results indicate that H3K56Ac facilitates efficient nucleosome assembly onto plasmid DNA. Consistent with this, when limiting, extracts prepared from cells with high levels of H3K56Ac (hst3Δ hst4Δ, Figure 6D) were able to assemble nucleosomes more efficiently than extracts from wild-type cells (Figure 6C). Importantly, although extracts from cac1Δ rtt106Δ hst3Δ hst4Δ quadruple mutant cells did contain elevated levels of H3K56Ac, this did not significantly alleviate the defect in nucleosome assembly observed in cac1Δ rtt106Δ mutant extracts (Figures 6C and 6D). Thus, the ability of H3K56Ac to stimulate nucleosome assembly depends on the presence of CAF-1 and Rtt106 in cell extracts.
Figure 6. H3K56Ac Facilitates CAF-1- and Rtt106-Dependent Nucleosome Assembly.
(A) Cell extracts prepared from rtt109Δ or H3K56R mutant cells assemble nucleosomes less efficiently than those prepared from wild-type cells. Buffer alone (−) or increasing amounts of extracts (μg) prepared from wild-type, H3K56R, and rtt109Δ mutant cells were used to assemble 32P-labeled plasmids relaxed by topoisomerase I into nucleosomes.
(B) Protein levels of Cac1 (the large subunit of CAF-1), Rtt106, PCNA, and H3 are not affected in H3K56R and rtt109Δ mutant cell extracts. Cell extracts used for nucleosome assembly in (A) were analyzed by western blotting using antibodies against the proteins indicated on the right.
(C) The enhancement of nucleosome assembly by H3K56Ac requires the presence of CAF-1 and Rtt106. Cell extracts were prepared from yeast strains with indicated genotype and used for supercoiling assays.
(D) H3K56Ac is higher in cac1Δ rtt106Δ hst3Δ hst4Δ mutant cell extracts than in wild-type or cac1Δ rtt106Δ extracts. Cell extracts used for plasmid supercoiling assays in (C) were analyzed by western blotting. * indicates a nonspecific protein detected by antibodies against Rtt106.
(E) H3K56Ac facilitates the formation of nucleosomes onto replicating DNA mediated by yeast CAF-1. Plasmids pSV011 were replicated in histone-depleted human S100 extracts in the presence of T antigen and [α-32P]-dATP. Recombinant CAF-1 and core histones purified from either hst3Δ hst4Δ (H3K56Ac) or rtt109Δ mutant cells (H3) were added to promote nucleosome assembly. Total DNA was visualized by ethidium bromide staining and replicating DNA by autoradiography.
Yeast CAF-1 promotes nucleosome assembly onto replicating DNA in a human cell extract for SV40 DNA replication (Kaufman et al., 1997). We used this assay to test whether H3K56Ac facilitates CAF-1-mediated nucleosome assembly onto replicating DNA in vitro. T antigen-driven SV40 DNA replications were performed in human S100 extracts with endogenous histones depleted. Recombinant yeast CAF-1 and core histones purified from either rtt109Δ or hst3Δ hst4Δ mutant cells were then added to the replication reactions to allow nucleosome assembly. As shown in Figure 6E, CAF-1 assembled nucleosomes onto replicating DNA more efficiently using core histones purified from hst3Δ hst4Δ cells than with core histones derived from rtt109Δ cells. These results strongly indicate that H3K56Ac also enhances replication-coupled nucleosome assembly by yeast CAF-1 in vitro.
CAF-1 and Rtt106 Are Involved in Deposition of H3K56Ac onto Replicating DNA In Vivo
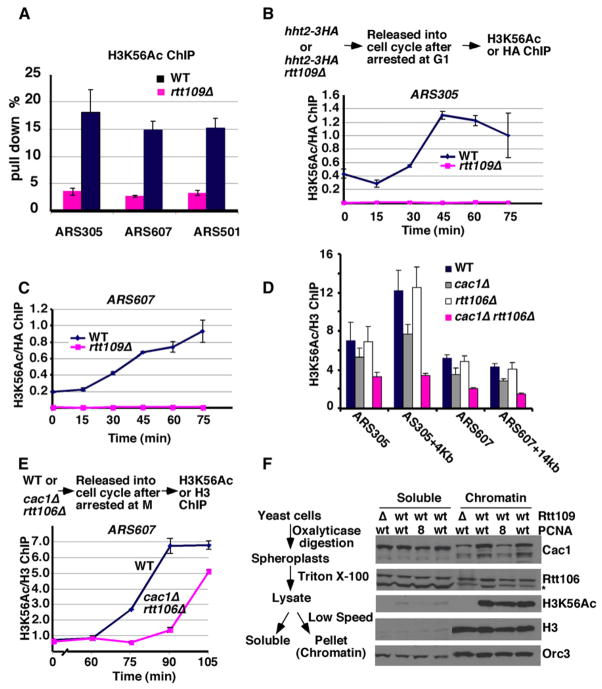
We then asked whether H3K56Ac was deposited onto replicating DNA during S phase of the cell cycle in yeast cells. Chromatin immunoprecipitation assays (ChIP) using antibodies specifically recognizing H3K56Ac were performed in wild-type and the rtt109Δ mutant cells (as a negative control). In wild-type cells, H3K56Ac was detected at two early replication origins (ARS305 and ARS607) as well as a late replication origin (ARS501) located on different chromosomes (Bell and Dutta, 2002) (Figure 7A). Next, we determined when H3K56Ac was localized at replication origins during cell-cycle progression. Although a low level of Rtt109-dependent H3K56Ac was detected at G1 phase of the cell cycle (Figure S7, Figures 7B–7C, and data not shown), considerably more H3K56Ac was detected at ARS305 (Figure 7B) and ARS607 (Figure 7C) during DNA replication. H3K56Ac was also detected on a DNA fragment located 14 Kb away from ARS607 (Figure S8A, ARS607+14 Kb) as well as a telomeric heterochromatin locus (Figures S8B–S8C), which is known replicate late in S phase, 15 min later than at the early replication origins. Moreover, when cells were released from G1 into fresh medium containing HU, very little H3K56Ac was incorporated at ARS607 + 14 Kb after 45 min in HU (Figure S9) compared with ARS607. HU allows initiation of DNA replication at ARS607 but prevents the replication fork from reaching ARS607 + 14 Kb (Han et al., 2007b). These results demonstrate that replication fork passage is required for H3K56Ac to be deposited onto DNA during S phase of the cell cycle.
Figure 7. CAF-1 and Rtt106 Are Involved in Deposition of H3K56Ac onto Replicating DNA during S Phase of the Cell Cycle.
(A) H3K56Ac is detected at DNA replication origins. ChIP assays using antibodies specific for H3K56Ac were performed from exponentially growing wild-type and rtt109Δ mutant cells. The percentage of immunoprecipitated DNA over the total input DNA was determined using primers amplifying two early replication origins (ARS305 and ARS607) and a late replication origin (ARS501).
(B and C) The association of H3K56Ac with replicating DNA increases during progression through S phase. Wild-type cells or rtt109Δ mutant cells in which the histone gene HHT2 was tagged with the HA epitope were synchronized in G1 using α factor and then released into the cell cycle. Every 15 min after release, cells were collected for ChIP assays using antibodies against H3K56Ac or against the HA epitope (B, upper panel). The immunoprecipitated DNA was analyzed by quantitative PCR using primers amplifying ARS305 (B) and ARS607 (C). The ratio of H3K56Ac ChIP signals over the corresponding H3-HA ChIP signals was calculated.
(D) The incorporation of H3K56Ac into chromatin is reduced in cells lacking CAF-1 and Rtt106. ChIP assays using antibodies against H3K56Ac or un-modified H3 were performed in wild-type, cac1Δ, rtt106Δ, or cac1Δ rtt106Δ double mutant cells. The ChIP DNAs were analyzed using four different primer pairs amplifying ARS305, ARS305+4Kb, ARS607, and ARS607+14Kb using quantitative PCR.
(E) Deposition of H3K56Ac into chromatin during S phase is reduced in cac1Δ rtt106Δ mutant cells. Wild-type and cac1Δ rtt106Δ double mutants synchronized in M phase using nocodazole were released into the cell cycle. H3K56Ac ChIP assays were performed as described in (D).
(F) Chromatin binding of Cac1 is partially reduced in rtt109Δ mutant cells. A chromatin-binding assay, as outlined on the left, was performed and proteins in the soluble and the chromatin fractions were detected by western blotting. Fractionation of an extract from a PCNA mutant strain (pol3-8) was included as a positive control for reduced CAF-1 association with chromatin. Data in (A)–(E) are represented as mean ± standard error of the mean (SEM).
Next we asked whether mutations in CAF-1 and Rtt106 affect deposition of H3K56Ac onto replicating DNA using ChIP assays. Neither cac1Δ nor rtt106Δ single mutations reduced the incorporation of H3K56Ac into replicating DNA. However, at four distinct DNA loci, the incorporation of H3K56Ac into chromatin was significantly reduced in cells lacking both CAF-1 and Rtt106 (Figure 7D). To ascertain that the reduced association of H3K56Ac with chromatin in cac1Δ rtt106Δ double mutant cells was indeed caused by a defect in the deposition of H3K56Ac onto replicating DNA, we used the H3K56Ac ChIP assay to monitor the deposition of H3K56Ac into chromatin when wild-type and cac1Δ rtt106Δ double mutant cells were released from arrest in mitosis into the cell cycle. Significantly more H3K56Ac was incorporated into replicating DNA during S phase in wild-type cells than in cac1Δ rtt106Δ mutant cells (Figures 7E and S10B). The reduced incorporation of H3K56Ac into replicating DNA was not due to lower levels of H3K56Ac in cac1Δ rtt106Δ double mutants (data not shown) or altered cell-cycle progression from mitosis to S phase (Figure S10A). Thus, CAF-1 and Rtt106 are required for efficient deposition of H3K56Ac onto replicating DNA during S phase of the cell cycle.
The Chromatin Binding of Cac1 Is Impaired in rtt109Δ Mutant Cells
In yeast and human cells, CAF-1 is recruited to replicating DNA through direct binding to PCNA (Shibahara and Stillman, 1999; Zhang et al., 2000). Moreover, the binding of yeast Cac1 to chromatin is significantly reduced in a PCNA mutant (Pol30-8) that cannot bind to CAF-1 (Zhang et al., 2000). How Rtt106 is recruited to the replication forks is not clear. Since H3K56Ac increases the binding of H3 to CAF-1 and Rtt106, it seems possible that this modification may serve as a “signal” for productive association of these two histone chaperones with chromatin. To test this idea, we utilized a chromatin fractionation assay to determine whether the chromatin binding of CAF-1 and Rtt106 is affected in rtt109Δ mutant cells. Proteins from early exponential phase cells were separated into a soluble fraction and a chromatin pellet and detected by western blotting (Figure 7F, left). As previously reported (Zhang et al., 2000), the majority of Orc3, a subunit of the Origin Recognition Complex (Bell and Dutta, 2002), was bound to chromatin. Remarkably, compared with wild-type cells, the association of Cac1 with chromatin was reduced in rtt109Δ mutant to a similar extent as in pol30-8 mutant cells. In contrast, chromatin binding of Rtt106 was not affected to a significant degree in either pol30-8 or rtt109Δ mutant cells (Figure 7F). Thus, H3K56Ac not only enhances the binding of H3 to CAF-1 but may also regulate the binding of CAF-1 to chromatin in yeast cells.
DISCUSSION
H3K56Ac Plays a Direct Role in Replication-Coupled Nucleosome Assembly
CAF-1, a protein conserved from yeast to human cells, deposits newly synthesized and acetylated forms of H3 and H4 onto replicating DNA to promote de novo nucleosome assembly during S phase of the cell cycle. However, the molecular function of the acetylation of new histones has, until now, remained largely unknown. We have presented several lines of evidence supporting the notion that a specific site of acetylation in new H3, namely H3K56Ac, regulates CAF-1- and Rtt106-dependent nucleosome assembly. First, H3K56Ac increases the binding affinity of CAF-1 and Rtt106 for H3 in vivo and in vitro. Second, H3K56Ac promotes nucleosome assembly in cell extracts in a CAF-1- and Rtt106-dependent manner. Third, H3K56Ac is incorporated into replicating DNA, and this deposition is compromised in cells lacking CAF-1 and Rtt106. These results indicate that modifications on newly synthesized histones play a direct role in determining how these histones are assembled into nucleosomes and extend the “histone code” hypothesis (Strahl and Allis, 2000) to the nucleosome assembly process.
In addition to H3K56Ac, H4 molecules that copurify with yeast and human CAF-1 are also acetylated at lysines 5, 8, and 12 (Verreault et al., 1996; Zhou et al., 2006). In contrast to mutations at Rtt109 that cripple histone H3 binding to CAF-1, mutations of Hat1, the enzyme that acetylates lysine residues 5 and 12 in newly synthesized histone H4 (Kleff et al., 1995), have no apparent effect on the binding of CAF-1 to histones (Figure S3D). Moreover, mutations that abolish H3K56Ac (_rtt109_Δ or H3K56R) synergistically enhance the slow growth and DNA-damage sensitivity caused by lysine-to-arginine mutations that eliminate acetylation of all the lysine residues in the N-terminal tails of either new H3 or H4 molecules (Figure 1). Thus, H3K56Ac plays a unique role in the nucleosome assembly pathway.
H3K56Ac Coordinates the Function of H3-H4 Chaperones in Nucleosome Assembly
In the yeast S. cerevisiae, three histone chaperones, CAF-1, Asf1, and Rtt106, have been implicated in the assembly of H3-H4 into nucleosomes (Huang et al., 2005; Kaufman et al., 1997; Tyler et al., 1999). However, how the roles of these histone chaperones are coordinated to promote nucleosome assembly is largely unknown. Our results suggest that these histone chaperones function in a hierarchical manner to promote nucleosome assembly. First, Asf1 binds to newly synthesized H3-H4 dimers (English et al., 2006) and presents those dimers for acetylation of H3K56 by the Rtt109-Vps75 complex (Han et al., 2007b, 2007c). H3K56Ac-H4 complexes are then transferred to Rtt106 and CAF-1 for deposition onto DNA and subsequent nucleo-some formation.
While the mechanism of parental histones segregation is still debated and it is not known whether this process requires CAF-1 (Groth et al., 2007a; Henikoff et al., 2004; Tagami et al., 2004), it is clear that deposition of newly synthesized histones requires CAF-1 and Asf1 in yeast cells. In the crystal structures of Asf1-H3-H4 complexes, Asf1 binds to a surface of H3 that is critical for formation of (H3-H4)2 tetramers (English et al., 2006). In vitro, Asf1 disrupts (H3-H4)2 tetramers and forms Asf1-H3-H4 heterotrimeric complexes (Natsume et al., 2007). Thus, it may not be energetically and/or kinetically possible for Asf1 alone to deposit histones onto replicating DNA for formation of (H3-H4)2 tetramers, the first building blocks needed for rapid de novo nucleosome assembly during S phase of the cell cycle. H3 lysine 56 is far away from the surface involved in the formation of (H3-H4)2 tetramers. Thus, the transfer of H3K56Ac-H4 dimers from Asf1 to CAF-1 and Rtt106, which bind preferentially to H3K56Ac, may ensure rapid formation of (H3-H4)2 tetramers and subsequent formation of nucleosomes.
How Is H3K56Ac Recognized by Rtt106 and CAF-1?
While several structural motifs, such as the PHD, Tudor, and chromodomains, all recognize methylated lysine residues, the bromodomain was the only known motif that bound acetylated lysine residues until now (Kouzarides, 2007). We have discovered a region of Rtt106 (amino acids 195–301) that recognizes H3K56Ac and, remarkably, this region is homologous to a domain of Pob3 that adopts a structure related to the PH domain. The PH-like domain of Pob3 does not belong to any of the six previously described subgroups of PH domain structures. While different subgroups of PH domain adopt similar structural folds, each subgroup of PH motifs recognizes different ligands ranging from inositol lipid, to phosphotyrosines and polyprolines (VanDemark et al., 2006). Since the region of Rtt106 that binds H3K56Ac is homologous to the PH domain of Pob3, these results suggest that the PH domain of Pob3 may also be involved in recognition of H3K56Ac and/or other histone modifications. Supporting this idea, FACT copurifies with H3K56Ac in yeast cells (Q.L. et al., unpublished data). Thus, the unique subgroup of PH domains that include those of Rtt106 and Pob3 may represent novel acetyl-lysine recognition modules.
We have shown that Cac1, but not the other two subunits of CAF-1 (Cac2 and Cac3), binds to H3 and H4 in vitro. These results suggest that Cac1 is a major determinant for CAF-1 binding to histones in vitro. Supporting this notion, no histones copurified with Cac2 from yeast cells in the absence of Cac1 (Zhou et al., 2006). Cac1 exhibits no sequence homology to the PH domain of Rtt106. It is possible that CAF-1 may adopt a distinct fold that binds preferentially to H3K56Ac. In this regard, we note that, while the association of Rtt106 with H3 is abolished in cells lacking H3K56Ac, CAF-1 binding to H3 is only partially reduced in yeast cells lacking H3K56Ac. Future work is needed to address how Cac1 recognizes H3K56Ac.
H3K56Ac Preserves Genome Integrity by Promoting Efficient Replication-Coupled Nucleosome Assembly
The major phenotypes reported in mutant cells that lack H3K56Ac (H3K56R, asf1Δ, or rtt109Δ mutants) include spontaneous chromosome breaks and sensitivity to genotoxic agents that impede DNA replication. (Collins et al., 2007; Driscoll et al., 2007; Han et al., 2007a; Masumoto et al., 2005). However, the exact molecular role of this modification in the DNA-damage response has remained enigmatic. We present two lines of evidence supporting the idea that defects in DNA replication-coupled nucleosome assembly are, at least in part, responsible for the acute DNA-damage sensitivity of cells that lack H3K56Ac. First, we have shown that _cac1_Δ _rtt106_Δ double mutant cells are sensitive to the same spectrum of genotoxic agents as the rtt109Δ mutant cells. Moreover, the DNA-damage sensitivity of the cac1Δ rtt106Δ double mutant cells approaches that of rtt109Δ mutant cells. Second, rtt106 mutants that cannot bind to H3K56Ac are quite sensitive to DNA-damaging agents in the absence of Cac1 (Figure 5F). However, cells lacking CAF-1 and Rtt106 are less sensitive than rtt109Δ mutant cells (Figures 5E and S5), suggesting that H3K56Ac may mediate interactions with nucleosome assembly factors other than CAF-1 and Rtt106. In support of this, the deposition of H3K56Ac onto replicating DNA is reduced but not abolished in cells lacking CAF-1 and Rtt106 (Figure 7E). Moreover, Pob3 also binds H3K56Ac in yeast cells (data not shown). Based on several lines of genetic evidence, FACT has been suggested to promote nucleosome assembly during DNA replication through an unknown mechanism (VanDemark et al., 2006). By analogy with our findings for CAF-1 and Rtt106, H3K56Ac may enhance the nucleosome assembly activity of FACT by increasing its association with new H3. Alternatively, the more pronounced DNA-damage sensitivity of rtt109Δ mutant cells, compared with the _cac1_Δ _rtt106_Δ mutant, may reflect an additional role of H3K56Ac following its deposition into chromatin. Despite the phenotypic differences between cells lacking Rtt109 and those lacking CAF-1 and Rtt106, our results indicate that H3K56Ac maintains genome stability by enhancing the efficiency of DNA replication-coupled nucleosome assembly.
Does H3K56Ac Enhance Nucleosome Assembly in Human Cells?
Recently, H3K56Ac has been reported in H3 purified from HeLa cells. In addition to H3K56Ac, human H3 is also methylated at lysine 56, whereas methylation of lysine 56 has not been detected in yeast H3 (Garcia et al., 2007; Masumoto et al., 2005). Compared with H3K56Ac in yeast cells, H3K56Ac is far less abundant in HeLa cells (Garcia et al., 2007). Moreover, sequence homologs of Rtt109 and Rtt106 have yet to be identified in human cells. These issues raise the question of whether H3K56Ac also promotes nucleosome assembly in human cells in a manner similar to its molecular function in yeast cells. Despite the lack of an apparent sequence homolog of Rtt109 in human cells, CAF-1, Asf1, Rtt106, and Rtt109 are clearly encoded in the genome of the fission yeast S. pombe. This yeast species diverged from S. cerevisiae around 400 million years ago and, in many aspects, its chromatin structure is more similar to that of human cells. Moreover, potential homologs of several proteins involved in the regulation of H3K56Ac in budding yeast can be identified in the human genome. For instance, there are two Asf1 homologs (ASF1a and ASF1b), five potential homologs of Vps75, which is a component of the Rtt109-Vps75 HAT complex in yeast cells, and seven sirtuins that share sequence similarity with the catalytic domains of the H3K56 deacetylases Hst3 and Hst4. In addition to the existence of sequence homologs, human Asf1 regulates the flow of the S phase histones during replication stress by unknown mechanisms (Groth et al., 2005). Thus, these proteins may function to regulate H3K56 acetylation and/or methylation or some other modifications of newly synthesized histones (Loyola et al., 2006), which in turn promote nucleosome assembly behind replication forks in higher eukaryotic cells.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Antibodies
Yeast strains are listed in Table S1 and were derived from the parental strain W303-1 using standard yeast media and genetic approaches. Cac2, the second subunit of CAF-1, Rtt106, and Asf1 were tagged at their C termini with the TAP tag according to published procedures (Puig et al., 2001). Antibody production and plasmid constructions are described in the Supplemental Experimental Procedures.
Fluorescence Microscopy
Haploid wild-type or rtt109Δ (S288C) cells were transformed with either pYES2-HHT1-GFP or pYES2-HHT1K56R-GFP and grown at 30°C in medium containing raffinose. After induction of H3 or H3K56R-GFP with 2% galactose for 1 hr, cells were fixed in 100 mM potassium phosphate buffer containing 3.7% formaldehyde for 10 min, washed in 100 mM potassium phosphate buffer (pH 7.4), and mounted on poly-lysine coated slides in mounting medium containing 50 ng/ml DAPI. Microscopy was performed using a Leica DM5500B epifluorescence microscope equipped with an X-Cite Series 120 light source and a Hamamatsu EM-CCD 512×512 camera.
In Vitro Binding of CAF-1 and Rtt106 to H3K56Ac
Recombinant GST-Rtt106 and CAF-1 were purified as described in the Supplemental Experimental Procedures and used for in vitro binding assays. To prepare H3 acetylated at lysine 56, recombinant Drosophila H3-H4 were mixed with Asf1 at 4°C overnight to allow formation of Asf1-H3-H4 complexes. These complexes were then used for histone acetylation, as previously described (Han et al., 2007a). As a negative control, reaction mixtures containing all of the components except acetyl-coenzyme A were also prepared and used for binding assays. Alternatively, core histones were purified from hst3Δ hst4Δ double mutant cells or from rtt109Δ mutant cells as described in the Supplemental Experimental Procedures and used for binding assays. To test whether Rtt106 binds H3K56Ac better than unacetylated H3, 5 μg of GST-Rtt106 and GST-REGα (a human proteasome activator that served as a control for nonspecific binding) were bound to glutathione-Sepharose beads and these beads were then incubated with different amounts of histone acetylation reaction at 4°C overnight. The beads were washed extensively, and the bound proteins were eluted with SDS-PAGE loading buffer, resolved by 15% SDS-PAGE, and detected by western blotting with antibodies against H3 or H3K56Ac. Band intensity was quantified using software Quantity One (Bio-Rad laboratories). Similar procedures were also performed to study how CAF-1 binds to H3 with or without H3K56 acetylation, except that anti-FLAG M2-Agarose beads (Sigma) were used to bind recombinant CAF-1.
In Vitro Plasmid Supercoiling Assay using Yeast Whole-Cell Extracts
Negatively supercoiled plasmid DNA molecules (pSV011), including those internally labeled with 32P as described in the Supplemental Experimental Procedures, were relaxed with DNA topoisomerase I (Promega) at 30°C for 1 hr. These plasmids were then incubated with different amounts of whole-cell extracts prepared as previously described (Ma et al., 1998) at 30°C for 1.5 hr. After digestion of RNA and proteins with RNAase A and Pronase, respectively, DNA products were purified, resolved in 1.25% native agarose gels, and detected by PhosphorImager or autoradiography.
Purification of Core Histones from Yeast Cells
To purify yeast core histones, we first prepared yeast chromatin according to published procedures (Altaf et al., 2007). The soluble chromatin fraction was then loaded onto a hydroxyapatite column. After extensive washes, histones were eluted with 1.4 M NaCl, dialyzed and stored at −80°C before use. A detailed protocol can be found in the Supplemental Experimental Procedures.
SV40 Replication-Coupled Nucleosome Assembly Assay
The assays were performed as previously described with the following modifications (Kaufman et al., 1997; Stillman, 1986). S100 extracts prepared from human 293 cells were first incubated with antibodies against H3 and H4 acetylated at lysines 5 and 12 to deplete endogenous H3-H4. The depleted S100 extracts were used to replicate pSVO11 DNA with SV40 T antigen (a kind gift from Dr. Bruce Stillman) and [α-32P]dATP at 30°C for 30 min. Core histones purified from _hst3_Δ _hst4_Δ or _rtt109_Δ yeast cells and recombinant yeast CAF-1 purified from Sf9 cells were then added to the reactions and incubation resumed for an additional 30 min. DNA was purified and resolved in 1.25% agarose gels. Total DNA was revealed by ethidium bromide staining and replicated DNA by autoradiography.
ChIP Assays
To determine if H3K56Ac was deposited onto replicating DNA, ChIP assays using antibodies against H3K56Ac were performed according to published procedures (Huang et al., 2007) in both asynchronous and synchronized cells. ChIP DNA was analyzed by quantitative PCR using primers amplifying replication origins as well as fragments downstream of replication origins. For normalization purposes, ChIP assays were also performed using antibodies against H3 in identical samples, and the ratios of H3K56Ac ChIP signals over the corresponding H3 ChIP signals were calculated.
Supplementary Material
Acknowledgments
We thank Jan van Deursen, Junjie Chen, Daniel Billadeau, and Rebecca Burgess for their critical reading of the manuscript. We thank Drs. Paul Kaufman and Jessica Tyler for plasmids and the Jacques Côté ’s laboratory for chromatin purification protocol. We thank Dr. Bruce Stillman for T-antigen, yeast strains, helpful discussions, and advice. We thank Junhong Han for performing experiments described in Figure 5E. This work is supported in part by grants from the CIHR to A.V. and by NIH grants to Z.Z. IRIC receives infrastructure support from FRSQ.
References
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Cote J. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell. 2007;28:1002–1014. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Blackwell JS, Jr, Wilkinson ST, Mosammaparast N, Pemberton LF. Mutational analysis of H3 and H4 N termini reveals distinct roles in nuclear import. J Biol Chem. 2007;282:20142–20150. doi: 10.1074/jbc.M701989200. [DOI] [PubMed] [Google Scholar]
- Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. The sirtuins hst3 and hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol. 2006;16:1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falbo KB, Shen X. Chromatin remodeling in DNA replication. J Cell Biochem. 2006;97:684–689. doi: 10.1002/jcb.20752. [DOI] [PubMed] [Google Scholar]
- Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, Shabanowitz J, Mishra N, Strahl BD, Allis CD, Hunt DF. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, Almouzni G. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol Cell. 2005;17:301–311. doi: 10.1016/j.molcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007a;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007b;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007a;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by Rtt109 and regulated by Asf1 is required for replisome integrity. J Biol Chem. 2007b;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Li Z, Xu RM, Zhang Z. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J Biol Chem. 2007c;282:14158–14164. doi: 10.1074/jbc.M700611200. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Furuyama T, Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004;20:320–326. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Huang S, Zhou H, Katzmann D, Hochstrasser M, Atanasova E, Zhang Z. Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing. Proc Natl Acad Sci USA. 2005;102:13410–13415. doi: 10.1073/pnas.0506176102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zhou H, Tarara J, Zhang Z. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. EMBO J. 2007;26:2274–2283. doi: 10.1038/sj.emboj.7601670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Luger K. Dynamic nucleosomes. Chromosome Res. 2006;14:5–16. doi: 10.1007/s10577-005-1026-1. [DOI] [PubMed] [Google Scholar]
- Ma XJ, Wu J, Altheim BA, Schultz MC, Grunstein M. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc Natl Acad Sci USA. 1998;95:6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci USA. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg D, Sims RJ., 3rd de FACTo nucleosome dynamics. J Biol Chem. 2006;281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- Shibahara K, Verreault A, Stillman B. The N-terminal domains of histones H3 and H4 are not necessary for chromatin assembly factor-1-mediated nucleosome assembly onto replicated DNA in vitro. Proc Natl Acad Sci USA. 2000;97:7766–7771. doi: 10.1073/pnas.97.14.7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986;45:555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3–K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- Tyler JK, Collins KA, Prasad-Sinha J, Amiott E, Bulger M, Harte PJ, Kobayashi R, Kadonaga JT. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol Cell Biol. 2001;21:6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDemark AP, Blanksma M, Ferris E, Heroux A, Hill CP, Formosa T. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol Cell. 2006;22:363–374. doi: 10.1016/j.molcel.2006.03.025. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- Zhou H, Madden BJ, Muddiman DC, Zhang Z. Chromatin assembly factor 1 interacts with histone h3 methylated at lysine 79 in the processes of epigenetic silencing and DNA repair. Biochemistry. 2006;45:2852–2861. doi: 10.1021/bi0521083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.