From individual Wnt pathways towards a Wnt signalling network (original) (raw)
Abstract
Wnt proteins play important roles during vertebrate and invertebrate development. They obviously have the ability to activate different intracellular signalling pathways. Based on the characteristic intracellular mediators used, these are commonly described as the Wnt/β-catenin, the Wnt/calcium and the Wnt/Jun N-terminal kinase pathways (also called planar cell polarity pathway). In the past, these different signalling events were mainly described as individual and independent signalling branches. Here, we discuss the possibility that Wnt proteins activate a complex intracellular signalling network rather than individual pathways and suggest a graph representation of this network. Furthermore, we discuss different ways of how to predict the specific outcome of an activation of this network in a particular cell type, which will require the use of mathematical models. We point out that the use of deterministic approaches via the application of differential equations is suitable to model only small aspects of the whole network and that more qualitative approaches are possibly a suitable starting point for the prediction of the global behaviour of such large protein interaction networks.
Keywords: Wnt, signal transduction, signalling networks, graphs
1. Introduction
Wnt proteins are important signalling molecules regulating diverse developmental processes such as differentiation, cell migration or proliferation. Misregulation of Wnt signalling can lead to tumour formation as well as metastasis. Wnt genes code for secreted glycoproteins approximately 350 amino acids in length (Logan & Nusse 2004; Clevers 2006). These proteins have a conserved pattern of cysteine residues and recent work indicates additional lipid modifications (Willert et al. 2003; Takada et al. 2006). Nineteen members of this family have been described in higher vertebrates. To activate intracellular signal transduction pathways, Wnt proteins interact with seven transmembrane receptors of the Frizzled family (Wodarz & Nusse 1998) of which 10 members were found in vertebrates. In addition, co-receptors of the LRP-5/6 type (Pinson et al. 2000; Tamai et al. 2000; Wehrli et al. 2000) and the receptor protein tyrosine kinases ror2 (Hikasa et al. 2002; Mikels & Nusse 2006) or ryk (Inoue et al. 2004; Lu et al. 2004) were indicated to be involved in Wnt signalling in certain conditions. These proteins may form heteromeric receptor complexes upon Wnt binding to elicit specific intracellular responses (Semenov et al. 2001; Cadigan & Liu 2006). Based on the rather high number of ligands belonging to the Wnt family and Frizzled receptors, the question arose whether all Wnts activate identical signal transduction pathways and thus are redundant or whether different signalling pathways can be activated by Wnt proteins pointing towards functional heterogeneity. Indeed, the work of many laboratories indicates that several signalling pathways can be activated by Wnt proteins as discussed below in more detail: the Wnt/β-catenin, the Wnt/Calcium and the Wnt/Jun N-terminal kinase (JNK) pathways, respectively. But if so, where does specificity arise? In the past, specificity has been assigned to the Wnt ligand or the Frizzled receptor type used; however, nowadays it seems rather probable that specificity comes from a particular combination of Wnt ligands, Frizzled receptors, co-receptors and even intracellular signalling components. Here, we first describe the idea of individual Wnt pathways as developed in the past by functional biological assays as well as experiments on a molecular level. We then discuss several recent findings suggesting an overlap of these different pathways resulting in the idea of a Wnt signalling network. Finally, we explore possibilities of how to predict the outcome of a Wnt stimulation for a particular cell type and thus come back to the question of specificity.
2. The historical idea of individual Wnt signalling pathways
Based on different biological activities, Wnt proteins were historically divided into two different subclasses. Whereas members of the Wnt-1/Wg class transform mammary epithelial C57mg cells and elicit secondary axis formation in Xenopus laevis embryos, members of the Wnt-5A class fail in both assays but rather regulate and influence cell adhesion and inhibit Wnt-1/Wg class members (Du et al. 1995; Torres et al. 1996). Although further experiments as discussed below indicate that this strict classification of Wnt proteins does not hold any more, it was the basis of the tremendous work of many laboratories that helped to identify at least three signalling pathways activated by Wnt proteins. Today, we know that certain biological effects of Wnt signalling, such as dorsal axis formation during embryogenesis, cell transformation or regulation of cell migration, indeed can mainly be assigned to either of these signalling pathways.
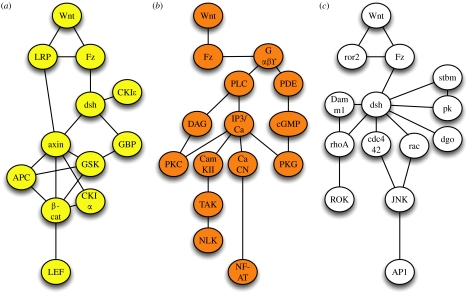
(a) The Wnt/β-catenin pathway
The major player of this pathway is the cytoplasmic protein β-catenin. In the absence of a Wnt ligand, the cytoplasmic concentration of free β-catenin is kept low due to the action of the destruction complex consisting of the proteins adenomatous polyposis coli (APC), axin and glycogen synthase kinase 3β (GSK3β). Upon binding to this complex, β-catenin is phosphorylated by casein kinase Iα (CKIα) and GSK-3β, which is a signal for ubiquitination and proteasomal degradation (Liu et al. 2002). Binding of a Wnt ligand to a Frizzled receptor and LRP-5/6 co-receptor leads to a signalling cascade that finally results in the disassembly of the destruction complex. In detail, LRP is phosphorylated by membrane-bound CKIγ and membrane-associated GSK-3β (Davidson et al. 2005; Zeng et al. 2005), which creates a docking site for axin. The cytoplasmic protein dishevelled can interact with the Frizzled receptor as well as with axin. During this process, dishevelled is modified itself, probably through CKIϵ, which increases its affinity to the GSK-3β inhibitor GBP/FRAT. As a result, the destruction complex disintegrates and cytoplasmic β-catenin can accumulate. Subsequently, β-catenin interacts with transcription factors of the TCF/LEF family of HMG box transcription factors turning them from a repressor into an activator of transcription. Within the nucleus, both LEF/TCF and β-catenin interact with additional proteins such as bcl9, pontin, SWI/SNF, PIASy, groucho or CtBP (Logan & Nusse 2004; Clevers 2006; Willert & Jones 2006).
As just exemplified for the Wnt/β-catenin pathway, intracellular signal transduction is based on the interaction of certain entities including proteins, lipids and ions. These interactions during signal transduction are widely presented by schematic cartoons that result—in a more mathematical sense—in a graph representation with nodes/vertices. Here, the participating entities are given as nodes, and edges drawn between these nodes represent interactions between the vertices. Such a graph can be drawn for the core components of the Wnt/β-catenin pathway as indicated in figure 1a. Within this static graph, potential interactions of entities are shown, whereas post-translational modifications of these components occurring during signal transduction, though possible, are not normally explicitly highlighted.
Figure 1.
The classical view of three independent Wnt signalling pathways. Graph representations are given, including the core components of the (a) canonical Wnt/β-catenin pathway (yellow), (b) the Wnt/calcium pathway (orange) and (c) the Wnt/JNK pathway (white, also called planar cell polarity pathway). Edges indicate direct protein–protein interactions or may relate to an epistatic relationship between two components within a signalling pathway.
(b) The Wnt/calcium pathway
Approximately 10 years ago, it was shown that injection of RNA coding for certain Wnts or Frizzleds into early zebrafish embryos triggered intracellular calcium release (Slusarski et al. 1997_a_,b). Loss of Wnt-11 or Wnt-5A function resulted in reduced intracellular calcium signalling (Eisenberg & Eisenberg 2002; Westfall et al. 2003). This finding was subsequently expanded by the observation that the Wnt-induced release of intracellular calcium is sufficient to activate different intracellular calcium-sensitive enzymes such as protein kinase C, PKC (Sheldahl et al. 1999; Winklbauer et al. 2001; Kinoshita et al. 2003; Tu et al. 2007), calcium–calmodulin-dependent kinase II, CamKII (Kuhl et al. 2000; Robitaille et al. 2002; Ishitani et al. 2003; Topol et al. 2003; Ouko et al. 2004; Kremenevskaja et al. 2005) and the calcium-sensitive phosphatase calcineurin that subsequently can activate the transcription factor NF-AT (Saneyoshi et al. 2002; Dejmek et al. 2006). Calcium signalling is a rapid event upon receptor stimulation. Within some of the above mentioned papers, however, assays were used and experimental designs were chosen that monitor cellular changes minutes or even hours after manipulation. The existence of such a calcium-mediated pathway directly activated by Wnt proteins was therefore debated over past years. Just recently, this issue has been resolved by the finding that purified Wnt-5A or Wnt-5A-derived peptides are able to trigger calcium release in several cell types (Kremenevskaja et al. 2005; Dejmek et al. 2006; Ma & Wang 2006; Safholm et al. 2006). As expected, the release of intracellular calcium is a rapid response of a cell within seconds upon ligand binding and depends on heterotrimeric G-proteins (Ma & Wang 2006) and the activation of the phosphatidyl inositol cycle (Slusarski et al. 1997_a_). Through the regulation of PDE6, a cGMP-specific phosphodiesterase, this pathway is also linked to the regulation of protein kinase G in some cell types (Ahumada et al. 2002; Ma & Wang 2006). The Wnt/calcium pathway has been linked to the activation of nemo-like kinase (NLK) that is able to phosphorylate TCF transcription factors and thereby inhibits canonical Wnt signalling (Ishitani et al. 1999, 2003; Meneghini et al. 1999). This pathway is involved in dorsoventral patterning of early X. laevis and zebrafish embryos (Kuhl et al. 2000; Saneyoshi et al. 2002; Westfall et al. 2003), regulation of the Wnt/β-catenin pathway (Ishitani et al. 2003), tumour formation (Weeraratna et al. 2002; Liang et al. 2003; Kremenevskaja et al. 2005) and more recently the regulation of epithelial–mesenchymal transitions (Garriock & Krieg 2007). The graph representation including the major components of this pathway is given in figure 1b.
(c) The Wnt/JNK or PCP pathway
Finally, the Wnt/JNK pathway forms the third signalling pathway activated by Wnts. This pathway was described earlier in Drosophila melanogaster as the planar cell polarity pathway. This pathway regulates the polarity of epithelial cells within the plane of the epithelium, e.g. orienting Drosophila wing hairs or regulating the organization of the ommatidia in the fly eye (Strutt 2003). In vertebrates, a similar pathway has been described regulating convergent extension movements during gastrulation or neurulation and cell migration of neural crest cells (Veeman et al. 2003_a_; De Calisto et al. 2005; Matsui et al. 2005). Whereas a distinct Wnt (wingless) ligand has not been assigned to this pathway in Drosophila, in vertebrates Wnt-11 or Wnt-5A is able to activate this signalling cascade. The core element of the Wnt/JNK pathway includes the activation of rho GTPases such as rhoA, rac or cdc42 downstream of dsh. The GTPases can activate more downstream mediators like JNK or rho kinase (ROK). In planar cell polarity signalling, this cascade of events is complemented by transmembrane proteins such as strabismus (stbm), diego (dgo) or prickle (pk) that also can bind dishevelled (Bastock et al. 2003; Strutt 2003; Jenny et al. 2005). It is thought that the asymmetric distribution of proteins such as stbm and pk within epithelial cells is involved in setting up planar polarity (Strutt 2003). In vertebrates, the receptor tyrosine kinase ror-2 has been shown to regulate convergent extension movements and bind to Wnt protein (Hikasa et al. 2002). Ror-2 was found to be upstream of cdc42 and to regulate JNK activity (Oishi et al. 2003). A graph representation including the major components of this pathway is indicated in figure 1c.
(d) Novel aspects: G-proteins in Wnt signalling
Whereas it has been thought for a long time that the Wnt/β-catenin signalling pathway does not involve heterotrimeric G-proteins, recent findings indicate their involvement in canonical signalling as well as planar cell polarity type of signalling. Activation of Frizzled-1 signalling through inducible Frizzled receptors in F9 teratocarcinoma cells required the function of Gαq and Gαo (Liu et al. 1999_a_), whereas triggering Frizzled-2 signalling through the corresponding inducible Frizzled receptor requires the action of Gαo and Gαt (Liu et al. 1999_b_). Further evidence for an involvement of G-proteins in canonical Wnt signalling was provided by the observation that overexpression of RGS4, a regulator of G-protein signalling, in early Xenopus embryos interferes with Wnt-8-induced secondary axis formation (Wu et al. 2000). TCF/β-catenin-mediated reporter gene activity as well as endogenous Wnt/β-catenin target genes can be induced by constitutively active Gαo and Gαq subunits (Liu et al. 2001). Depletion of G-proteins by the use of antisense oligonucleotides or siRNA interferes with Wnt- and Frizzled-induced activation of the Wnt/β-catenin pathway (Liu et al. 2001). In Drosophila, genetic epistasis experiments are indicative of the involvement of G-proteins downstream of Frizzled but upstream of dishevelled in both canonical and non-canonical Wnt signalling (Katanaev et al. 2005). These findings were complemented by the observation of a ligand-dependent association/dissociation of Gαo and Gαq subunits with Frizzled receptors (Liu et al. 2005). Moreover, also dsh and axin have been linked directly to G-proteins. The βγ subunits were found to associate with dishevelled (Angers et al. 2006) whereas Gα12 binds to axin (Stemmle et al. 2006). Interestingly, Gα12 and Gα13 have also been implicated in regulating convergent extension movements (Lin et al. 2005). This list of G-protein subunits involved in Wnt signalling becomes even longer if one takes into account the finding that some Wnts probably activate protein kinase A through the classical Gαs pathway (Chen et al. 2005; Pourquie 2005) whereas a non-canonical Wnt ligand-like Wnt-11 reduces PKA activity through Gαi (Park et al. 2006). PKA has been correlated to the regulation of β-catenin stability as well as JNK activity (Hino et al. 2005; Park et al. 2006). Considering the fact that the Wnt/calcium pathway also involves G-proteins as outlined above, it seems very probable that all three Wnt signalling branches involve heterotrimeric G-proteins. Currently, a total of 16 Gα subunits with specific signalling activities are known in addition to 4β and 7β subunits (Birnbaumer 2007_a_,b). The latter subunits form different βγ heterodimers with obvious identical signalling activities. As outlined, several Gα subtypes as well as βγ heterodimers have been linked to different biochemical downstream events during Wnt signalling as summarized in table 1. Considering Wnt signalling to be G-protein coupled, indeed leaves us with a plethora of different Wnt/Frizzled/G-Protein and co-receptor combinations for potential downstream effects.
Table 1.
G-proteins implicated in Wnt and Frizzled signalling. (PCP, planar cell polarity; PDE, cGMP-specific phosphodiesterase; PKC, protein kinase C; PLC, phospholipase Cβ; AC, adenylate cyclase; PKA, protein kinase A; CamKII, calcium–calmodulin-dependent kinase II.)
| G-protein subunit | downstream mediators | references |
|---|---|---|
| αo | β-catenin | Katanaev et al. (2005) |
| β-catenin | Liu et al. (2001) | |
| β-catenin | Liu et al. (2005) | |
| PCP signalling | Katanaev et al. (2005) | |
| PDE | Ma & Wang (2006) | |
| PKC | Liu et al. (1999_a_) | |
| differentiation assay | Liu et al. (1999_b_) | |
| αt2 | PDE | Ma & Wang (2006) |
| PDE | Ahumada et al. (2002) | |
| differentiation assay | Liu et al. (1999_b_) | |
| αq | β-catenin | Liu et al. (2001) |
| β-catenin | Liu et al. (2005) | |
| PLC/PKCδ | Tu et al. (2007) | |
| PKC | Liu et al. (2001) | |
| α11 | PLC/PKCδ | Tu et al. (2007) |
| αs | AC/PKA | Chen et al. (2005) |
| αi | AC/PKA | Park et al. (2006) |
| βγ | PLC/PKC/cdc42 | Penzo-Mendèz et al. (2003) |
| PLC | Slusarski et al. (1997_a_) | |
| PLC/CamKII | Kuhl et al. (2000) | |
| PLC/PKC | Sheldahl et al. (1999) | |
| differentiation assay | Liu et al. (1999_b_) |
3. Towards a Wnt signalling network: overlapping activities of individual components
(a) Evolving a novel concept of Wnt signalling as a network
As mentioned before, the current model of Wnt signalling is based on the assumption that different independent Wnt signalling pathways exist, e.g. canonical and non-canonical pathways, which represent the underlying molecular mechanism of certain biological effects. However, in some cases, as listed and discussed below, it has been observed that (i) particular proteins are involved in more than one of these signalling pathways, (ii) individual proteins have been shown to be involved in regulating different biological processes that were originally assigned to either Wnt pathways and finally, (iii) different Wnt pathways seem to act in vivo in one particular cell type to regulate each other at the same time. In an expansion of the individual pathway model, we would like to suggest instead that Wnt proteins activate a signalling network. The outcome of a Wnt stimulation in this model would depend on the molecular signature of the system used as well as the recent history of the system (i.e. presence and influence of other growth factors that might interfere with Wnt signal transduction or regulate mediators of Wnt signalling transcriptionally). The points for this line of argument in favour of a Wnt signalling network can be listed as follows:
- The original classification of Wnts into different functional classes cannot hold any more. Wnt-5A is able to activate not only calcium signalling (Slusarski et al. 1997_a_; Dejmek et al. 2006; Ma & Wang 2006; Safholm et al. 2006) and JNK (Yamanaka et al. 2002), but also functions in the β-catenin pathway if the correct receptor is present, i.e. Fz-5 (He et al. 1997). Also, Frizzled receptors have the ability to activate different signalling pathways. Fz-5 has been shown to activate β-catenin signalling (He et al. 1997) and also is involved in PKC activation (Weeraratna et al. 2002). Furthermore, Fz-4 has been shown to activate different signalling pathways (Robitaille et al. 2002; Mikels & Nusse 2006). Finally, Fz-7 can also activate canonical as well as non-canonical signalling branches (Medina et al. 2000). Thus, activation of different Wnt signalling aspects by Wnts or Frizzled receptors seems to be highly context dependent.
- The phosphoprotein dishevelled has been involved in canonical β-catenin and non-canonical JNK activations (Boutros et al. 1998) as well as in the Wnt/calcium pathway (Sheldahl et al. 2003) indicating an overlap of intracellular signalling pathway mediators.
- As discussed above, it is reasonable to argue that heterotrimeric G-proteins are involved in some, if not all, aspects of Wnt signalling. As some Gα proteins are involved in different Wnt branches (Katanaev et al. 2005), for example, Gαo in canonical as well as planar cell polarity signalling, there is again an overlap of pathway activities.
- Wnt-mediated activation of JNK within planar cell polarity type of signalling has been shown to require the action of PKC isoforms, in particular PKCδ (Pandur et al. 2002; Kinoshita et al. 2003). Activation of PKC requires DAG (and in some cases also calcium ions), an aspect so far assigned to the Wnt/calcium pathway. Furthermore, it has been demonstrated that during Xenopus gastrulation movements, cdc42 is activated by PKC downstream of βγ subunits of G-proteins (and thus probably downstream of PLCβ). Cdc42 may also activate JNK (Tao et al. 2001). Thus, there is an overlap of Wnt/calcium and Wnt/JNK signalling in this context.
- The calcium-sensitive transcription factor NF-AT has been linked to Wnt-mediated dorsal–ventral patterning in Xenopus embryos, probably due to an inhibitory role onto the Wnt/β-catenin pathway (Saneyoshi et al. 2002). Interestingly, NF-AT is also required for regulating neural convergent extension movements, a process that depends on a planar cell polarity type signalling pathway (Borchers et al. 2006).
- CamKII, activated by the Wnt/calcium pathway, has been shown to be involved in dorsoventral patterning in Xenopus and zebrafish, probably through the inhibition of the Wnt/β-catenin signalling pathway (Kuhl et al. 2000; Ishitani et al. 2003; Westfall et al. 2003), as well as in regulating epithelial–mesenchymal transitions, a prerequisite for cell migration processes, i.e. neural crest cell migration, normally linked to the PCP pathway (De Calisto et al. 2005). Also, PKC activated by the Wnt/calcium pathway has some ventralizing activity (Busa & Gimlich 1989).
- The PCP pathway component prickle has been shown to modulate intracellular calcium signalling (Veeman et al. 2003_b_).
- The atypical receptor tyrosine kinase ror2 is activated by CKIϵ (Kani et al. 2004) which itself is regulated through canonical Wnt signalling and has been shown to be involved in the Wnt/β-catenin pathway (Cong et al. 2004; Swiatek et al. 2004). CKIϵ also seems to be required for the planar cell polarity pathway (Klein et al. 2006; Strutt et al. 2006). In line with this is the observation that the protein diversin (the vertebrate homologue to Drosophila diego) not only activates JNK but also recruits CKIϵ to the destruction complex of the β-catenin pathway (Schwarz-Romond et al. 2002).
- JNK is involved not only in regulating cell movements through the Wnt/JNK/planar cell polarity pathway, but also in inhibiting the action of the Wnt/β-catenin pathway (Liao et al. 2006). Here, it is noteworthy to mention that Wnt-mediated activation of JNK can regulate target gene expression through AP-1 (Jun/fos; Li et al. 1999; Weber et al. 2000).
- Inhibition of the canonical Wnt branch through the extracellular inhibitor dickkopf (dkk; Glinka et al. 1998) results in an activation of non-canonical Wnt signalling as indicated by the activation of JNK (Pandur et al. 2002; Lee et al. 2004; Caneparo et al. 2007), indicating a balance between canonical and non-canonical branches through extracellular regulation, probably through modifying receptor complex compositions.
- Just recently, it has been shown that the src kinase family member yes is activated by Wnt-5A signalling (Dejmek et al. 2006), whereas it was reported earlier that yes and fyn are also involved in regulating convergent extension movements (Jopling & den Hertog 2005). Interestingly, src kinases are regulated through G-proteins (Ma et al. 2000) and tyrosine phosphorylation by integrating these inputs (Roskoski 2005). In future, it will be of high interest to analyse whether src kinase integrates information from G-protein coupled Frizzled receptors and ror-2 tyrosine kinase in this setting. Of interest in this context is the observation that the src kinase member fyn also modulates calcium signalling (Sharma et al. 2005) during zebrafish development through an unknown mechanism. The Wnt-5A-mediated activation of yes was shown to regulate cdc42 (Dejmek et al. 2006). Activation of cdc42 through src kinase members has been shown before (Yokoyama & Miller 2003) in a different context. More downstream, the Wnt-5A–yes–cdc42 axis results in binding of CKIα to NF-AT transcription factor (Dejmek et al. 2006). CKIα binding to NF-AT prevents nuclear localization and accumulation of NF-AT (Zhu et al. 1998; Okamura et al. 2004). With respect to the demonstration that Wnt proteins activate a signalling network, this observation is of particular interest as it involves proteins previously linked to planar cell polarity signalling (cdc42), Wnt/calcium signalling (NF-AT) or Wnt/β-catenin signalling (CKIα).
- Wnt-3A has been shown to simultaneously activate canonical as well as non-canonical signalling, clearly demonstrating that one particular Wnt ligand can activate different branches at the same time within the same cell type (Tu et al. 2007).
- Finally, it is of interest to come back to the former observation that Wnt-5A class members are able to inhibit the function of Wnt-1/Wg class members, that is they inhibit the canonical Wnt/β-catenin pathway. Multiple possibilities to explain this behaviour on a molecular level are suggested above, i.e. involving JNK, CamKII, PKC or CaCN/NF-AT. Furthermore, calcium can bind to Naked which interacts with and regulates dishevelled (Zeng et al. 2000; Wharton et al. 2001; Yan et al. 2001). One question of interest, however, is whether this also occurs in vivo and not only under gain of function situations. Interestingly, there are some examples described in the literature. In Wnt-5A−/− mice, canonical Wnt signalling is upregulated in the limb buds (Topol et al. 2003). In fish embryos deficient for maternal as well as zygotic Wnt-5A, expression levels of β-catenin are elevated (Westfall et al. 2003) and this can be rescued by a constitutively active CamKII mutant. Depleting Wnt-11 in P19 teratocarcinoma cells by siRNA results in an upregulation of β-catenin signalling (Maye et al. 2004) and finally, during appendage regeneration in fishes distinct Wnt signalling pathways have opposing effects (Stoick-Cooper et al. 2007).
Taken together, these findings clearly indicate that different Wnt signalling pathways are simultaneously active within the same cell type, further supporting the idea that Wnt pathways are highly connected to form a Wnt signalling network. This network seems to be activated by either one or more ligands acting on a certain cell type.
(b) Balanced Wnt signalling networks during eye formation and cardiac development
In this section, we would like to discuss two biological examples from developmental biology for which it seems to be appropriate to follow the idea of a Wnt signalling network in more detail, in particular early steps of eye formation in the forebrain as well as induction of cardiac progenitor cells.
Induction of eye progenitor cells takes place in the anterior-most part of the neural plate, the forebrain anlage. This region is characterized through a low β-catenin signalling activity, and an increase in canonical Wnt/β-catenin activity results in a posteriorization of neural tissue and a loss of eye structures (Heisenberg et al. 2001). In line with this finding, canonical Wnt activity in the developing eye could not be detected before stage 20 in Xenopus embryos (Van Raay et al. 2005). Recently, we demonstrated that the activation of a non-canonical Wnt pathway by Wnt-4 is, however, required for eye formation in X. laevis (Maurus et al. 2005). The probable receptor for Wnt-4 in this setting is Fz-3 which was previously reported to activate non-canonical Wnt signalling and to be required for eye development (Rasmussen et al. 2001). Wnt-4 and Fz-3 were postulated to be a ligand–receptor combination in other settings as well (Lyuksyutova et al. 2003). Similar to these findings, it was reported that Wnt-11/Fz-5 has a comparable role in eye development in zebrafish (Cavodeassi et al. 2005). Here, the authors demonstrated that activation of a non-canonical Wnt pathway inhibits β-catenin signalling and regulates coherence of cells as well as cell movements within the eye field. In agreement with both studies, it was demonstrated recently that a planar cell polarity type of signalling pathway involving dsh, PKC and JNK is important for eye formation, cell movements and marker gene expression in the developing Xenopus eye (Lee et al. 2006). In summary, in the eye-forming region, non-canonical Wnt signalling branches negatively influence the Wnt/β-catenin branch and regulate cell adhesion, cell migration and gene expression obviously at the same time and thus represent a Wnt signalling network.
A comparable situation can be found during early heart development in Xenopus and zebrafish. In this context, Wnt-11 or the close homologue Wnt-11R activate a non-canonical Wnt signalling pathway (Pandur et al. 2002; Garriock et al. 2005; Matsui et al. 2005). Loss of non-canonical Wnt function results in morphological defects such as failure of heart precursor cells to fuse at the ventral midline of the embryo, resulting cardia bifida or failure in heart tube formation (Pandur et al. 2002; Matsui et al. 2005). Activation of non-canonical signalling by Wnt-11 promotes cardiomyocyte formation in different cellular setting such as Xenopus mesodermal cells (Pandur et al. 2002; Garriock et al. 2005), quail mesoderm (Eisenberg et al. 1997; Eisenberg & Eisenberg 1999), murine embryonic stem cells as well as teratocarcinoma cells (Pandur et al. 2002; Terami et al. 2004), murine circulating human endothelial progenitor cells (Koyanagi et al. 2005) and in murine bone marrow and mesenchymal stem cells (Belema Bedada et al. 2005; Schulze et al. 2005). Interestingly, at the same time, activation of canonical Wnt signalling inhibits cardiogenesis in vivo, thus indicating that canonical Wnt signalling needs to be off for early cardiac specification (Marvin et al. 2001; Schneider & Mercola 2001; Tzahor & Lassar 2001). Furthermore, this requirement for inhibition of canonical Wnt signalling needs to be regulated in a time-dependent manner (Nakamura et al. 2003; Naito et al. 2007), suggesting that β-catenin signalling needs to be tightly regulated and fine tuned. Thus, in both cellular settings, early cardiac and eye development, a Wnt network needs to be activated in which non-canonical signalling branches inhibit β-catenin signalling and regulate the cell cohesion and migration required for proper tissue morphogenesis.
4. Models to predict the behaviour of the Wnt signalling network
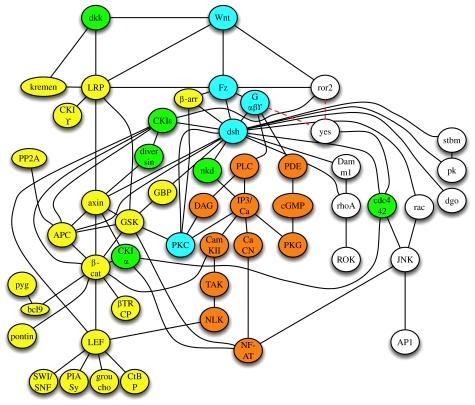
All these issues discussed above strongly argue in favour of a highly connected Wnt signalling network. An extended version of the interactions discussed is given in figure 2. This hypothesis can be further extended by recent whole-genome yeast two-hybrid screens in humans (Stelzl et al. 2005) depicting a Wnt signalling network. Furthermore, functional large-scale screening methods indicate that the number of molecules involved in Wnt signalling might be markedly higher. In an RNAi screen in Drosophila, a total of 238 potential regulators of the Wnt/β-catenin pathway were identified (DasGupta et al. 2005). Thus, it becomes clear that a network representing the discussed interactions, as depicted in figure 2, still clearly represents a simplification of reality. A question that comes to mind immediately is whether it will ever be possible to predict the specificity or the outcome of such a signalling network by appropriate theoretical and mathematical methods.
Figure 2.
The integrative approach of analysing Wnt signalling results in a Wnt signalling network. Several components have been linked to all signalling branches such as Wnt, Fz, heterotrimeric G-proteins, dsh or PKC (indicated in light blue); others were linked to at least two branches such as CKIϵ, diversin, dkk, cdc 42 or CKIα (indicated in green). Please note that this network does not represent all interactions identified in Wnt signalling so far for reasons of clarity. Most of the edges represent direct interactions of entities indicated and some of them however represent the epistatic relationship of these components within the network as discussed in the main text. Dashed lines for the upstream molecules of yes indicate that the upstream receptor for yes activation through Wnts is unknown so far.
Modelling and simulation of complex systems is widely used to predict their behaviour (for a paper-based review see De Jong 2002). The term modelling represents the description of a system with mathematical methods, whereas the term simulation describes the computation of solutions for the model chosen. In principle, several modelling methods can be distinguished based on their underlying theoretical and mathematical approaches (i.e. quantitative versus qualitative, deterministic versus stochastic, static versus dynamic and discrete versus continuous). In a particular case, use of these different approaches depends on the data available and the purpose of the model. In the following section, we will briefly discuss two different approaches that have been used to model parts of the Wnt signalling network by the use of differential equations.
(a) Ordinary differential equations
Ordinary differential equations have been widely used to model genetic regulatory networks, metabolism and signalling pathways (Heinrich & Schuster 1996; Fell 1997; Collado-Vides & Hofestädt 2002; Fall et al. 2002; Szallasi et al. 2006). Several assumptions have been made for the usage of this approach. It is assumed that all reactions within a signal transduction pathway can be described by the use of laws used in physical chemistry. This kinetic model requires detailed knowledge of the underlying reactions, e.g. the concentration of reaction partners, the reaction rates of individual reaction steps, association and dissociation rates. Furthermore, it is assumed that all reaction steps occur in a well-mixed solution and that all reaction partners are available immediately for the next reaction step (that means the reaction is not limited through diffusion processes). Within this physicochemical modelling approach, the system of interest is described with a set of coupled differential equations. This approach is termed a deterministic approach as the behaviour of the system can be predicted for every time point of the reaction if all reactions of the system have been described and all parameters are known. Unfortunately, all variables of the system have to be known exactly. For the simple example of modelling the rate of change for a certain cellular property dX/dt=−X/τ, we seek a function X(t) whose derivative is proportional to itself with proportionality constant 1/τ. Remembering that the derivative of an exponential function is still the exponential function, we expect the exact solution to be of the form X=Ce−t/τ. That means we have to specify exactly two parameters, one for the rate and the other for the initial solution. This gets even more complicated with the nonlinear coupling of these differential equations. Unless they are stable (robust), parameters have to be known precisely, which is rarely the case, as small perturbations may have large impacts.
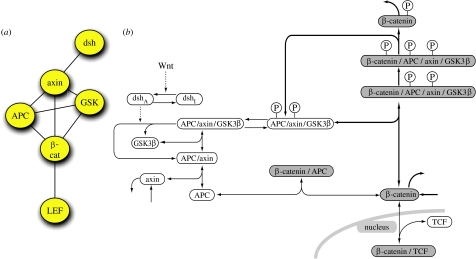
A quantitative and moderately complex model of part of the Wnt/β-catenin pathway including its core components has been established (Lee et al. 2003) and just recently been reused to predict the behaviour of the canonical Wnt pathway branch in a tumour cell (Kim et al. 2007). Based on 15 linear ordinary differential equations that are coupled with nonlinear algebraic equations, this Lee–Heinrich model includes the following main proteins and their interactions of the canonical Wnt branch: dishevelled (dsh), GSK3β, APC, axin, β-catenin and TCF (figure 3). The model is mainly based on data obtained from quantitative measurements of protein concentrations in Xenopus egg extracts and predicts the behaviour of this core element of the Wnt/β-catenin signalling branch. Data obtained from this simulation have been nicely confirmed in the Xenopus egg extract system (which represents a well-mixed reaction system of course). However, because this model was established using quantitative measurements from Xenopus egg extracts and, in addition, some of the parameters used were derived from estimations, it is probable that these concentrations differ in other organisms or cellular systems. Additional variability is introduced by unavoidable measurement errors. Thus, it is questionable whether such a model will also be correct in other systems. This question can at least in part be evaluated by robustness analyses for which one or more parameters are changed and the model tested for its predicted outcome thereafter. Furthermore, one should keep in mind that the original Lee–Heinrich model does not include an activation with a Wnt ligand but relies on an artificial activation using dishevelled to be inactive (dsh=0) or to be active (dsh=1). Furthermore, most of the components indicated and discussed before for canonical as well as non-canonical Wnt branches were not considered. This shows that modelling by ordinary differential equations requires data that are usually not easily obtainable for moderately large pathways or even more complex networks. This is even more the case both for models that lack the assumptions of a spatially homogeneous system and reflect cellular asymmetries of participating components, and for reactions that occur deterministically and continuously. Both cases would require different modelling approaches, i.e. the use of partial differential equations, compartment models or stochastic differential equations. Interestingly, the asymmetric cellular distribution of planar cell polarity pathway components has been modelled by the use of partial differential equations just recently (Amonlirdviman et al. 2005). Furthermore, as the model gets more complex, simulation time rises with the increase of the number of parameters involved, which in turn limits the inferential capabilities of these models. Thus, although quantitative approaches using differential equations were used to describe and predict the behaviour parts of the Wnt network, it is rather unlikely that these approaches will be currently suitable for predicting the outcome of the Wnt network. Therefore, different approaches using more qualitative knowledge seem more promising.
Figure 3.
Modelling aspects of Wnt signalling by the use of ordinary differential equations. Part of the canonical Wnt signalling branch has been modelled in the Lee–Heinrich model (Lee et al. 2003). (a) Components of the network as indicated in figure 1a, which were included in the Lee–Heinrich model. In addition, dephosphorylation events, de novo synthesis and degradation of some components are included. (b) Reaction scheme included in the Lee–Heinrich model. Note that just a very small aspect of the Wnt signalling network has been implemented in the Lee–Heinrich model.
(b) Qualitative approaches
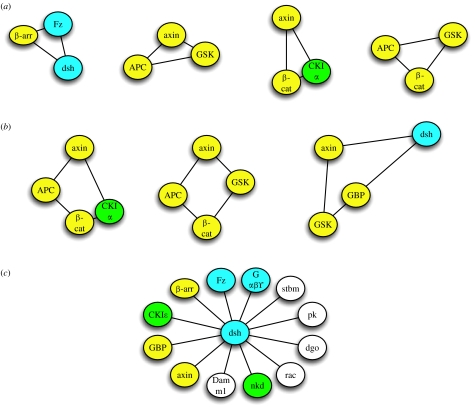
Different more qualitative models have been used to describe the behaviour of complex systems including graph methods, Bayesian networks, Boolean networks or Petri networks, to name just a few (Hornberg et al. 2005; Klipp et al. 2005; Szallasi et al. 2006; Alon 2007). Graph methods are widely used to get an overview of a complex network as done in §§1–3 in this paper. Components of the network, i.e. a signal transduction pathway in a particular cell type, are identified. This can be done, for example, by extracting the required information from the literature, the use of appropriate public databases and by experimental screening methods such as DNA microarrays or proteomic approaches. Individual entities of the network can be represented as nodes/vertices. Interaction between these components or their relationship can be drawn as edges between these vertices. Such a representation of the system is an ‘undirected graph’ of the network (see figure 2, for example). However, it provides just a minimum of information as it is a static representation of the network. These graphs can be extended by including additional features such as the direction of flow of information (directed graphs), including different weights for the interaction (directed and weighted) as well as spatial information (Ma'ayan et al. 2005). These networks can be used to elucidate the topology of signal transduction pathways and can be described by using different parameters such as mean connectivity of nodes, local connectivity of nodes (by calculating so-called clustering coefficients and grid coefficients) and the characteristic path length to connect two individual nodes with the shortest path possible. A high clustering coefficient and a short path length distinguish real networks from random networks and characterize them as (ultra) small world networks. Interestingly, it was found by Barabasi & Oltvai (2004) that cellular networks have certain features that characterize them as scale-free networks, in particular that they have a power-law degree distribution. Thus, within these networks, few vertices have many links characterizing them as hubs and many vertices have few links. Intensive analysis of real networks furthermore identified several design motifs that occurred more often than just randomly in these networks, such as triads or tetrads of interactions (Milo et al. 2002; Li et al. 2004; Yook et al. 2004). The destruction complex within the β-catenin branch of Wnt signalling for example, consisting of axin, APC, GSK-3β, CKIα and β-catenin, represents several overlapping triads and tetrads of interactions, and a close inspection identifies many more (figure 4a,b). Re-examination of figure 2 also reveals that the major components of the Wnt/β-catenin path, such as dsh, axin, β-catenin or LEF, have more links than other proteins, thereby indicating that they are hubs. A more extended representation of the Wnt network including 111 nodes (data not shown) has a mean shortest path length of 3.5 and a mean node degree of 2.9 with several hubs such as dsh, axin or β-catenin with more than 15 edges. Keeping in mind that cellular networks are small-world and scale-free networks that follow certain design principles, it may be possible to predict additional vertices or interactions between them within the network as shown recently (Albert & Albert 2004). Thus, network theories will be a valuable to tool to predict the final design of a network leading to experimentally testable hypotheses.
Figure 4.
Network motifs of direct protein–protein interactions found within the Wnt signalling network. Examples for (a) triads and (b) tetrads of interactions identified. (c) Major components of the network, i.e. dishevelled represent hubs. Not all known interactions of dsh are shown. Additional interaction partners include Frodo/Dapper, Iday, Naked, MuSK, Eps8, Par-1, PAK and PP2C. Other hubs are LEF/TCF, β-catenin, axin or Frizzled.
Furthermore, methods will be required to predict the dynamic behaviour of such static networks. In Boolean networks, for example, nodes are connected through logical functions (AND, OR and NOT) and can either be ON or OFF. These models can predict in a deterministic and discrete simulation the state of each node at each time point. Such a model has been used to model genetic regulatory networks in which genes can be either ON or OFF, thereby transferring a static network of genetic interactions into a dynamic model. These Boolean models can be learned from data via specifying complete truth tables, but typically this is not feasible in biological systems. The computational learning community has developed methods for learning these Boolean functions from input–output observations (Kearns & Vazirani 1994; Liang et al. 1998). A Boolean model of the segment polarity gene network in the fly has recently been shown to predict the behaviour of the network fairly well, as did a model based on differential equations (von Dassow et al. 2000; Albert & Othmer 2003). Extensions of these Boolean networks are generalized logical networks that also allow values other than 0 and 1 for the individual components.
The so-called Petri nets (Adam Petri's PhD thesis 1962, or place/transition network) are another way of modelling dynamic behaviour and have been developed for different simulation purposes (Bernadinello & De Cindio 1992). Petri nets consist of two types of nodes, namely place nodes and transition nodes, and directed arcs connecting places with transitions. Places represent objects, e.g. molecules, and contain tokens indicating the currently existing number of these objects. A transition is performed if the incoming places contain enough tokens, and consists of removing tokens from the incoming places and adding tokens to the output places. The execution is non-deterministic. Several extensions of this basic methodology exist, e.g. timed Petri nets and stochastic Petri nets, which make them suitable for modelling biochemical pathways. Petri nets just recently were applied to analyse intracellular signal transduction pathways (Heiner et al. 2004).
5. Conclusions
These few examples indicate that more qualitative approaches are capable of predicting the outcome of complex networks and thus might be a valuable tool to transfer a static graph representation of highly connected signalling networks, such as the Wnt signalling network, into a dynamic network of interactions. In the future, methods will surely be developed that allow us to translate our current knowledge of interactions and their interdependence into dynamic modelling tools that probably will also allow prediction of the global outcome of a stimulated Wnt signalling network in a particular cell. Of course, such qualitative models also have disadvantages, such as quantitative predictions not being possible. Also, settings in which the characteristics of an entity highly depend on quantitative parameters may be hard to cover. The outcome of calcium signalling seems to depend on the frequency as well as the amplitude of calcium transients (Berridge 1997). The selected examples of the Wnt signalling network, however, indicate that this network can be described by the use of mathematical network theory which will allow analyses of this network by the use of appropriate network models.
Acknowledgments
This work was supported by grants of the DFG to M.K. (SFB 497, A6) and H.K. (SFB 518, C5) and Stifterverband für die Deutsche Wissenschaft to H.K. We thank Christian Wawra for drawing figure 3b and Christian Wawra, André Müller and Barbara Kracher for their discussions.
Footnotes
One contribution of 11 to a Discussion Meeting Issue ‘Calcium signals and developmental patterning’.
References
- Ahumada A, Slusarski D.C, Liu X, Moon R.T, Malbon C.C, Wang H.Y. Signaling of rat Frizzled-2 through phosphodiesterase and cyclic GMP. Science. 2002;298:2006–2010. doi: 10.1126/science.1073776. doi:10.1126/science.1073776 [DOI] [PubMed] [Google Scholar]
- Albert I, Albert R. Conserved network motifs allow protein–protein interaction prediction. Bioinformatics. 2004;20:3346–3352. doi: 10.1093/bioinformatics/bth402. doi:10.1093/bioinformatics/bth402 [DOI] [PubMed] [Google Scholar]
- Albert R, Othmer H.G. The topology of the regulatory interactions predicts the expression pattern of the segment polarity genes in Drosophila melanogaster. J. Theor. Biol. 2003;223:1–18. doi: 10.1016/s0022-5193(03)00035-3. doi:10.1016/S0022-5193(03)00035-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. Chapman & Hall/CRC; London, UK: 2007. An introduction to systems biology: design principles of biological circuits. [Google Scholar]
- Amonlirdviman K, Khare N.A, Tree D.R, Chen W.S, Axelrod J.D, Tomlin C.J. Mathematical modeling of planar cell polarity to understand domineering nonautonomy. Science. 2005;307:423–426. doi: 10.1126/science.1105471. doi:10.1126/science.1105471 [DOI] [PubMed] [Google Scholar]
- Angers S, Thorpe C.J, Biechele T.L, Goldenberg S.J, Zheng N, MacCoss M.J, Moon R.T. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat. Cell Biol. 2006;8:348–357. doi: 10.1038/ncb1381. doi:10.1038/ncb1381 [DOI] [PubMed] [Google Scholar]
- Barabasi A.L, Oltvai Z.N. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. doi:10.1038/nrg1272 [DOI] [PubMed] [Google Scholar]
- Bastock R, Strutt H, Strutt D. Strabismus is asymmetrically localised and binds to Prickle and Dishevelled during Drosophila planar polarity patterning. Development. 2003;130:3007–3014. doi: 10.1242/dev.00526. doi:10.1242/dev.00526 [DOI] [PubMed] [Google Scholar]
- Belema Bedada F, Technau A, Ebelt H, Schulze M, Braun T. Activation of myogenic differentiation pathways in adult bone marrow-derived stem cells. Mol. Cell Biol. 2005;25:9509–9519. doi: 10.1128/MCB.25.21.9509-9519.2005. doi:10.1128/MCB.25.21.9509-9519.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernadinello L, De Cindio F. A survey of basic net models and modular net classes. Lect. Notes Comput. Sci. 1992;609:304–351. [Google Scholar]
- Berridge M.J. The AM and FM of calcium signalling. Nature. 1997;386:759–760. doi: 10.1038/386759a0. doi:10.1038/386759a0 [DOI] [PubMed] [Google Scholar]
- Birnbaumer, L. 2007_a_ Expansion of signal transduction by G proteins. The second 15 years or so: from 3 to 16 α subunits plus βγ dimers. Biochim. Biophys. Acta1768, 772–793. (doi:10.1016/j.bbamem.2006.12.002) [DOI] [PMC free article] [PubMed]
- Birnbaumer, L. 2007_b_ The discovery of signal transduction by G proteins. A personal account and an overview of the initial findings and contributions that led to our present understanding. Biochim. Biophys. Acta1768, 756–771. (doi:10.1016/j.bbamem.2006.09.027) [DOI] [PMC free article] [PubMed]
- Borchers A, Fonar Y, Frank D, Baker J.C. XNF-ATc3 affects neural convergent extension. Development. 2006;133:1745–1755. doi: 10.1242/dev.02343. doi:10.1242/dev.02343 [DOI] [PubMed] [Google Scholar]
- Boutros M, Paricio N, Strutt D.I, Mlodzik M. Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell. 1998;94:109–118. doi: 10.1016/s0092-8674(00)81226-x. doi:10.1016/S0092-8674(00)81226-X [DOI] [PubMed] [Google Scholar]
- Busa W.B, Gimlich R.L. Lithium-induced teratogenesis in frog embryos prevented by a polyphosphoinositide cycle intermediate or a diacylglycerol analog. Dev. Biol. 1989;132:315–324. doi: 10.1016/0012-1606(89)90228-5. doi:10.1016/0012-1606(89)90228-5 [DOI] [PubMed] [Google Scholar]
- Cadigan K.M, Liu Y.I. Wnt signaling: complexity at the surface. J. Cell Sci. 2006;119:395–402. doi: 10.1242/jcs.02826. doi:10.1242/jcs.02826 [DOI] [PubMed] [Google Scholar]
- Caneparo, L., Huang, Y.-L., Staudt, N., Masasumi, T., Ahrendt, R., Kazanskaya, O., Niehrs, C. & Houart, C. 2007 Dickkopf-1 regulates gastrulation movements by coordinated modulation of Wnt/β-catenin and Wnt/PCP activities, through interaction with the Dally-like homolog Knypek. Genes Dev 21, 465–480. (doi:10.1101/gad.406007) [DOI] [PMC free article] [PubMed]
- Cavodeassi F, Carreira-Barbosa F, Young R.M, Concha M.L, Allende M.L, Houart C, Tada M, Wilson S.W. Early stages of zebrafish eye formation require the coordinated activity of Wnt11, Fz5, and the Wnt/beta-catenin pathway. Neuron. 2005;47:43–56. doi: 10.1016/j.neuron.2005.05.026. doi:10.1016/j.neuron.2005.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.E, Ginty D.D, Fan C.M. Protein kinase a signalling via CREB controls myogenesis induced by Wnt proteins. Nature. 2005;433:317–322. doi: 10.1038/nature03126. doi:10.1038/nature03126 [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. doi:10.1016/j.cell.2006.10.018 [DOI] [PubMed] [Google Scholar]
- Collado-Vides J, Hofestädt R. MIT Press; Cambridge, MA: 2002. Gene regulation and metabolism: postgenomic computational approaches. [Google Scholar]
- Cong F, Schweizer L, Varmus H. Casein kinase Iepsilon modulates the signaling specificities of Dishevelled. Mol. Cell Biol. 2004;24:2000–2011. doi: 10.1128/MCB.24.5.2000-2011.2004. doi:10.1128/MCB.24.5.2000-2011.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta R, Kaykas A, Moon R.T, Perrimon N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science. 2005;308:826–833. doi: 10.1126/science.1109374. doi:10.1126/science.1109374 [DOI] [PubMed] [Google Scholar]
- Davidson G, Wu W, Shen J, Bilic J, Fenger U, Stannek P, Glinka A, Niehrs C. Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature. 2005;438:867–872. doi: 10.1038/nature04170. doi:10.1038/nature04170 [DOI] [PubMed] [Google Scholar]
- De Calisto J, Araya C, Marchant L, Riaz C.F, Mayor R. Essential role of non-canonical Wnt signalling in neural crest migration. Development. 2005;132:2587–2597. doi: 10.1242/dev.01857. doi:10.1242/dev.01857 [DOI] [PubMed] [Google Scholar]
- De Jong H. Modeling and simulation of genetic regulators systems: a literature review. J. Comput. Biol. 2002;9:67–103. doi: 10.1089/10665270252833208. doi:10.1089/10665270252833208 [DOI] [PubMed] [Google Scholar]
- Dejmek J, Safholm A, Kamp Nielsen C, Andersson T, Leandersson K. Wnt-5a/Ca2+-induced NFAT activity is counteracted by Wnt-5a/Yes-Cdc42-casein kinase 1α signaling in human mammary epithelial cells. Mol. Cell Biol. 2006;26:6024–6036. doi: 10.1128/MCB.02354-05. doi:10.1128/MCB.02354-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S.J, Purcell S.M, Christian J.L, McGrew L.L, Moon R.T. Identification of distinct classes and functional domains of Wnts through expression of wild-type and chimeric proteins in Xenopus embryos. Mol. Cell Biol. 1995;15:2625–2634. doi: 10.1128/mcb.15.5.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg C.A, Eisenberg L.M. WNT11 promotes cardiac tissue formation of early mesoderm. Dev. Dyn. 1999;216:45–58. doi: 10.1002/(SICI)1097-0177(199909)216:1<45::AID-DVDY7>3.0.CO;2-L. doi:10.1002/(SICI)1097-0177(199909)216:1<45::AID-DVDY7>3.0.CO;2-L [DOI] [PubMed] [Google Scholar]
- Eisenberg L.M, Eisenberg C.A. Cardiovascular molecular morphogenesis: myofibrillogenesis. Springer; New York, NY: 2002. Onset of a cardiac phenotype in the early embryo. [Google Scholar]
- Eisenberg C.A, Gourdie R.G, Eisenberg L.M. Wnt-11 is expressed in early avian mesoderm and required for the differentiation of the quail mesoderm cell line QCE-6. Development. 1997;124:525–536. doi: 10.1242/dev.124.2.525. [DOI] [PubMed] [Google Scholar]
- Fall C.P, Marland E.S, Wagner J.M, Tysson J.J. Springer; New York, NY: 2002. Computational cell biology. [Google Scholar]
- Fell D. Frontiers in metabolism 2. Portland Press; London, UK; Miami, FL: 1997. Understanding the control of metabolism. [Google Scholar]
- Garriock, R. J. & Krieg, P. A. 2007 Wnt11-R signaling regulates a calcium sensitive EMT event essential for dorsal fin development of Xenopus Dev. Biol 304, 127–140. (doi:10.1016/j.ydbio.2006.12.020) [DOI] [PMC free article] [PubMed]
- Garriock R.J, D'Agostino S.L, Pilcher K.C, Krieg P.A. Wnt11-R, a protein closely related to mammalian Wnt11, is required for heart morphogenesis in Xenopus. Dev. Biol. 2005;279:179–192. doi: 10.1016/j.ydbio.2004.12.013. doi:10.1016/j.ydbio.2004.12.013 [DOI] [PubMed] [Google Scholar]
- Glinka A, Wu W, Delius H, Monaghan A.P, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–362. doi: 10.1038/34848. doi:10.1038/34848 [DOI] [PubMed] [Google Scholar]
- He X, Saint-Jeannet J.P, Wang Y, Nathans J, Dawid I, Varmus H. A member of the Frizzled protein family mediating axis induction by Wnt-5A. Science. 1997;275:1652–1654. doi: 10.1126/science.275.5306.1652. doi:10.1126/science.275.5306.1652 [DOI] [PubMed] [Google Scholar]
- Heiner M, Koch I, Will J. Model validation of biological pathways using Petri nets—demonstrated for apoptosis. BioSystems. 2004;75:15–28. doi: 10.1016/j.biosystems.2004.03.003. doi:10.1016/j.biosystems.2004.03.003 [DOI] [PubMed] [Google Scholar]
- Heinrich R, Schuster S. Chapman and Hall; New York, NY: 1996. The regulation of cellular systems. [Google Scholar]
- Heisenberg C.P, et al. A mutation in the Gsk3-binding domain of zebrafish Masterblind/Axin1 leads to a fate transformation of telencephalon and eyes to diencephalon. Genes Dev. 2001;15:1427–1434. doi: 10.1101/gad.194301. doi:10.1101/gad.194301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikasa H, Shibata M, Hiratani I, Taira M. The Xenopus receptor tyrosine kinase Xror2 modulates morphogenetic movements of the axial mesoderm and neuroectoderm via Wnt signaling. Development. 2002;129:5227–5239. doi: 10.1242/dev.129.22.5227. [DOI] [PubMed] [Google Scholar]
- Hino S, Tanji C, Nakayama K.I, Kikuchi A. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination. Mol. Cell Biol. 2005;25:9063–9072. doi: 10.1128/MCB.25.20.9063-9072.2005. doi:10.1128/MCB.25.20.9063-9072.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberg J.J, Binder B, Bruggeman F.J, Schoeberl B, Heinrich R, Westerhoff H.V. Control of MAPK signalling: from complexity to what really matters. Oncogene. 2005;24:5533–5542. doi: 10.1038/sj.onc.1208817. doi:10.1038/sj.onc.1208817 [DOI] [PubMed] [Google Scholar]
- Inoue T, Oz H.S, Wiland D, Gharib S, Deshpande R, Hill R.J, Katz W.S, Sternberg P.W. C. elegans LIN-18 is a Ryk ortholog and functions in parallel to LIN-17/Frizzled in Wnt signaling. Cell. 2004;118:795–806. doi: 10.1016/j.cell.2004.09.001. doi:10.1016/j.cell.2004.09.001 [DOI] [PubMed] [Google Scholar]
- Ishitani T, et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature. 1999;399:798–802. doi: 10.1038/21674. doi:10.1038/21674 [DOI] [PubMed] [Google Scholar]
- Ishitani T, et al. The TAK1-NLK mitogen-activated protein kinase cascade functions in the Wnt-5a/Ca2+ pathway to antagonize Wnt/beta-catenin signaling. Mol. Cell Biol. 2003;23:131–139. doi: 10.1128/MCB.23.1.131-139.2003. doi:10.1128/MCB.23.1.131-139.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Reynolds-Kenneally J, Das G, Burnett M, Mlodzik M. Diego and Prickle regulate Frizzled planar cell polarity signalling by competing for Dishevelled binding. Nat. Cell Biol. 2005;7:691–697. doi: 10.1038/ncb1271. doi:10.1038/ncb1271 [DOI] [PubMed] [Google Scholar]
- Jopling C, den Hertog J. Fyn/Yes and non-canonical Wnt signalling converge on RhoA in vertebrate gastrulation cell movements. EMBO Rep. 2005;6:426–431. doi: 10.1038/sj.embor.7400386. doi:10.1038/sj.embor.7400386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kani S, et al. The receptor tyrosine kinase Ror2 associates with and is activated by casein kinase Iepsilon. J. Biol. Chem. 2004;279:50 102–50 109. doi: 10.1074/jbc.M409039200. doi:10.1074/jbc.M409039200 [DOI] [PubMed] [Google Scholar]
- Katanaev V.L, Ponzielli R, Semeriva M, Tomlinson A. Trimeric G protein-dependent frizzled signaling in Drosophila. Cell. 2005;120:111–122. doi: 10.1016/j.cell.2004.11.014. doi:10.1016/j.cell.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Kearns M.J, Vazirani U.V. MIT Press; Cambridge, MA: 1994. An introduction to computational learning theory. [Google Scholar]
- Kim D, Rath O, Kolch W, Cho K.-H. A hidden oncogenic positive feedback loop caused by crosstalk between Wnt and ERK pathways. Oncogene. 2007;26:4571–4579. doi: 10.1038/sj.onc.1210230. doi:10.1038/sj.onc.1210230 [DOI] [PubMed] [Google Scholar]
- Kinoshita N, Iioka H, Miyakoshi A, Ueno N. PKC delta is essential for Dishevelled function in a noncanonical Wnt pathway that regulates Xenopus convergent extension movements. Genes. Dev. 2003;17:1663–1676. doi: 10.1101/gad.1101303. doi:10.1101/gad.1101303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein T.J, Jenny A, Djiane A, Mlodzik M. CKIepsilon/discs overgrown promotes both Wnt-Fz/beta-catenin and Fz/PCP signaling in Drosophila. Curr. Biol. 2006;16:1337–1343. doi: 10.1016/j.cub.2006.06.030. doi:10.1016/j.cub.2006.06.030 [DOI] [PubMed] [Google Scholar]
- Klipp E, Herwig R, Kowald A, Wierling C, Lehrach H. Wiley-VCH; Weinheim, Germany: 2005. Systems biology in practice. [Google Scholar]
- Koyanagi M, Haendeler J, Badorff C, Brandes R.P, Hoffmann J, Pandur P, Zeiher A.M, Kuhl M, Dimmeler S. Non-canonical Wnt signaling enhances differentiation of human circulating progenitor cells to cardiomyogenic cells. J. Biol. Chem. 2005;280:16 838–16 842. doi: 10.1074/jbc.M500323200. doi:10.1074/jbc.M500323200 [DOI] [PubMed] [Google Scholar]
- Kremenevskaja N, von Wasielewski R, Rao A.S, Schofl C, Andersson T, Brabant G. Wnt-5a has tumor suppressor activity in thyroid carcinoma. Oncogene. 2005;24:2144–2154. doi: 10.1038/sj.onc.1208370. doi:10.1038/sj.onc.1208370 [DOI] [PubMed] [Google Scholar]
- Kuhl M, Sheldahl L.C, Malbon C.C, Moon R.T. Ca2+/calmodulin-dependent protein kinase II is stimulated by Wnt and Frizzled homologs and promotes ventral cell fates in Xenopus. J. Biol. Chem. 2000;275:12 701–12 711. doi: 10.1074/jbc.275.17.12701. doi:10.1074/jbc.275.17.12701 [DOI] [PubMed] [Google Scholar]
- Lee E, Salic A, Kruger R, Heinrich R, Kirschner M.W. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol. 2003;1:E10. doi: 10.1371/journal.pbio.0000010. doi:10.1371/journal.pbio.0000010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.Y, He B, You L, Xu Z, Mazieres J, Reguart N, Mikami I, Batra S, Jablons D.M. Dickkopf-1 antagonizes Wnt signaling independent of beta-catenin in human mesothelioma. Biochem. Biophys. Res. Commun. 2004;323:1246–1250. doi: 10.1016/j.bbrc.2004.09.001. doi:10.1016/j.bbrc.2004.09.001 [DOI] [PubMed] [Google Scholar]
- Lee H.S, Bong Y.S, Moore K.B, Soria K, Moody S.A, Daar I.O. Dishevelled mediates ephrinB1 signalling in the eye field through the planar cell polarity pathway. Nat. Cell Biol. 2006;8:55–63. doi: 10.1038/ncb1344. doi:10.1038/ncb1344 [DOI] [PubMed] [Google Scholar]
- Li L, Yuan H, Xie W, Mao J, Caruso A.M, McMahon A, Sussman D.J, Wu D. Dishevelled proteins lead to two signaling pathways. Regulation of LEF-1 and c-Jun N-terminal kinase in mammalian cells. J. Biol. Chem. 1999;274:129–134. doi: 10.1074/jbc.274.1.129. doi:10.1074/jbc.274.1.129 [DOI] [PubMed] [Google Scholar]
- Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. doi:10.1126/science.1091403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Fuhrman S, Somogyi R. Reveal, a general reverse engineering algorithm for inference of genetic network architecture. Pacific Symp. Biocomput. 1998;3:18–29. [PubMed] [Google Scholar]
- Liang H, Chen Q, Coles A.H, Anderson S.J, Pihan G, Bradley A, Gerstein R, Jurecic R, Jones S.N. Wnt5a inhibits B cell proliferation and functions as a tumor suppressor in hematopoietic tissue. Cancer Cell. 2003;4:349–360. doi: 10.1016/s1535-6108(03)00268-x. doi:10.1016/S1535-6108(03)00268-X [DOI] [PubMed] [Google Scholar]
- Liao G, Tao Q, Kofron M, Chen J.S, Schloemer A, Davis R.J, Hsieh J.C, Wylie C, Heasman J, Kuan C.Y. Jun NH2-terminal kinase (JNK) prevents nuclear beta-catenin accumulation and regulates axis formation in Xenopus embryos. Proc. Natl Acad. Sci. USA. 2006;103:16 313–16 318. doi: 10.1073/pnas.0602557103. doi:10.1073/pnas.0602557103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Sepich D.S, Chen S, Topczewski J, Yin C, Solnica-Krezel L, Hamm H. Essential roles of Gα12/13 signaling in distinct cell behaviors driving zebrafish convergence and extension gastrulation movements. J. Cell Biol. 2005;169:777–787. doi: 10.1083/jcb.200501104. doi:10.1083/jcb.200501104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Liu X, Wang H, Moon R.T, Malbon C.C. Activation of rat frizzled-1 promotes Wnt signaling and differentiation of mouse F9 teratocarcinoma cells via pathways that require Gαq and Gαo function. J. Biol. Chem. 1999a;274:33 539–33 544. doi: 10.1074/jbc.274.47.33539. doi:10.1074/jbc.274.47.33539 [DOI] [PubMed] [Google Scholar]
- Liu X, Liu T, Slusarski D.C, Yang-Snyder J, Malbon C.C, Moon R.T, Wang H. Activation of a frizzled-2/beta-adrenergic receptor chimera promotes Wnt signaling and differentiation of mouse F9 teratocarcinoma cells via Galphao and Galphat. Proc. Natl Acad. Sci. USA. 1999b;96:14 383–14 388. doi: 10.1073/pnas.96.25.14383. doi:10.1073/pnas.96.25.14383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, DeCostanzo A.J, Liu X, Wang H, Hallagan S, Moon R.T, Malbon C.C. G protein signaling from activated rat frizzled-1 to the beta-catenin-Lef-Tcf pathway. Science. 2001;292:1718–1722. doi: 10.1126/science.1060100. doi:10.1126/science.1060100 [DOI] [PubMed] [Google Scholar]
- Liu C, Li Y, Semenov M, Han C, Baeg G.H, Tan Y, Zhang Z, Lin X, He X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. doi:10.1016/S0092-8674(02)00685-2 [DOI] [PubMed] [Google Scholar]
- Liu X, Rubin J.S, Kimmel A.R. Rapid, Wnt-induced changes in GSK3beta associations that regulate beta-catenin stabilization are mediated by Galpha proteins. Curr. Biol. 2005;15:1989–1997. doi: 10.1016/j.cub.2005.10.050. doi:10.1016/j.cub.2005.10.050 [DOI] [PubMed] [Google Scholar]
- Logan C.Y, Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. doi:10.1146/annurev.cellbio.20.010403.113126 [DOI] [PubMed] [Google Scholar]
- Lu W, Yamamoto V, Ortega B, Baltimore D. Mammalian Ryk is a Wnt coreceptor required for stimulation of neurite outgrowth. Cell. 2004;119:97–108. doi: 10.1016/j.cell.2004.09.019. doi:10.1016/j.cell.2004.09.019 [DOI] [PubMed] [Google Scholar]
- Lyuksyutova A.I, Lu C.C, Milanesio N, King L.A, Guo N, Wang Y, Nathans J, Tessier-Lavigne M, Zou Y. Anterior–posterior guidance of commissural axons by Wnt-frizzled signaling. Science. 2003;302:1984–1988. doi: 10.1126/science.1089610. doi:10.1126/science.1089610 [DOI] [PubMed] [Google Scholar]
- Ma L, Wang H.Y. Suppression of cyclic GMP-dependent protein kinase is essential to the Wnt/cGMP/Ca2+ pathway. J. Biol. Chem. 2006;281:30 990–31 001. doi: 10.1074/jbc.M603603200. doi:10.1074/jbc.M603603200 [DOI] [PubMed] [Google Scholar]
- Ma Y.C, Huang J, Ali S, Lowry W, Huang X.Y. Src tyrosine kinase is a novel direct effector of G proteins. Cell. 2000;102:635–646. doi: 10.1016/s0092-8674(00)00086-6. doi:10.1016/S0092-8674(00)00086-6 [DOI] [PubMed] [Google Scholar]
- Ma'ayan A, Blitzer R.D, Iyengar R. Toward predictive models of mammalian cells. Annu. Rev. Biophys. Biomol. Struct. 2005;34:319–349. doi: 10.1146/annurev.biophys.34.040204.144415. doi:10.1146/annurev.biophys.34.040204.144415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin M.J, Di Rocco G, Gardiner A, Bush S.M, Lassar A.B. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 2001;15:316–327. doi: 10.1101/gad.855501. doi:10.1101/gad.855501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Raya A, Kawakami Y, Callol-Massot C, Capdevila J, Rodriguez-Esteban C, Izpisua Belmonte J.C. Noncanonical Wnt signaling regulates midline convergence of organ primordia during zebrafish development. Genes Dev. 2005;19:164–175. doi: 10.1101/gad.1253605. doi:10.1101/gad.1253605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurus D, Heligon C, Burger-Schwarzler A, Brandli A.W, Kuhl M. Noncanonical Wnt-4 signaling and EAF2 are required for eye development in Xenopus laevis. EMBO J. 2005;24:1181–1191. doi: 10.1038/sj.emboj.7600603. doi:10.1038/sj.emboj.7600603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maye P, Zheng J, Li L, Wu D. Multiple mechanisms for Wnt11-mediated repression of the canonical Wnt signaling pathway. J. Biol. Chem. 2004;279:24 659–24 665. doi: 10.1074/jbc.M311724200. doi:10.1074/jbc.M311724200 [DOI] [PubMed] [Google Scholar]
- Medina A, Reintsch W, Steinbeisser H. Xenopus frizzled 7 can act in canonical and non-canonical Wnt signaling pathways: implications on early patterning and morphogenesis. Mech. Dev. 2000;92:227–237. doi: 10.1016/s0925-4773(00)00240-9. doi:10.1016/S0925-4773(00)00240-9 [DOI] [PubMed] [Google Scholar]
- Meneghini M.D, Ishitani T, Carter J.C, Hisamoto N, Ninomiya-Tsuji J, Thorpe C.J, Hamill D.R, Matsumoto K, Bowerman B. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature. 1999;399:793–797. doi: 10.1038/21666. doi:10.1038/21666 [DOI] [PubMed] [Google Scholar]
- Mikels A.J, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. doi:10.1371/journal.pbio.0040115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. doi:10.1126/science.298.5594.824 [DOI] [PubMed] [Google Scholar]
- Naito, A. T., Shiojima, I., Akazawa, H., Hidaka, K., Morisaki, T., Kikuchi, A. & Komuro, I. 2007 Developmental stage-specific biphasic roles of Wnt/β-catenin signaling in cardiomyogenesis and hematopoiesis. Proc. Natl Acad. Sci. USA103, 19 812–19 817. (doi:10.1073/pnas.0605768103) [DOI] [PMC free article] [PubMed]
- Nakamura T, Sano M, Songyang Z, Schneider M.D. A Wnt- and beta-catenin-dependent pathway for mammalian cardiac myogenesis. Proc. Natl Acad. Sci. USA. 2003;100:5834–5839. doi: 10.1073/pnas.0935626100. doi:10.1073/pnas.0935626100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi I, et al. The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes Cells. 2003;8:645–654. doi: 10.1046/j.1365-2443.2003.00662.x. doi:10.1046/j.1365-2443.2003.00662.x [DOI] [PubMed] [Google Scholar]
- Okamura H, Garcia-Rodriguez C, Martinson H, Qin J, Virshup D.M, Rao A. A conserved docking motif for CK1 binding controls the nuclear localization of NFAT1. Mol. Cell Biol. 2004;24:4184–4195. doi: 10.1128/MCB.24.10.4184-4195.2004. doi:10.1128/MCB.24.10.4184-4195.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouko L, Ziegler T.R, Gu L.H, Eisenberg L.M, Yang V.W. Wnt11 signaling promotes proliferation, transformation, and migration of IEC6 intestinal epithelial cells. J. Biol. Chem. 2004;279:26 707–26 715. doi: 10.1074/jbc.M402877200. doi:10.1074/jbc.M402877200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandur P, Lasche M, Eisenberg L.M, Kuhl M. Wnt-11 activation of a non-canonical Wnt signalling pathway is required for cardiogenesis. Nature. 2002;418:636–641. doi: 10.1038/nature00921. doi:10.1038/nature00921 [DOI] [PubMed] [Google Scholar]
- Park E, Kim G.H, Choi S.C, Han J.K. Role of PKA as a negative regulator of PCP signaling pathway during Xenopus gastrulation movements. Dev. Biol. 2006;292:344–357. doi: 10.1016/j.ydbio.2006.01.011. doi:10.1016/j.ydbio.2006.01.011 [DOI] [PubMed] [Google Scholar]
- Penzo-Mendèz A, Umbhauer M, Djiane A, Boucaut J.-C, Riou J.-F. Activation of Gβγ signaling downstream of Wnt-11/Xfz7 regulates Cdc42 activity during Xenopus gastrulation. Dev. Biol. 2003;257:302–314. doi: 10.1016/s0012-1606(03)00067-8. doi:10.1016/s0012-1606(03)00067-8 [DOI] [PubMed] [Google Scholar]
- Pinson K.I, Brennan J, Monkley S, Avery B.J, Skarnes W.C. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407:535–538. doi: 10.1038/35035124. doi:10.1038/35035124 [DOI] [PubMed] [Google Scholar]
- Pourquie O. Signal transduction: a new canon. Nature. 2005;433:208–209. doi: 10.1038/433208a. doi:10.1038/433208a [DOI] [PubMed] [Google Scholar]
- Rasmussen J.T, Deardorff M.A, Tan C, Rao M.S, Klein P.S, Vetter M.L. Regulation of eye development by frizzled signaling in Xenopus. Proc. Natl Acad. Sci. USA. 2001;98:3861–3866. doi: 10.1073/pnas.071586298. doi:10.1073/pnas.071586298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille J, et al. Mutant frizzled-4 disrupts retinal angiogenesis in familial exudative vitreoretinopathy. Nat. Genet. 2002;32:326–330. doi: 10.1038/ng957. doi:10.1038/ng957 [DOI] [PubMed] [Google Scholar]
- Roskoski R., Jr Src kinase regulation by phosphorylation and dephosphorylation. Biochem. Biophys. Res. Commun. 2005;331:1–14. doi: 10.1016/j.bbrc.2005.03.012. doi:10.1016/j.bbrc.2005.03.012 [DOI] [PubMed] [Google Scholar]
- Safholm A, Leandersson K, Dejmek J, Nielsen C.K, Villoutreix B.O, Andersson T. A formylated hexapeptide ligand mimics the ability of Wnt-5a to impair migration of human breast epithelial cells. J. Biol. Chem. 2006;281:2740–2749. doi: 10.1074/jbc.M508386200. doi:10.1074/jbc.M508386200 [DOI] [PubMed] [Google Scholar]
- Saneyoshi T, Kume S, Amasaki Y, Mikoshiba K. The Wnt/calcium pathway activates NF-AT and promotes ventral cell fate in Xenopus embryos. Nature. 2002;417:295–299. doi: 10.1038/417295a. doi:10.1038/417295a [DOI] [PubMed] [Google Scholar]
- Schneider V.A, Mercola M. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev. 2001;15:304–315. doi: 10.1101/gad.855601. doi:10.1101/gad.855601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze M, Belema-Bedada F, Technau A, Braun T. Mesenchymal stem cells are recruited to striated muscle by NFAT/IL-4-mediated cell fusion. Genes Dev. 2005;19:1787–1798. doi: 10.1101/gad.339305. doi:10.1101/gad.339305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Romond T, Asbrand C, Bakkers J, Kuhl M, Schaeffer H.J, Huelsken J, Behrens J, Hammerschmidt M, Birchmeier W. The ankyrin repeat protein Diversin recruits casein kinase Iepsilon to the beta-catenin degradation complex and acts in both canonical Wnt and Wnt/JNK signaling. Genes Dev. 2002;16:2073–2084. doi: 10.1101/gad.230402. doi:10.1101/gad.230402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenov M.V, Tamai K, Brott B.K, Kuhl M, Sokol S, He X. Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Curr. Biol. 2001;11:951–961. doi: 10.1016/s0960-9822(01)00290-1. doi:10.1016/S0960-9822(01)00290-1 [DOI] [PubMed] [Google Scholar]
- Sharma D, Holets L, Zhang X, Kinsey W.H. Role of Fyn kinase in signaling associated with epiboly during zebrafish development. Dev. Biol. 2005;285:462–476. doi: 10.1016/j.ydbio.2005.07.018. doi:10.1016/j.ydbio.2005.07.018 [DOI] [PubMed] [Google Scholar]
- Sheldahl L.C, Park M, Malbon C.C, Moon R.T. Protein kinase C is differentially stimulated by Wnt and Frizzled homologs in a G-protein-dependent manner. Curr. Biol. 1999;9:695–698. doi: 10.1016/s0960-9822(99)80310-8. doi:10.1016/S0960-9822(99)80310-8 [DOI] [PubMed] [Google Scholar]
- Sheldahl L.C, Slusarski D.C, Pandur P, Miller J.R, Kuhl M, Moon R.T. Dishevelled activates Ca2+ flux, PKC, and CamKII in vertebrate embryos. J. Cell Biol. 2003;161:769–777. doi: 10.1083/jcb.200211094. doi:10.1083/jcb.200211094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusarski D.C, Corces V.G, Moon R.T. Interaction of Wnt and a Frizzled homologue triggers G-protein-linked phosphatidylinositol signalling. Nature. 1997a;390:410–413. doi: 10.1038/37138. doi:10.1038/37138 [DOI] [PubMed] [Google Scholar]
- Slusarski D.C, Yang-Snyder J, Busa W.B, Moon R.T. Modulation of embryonic intracellular Ca2+ signaling by Wnt-5A. Dev. Biol. 1997b;182:114–120. doi: 10.1006/dbio.1996.8463. doi:10.1006/dbio.1996.8463 [DOI] [PubMed] [Google Scholar]
- Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. doi:10.1016/j.cell.2005.08.029 [DOI] [PubMed] [Google Scholar]
- Stemmle L.N, Fields T.A, Casey P.J. The regulator of G protein signaling domain of axin selectively interacts with Galpha12 but not Galpha13. Mol. Pharmacol. 2006;70:1461–1468. doi: 10.1124/mol.106.023705. doi:10.1124/mol.106.023705 [DOI] [PubMed] [Google Scholar]
- Stoick-Cooper C.L, Weidinger G, Riehle K.J, Hubbert C, Major M.B, Fausto N, Moon R.T. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–489. doi: 10.1242/dev.001123. doi:10.1242/dev.001123 [DOI] [PubMed] [Google Scholar]
- Strutt D. Frizzled signalling and cell polarisation in Drosophila and vertebrates. Development. 2003;130:4501–4513. doi: 10.1242/dev.00695. doi:10.1242/dev.00695 [DOI] [PubMed] [Google Scholar]
- Strutt H, Price M.A, Strutt D. Planar polarity is positively regulated by casein kinase Iepsilon in Drosophila. Curr. Biol. 2006;16:1329–1336. doi: 10.1016/j.cub.2006.04.041. doi:10.1016/j.cub.2006.04.041 [DOI] [PubMed] [Google Scholar]
- Swiatek W, Tsai I.C, Klimowski L, Pepler A, Barnette J, Yost H.J, Virshup D.M. Regulation of casein kinase I epsilon activity by Wnt signaling. J. Biol. Chem. 2004;279:13 011–13 017. doi: 10.1074/jbc.M304682200. doi:10.1074/jbc.M304682200 [DOI] [PubMed] [Google Scholar]
- Szallasi Z, Stelling J, Periwal V. MIT Press; Cambridge, MA: 2006. System modeling in cellular biology. [Google Scholar]
- Takada R, Satomi Y, Kurata T, Ueno N, Norioka S, Kondoh H, Takao T, Takada S. Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev. Cell. 2006;11:791–801. doi: 10.1016/j.devcel.2006.10.003. doi:10.1016/j.devcel.2006.10.003 [DOI] [PubMed] [Google Scholar]
- Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet J.P, He X. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. doi:10.1038/35035117 [DOI] [PubMed] [Google Scholar]
- Tao W, Pennica D, Xu L, Kalejta R.F, Levine A.J. Wrch-1, a novel member of the Rho gene family that is regulated by Wnt-1. Genes Dev. 2001;15:1796–1807. doi: 10.1101/gad.894301. doi:10.1101/gad.894301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terami H, Hidaka K, Katsumata T, Iio A, Morisaki T. Wnt-11 facilitates embryonic stem cell differentiation to Nkx2.5-positive cardiomyocytes. Biochem. Biophys. Res. Commun. 2004;325:968–975. doi: 10.1016/j.bbrc.2004.10.103. doi:10.1016/j.bbrc.2004.10.103 [DOI] [PubMed] [Google Scholar]
- Topol L, Jiang X, Choi H, Garrett-Beal L, Carolan P.J, Yang Y. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent beta-catenin degradation. J. Cell Biol. 2003;162:899–908. doi: 10.1083/jcb.200303158. doi:10.1083/jcb.200303158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M.A, Yang-Snyder J.A, Purcell S.M, DeMarais A.A, McGrew L.L, Moon R.T. Activities of the Wnt-1 class of secreted signaling factors are antagonized by the Wnt-5A class and by a dominant negative cadherin in early Xenopus development. J. Cell Biol. 1996;133:1123–1137. doi: 10.1083/jcb.133.5.1123. doi:10.1083/jcb.133.5.1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X, Joeng K.S, Nakayama K.I, Nakayama K, Rajagopal J, Carroll T.J, McMahon A.P, Long F. Noncanonical Wnt signaling through G protein-linked PKCdelta activation promotes bone formation. Dev. Cell. 2007;12:113–127. doi: 10.1016/j.devcel.2006.11.00. doi:10.1016/j.devcel.2006.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzahor E, Lassar A.B. Wnt signals from the neural tube block ectopic cardiogenesis. Genes Dev. 2001;15:255–260. doi: 10.1101/gad.871501. doi:10.1101/gad.871501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raay T.J, Moore K.B, Iordanova I, Steele M, Jamrich M, Harris W.A, Vetter M.L. Frizzled 5 signaling governs the neural potential of progenitors in the developing Xenopus retina. Neuron. 2005;46:23–36. doi: 10.1016/j.neuron.2005.02.023. doi:10.1016/j.neuron.2005.02.023 [DOI] [PubMed] [Google Scholar]
- Veeman M.T, Axelrod J.D, Moon R.T. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev. Cell. 2003a;5:367–377. doi: 10.1016/s1534-5807(03)00266-1. doi:10.1016/S1534-5807(03)00266-1 [DOI] [PubMed] [Google Scholar]
- Veeman M.T, Slusarski D.C, Kaykas A, Louie S.H, Moon R.T. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 2003b;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. doi:10.1016/S0960-9822(03)00240-9 [DOI] [PubMed] [Google Scholar]
- von Dassow G, Meir E, Munro E.M, Odell G.M. The segment polarity network is a robust developmental module. Nature. 2000;406:188–192. doi: 10.1038/35018085. doi:10.1038/35018085 [DOI] [PubMed] [Google Scholar]
- Weber U, Paricio N, Mlodzik M. Jun mediates Frizzled-induced R3/R4 cell fate distinction and planar polarity determination in the Drosophila eye. Development. 2000;127:3619–3629. doi: 10.1242/dev.127.16.3619. [DOI] [PubMed] [Google Scholar]
- Weeraratna A.T, Jiang Y, Hostetter G, Rosenblatt K, Duray P, Bittner M, Trent J.M. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1:279–288. doi: 10.1016/s1535-6108(02)00045-4. doi:10.1016/S1535-6108(02)00045-4 [DOI] [PubMed] [Google Scholar]
- Wehrli M, Dougan S.T, Caldwell K, O'Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, DiNardo S. Arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature. 2000;407:527–530. doi: 10.1038/35035110. doi:10.1038/35035110 [DOI] [PubMed] [Google Scholar]
- Westfall T.A, Brimeyer R, Twedt J, Gladon J, Olberding A, Furutani-Seiki M, Slusarski D.C. Wnt-5/pipetail functions in vertebrate axis formation as a negative regulator of Wnt/beta-catenin activity. J. Cell Biol. 2003;162:889–898. doi: 10.1083/jcb.200303107. doi:10.1083/jcb.200303107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton K.A, Jr, Zimmermann G, Rousset R, Scott M.P. Vertebrate proteins related to Drosophila naked cuticle bind Dishevelled and antagonize Wnt signaling. Dev. Biol. 2001;234:93–106. doi: 10.1006/dbio.2001.0238. doi:10.1006/dbio.2001.0238 [DOI] [PubMed] [Google Scholar]
- Willert K, Jones K.A. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. doi:10.1101/gad.1424006 [DOI] [PubMed] [Google Scholar]
- Willert K, Brown J.D, Danenberg E, Duncan A.W, Weissman I.L, Reya T, Yates J.R, III, Nusse R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–452. doi: 10.1038/nature01611. doi:10.1038/nature01611 [DOI] [PubMed] [Google Scholar]
- Winklbauer R, Medina A, Swain R.K, Steinbeisser H. Frizzled-7 signalling controls tissue separation during Xenopus gastrulation. Nature. 2001;413:856–860. doi: 10.1038/35101621. doi:10.1038/35101621 [DOI] [PubMed] [Google Scholar]
- Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. doi:10.1146/annurev.cellbio.14.1.59 [DOI] [PubMed] [Google Scholar]
- Wu C, Zeng Q, Blumer K.J, Muslin A.J. RGS proteins inhibit Xwnt-8 signaling in Xenopus embryonic development. Development. 2000;127:2773–2784. doi: 10.1242/dev.127.13.2773. [DOI] [PubMed] [Google Scholar]
- Yamanaka H, Moriguchi T, Masuyama N, Kusakabe M, Hanafusa H, Takada R, Takada S, Nishida E. JNK functions in the non-canonical Wnt pathway to regulate convergent extension movements in vertebrates. EMBO Rep. 2002;3:69–75. doi: 10.1093/embo-reports/kvf008. doi:10.1093/embo-reports/kvf008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Wallingford J.B, Sun T.Q, Nelson A.M, Sakanaka C, Reinhard C, Harland R.M, Fantl W.J, Williams L.T. Cell autonomous regulation of multiple Dishevelled-dependent pathways by mammalian Nkd. Proc. Natl Acad. Sci. USA. 2001;98:3802–3807. doi: 10.1073/pnas.071041898. doi:10.1073/pnas.071041898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama N, Miller W.T. Biochemical properties of the Cdc42-associated tyrosine kinase ACK1. Substrate specificity, authphosphorylation, and interaction with Hck. J. Biol. Chem. 2003;278:47 713–47 723. doi: 10.1074/jbc.M306716200. doi:10.1074/jbc.M306716200 [DOI] [PubMed] [Google Scholar]
- Yook S.H, Oltvai Z.N, Barabasi A.L. Functional and topological characterization of protein interaction networks. Proteomics. 2004;4:928–942. doi: 10.1002/pmic.200300636. doi:10.1002/pmic.200300636 [DOI] [PubMed] [Google Scholar]
- Zeng W, Wharton K.A, Jr, Mack J.A, Wang K, Gadbaw M, Suyama K, Klein P.S, Scott M.P. Naked cuticle encodes an inducible antagonist of Wnt signalling. Nature. 2000;403:789–795. doi: 10.1038/35001615. doi:10.1038/35001615 [DOI] [PubMed] [Google Scholar]
- Zeng X, Tamai K, Doble B, Li S, Huang H, Habas R, Okamura H, Woodgett J, He X. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature. 2005;438:873–877. doi: 10.1038/nature04185. doi:10.1038/nature04185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Shibasaki F, Price R, Guillemot J.C, Yano T, Dotsch V, Wagner G, Ferrara P, McKeon F. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell. 1998;93:851–861. doi: 10.1016/s0092-8674(00)81445-2. doi:10.1016/S0092-8674(00)81445-2 [DOI] [PubMed] [Google Scholar]