Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development (original) (raw)
Abstract
The onset of pancreas development in the foregut endoderm is marked by activation of the homeobox gene Pdx1 (IPF1). Pdx1 is essential for the expansion of the pancreatic primordium and the development of endocrine islets. The control of Pdx1 expression has been only partially elucidated. We demonstrate here that the winged-helix transcription factors Foxa1 and Foxa2 co-occupy multiple regulatory domains in the Pdx1 gene. Compound conditional ablation of both Foxa1 and Foxa2 in the pancreatic primordium results in complete loss of Pdx1 expression and severe pancreatic hypoplasia. Mutant mice exhibit hyperglycemia with severely disrupted acinar and islet development, and die shortly after birth. Assessment of developmental markers in the mutant pancreas revealed a failure in the expansion of the pancreatic anlage, a blockage of exocrine and endocrine cell differentiation, and an arrest at the primitive duct stage. Comparing their relative developmental activity, we find that Foxa2 is the major regulator in promoting pancreas development and cell differentiation. Using chromatin immunoprecipitations (ChIP) and ChIP sequencing (ChIPSeq) of fetal pancreas and islet chromatin, we demonstrate that Foxa1 and Foxa2 predominantly occupy a distal enhancer at −6.4 kb relative to the transcriptional start site in the Pdx1 gene. In addition, occupancy of the well-characterized proximal Pdx1 enhancer by Foxa1 and Foxa2 is developmental stage-dependent. Thus, the regulation of Pdx1 expression by Foxa1 and Foxa2 is a key early event controlling the expansion and differentiation of the pancreatic primordia.
Keywords: Pancreas, development, Pdx1, Foxa1, Foxa2, conditional gene ablation
Mouse pancreas development begins on embryonic day 8.5–9.0 (E8.5–E9.0) when two epithelial buds emerge from the ventral and dorsal surfaces of the posterior foregut endoderm. These rudimentary buds undergo branching morphogenesis to form a ductal tree consisting primarily of undifferentiated ductal epithelia or pancreatic cords. By E13.5–E14.5, intensive epithelial cell proliferation and differentiation initiates in what has been termed the “secondary transition,” and by E16.5, exocrine acinar cells separate from the central ducts while endocrine cells begin to cluster into islet-like structures. Additional islet remodeling and maturation is completed 3 wk after birth, resulting in a mature functional pancreas (Jorgensen et al. 2007).
These early morphogenetic processes coincide with extensive migration and differentiation of epithelial cell lineages, whose fate can be traced through the dynamic expression of several key transcription factors. For instance, the initial expression of Pdx1 (E8.5–E9.0) highlights the prospective pancreatic domain even before the pancreatic buds can be distinguished morphologically. The initial broad expression of Pdx1 becomes restricted to differentiated β- and δ-cells upon the completion of the secondary transition (Guz et al. 1995). Lineage tracing studies have shown that Pdx1Cre+ precursors give rise to all three epithelial cell lineages (ductal, exocrine, and endocrine) of the pancreas (Gu et al. 2003), consistent with the finding that Pdx1 ablation causes pancreatic agenesis (Jonsson et al. 1994; Offield et al. 1996).
Like Pdx1, Ptf1a+ precursor cells contribute to all three cell lineages; however, its expression becomes restricted to acinar progenitor cells by E13.5 (Kawaguchi et al. 2002), consistent with the total absence of acinar cells in the _Ptf1a_-deficient pancreas (Krapp et al. 1998). In contrast, Ngn3 specifically marks islet precursor cells (Apelqvist et al. 1999; Schwitzgebel et al. 2000; Gu et al. 2002), and the _Ngn3_-null pancreas does not develop endocrine cells (Gradwohl et al. 2000). Once committed to the endocrine fate, the final differentiation of individual hormone-producing cells (α, β, δ, ε, and PP cells) is dependent on a number of transcription factors, including members of the paired-box transcription factor family (Pax), the NK2 homeoprotein transcription factor (Nkx), the forkhead box transcription factors, and the Maf transcription factors (Lee et al. 2002, 2005; Habener et al. 2005;Vatamaniuk et al. 2006; Murtaugh 2007).
Although multiple DNA-binding proteins have been shown to participate in the elaboration of pancreatic cell types, the transcriptional events that govern the earliest steps in organ development remain to be fully elucidated. Recent studies have defined the _cis_-regulatory elements of Pdx1 that are critical for the maintenance of precise Pdx1 expression levels and thus for pancreas development (Fujitani et al. 2006). This essential enhancer of the Pdx1 gene, termed “Area I–II–III,” harbors binding sites for multiple _trans_-activators including Foxa2 (Wu et al. 1997; Gerrish et al. 2000, 2001; Samaras et al. 2002, 2003; Fujitani et al. 2006; Wiebe et al. 2007; Vanhoose et al. 2008). Transgenic rescue further confirmed that this enhancer element is sufficient to direct Pdx1 expression and restore pancreas development and islet maturation in Pdx1−/− mice (Gannon et al. 2001; Boyer et al. 2006). In contrast to this principal control region located between −2800 and −1600 base pairs (bp), the contribution of a highly conserved distal enhancer element between −6530 and −6045 bp, termed “Area IV,” is less clear (Gerrish et al. 2004). Independent of Area I–II–III, this distal enhancer is capable of directing pancreatic β-cell-selective reporter gene expression and potentiating the proximal enhancer activity (Sharma et al. 1997; Gerrish et al. 2004).
In vitro Foxa2 binding to these Pdx1 enhancers has been studied by several groups (Sharma et al. 1997; Wu et al. 1997; Gerrish et al. 2000, 2004; Ben-Shushan et al. 2001). To evaluate the importance of the suggested regulatory hierarchy between Foxa2 and Pdx1 in vivo, we previously used endoderm-specific or β-cell-specific Foxa2 mutants (Foxa2loxP/loxP,Foxa3Cre; Foxa2loxP/loxP, InsCre) mice. Foxa2 deficiency resulted in the absence of mature α cells and a reduction of Pdx1 expression and β-cell differentiation (Lee et al. 2002, 2005). Given that Foxa1 is a close homolog to Foxa2 and contains an almost identical DNA-binding domain (Lai et al. 1991; Kaestner et al. 1994), we hypothesized that this factor may also participate in the regulation of Pdx1, and thus control pancreas development.
Here we report that, using a novel Foxa1 loxP allele, removal of both Foxa1 and Foxa2 from the pancreatic primordia causes near total pancreatic agenesis and loss of Pdx1 expression. Both Foxa factors predominantly occupy the distal Pdx1 enhancer during pancreas development, with an increase in Foxa2 binding to both enhancers during development. These data establish that Foxa1 and Foxa2 act upstream of Pdx1 in the regulatory hierarchy governing pancreatic development.
Results
The Pdx1 enhancer is bound by both Foxa1 and Foxa2 in vivo
Foxa1 is coexpressed with Foxa2 in the foregut endoderm from which the pancreatic buds are derived (Monaghan et al. 1993). Both genes are activated during early pancreas development, with highest levels in mature islets (Supplemental Fig. 1A). Given the high degree of sequence similarity between mouse Foxa1 and Foxa2, especially in the DNA-binding domain (Lai et al. 1991; Kaestner et al. 1994), we hypothesized that Foxa1 binds to the previously described Foxa2 sites within the regulatory regions of the Pdx1 gene (Area I–II–III; Supplemental Fig. 1B) (Wu et al. 1997; Gerrish et al. 2000; Ben-Shushan et al. 2001), and both factors participate in regulating Pdx1 expression in the pancreatic anlage. We performed chromatin immunoprecipitation (ChIP) assays with antibodies specific to either Foxa1 or Foxa2 on chromatin isolated from primary mouse islets. Both Foxa1 and Foxa2 bound to the Area I–II–III enhancer of Pdx1 in vivo (Supplemental Fig. 1C).
Derivation of FoxalloxP mice
Foxa1−/− mice elaborate a morphologically normal pancreas, but exhibit reduced Gcg (glucagon) transcript levels and die shortly after birth (Kaestner et al. 1999; Shih et al. 1999). These Foxa1−/− mice also have defects in β-cell function (Vatamaniuk et al. 2006) and in nonpancreatic tissues (Behr et al. 2004; Gao et al. 2005; Wan et al. 2005; Ferri et al. 2007). In order to study pancreas development without confounding effects from other tissues, we designed a novel loxP conditional allele for Foxa1. A targeting vector, engineered to flank exon 2 of Foxa1 with a single loxP site and a FRT-PGKneo-FRT-loxP cassette (Supplemental Fig. 2A), was electroporated into embryonic stem (ES) cells. Correctly targeted ES clones were identified by Southern blot with a 5′ external probe (Supplemental Fig. 2B) and expanded for blastocyst injection. One of the resulting chimeric males transmitted the loxP allele through the germ line. F1 offspring were bred to Flp1 “deleter” mice (Rodriguez et al. 2000), which express the Flp recombinase enzyme ubiquitously, for removal of the FRT-flanked PGKneo cassette (Supplemental Fig. 2A). Foxa1loxP/loxP mice (Supplemental Fig. 2C), obtained by intercrossing of Foxa1loxP/+ mice, were healthy and fertile, confirming that the Foxa1loxP allele is functionally wild type. The subsequent Cre-mediated excision of the loxP-flanked target resulted in a _Foxa1_-null allele that lacks the coding sequence for the entire DNA-binding domain.
Severe pancreatic hypoplasia in compound Foxa1 and Foxa2 mutants
To investigate whether Foxa1 and Foxa2 cooperate in early pancreas development, we derived Foxa1loxP/loxP, Foxa2loxP/loxP,Pdx1CreE mice using the Foxa1loxP allele described above and the previously reported Foxa2loxP (Sund et al. 2000) and Pdx1CreEarly mice (referred to as Pdx1CreE throughout) (Gu et al. 2002; Heiser et al. 2006). Surprisingly, Foxa1loxP/loxP,Pdx1CreE mice (n = 20) and Foxa1loxP/loxP,Foxa2loxP/+,Pdx1CreE mice (n = 61) were not only viable and fertile, but also euglycemic (Supplemental Fig. 2D). Given that the Pdx1CreE transgene mediates gene ablation in the entire endocrine pancreas including α cells (Gu et al. 2002), this finding demonstrates that the neonatal lethality of Foxa1−/− mice is the consequence of nonpancreatic defects (Kaestner et al. 1999; Behr et al. 2004). In contrast, Foxa1loxP/loxP, Foxa2loxP/loxP,Pdx1CreE mice (n = 37) were severely hyperglycemic (Supplemental Fig. 2D) and died within the first 2 d of life. Interestingly, Foxa1loxP/+,Foxa2loxP/loxP, Pdx1CreE mice (n = 8) also died before postnatal day 5.
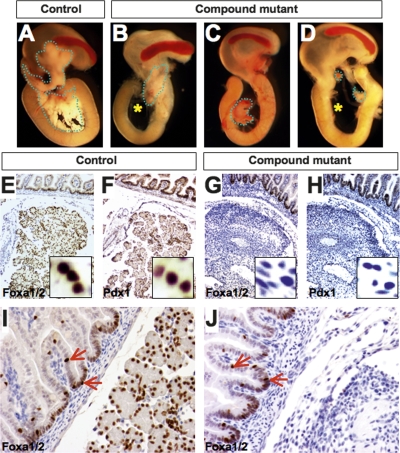
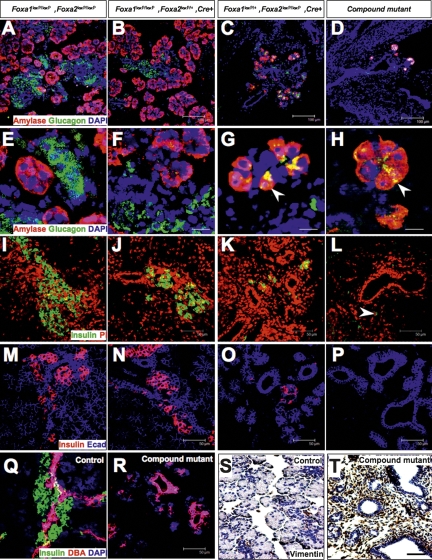
In Foxa1loxP/loxP,Foxa2loxP/loxP,Pdx1CreE mice (referred to as “compound mutants” below), Cre recombinase initiated deletion of both genes on E9.0. When we analyzed embryos at late gestation (E18.5), only minimal pancreatic tissue remained in the compound mutant mice, in contrast to their control littermates (Fig. 1A–D). This dramatic phenotype exhibited variable penetrance in mutant mice, affecting either the ventral or dorsal pancreas (Fig. 1B,C), or both (Fig. 1D). This severe pancreatic hypoplasia explains the early postnatal death of Foxa1/Foxa2 compound mutant mice. As expected, no Foxa1 or Foxa2 protein was detected by immunohistochemistry in the remaining pancreatic tissue recovered from compound mutants at E18.5 (Fig. 1E,G), while a concomitant loss of Pdx1 protein in these mutant tissues was revealed in adjacent sections (Fig. 1F,H). In contrast to the pancreas, the duodenum developed normally in compound mutants. Equivalent levels of Foxa1/Foxa2 proteins were detected by immunohistochemistry in control and mutant duodenal epithelia using an antibody that recognizes both Foxa1 and Foxa2, and the distribution of Foxa1/Foxa2+ cells within the duodenal epithelia was also identical (Fig. 1I,J). Because Pdx1CreE expression in the duodenum is mosaic, this is either because the transgene did not result in Foxa1/Foxa2 deletion in this tissue or because _Foxa1/Foxa2_-null cells were replaced by wild-type cells.
Figure 1.
Conditional ablation of Foxa1 and Foxa2 in the pancreatic primordia causes pancreatic hypoplasia. (A) Control E18.5 pancreas, outlined by the dotted line, is located between the stomach and duodenum. (B–D) Foxa1 and Foxa2 compound mutant pancreata were severely hypoplastic, with variable penetrance of the affected pancreatic lobes. Asterisks indicate the complete absence of the ventral pancreas. (E–H) Adjacent sections of control and compound mutant pancreas (E18.5) were stained for Foxa1/2 and Pdx1, with all three proteins being absent in the mutant pancreas. (I,J) Foxa1/Foxa2 expression in duodenal epithelia of the compound mutants was unaffected.
Decreased Pdx1 in the pancreatic buds of Foxa1/Foxa2 compound mutants
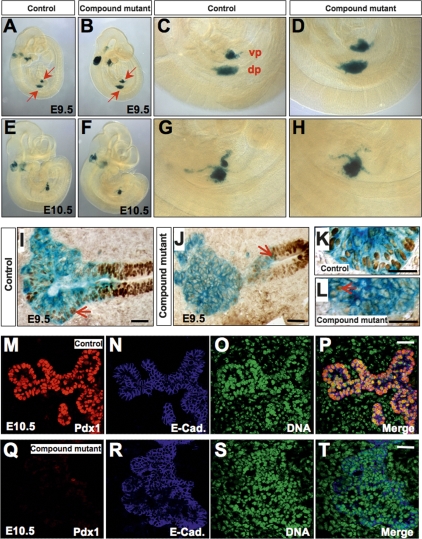
Even though Pdx1 is a master regulator of pancreas formation, and Pdx1 mutant mice or people lack a mature pancreas (Jonsson et al. 1994; Stoffers et al. 1997), pancreas development is initiated in Pdx1−/− mice, and a small pancreatic rudiment is present (Offield et al. 1996). We examined pancreatic bud formation in Foxa1/Foxa2 compound mutant animals utilizing the Rosa26R reporter line, which marks those cells in which the Pdx1CreE has been active. Both dorsal and ventral pancreatic buds formed (Fig. 2A–D, E9.5) and initially expanded (Fig. 2E–H, E10.5) in compound mutant mice. Sections of these β-gal-stained embryos were analyzed by immunohistochemistry for Foxa1/2. In contrast to control pancreatic buds (Fig. 2I), where nuclear Foxa1/Foxa2 was detected in both the foregut epithelium and the growing tips of the pancreatic primordia at E9.5 (Fig. 2I,K), a vast majority of the cells in the compound mutant pancreas were Foxa1/Foxa2-negative (Fig. 2J,L), except for a few scattered cells in the pancreatic anlage that retained nuclear staining (Fig. 2L, arrow). The gut epithelium of the mutant embryos was clearly Foxa1/Foxa2-positive (Fig. 2J).
Figure 2.
Loss of Pdx1 expression in the pancreatic buds of compound mutant mice. (A–D) The pancreatic domain marked by the Pdx1CreE transgene was detected by β-galactosidase staining after introgression of the Rosa26R reporter allele into our Foxa1loxP/loxP,Foxa2loxP/loxP,Pdx1CreE mice. Both ventral and dorsal pancreatic buds (blue staining, arrows) were visible in compound mutant mice at E9.5. (E–H) These mutant buds expanded initially at E10.5. (I) As the pancreatic buds evaginated from the gut epithelium, Foxa1/2 expression was seen in a majority of control pancreatic precursor cells as well as in the gut epithelium. The arrow points to the pancreatic bud, which is positive for nuclear Foxa1/2. (J) Foxa1/2 expression was decreased in the pancreatic anlage of compound mutant mice at E9.5. (K) Higher magnification of I. (L) In compound mutant pancreatic buds, only scattered single cells remained Foxa1/a2-positive (arrow). (M–T) Confocal fluorescent staining demonstrates that Pdx1 expression is extinguished in the compound mutant pancreatic anlage on day E10.5. Control pancreatic epithelial cells strongly express Pdx1. E-cadherin staining indicates the extent of the pancreatic epithelium. Bars, 25 μm.
Pdx1 expression was reduced in compound mutant pancreatic anlagen compared with control sections as early as E9.5 (Supplemental Fig. 3A,B). At E10.5, the growing pancreatic epithelial buds in control mice strongly expressed Pdx1 (Fig. 2M–P), while Pdx1 expression in the mutant epithelial cords was almost completely extinguished (Fig. 2Q–T). Thus, early pancreatic primordia in compound mutant mice formed with a normal number of Pdx1+ precursor cells in which Foxa1 and Foxa2 had not yet been deleted, but by E10.5 the Pdx1CreE transgene efficiently removed both Foxa genes, resulting in a loss of Pdx1 expression in the developing pancreas.
Pancreatic precursor cells fail to expand in _Foxa1/Foxa2-_deficient buds
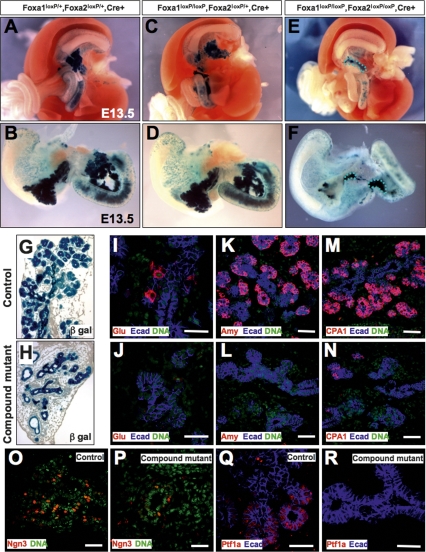
During early pancreas development, Pdx1-expressing precursor cells expand to form a branched system of epithelial cords. Outgrowth and differentiation of exocrine and endocrine progenitors from these primitive cords initiates around E13.5. We observed near normal amounts of pancreas tissue, as indicated by β-gal activity, in Foxa1loxP/+,Foxa2loxP/+,Pdx1CreE,Rosa26R mice at E13.5 (Fig. 3A,B). Interestingly, mice with only one remaining wild-type Foxa2 allele (Foxa1loxP/loxP, Foxa2loxP/+,Pdx1CreE) developed normal dorsal and ventral pancreata (Fig. 3C,D). In contrast, the pancreas anlage of compound mutant mice failed to expand (Fig. 3E,F).
Figure 3.
Pancreatic precursors fail to expand in Foxa1 and Foxa2 compound mutants. (A–F) Whole-mount gut tissues were dissected from E13.5 embryos of the indicated genotypes. Dorsal and ventral pancreata developed in Foxa2loxP/+,Foxa2loxP/+, Pdx1CreE and Foxa2loxP/loxP,Foxa2loxP/+, Pdx1CreE mice as indicated by β-galactosidase activity, but failed to grow in compound mutant mice (dotted lines). (G,H) E13.5 control pancreas formed a ductal epithelial tree with abundant exocrine structures visible at the ductal tips. Compound mutant pancreata consisted primarily of ducts. (I–R) Control and compound mutant sections were stained with glucagon (I,J), amylase (K,L), CPA1 (M,N), Ngn3 (O,P), and Ptf1a (Q,R) antibodies in red, and counterstained for E-cadherin (blue) or DNA (green). Bars, 25 μm.
At E13.5, the control pancreas formed a ductal tree (Fig. 3G) concomitant with epithelial cell differentiation and proliferation. In contrast, the compound mutant pancreas contained epithelial cords without clear exocrine structure (Fig. 3H). This lack of morphological transformation in mutant tissue was supported by the scarcity of immature endocrine and exocrine cells marked by glucagon (Fig. 3J) and amylase (Fig. 3L) antibodies, while an abundance of glucagon+ and amylase+ cells were detected in the control pancreas (Fig. 3I,K). In addition, Cpa1+ cells, which form the multipotent progenitor domains at the tips of the ductal tree at this stage (Fig. 3M; Zhou et al. 2007), were very rare in the compound mutant pancreas (Fig. 3N).
Using antibodies against Ngn3 and Ptf1a, known lineage determinants for endocrine and exocrine cells, respectively, we observed abundant Ngn3+ cells in the control ductal trunk (Fig. 3O), and Ptf1a+ cells in emerging acini (Fig. 3Q). In sharp contrast, compound mutant pancreata contained only a few cells expressing either factor (Fig. 3P,R). To quantify this reduction, we counted Ngn3+ and Ptf1a+ cells that were colabeled by β-gal staining in the control and mutant pancreas (Supplemental Fig. 3). This analysis revealed an 84% and 93% reduction of Ngn3+ and Ptf1a+ cells, respectively, in compound mutants (Supplemental Fig. 3M). Thus, the compound mutant pancreatic epithelial cords failed to elaborate sufficient numbers of endocrine and exocrine progenitors, resulting in severely reduced numbers of glucagon+ and amylase+ cells.
Foxa1 and Foxa2 are not equivalent in promoting pancreas development
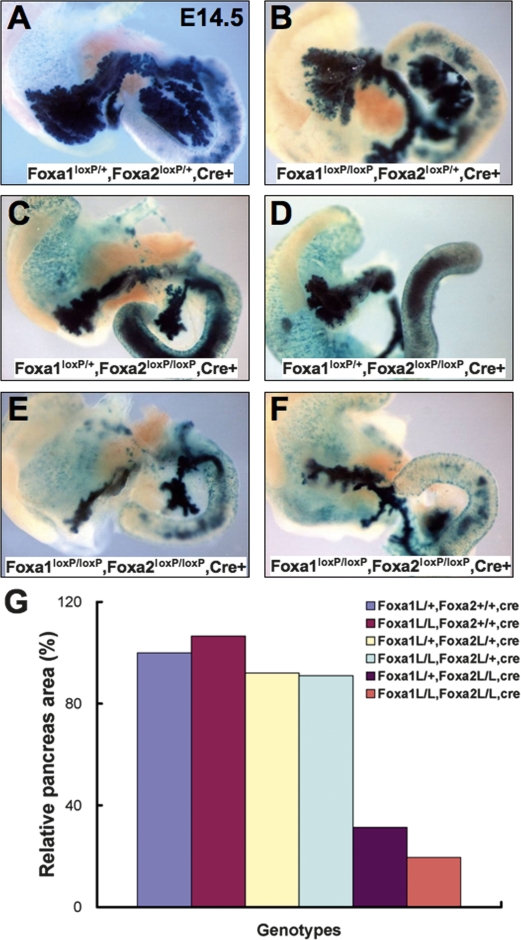
In order to determine whether Foxa1 and Foxa2 are completely interchangeable in pancreas formation, we analyzed E14.5 mouse embryos with various combinations of Foxa1 and Foxa2 alleles. Interestingly, when one or both Foxa2 wild-type alleles were present (Fig. 4A,B), the number of wild-type Foxa1 alleles did not have an impact on pancreatic size. In other words, one copy of the Foxa2 gene was sufficient to induce pancreas formation. When both Foxa2 alleles were removed, however, a clear reduction in pancreatic mass was seen (Fig. 4C–F). Importantly, in this scenario, Foxa1 gene copy number became a determinant of the size of pancreatic lobes (Fig. 4, cf. C and E, and D and F). This was reflected by the relative area of the pancreas: 31% of control in Foxa1loxP/+,Foxa2loxP/loxP,Pdx1CreE mice, compared with 19% in compound mutant mice (Fig. 4G). While these data demonstrate that Foxa2 is more potent in promoting pancreas growth than Foxa1, Foxa1 is also a positive regulator of pancreas development. In addition, although both pancreatic lobes were affected in these mutant mice, the ventral pancreas was more sensitive to Foxa deficiency than the dorsal pancreas (Fig. 4D,F).
Figure 4.
Foxa1 and Foxa2 regulate pancreas growth with different potency. (A–F) Pancreas development in E14.5 mice with various combinations of Foxa1 and Foxa2 alleles, in mice with introgressed Rosa26R reporter allele. (A,B) One wild-type Foxa2 allele is sufficient to specify a nearly normal-size pancreas. Severe hypoplasia was seen in Foxa2loxP/+,Foxa2loxP/loxP,Pdx1CreE mice (C,D) and compound mutants (E,F), with the ventral pancreas being affected more severely (D,F). (G) The extent of the pancreas, as marked by β-galactosidase activity of the Rosa26R reporter present in all embryos, was quantified in mice with indicated Foxa1/Foxa2 genotypes.
The notion of differential competency of the two Foxa genes was supported by our analysis of late gestation pancreata. E17.5 mice with only one remaining wild-type _Foxa_2 allele (Foxa1loxP/loxP,Foxa2loxP/+,Pdx1CreE) specified similar numbers of exocrine and endocrine cells as control pancreata (Fig. 5A,B). In contrast, a dramatic reduction in both cell types was seen in the Foxa1loxP/+,Foxa2loxP/loxP,Pdx1CreE pancreas (Fig. 5C). However, when compared with the Foxa1/Foxa2-devoid pancreas, where amylase+ or glucagon+ cells were rarely seen (Fig. 5D), one copy of Foxa1 was sufficient to specify a limited number of either cell type (Fig. 5C). Interestingly, while amylase+ and glucagon+ cells were clearly separated from each other in control pancreata, with amylase+ cells spread peripherally and glucagon+ cells occupying the central core (Fig. 5E,F), in Foxa1loxP/+,Foxa2loxP/loxP,Pdx1CreE and compound mutants, some double-positive (amylase+/glucagon+) cells were observed (Fig. 5G,H; Supplemental Fig. 4).
Figure 5.
Disrupted pancreatic differentiation in compound mutants. (A–R) Indirect immunofluorescent staining of E17.5 pancreas sections from mice with the indicated genotypes for amylase and glucagon (A–H), insulin (I–L), E-cadherin and insulin (M–P), DBA and insulin (Q–R), or vimentin (S–T) antibodies and visualized by confocal microscopy. Arrows in G and H point to apparently double-positive amylase+/glucagon+ cells, and the optical section depth in these micrographs is 0.7 μm. Arrow in L points to a single insulin+ cell in the compound mutant pancreas. Bars: A–D, 100 μm; E–H, 17.5 μm; I–T, 50 μm.
Likewise, one copy of Foxa2 was sufficient to specify a nearly normal complement of insulin+ β cells as compared with control mice (Fig. 5I,J). In contrast, the number of β cells was greatly reduced in Foxa1loxP/+, Foxa2loxP/loxP,Pdx1CreE mice (Fig. 5K,O). Very few β cells were present in compound Foxa1/Foxa2 mutant pancreata from E17.5 (Fig. 5L, arrow) through postnatal day 1 (data not shown). Accompanying the reduced endocrine cell mass, we noted a loss of total epithelial tissue in compound mutant pancreata (Fig. 5O,P), as indicated by E-cadherin staining (Fig. 5M–P). In fact, the remaining epithelial cells in the compound mutant pancreas were almost exclusively ductal, as they were strongly positive for DBA lectin (Fig. 5R). We detected only scattered single δ cells and ε cells in sections of compound mutant pancreas, demonstrating that α and β cells had not been reallocated to other lineages in Foxa1/_Foxa2-_deficient pancreas (data not shown). Finally, we found that the spaces between the remaining ductal epithelia in the compound mutant pancreas were filled with stromal tissue, including fibroblasts and smooth muscle cells, as identified by vimentin (Fig. 5S,T) and smooth muscle actin (data not shown) staining. Thus, compound loss of Foxa1 and Foxa2 arrested pancreas growth at the early pancreatic cord stage.
Foxa1/Foxa2 are required for the expression of several endocrine transcriptional regulators
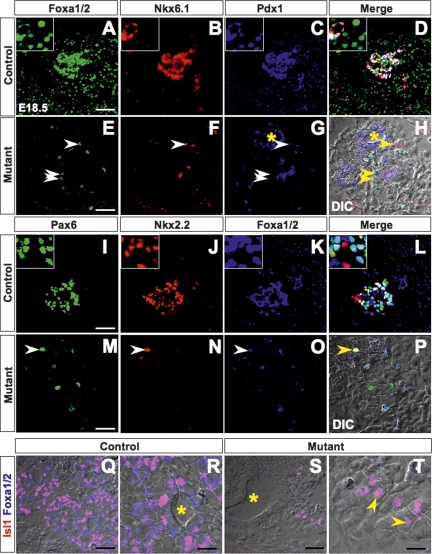
Disrupting either Nkx6.1 or Nkx2.2 affects β-cell development (Sussel et al. 1998; Sander et al. 2000; Wang et al. 2004), while abolishing Pax6 primarily inhibits α-cell differentiation (Sander et al. 1997). None of these mutants demonstrates a phenotype with a severity comparable with that of _Pdx1_-null (Jonsson et al. 1994; Offield et al. 1996), or Foxa1/2 compound mutant pancreata. We examined the impact of Foxa1 and Foxa2 compound mutation on several early transcription factors shown to be critical for islet development. Although these key regulators were present in control endocrine compartments with expression patterns consistent with the literature (Fig. 6A–D, I–L), they were largely depleted in the Foxa1/Foxa2 compound mutant pancreas (Fig. 6E–H, M–P). This was illustrated by costaining for these factors with Foxa1/2, which were expressed in all control pancreatic epithelial cells (Fig. 6A,K) but only in scattered compound mutant cells (Fig. 6E,O, arrows). These single Foxa1/2-positive cells had apparently escaped Cre-mediated gene excision, and coexpressed Nkx6.1 (Fig. 6F), Pdx1 (Fig. 6G), Pax6 (Fig. 6M), and Nkx2.2 (Fig. 6N). In addition, we analyzed the expression of Isl1, which has been shown to be essential for formation of the dorsal pancreas (Ahlgren et al. 1997). In contrast to controls (Fig. 6Q,R), we found a dramatic reduction in the number of Isl1-expressing cells in compound mutant tissues (Fig. 6S,T), where the majority of the remaining Isl1-positive cells coexpressed Foxa1/2 protein in their nuclei. Thus, disruption of Foxa1 and Foxa2 caused pancreatic tissue loss with a concomitant reduction of cells expressing key transcriptional regulators.
Figure 6.
The impact of Foxa1 and Foxa2 ablation on the pancreatic transcription factor network. E18.5 pancreas sections from control and compound mutant mice were stained with Foxa1/2, Nkx6.1, and Pdx1 (A–H); Pax6, Nkx2.2, and Foxa1/2 (I–P); or Isl1 and Foxa1/2 (Q–T) antibodies by indirect immunofluorescence and visualized by confocal microscopy. Arrows, in compound mutant sections, point to remaining Foxa1/2-positive cells that coexpress additional pancreatic transcription factors. Asterisks in G, H, R, and S mark the ducts. H, P, and Q–T are bright-field images or merged images including bright-field images. Bars: R,T, 17.5 μm; all others, 30 μm.
Foxa occupancy of the distal and proximal Pdx1 enhancers
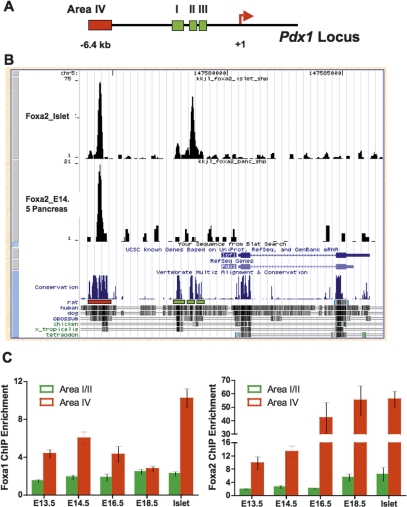
Both Foxa1 and Foxa2 bind to the proximal enhancer Area I–II–III, in the Pdx1 gene in mature pancreatic islets (Supplemental Fig. 1). When Area I–II–III is deleted by gene targeting, the resulting mice display severe pancreatic hypoplasia; however, they develop a larger pancreas than our Foxa1/Foxa2 compound mutants, with ample exocrine tissue and even some endocrine cells remaining (Fujitani et al. 2006). Combined with our finding that Foxa1/_Foxa2-_deficient cells extinguish Pdx1 expression completely, we hypothesized that Foxa1 and Foxa2 may bind to additional enhancer elements in the regulatory regions of Pdx1 that were not deleted by Fujitani et al. (2006). To address this question, we performed ChIP sequencing (ChipSeq) experiments using a Foxa2-specific antibody on chromatin isolated from E14.5 wild-type fetal pancreas and primary islets, respectively. We chose E14.5 pancreas, since at this stage all acinar cells continue to express Pdx1, and Pdx1-expressing progenitor cells are still present in large numbers in the central epithelial cords. Alignment of the sequence tags obtained to the Pdx1 locus revealed that in the fetal pancreas, Foxa2 does not bind effectively to the well-characterized Area I–II–III enhancer, but rather predominantly to Area IV farther upstream (Fig. 7A,B). This enhancer element, located 6.4 kb upstream of the transcription start site, is highly conserved among different animal species (Fig. 7B; Gerrish et al. 2004). Notably, the binding of Foxa2 to Area IV or Area I–II–III is significantly enhanced in pancreatic islets compared with fetal pancreas (Fig. 7B).
Figure 7.
Occupancy of Pdx1 enhancers by Foxa1/Foxa2 increases with developmental time. (A) Schematic diagram of the Pdx1 locus indicating the distal (Area IV), and proximal (Area I–II–III) enhancer. (B) Foxa2 ChipSeq demonstrates significant occupancy of Area IV in E14.5 mouse fetal pancreas, while increased occupancy of Area I–II–III is seen in pancreatic islets. Area IV (red box) is evolutionarily conserved as illustrated by multispecies sequence alignment. (C) ChIP experiments using chromatin from the developing pancreas or mature islets demonstrate preferential Foxa1/Foxa2 occupancy of Area IV. Error bars represent SEM. Each ChIP was performed on chromatin samples from two to six biological replicates, and enrichment of both enhancers in the ChIP samples compared with input chromatin was statistically significant (P < 0.05) for all time points and both antibodies analyzed, with the exception of binding of either Foxa1 or Foxa2 to Area I–II in mature islets, where the _P_-value was <0.06 (Student’s _t_-test).
Real-time PCR with primers specific for Area IV confirmed our ChipSeq data, demonstrating an average enrichment of 13-fold and 56-fold in Foxa2 immunoprecipitated DNA compared with input DNA in E14.5 pancreas and adult pancreatic islets, resepectively (Fig. 7C). In contrast, the average ChIP enrichments for Area I–II were only 2.3-fold and 6.4-fold in E14.5 pancreas and islet, respectively (Fig. 7C). Foxa2 ChIP of pancreatic chromatin samples from various developmental stages demonstrated that Foxa2 occupancy of Area I–II increases with age (Fig. 7C). Although the binding of Foxa2 to Area IV is always greater than to Area I–II (Fig. 7C), it is important to note that the Foxa2 ChIP enrichment for Area I–II is statistically significant compared with input throughout all fetal stages tested (P < 0.05).
ChIP assay using a Foxa1-specific antibody revealed that the binding affinity of this protein for Area IV is also greater than that for Area I–II, with maximal occupancy in mature islets (Fig. 7C). In contrast, Foxa1 shows near constant binding to Area I–II throughout pancreas development (Fig. 7C). An overall lower ChIP enrichment was seen from Foxa1ChIP experiment compared with Foxa2 ChIP, which could either reflect the relative abundance or intrinsic affinity of the two proteins, or differences in the efficacy of the two antibodies used. Thus, while we are able to analyze the developmental time course of enhancer occupancy for each factor individually during pancreatic development, we cannot assess which of the two proteins occupies more binding sites in vivo.
The strong binding of both Foxa1 and Foxa2 to the Area IV enhancer in early pancreas development supports the observation that this enhancer alone is capable of directing Pdx1 expression (Sharma et al. 1997; Gerrish et al. 2004). This finding also explains why the phenotype of the Area I–II–III enhancer mutant described by Fujitani et al. (2006) is milder than the one we describe in our compound mutants. Our data demonstrate that differential occupancy of various _cis_-regulatory elements in the Pdx1 gene by Foxa1 and Foxa2 is dependent on developmental time, adding a further dimension to the regulatory properties of these transcription factors.
Discussion
Signals from the adjacent mesoderm initiate pancreas formation (Moriya et al. 2000; Stafford and Prince 2002; Chen et al. 2004; Martin et al. 2005; Molotkov et al. 2005), while signals from endothelial cells promote pancreatic expansion (Lammert et al. 2001, 2003). In addition to these extrinsic signals, we demonstrated here that the involvement of early intrinsic lineage determinants, inherited by the pancreas from early endoderm, is essential in orchestrating pancreatic organogenesis. Our data extend and complement the finding that targeted deletion of the proximal Pdx1 enhancer (Area I–II–III) causes pancreatic hypoplasia (Fujitani et al. 2006). We demonstrate that Foxa1 and Foxa2 are the essential transcription factors required for Pdx1 expression and pancreas growth in vivo.
Foxa proteins regulate Pdx1 expression and pancreas development
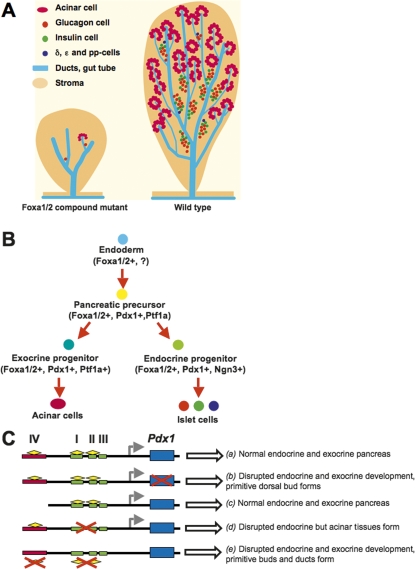
We used the Pdx1CreE mouse line to inactivate Foxa genes in early pancreatic primordia. The endogenous Pdx1 gene is initially activated along with the Cre transgene in the prospective pancreatic domains of Foxa1loxP/loxP, Foxa2loxP/loxP,Pdx1CreE mice. This was evident, as both ventral and dorsal pancreatic buds formed in the compound mutant mice and transiently expressed Pdx1. During the primary transition these precursor cells gave rise to daughter cells in the primitive pancreatic ducts. The remaining Pdx1 protein in these early ductal epithelia likely reflects the time required for the deletion of the four Foxa1/2 gene alleles, the degradation of Foxa1/2 proteins carried over from the endoderm parental cells, the loss of Pdx1 gene transcription, and the decay of existing Pdx1 protein. Thus, the variable penetrance of the mutant phenotype described above may reflect slight variations in the timing of Foxa1/Foxa2 deletion. Nevertheless, Pdx1 expression in exocrine and endocrine cells, as well as the differentiation of these tissues from the ductal epithelia, was completely dependent on Foxa1 and Foxa2. This conclusion is supported by the overall disruption of acinar and endocrine differentiation in Foxa1/Foxa2 compound mutant mice. The immediate consequences of this disrupted differentiation program are the severely reduced exocrine and endocrine progenitor cell populations (Ngn3+ and Ptf1+ cells), which determine the ultimate pancreatic mass. The attenuation of the multipotent progenitor domain was pronounced, consistent with the drastically reduced ductal tip outgrowth and differentiation in the mutant pancreas. Our results are summarized schematically in Figure 8. The near total absence of β cells and other mature exocrine and endocrine cell types (Fig. 8A) represents the cumulative blockage of pancreatic differentiation at multiple transitional steps where Foxa1 and Foxa2 are needed (Fig. 8B). These changes are consistent with the decrease in cells expressing various endocrine transcriptional regulators. Further, the expression of these factors, including Nkx6.1, Nkx2.2, Pax6, Isl1, and Pdx1, in the few cells that retained Foxa1/2 in the mutant pancreas is consistent with a cell-autonomous regulatory effect of the Foxa factors in pancreas differentiation.
Figure 8.
Occupancy of two Pdx1 enhancers by Foxa1 and Foxa2 controls pancreas development. (A) Loss of Foxa1 and Foxa2 prevents the expansion and differentiation of pancreatic progenitor cells, leading to disrupted exocrine and endocrine development. (B) Foxa1 and Foxa2 are among the earliest factors required for the expansion of pancreatic progenitors, the differentiation of endocrine and exocrine compartments, and the maturation of islet cells. (C) Diagram illustrating the cis_-regulatory elements of Pdx1, including the distal enhancer (Area IV; red box), and the proximal enhancer (Area I–II–III; green boxes). (a) In the wild-type embryo, both enhancers, together with key transactivators including Foxa1/Foxa2 (yellow diamonds) direct Pdx1 gene (blue box) expression and normal pancreas development. (b) Ablation of Pdx1 causes abrogation of pancreas development, with only a small dorsal ductule remaining (Jonsson et al. 1994; Offield et al. 1996). (c) A Pdx1 transgene directed by either a 6.2-kb fragment including Area I–II–III and half of Area IV, or a 4.5-kb fragment including only Area I–II–III is capable of rescuing the Pdx1_−/− pancreas phenotype (Gannon et al. 2001; Boyer et al. 2006). (d) Targeted deletion of Area I–II–III leads to severe pancreatic hypoplasia, affecting in particular the endocrine compartment, while acinar tissue does form (Fujitani et al. 2006). (e) Removal of Foxa1 and Foxa2 in the early pancreatic buds, as shown in this study, causes elimination of Pdx1 expression and disrupted exocrine and endocrine pancreas development.
Developmental occupancy of Pdx1 enhancers
Foxa1/Foxa2 compound mutants display a more severe phenotype in exocrine pancreas development than Pdx1ΔI–II–III/ΔI–II–III mice, as acinar cells are abundantly present the latter model (Fujitani et al. 2006). This discrepancy is explained by our discovery that throughout early pancreas development, Foxa1 and Foxa2 predominantly bind to Area IV, a region left intact in the Pdx1ΔI–II–III/ΔI–II–III model. Previous experiments have shown that key pancreatic transcription factors, including Foxa2, Pdx1, and Nkx2.2 are capable of transactivating this enhancer region in vitro (Gerrish et al. 2004). Furthermore, this distal enhancer can independently direct Pdx1 expression and potentiate proximal enhancer activity (Sharma et al. 1997; Gerrish et al. 2004). Thus, in the absence of Area I–II–III, Area IV plus Foxa1/Foxa2 are sufficient to specify some acinar development, while endocrine differentiation is blocked (Fig. 8C; Fujitani et al. 2006).
Although Foxa1 and Foxa2 bind to Area I–II–III with relatively low affinity, both factors occupy this core enhancer element throughout pancreas development and in mature islets. Our data support the importance of Foxa1/Foxa2 occupancy of the Area I–II–III enhancer, as previously demonstrated by in vitro experiments (Gerrish et al. 2000). The transactivation of Area I–II–III by Foxa1/Foxa2 and other factors such as Ptf1a (Wiebe et al. 2007) is sufficient for some level of Pdx1 expression, since a Pdx1 transgene driven by Area I–II–III alone rescued the Pdx1−/− pancreas phenotype (Fig. 8C; Boyer et al. 2006). Our data and previous work (Fujitani et al. 2006) further suggest that endocrine differentiation requires higher levels of Pdx1 expression than acinar development.
Differential contributions of Foxa factors to pancreas development
Our study, utilizing a novel Foxa1 conditional allele, overcomes a major drawback of experiments using Foxa1−/− mice that, in addition to endoderm defects, have neurological and renal phenotypes as well (Behr et al. 2004; Ferri et al. 2007). Unexpectedly, Foxa1loxP/loxP, Pdx1CreE mice did not demonstrate major defects in pancreas development. Most strikingly, Foxa1loxP/loxP, Foxa2loxP/+,Pdx1CreE mice, with only one copy of wild-type Foxa2, developed a relatively normal pancreas with all mature cell types, and were metabolically healthy. In contrast, mice with one allele of wild-type Foxa1 alone failed to thrive. Thus, one allele of Foxa2, but not Foxa1, was sufficient for normal pancreas development and function, suggesting that the previously reported postnatal death of Foxa1−/− mice (Kaestner et al. 1999; Shih et al. 1999) is due to nonpancreatic defects.
The notion that Foxa1 is a positive regulator of pancreas development is supported by the findings that (1) in the absence of both Foxa1 and Foxa2 genes, very few amylase+ or glucagon+ cells, and no β cells were specified; (2) in the absence of Foxa2, one allele of Foxa1 specified more amylase+, glucagon+, and β cells than no Foxa1 alleles; and (3) in the absence of Foxa2, two alleles of Foxa1 specified a pancreas with exocrine and endocrine compartments, albeit with reduced α-cell number (Lee et al. 2005).
We were also able to directly compare the “developmental potency” of Foxa1 and Foxa2 in vivo. While both Foxa proteins can occupy the Pdx1 enhancers, binding by Foxa2 appears to predominate, if one assumes that the efficacy of ChIP with both antibodies is equivalent. This relative comparison to a large extent agrees with our in vivo data as well as previous observations that two copies of the Foxa1 (as in Foxa2loxP/loxP,Foxa3Cre mice) (Lee et al. 2005) promote pancreatic development with fewer α cells than one copy of the Foxa2 gene alone (in Foxa1loxP/loxP, Foxa2loxP/+,Pdx1CreE mice). We conclude that the dynamic regulation of Pdx1 enhancers by Foxa2 and Foxa1 plays a critical role in pancreas development.
Materials and methods
Mice
The Foxa1 loxP targeting vector was constructed by retrieval of a 7.9-kb genomic DNA sequence encompassing the Foxa1 gene from a bacterial artificial chromosome (University of California at Santa Cruz), followed by insertion of a loxP sequence 273 bp upstream, and a FRT-PGKneo-FRT-loxP fragment immediately downstream from exon 2. Homologous recombination in the R1 ES cell line was screened by Southern blot with a 5′-external probe. Wild-type and targeted alleles were identified as 7.4-kb HindIII and 4.0-kb HindIII/SpeI fragments, respectively. Targeted ES cells were injected into C57/BL6 blastocysts, and one of the resulting chimeric males transmitted the loxP allele through the germ line. The offspring were bred to Flp1 deleter mice (Rodriguez et al. 2000) for removal of the FRT-flanked PGKneo cassette, resulting in Foxa1loxP/+ mice identified by genomic PCR with the following primers: forward, 5′-CTGTG GATTATGTTCCTGAT-3′; and reverse, 5′-GTGTCAGGATG CCTATCTGGT-3′. Foxa2loxP/+(Sund et al. 2001), Pdx1CreE (Gu et al. 2002; Heiser et al. 2006), and Rosa26R (Soriano 1999) mice have been described previously. Blood glucose levels were measured as previously described (Gao et al. 2007).
β-galactosidase staining and immunohistochemistry
Embryos or whole-mount pancreata were dissected into fresh PBS, fixed for 1 h in PBS with 1% formaldehyde, 0.2% gluteraldehyde, 2 mM MgCl2, 5 mM EGTA, 0.2% NP-40, and washed in PBS with 0.2% NP-40 three times. Embryos or tissues were then stained overnight at 37°C in a solution containing 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 2 mM MgCl, 0.01% sodium deoxycholate, 0.02% NP-40, and 1 mg/mL X-gal. Embryos were washed three times in PBS with 0.02% NP-40 and post-fixed in 4% paraformaldehyde. Whole embryos or tissues were imaged using a Leica dissecting microscope. To quantify the pancreatic tissue, one representative image was taken for each β-galactosidase-stained whole-mount tissue, and two-dimensional areas of β-galactosidase-positive pancreases were measured by ImageJ 1.38x (National Institutes of Health).
For immunohistochemistry after β-galactosidase staining, embryos or tissues were incubated in 30% sucrose in PBS overnight, embedded in OCT, and cryosectioned. Immunohistochemistry was performed using the Vectastain Elite ABC kit (Vector Laboratories). Primary antibodies used in immunohistochemistry included mouse anti-Ngn3 (1:1000; Developmental Studies Hybridoma Bank at the University of Iowa, F25A1B3), rabbit anti-Ptf1a (1:50000; gift from C. Wright, Vanderbilt University), guinea pig anti-Pdx1 (1:10000; gift from C. Wright), goat anti-Foxa1/2 (1:1000; Santa Cruz Biotechnologies, sc-6553), rabbit anti-carboxypeptidase A (1:1500; AbD Serotec, 1810-0006), rabbit anti-glucagon (predilute; Invitrogen, 080064), and rabbit anti-amylase (1:1000; Sigma, A8273). For quantification, Ngn3+ or Ptf1a+ cells from one section of each five control and five compound mutant embryos were manually counted under a light microscope.
Immunofluorescent analysis
Procedures of confocal immunofluorescent analysis have been described previously (Gao et al. 2007). Primary antibodies not mentioned above included guinea pig anti-insulin (1:1000; Linco), mouse anti-E-cadherin (1:500; BD Biosciences), biotinylated dolichos biflorus agglutinin (1:1000; Vector Laboratories), rabbit anti-Pax6 (1:500; Covance, PRB-278P), mouse anti-Nkx2.2 (1:50; Developmental Studies Hybridoma Bank), mouse anti-Nkx6.1 (1:500; Developmental Studies Hybridoma Bank), and mouse anti-Isl1 (1:50; Developmental Studies Hybridoma Bank).
ChIP
Foxa1 and Foxa2 ChIP were performed as described (Friedman et al. 2004), using antibodies against Foxa1 (Santa Cruz Biotechnologies, sc-22841) and Foxa2 (Santa Cruz Biotechnologies, sc-6554), or Foxa1-specific antiserum (kind gift of G. Schütz, Heidelberg, Germany), and Foxa2-specific antiserum (kind gift of J.A. Whitsett). The ChIP PCR primers spanned the Foxa2-binding sites within Area I–II and IV of the mouse Pdx1 5′ upstream sequence (Wu et al. 1997; Gerrish et al. 2000, 2004) Area1F, TGCCTGCAAAACCACTAAGA; Area1R, GAG GTACCCTGCCTCCTCTC; Area2F, ATGAAGCGTCGAGAT GGAAG; Area2R, CACCCCAGGATGTTTGCTTA; Area 4F, TGCCTCAGTGCCCTTTACTC; Area4R, CTAAGAGTGCT CTGGGCTCTG.
ChIPSeq
Each biological sample was generated from pooled pancreata isolated from 10–15 E14.5 CD1 embryos, or 3000–5000 pancreatic islets. The dissected tissues were cross-linked as described previously (Tuteja et al. 2008). After washing in PBS to remove excess formaldehyde and glycine, the fixed tissues were homogenized in 200 μL of cold whole-cell lysis buffer (10 mM Tris-HCl at pH 8.0, 10 mM NaCl, 3 mM MgCl2, 1% NP-40, 0.1% SDS, 0.5% deoxycholic acid) and protease inhibitors. After incubating on ice for 10 min, lysates were sonicated using a Diagenode Bioruptor (30-sec on/off pulses for 10 min, on high setting). Debris was removed by centrifugation at 13,000_g_ for 10 min, and the supernatant was collected and snap frozen in liquid nitrogen. As input, 10 μL of sonicated chromatin was incubated in PBS with 200 mM NaCl overnight at 65°C, treated with proteinase K and purified with the QIAquick PCR Purification Kit (Qiagen).
Immunoprecipitations were performed as previously described (Tuteja et al. 2008), except that the herring sperm DNA was excluded from the agarose bead-blocking step. For ChIPSeq experiments, the immunoprecipitated DNA was modified for sequencing following the manufacturer’s protocol (Illumina). Briefly, DNAs were blunted with a combination of T4 DNA polymerase, Klenow polymerase, and T4 PNK, then a single 3′-end “A” base was added using Klenow exo (3′-to-5′ exo minus). Adapters provided by Illumina were then ligated to the ends of the modified DNA before size selection of ∼200-bp fragments via PAGE extraction. The isolated DNA samples were used as the template for amplification by 18 cycles of PCR. Amplified products were column purified with the QIAquick PCR Purification Kit (Qiagen) and assayed for quantity and quality with the Agilent 2100 Bioanalyzer (Agilent Technologies). Cluster generation and sequence alignment to the mouse genome (mm8) following pipeline processing were performed following Illumina’s protocol. Only sequence tags uniquely mapping to the Pdx1 locus were considered for this analysis.
Immunoprecipitated DNA was also used to confirm enrichment of target DNA fragments via qPCR. PCR reactions were assembled in triplicate with SYBR GreenER qPCR Supermix (Invitrogen) and run using the SYBR Green (with dissociation curve) program on the Mx3000 Multiplex Quantitative PCR System (Stratagene). The enrichment of target sequences in ChIP material was calculated relative to the myelin basic protein (MBP) locus as a reference for nonspecific binding, and normalized to their relative amplification in input DNA.
Acknowledgments
We thank Dr. Catherine Lee for critical reading the manuscript, Dr. Hong Fu for ES cell work, Sophia Hammani and Elizabeth Helmbrecht for maintaining our mouse colony, Dr. Douglas Melton for kindly providing us with the Pdx1CreE mice, and Dr. Christopher Wright for the Pdx1 and Ptf1a antibodies. We thank Alan Fox, Olga Smirnova, Geetu Tuteja, and Dr. Peter White for help with the ChIPSeq experiment. We are grateful to the Penn Morphology Core (P30DK50306) for tissue embedding and sectioning. This work was supported by NIDDK grants R01-DK055342 and P01-DK049210 to K.H.K. N.G. is supported by a JDRF fellowship (3-2007-521).
Footnotes
References
- Ahlgren U., Pfaff S.L., Jessell T.M., Edlund T., Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- Apelqvist A., Li H., Sommer L., Beatus P., Anderson D.J., Honjo T., de Hrabe Angelis M., Lendahl U., Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Behr R., Brestelli J., Fulmer J.T., Miyawaki N., Kleyman T.R., Kaestner K.H. Mild nephrogenic diabetes insipidus caused by Foxa1 deficiency. J. Biol. Chem. 2004;279:41936–41941. doi: 10.1074/jbc.M403354200. [DOI] [PubMed] [Google Scholar]
- Ben-Shushan E., Marshak S., Shoshkes M., Cerasi E., Melloul D. A pancreatic β-cell-specific enhancer in the human PDX-1 gene is regulated by hepatocyte nuclear factor 3β (HNF-3β), HNF-1α, and SPs transcription factors. J. Biol. Chem. 2001;276:17533–17540. doi: 10.1074/jbc.M009088200. [DOI] [PubMed] [Google Scholar]
- Boyer D.F., Fujitani Y., Gannon M., Powers A.C., Stein R.W., Wright C.V. Complementation rescue of Pdx1 null phenotype demonstrates distinct roles of proximal and distal cis-regulatory sequences in pancreatic and duodenal expression. Dev. Biol. 2006;298:616–631. doi: 10.1016/j.ydbio.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Chen Y., Pan F.C., Brandes N., Afelik S., Solter M., Pieler T. Retinoic acid signaling is essential for pancreas development and promotes endocrine at the expense of exocrine cell differentiation in Xenopus. Dev. Biol. 2004;271:144–160. doi: 10.1016/j.ydbio.2004.03.030. [DOI] [PubMed] [Google Scholar]
- Ferri A.L., Lin W., Mavromatakis Y.E., Wang J.C., Sasaki H., Whitsett J.A., Ang S.L. Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development. 2007;134:2761–2769. doi: 10.1242/dev.000141. [DOI] [PubMed] [Google Scholar]
- Friedman J.R., Larris B., Le P.P., Peiris T.H., Arsenlis A., Schug J., Tobias J.W., Kaestner K.H., Greenbaum L.E. Orthogonal analysis of C/EBPβ targets in vivo during liver proliferation. Proc. Natl. Acad. Sci. 2004;101:12986–12991. doi: 10.1073/pnas.0402875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y., Fujitani S., Boyer D.F., Gannon M., Kawaguchi Y., Ray M., Shiota M., Stein R.W., Magnuson M.A., Wright C.V. Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes & Dev. 2006;20:253–266. doi: 10.1101/gad.1360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon M., Gamer L.W., Wright C.V. Regulatory regions driving developmental and tissue-specific expression of the essential pancreatic gene pdx1. Dev. Biol. 2001;238:185–201. doi: 10.1006/dbio.2001.0359. [DOI] [PubMed] [Google Scholar]
- Gao N., Ishii K., Mirosevich J., Kuwajima S., Oppenheimer S.R., Roberts R.L., Jiang M., Yu X., Shappell S.B., Caprioli R.M., et al. Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development. 2005;132:3431–3443. doi: 10.1242/dev.01917. [DOI] [PubMed] [Google Scholar]
- Gao N., White P., Doliba N., Golson M.L., Matschinsky F.M., Kaestner K.H. Foxa2 controls vesicle docking and insulin secretion in mature β cells. Cell Metab. 2007;6:267–279. doi: 10.1016/j.cmet.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Gerrish K., Gannon M., Shih D., Henderson E., Stoffel M., Wright C.V., Stein R. Pancreatic β cell-specific transcription of the pdx-1 gene. The role of conserved upstream control regions and their hepatic nuclear factor 3β sites. J. Biol. Chem. 2000;275:3485–3492. doi: 10.1074/jbc.275.5.3485. [DOI] [PubMed] [Google Scholar]
- Gerrish K., Cissell M.A., Stein R. The role of hepatic nuclear factor 1 α and PDX-1 in transcriptional regulation of the pdx-1 gene. J. Biol. Chem. 2001;276:47775–47784. doi: 10.1074/jbc.M109244200. [DOI] [PubMed] [Google Scholar]
- Gerrish K., Van Velkinburgh J.C., Stein R. Conserved transcriptional regulatory domains of the pdx-1 gene. Mol. Endocrinol. 2004;18:533–548. doi: 10.1210/me.2003-0371. [DOI] [PubMed] [Google Scholar]
- Gradwohl G., Dierich A., LeMeur M., Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G., Dubauskaite J., Melton D.A. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Gu G., Brown J.R., Melton D.A. Direct lineage tracing reveals the ontogeny of pancreatic cell fates during mouse embryogenesis. Mech. Dev. 2003;120:35–43. doi: 10.1016/s0925-4773(02)00330-1. [DOI] [PubMed] [Google Scholar]
- Guz Y., Montminy M.R., Stein R., Leonard J., Gamer L.W., Wright C.V., Teitelman G. Expression of murine STF-1, a putative insulin gene transcription factor, in β cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development. 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- Habener J.F., Kemp D.M., Thomas M.K. Minireview: Transcriptional regulation in pancreatic development. Endocrinology. 2005;146:1025–1034. doi: 10.1210/en.2004-1576. [DOI] [PubMed] [Google Scholar]
- Heiser P.W., Lau J., Taketo M.M., Herrera P.L., Hebrok M. Stabilization of β-catenin impacts pancreas growth. Development. 2006;133:2023–2032. doi: 10.1242/dev.02366. [DOI] [PubMed] [Google Scholar]
- Jonsson J., Carlsson L., Edlund T., Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Jorgensen M.C., Ahnfelt-Ronne J., Hald J., Madsen O.D., Serup P., Hecksher-Sorensen J. An illustrated review of early pancreas development in the mouse. Endocr. Rev. 2007;28:685–705. doi: 10.1210/er.2007-0016. [DOI] [PubMed] [Google Scholar]
- Kaestner K.H., Hiemisch H., Luckow B., Schutz G. The HNF-3 gene family of transcription factors in mice: Gene structure, cDNA sequence, and mRNA distribution. Genomics. 1994;20:377–385. doi: 10.1006/geno.1994.1191. [DOI] [PubMed] [Google Scholar]
- Kaestner K.H., Katz J., Liu Y., Drucker D.J., Schutz G. Inactivation of the winged helix transcription factor HNF3α affects glucose homeostasis and islet glucagon gene expression in vivo. Genes & Dev. 1999;13:495–504. doi: 10.1101/gad.13.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y., Cooper B., Gannon M., Ray M., MacDonald R.J., Wright C.V. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Krapp A., Knofler M., Ledermann B., Burki K., Berney C., Zoerkler N., Hagenbuchle O., Wellauer P.K. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes & Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E., Prezioso V.R., Tao W.F., Chen W.S., Darnell J.E. Hepatocyte nuclear factor 3 α belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes & Dev. 1991;5:416–427. doi: 10.1101/gad.5.3.416. [DOI] [PubMed] [Google Scholar]
- Lammert E., Cleaver O., Melton D. Induction of pancreatic differentiation by signals from blood vessels. Science. 2001;294:564–567. doi: 10.1126/science.1064344. [DOI] [PubMed] [Google Scholar]
- Lammert E., Cleaver O., Melton D. Role of endothelial cells in early pancreas and liver development. Mech. Dev. 2003;120:59–64. doi: 10.1016/s0925-4773(02)00332-5. [DOI] [PubMed] [Google Scholar]
- Lee C.S., Sund N.J., Vatamaniuk M.Z., Matschinsky F.M., Stoffers D.A., Kaestner K.H. Foxa2 controls Pdx1 gene expression in pancreatic β-cells in vivo. Diabetes. 2002;51:2546–2551. doi: 10.2337/diabetes.51.8.2546. [DOI] [PubMed] [Google Scholar]
- Lee C.S., Sund N.J., Behr R., Herrera P.L., Kaestner K.H. Foxa2 is required for the differentiation of pancreatic α-cells. Dev. Biol. 2005;278:484–495. doi: 10.1016/j.ydbio.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Martin M., Gallego-Llamas J., Ribes V., Kedinger M., Niederreither K., Chambon P., Dolle P., Gradwohl G. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev. Biol. 2005;284:399–411. doi: 10.1016/j.ydbio.2005.05.035. [DOI] [PubMed] [Google Scholar]
- Molotkov A., Molotkova N., Duester G. Retinoic acid generated by Raldh2 in mesoderm is required for mouse dorsal endodermal pancreas development. Dev. Dyn. 2005;232:950–957. doi: 10.1002/dvdy.20256. [DOI] [PubMed] [Google Scholar]
- Monaghan A.P., Kaestner K.H., Grau E., Schutz G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 α, β and γ genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development. 1993;119:567–578. doi: 10.1242/dev.119.3.567. [DOI] [PubMed] [Google Scholar]
- Moriya N., Komazaki S., Takahashi S., Yokota C., Asashima M. In vitro pancreas formation from Xenopus ectoderm treated with activin and retinoic acid. Dev. Growth Differ. 2000;42:593–602. doi: 10.1046/j.1440-169x.2000.00542.x. [DOI] [PubMed] [Google Scholar]
- Murtaugh L.C. Pancreas and β-cell development: From the actual to the possible. Development. 2007;134:427–438. doi: 10.1242/dev.02770. [DOI] [PubMed] [Google Scholar]
- Offield M.F., Jetton T.L., Labosky P.A., Ray M., Stein R.W., Magnuson M.A., Hogan B.L., Wright C.V. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Rodriguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Samaras S.E., Cissell M.A., Gerrish K., Wright C.V., Gannon M., Stein R. Conserved sequences in a tissue-specific regulatory region of the pdx-1 gene mediate transcription in Pancreatic β cells: Role for hepatocyte nuclear factor 3 β and Pax6. Mol. Cell. Biol. 2002;22:4702–4713. doi: 10.1128/MCB.22.13.4702-4713.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaras S.E., Zhao L., Means A., Henderson E., Matsuoka T.A., Stein R. The islet β cell-enriched RIPE3b1/Maf transcription factor regulates pdx-1 expression. J. Biol. Chem. 2003;278:12263–12270. doi: 10.1074/jbc.M210801200. [DOI] [PubMed] [Google Scholar]
- Sander M., Neubuser A., Kalamaras J., Ee H.C., Martin G.R., German M.S. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes & Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- Sander M., Sussel L., Conners J., Scheel D., Kalamaras J., Dela Cruz F., Schwitzgebel V., Hayes-Jordan A., German M. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of β-cell formation in the pancreas. Development. 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- Schwitzgebel V.M., Scheel D.W., Conners J.R., Kalamaras J., Lee J.E., Anderson D.J., Sussel L., Johnson J.D., German M.S. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- Sharma S., Jhala U.S., Johnson T., Ferreri K., Leonard J., Montminy M. Hormonal regulation of an islet-specific enhancer in the pancreatic homeobox gene STF-1. Mol. Cell. Biol. 1997;17:2598–2604. doi: 10.1128/mcb.17.5.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih D.Q., Navas M.A., Kuwajima S., Duncan S.A., Stoffel M. Impaired glucose homeostasis and neonatal mortality in hepatocyte nuclear factor 3α-deficient mice. Proc. Natl. Acad. Sci. 1999;96:10152–10157. doi: 10.1073/pnas.96.18.10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stafford D., Prince V.E. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr. Biol. 2002;12:1215–1220. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- Stoffers D.A., Zinkin N.T., Stanojevic V., Clarke W.L., Habener J.F. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- Sund N.J., Ang S.L., Sackett S.D., Shen W., Daigle N., Magnuson M.A., Kaestner K.H. Hepatocyte nuclear factor 3β (Foxa2) is dispensable for maintaining the differentiated state of the adult hepatocyte. Mol. Cell. Biol. 2000;20:5175–5183. doi: 10.1128/mcb.20.14.5175-5183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sund N.J., Vatamaniuk M.Z., Casey M., Ang S.L., Magnuson M.A., Stoffers D.A., Matschinsky F.M., Kaestner K.H. Tissue-specific deletion of Foxa2 in pancreatic β cells results in hyperinsulinemic hypoglycemia. Genes & Dev. 2001;15:1706–1715. doi: 10.1101/gad.901601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L., Kalamaras J., Hartigan-O’Connor D.J., Meneses J.J., Pedersen R.A., Rubenstein J.L., German M.S. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic β cells. Development. 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- Tuteja G., Jensen S.T., White P., Kaestner K.H. Cis-regulatory modules in the mammalian liver: Composition depends on strength of Foxa2 consensus site. Nucleic Acids Res. 2008;36:4149–4167. doi: 10.1093/nar/gkn366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhoose A.M., Samaras S., Artner I., Henderson E., Hang Y., Stein R. MafA and MafB regulate Pdx1 transcription through the Area II control region in pancreatic β cells. J. Biol. Chem. 2008;283:22612–22619. doi: 10.1074/jbc.M802902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatamaniuk M.Z., Gupta R.K., Lantz K.A., Doliba N.M., Matschinsky F.M., Kaestner K.H. Foxa1-deficient mice exhibit impaired insulin secretion due to uncoupled oxidative phosphorylation. Diabetes. 2006;55:2730–2736. doi: 10.2337/db05-0470. [DOI] [PubMed] [Google Scholar]
- Wan H., Dingle S., Xu Y., Besnard V., Kaestner K.H., Ang S.L., Wert S., Stahlman M.T., Whitsett J.A. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J. Biol. Chem. 2005;280:13809–13816. doi: 10.1074/jbc.M414122200. [DOI] [PubMed] [Google Scholar]
- Wang J., Elghazi L., Parker S.E., Kizilocak H., Asano M., Sussel L., Sosa-Pineda B. The concerted activities of Pax4 and Nkx2.2 are essential to initiate pancreatic β-cell differentiation. Dev. Biol. 2004;266:178–189. doi: 10.1016/j.ydbio.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Wiebe P.O., Kormish J.D., Roper V.T., Fujitani Y., Alston N.I., Zaret K.S., Wright C.V., Stein R.W., Gannon M. Ptf1a binds to and activates area III, a highly conserved region of the Pdx1 promoter that mediates early pancreas-wide Pdx1 expression. Mol. Cell. Biol. 2007;27:4093–4104. doi: 10.1128/MCB.01978-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.L., Gannon M., Peshavaria M., Offield M.F., Henderson E., Ray M., Marks A., Gamer L.W., Wright C.V., Stein R. Hepatocyte nuclear factor 3β is involved in pancreatic β-cell-specific transcription of the pdx-1 gene. Mol. Cell. Biol. 1997;17:6002–6013. doi: 10.1128/mcb.17.10.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Law A.C., Rajagopal J., Anderson W.J., Gray P.A., Melton D.A. A multipotent progenitor domain guides pancreatic organogenesis. Dev. Cell. 2007;13:103–114. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]