Global analysis of host-pathogen interactions that regulate early stage HIV-1 replication (original) (raw)
. Author manuscript; available in PMC: 2009 Oct 3.
Abstract
Human Immunodeficiency Viruses (HIV-1 and HIV-2) rely upon host-encoded proteins to facilitate their replication. Here we combined genome-wide siRNA analyses with interrogation of human interactome databases to assemble a host-pathogen biochemical network containing 213 confirmed host cellular factors and 11 HIV-1-encoded proteins. Protein complexes that regulate ubiquitin conjugation, proteolysis, DNA damage response and RNA splicing were identified as important modulators of early stage HIV-1 infection. Additionally, over 40 new factors were shown to specifically influence initiation and/or kinetics of HIV-1 DNA synthesis, including cytoskeletal regulatory proteins, modulators of post-translational modification, and nucleic acid binding proteins. Finally, fifteen proteins with diverse functional roles, including nuclear transport, prostaglandin synthesis, ubiquitination, and transcription, were found to influence nuclear import or viral DNA integration. Taken together, the multi-scale approach described here has uncovered multiprotein virus-host interactions that likely act in concert to facilitate early steps of HIV-1 infection.
Introduction
Over the course of the last several decades a number of host cell proteins that influence early steps of retroviral replication have been defined (Goff, 2007). However, it is likely that many other cellular factors and processes are exploited by the virus during these stages, which involves uncoating steps that give rise to an active reverse transcription complex (RTC), movement of the viral preintegration complex (PIC) to the cell nucleus, and then integration of the linear viral DNA into a host cell chromosome to generate the provirus (Nisole and Saib, 2004). Recently, a genome-wide siRNA analysis revealed over 250 host cellular factors that influence HIV-1 infection (Brass et al., 2008). Notably, only a small fraction of these factors were proposed to influence the early stages of HIV-1 replication, making it likely that additional cellular factors that regulate these steps remain to be identified. Here we present a genome-wide analysis of viral-host interactions affecting early steps of HIV-1 infection.
Results
In this study, we combined several genome-wide analytical methods to characterize host factors required in the early steps of HIV infection. We performed genome-wide RNAi screens for genes required for infection utilizing a single cycle HIV-1 reporter virus engineered to encode luciferase and bearing the Vesicular Stomatitis Virus Glycoprotein (VSV-G) on its surface to facilitate efficient infection (Figure 1a). As controls, parallel screens were performed with other viral vectors encoding luciferase; 1) Moloney murine leukemia virus (MuLV) vector pseudotyped with VSV-G, and 2) an adeno-associated virus (AAV) vector (Figure S1). Prior to infection, human 293T cells were transfected with an arrayed genome-wide siRNA library, which targets approximately 20,000 human genes. Typically, six unique siRNAs were used to interrogate each gene, with two siRNAs targeting the same gene arrayed in a single well (3 wells/gene) (Figure 1b). The toxicity associated with each pair of siRNAs was also measured by assaying viable cell number (Figure 1b).
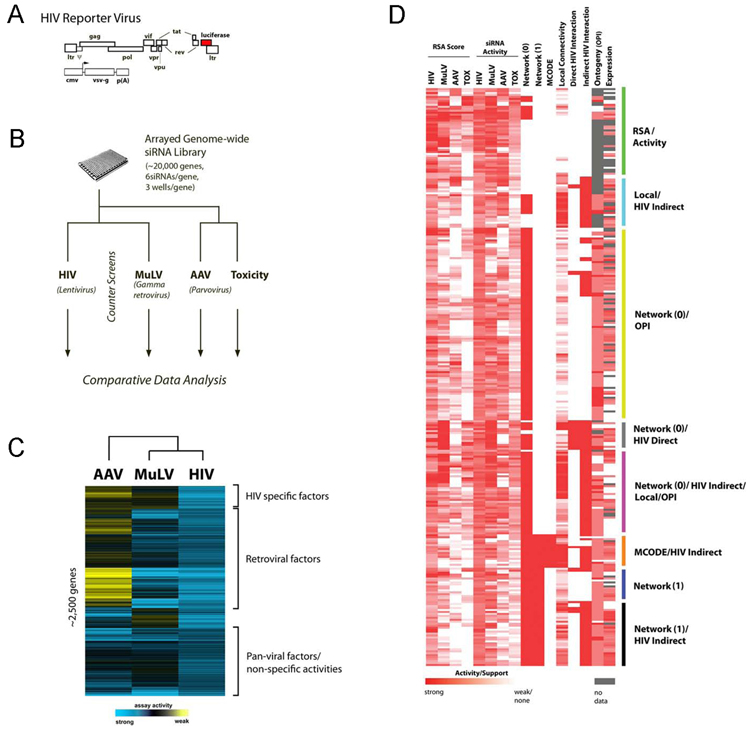
Figure 1. Integrative analysis of HIV-host interactions.
(a) To monitor early stages of HIV replication, we carried out infections using a single cycle, VSV-G pseudotyped, HIV-1 vector encoding luciferase. (b) Human 293T cells were transfected with an arrayed genome-wide siRNA library and then challenged with pNL43R+E− luc [VSV-G]. Infection was monitored by measuring luciferase activity. Additionally, counterscreens were performed to identify those host factors affecting gamma retrovirus (MuLV) or parvovirus (AAV) infection, as well as cellular viability (toxicity). (c) Inhibition data for each gene was normalized, scaled, and enriched for genes which have at least 2 siRNAs supporting their activity (RSA analysis). Comparative analysis of all viral screens, after filtering for toxicity, was visualized through hierarchical clustering. (d) To facilitate the validation of biologically relevant functional and biochemical activities, we employed a multi-scale approach to select candidate genes for further study. In addition to identifying genes based on either gene (RSA) or single siRNA activity in the genome-wide assay, an ‘evidence score’ was compiled for each gene based upon a variety of other supporting criteria. Specifically, proteins, which were encompassed in statistically significant host protein interaction networks (Network 0, Network 1, MCODE), or had significant network connectivity with other putative host factors (local connectivity) or HIV encoded proteins (Direct/Indirect HIV interaction), were given additional consideration. Furthermore, genes identified through the genome-wide assay, which were members of over-represented functional groups (ontology/OPI), or had coincident expression with CD4/CXCR4 or CD4/CXCR5, were also given additional weight. The plot depicts the activities and evidence support of 295 genes for which activities were subsequently confirmed by two or more siRNAs (Table S3).
Since the primary screen was executed in an arrayed format, we were able to employ Redundant siRNA Analysis (RSA) to identify genes that were significantly inhibited by at least two independent siRNAs, many of which possessed reproducible, but moderate activities (45% or greater reduction of HIV infectivity) (Konig et al., 2007). This analysis revealed that a significant fraction of host proteins required by HIV-1 are also required by MuLV (80%; Figure 1c). These proteins included a number of factors that are already known to affect retroviral replication (Brass et al., 2008; Goff, 2007) (Table S1).
To characterize the interactions between HIV-1 and these host cell proteins, we developed an integrative approach that considered additional lines of functional, biochemical, or transcriptional data (collectively called ‘evidence score’), for candidate gene verification. This approach is based on the assumption that a cellular factor is more likely to be a proximal regulator of viral replication if its activity is supported by multiple independent lines of evidence.
Using a yeast two-hybrid (Y2H) human protein-protein interaction database (Hynet) (Mukherji et al., 2006), we identified an extended interactome network, which contained 2458 putative host cellular factors affecting HIV infection, which form 120,211 direct or indirect interactions (network 0; data not shown). This network map was further refined by removing protein clusters, which influenced infection by the AAV vector or were associated with cellular toxicity (network 1; p<0.001; Figure S2a). Additionally, we applied a graph theoretic clustering algorithm (MCODE) to network 1 to identify regions of locally enhanced connectivity (Figure S2b-g)(Bader and Hogue, 2003). Finally, since the Hynet Y2H dataset employed in these analyses did not contain viral/host protein-protein interactions, we integrated protein interaction relationships contained in the NCBI HIV-1 Protein Interaction database (http://www.ncbi.nlm.nih.gov/RefSeq/HIVInteractions/), as well as a newly generated HIV/host Y2H interaction data in which each of the HIV-encoded proteins was tested individually against a library of all human proteins (see Experimental Procedure). The resulting host-pathogen interaction map between network 0 proteins and HIV-encoded proteins was determined to be of high statistical significance (direct interactions, p=0.005; indirect interactions, p=10−103, see supporting online text).
In an effort to identify those genes most relevant to HIV-1 infection, the mRNA expression profiles of the identified host genes were also correlated to those of the viral receptor CD4 and coreceptors CXCR4 and CCR5. The majority of host proteins in network 0 displayed strong expression patterns in tissues of lymphoid and neuronal origin (Figure S3). In a subsequent analysis for each candidate host factor, we calculated correlation coefficients with CD4/CXCR4 and CD4/CCR5 co-expression, and incorporated this value into the evidence score (see Experimental Procedure). Finally, to correlate gene functions, processes, components, or domains with gene activities across the four assays previously described (siRNA screens against the three viral vectors, and cytotoxicity), we employed an ontogeny-based pattern identification algorithm (OPI), and identified over 100 enriched activity signatures across the four genome-wide screens (Figure S4, Table S2). Since OPI activity profiles are supported by multiple functionally related genes and are statistically reliable (p<0.05), they were also considered as favorable evidence for the elucidation of physiologically relevant host cellular factors.
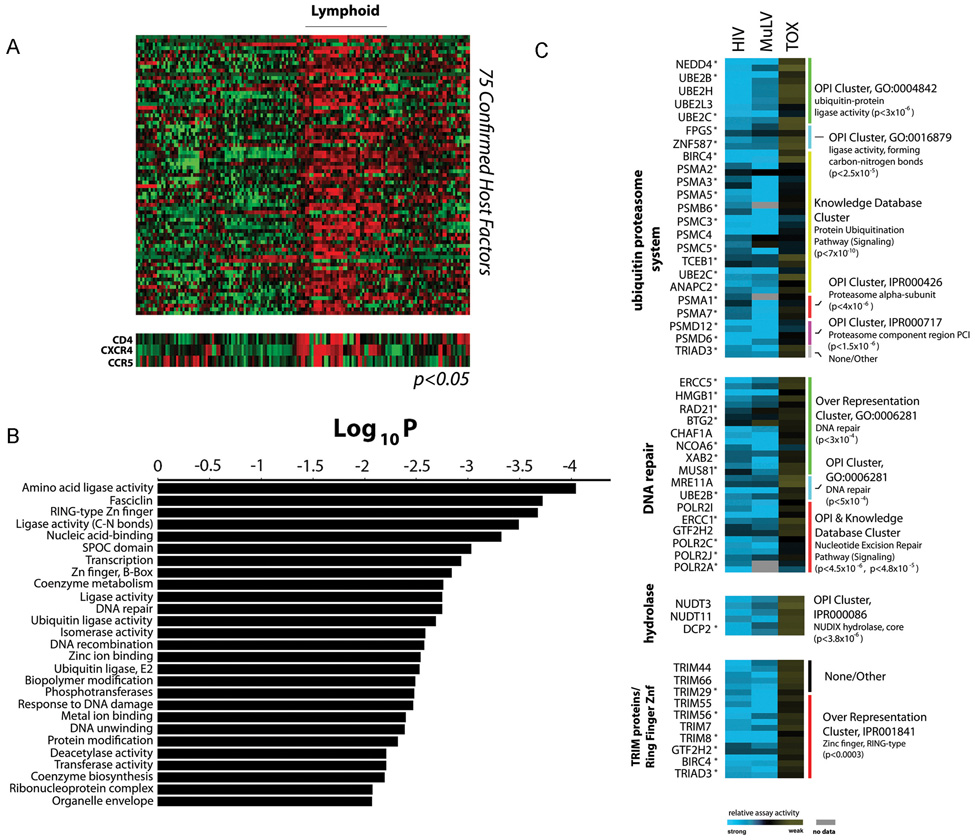
We combined these lines of evidence into a decision matrix to prioritize those genes that are likely regulators of HIV-1 replication. Criteria included gene-based (RSA score) and siRNA-based activity, gene expression signatures, gene ontology data (OPI), and cellular protein-protein interaction (using Network 0, Network 1, MCODE, and local connectivity) and viral-host interaction data (HIV Direct/Indirect) (Figure 1d, Figure S2, Table S2, data not shown). This enabled us to identify approximately 800 strongly supported genes based on evidence scores, of which 295 have been confirmed to inhibit HIV (VSV-G) infection more than 45% with at least 2 siRNAs, while not significantly affecting cellular viability (Table S3). Most confirmed factors showed broad expression in cell types relevant to HIV pathogenesis (Figure S3), and in addition, 75 of the confirmed factors share highly coincident expression patterns with CD4/CXCR4 or CD4/CCR5 (Figure 2a and Figure 3).
Figure 2. Characterization of confirmed factors required for infection by the VSV-G pseudotyped HIV-1 vector.
(a) Tissue expression of 75 confirmed host cellular factors with statistically coincident expression profiles to HIV receptor/co-receptors CD4/CXCR4 or CD4/CXCR5 (p<0.05, standard χ2-Chi-square test, _R_CD4,CXCR4 ≥ 0.18, _R_CD4,CCR5 ≥ 0.13). (b) Statistically significant over-representation of functional classes and protein families based upon gene ontology (GO) and interpro (IPR) domain mapping of 295 confirmed host factors required for HIV infection (Table S4). P values were calculated by an accumulated hypergeometric distribution function (Zar, 1999) (c) Relative activities of confirmed genes, each represented by two active siRNAs, across HIV, MuLV, and cytotoxicity assays (TOX) is shown from low (blue) to high (yellow) (Table S2, Figure S4). Functional classes are derived from ontogeny-based pattern identification algorithm (OPI & Knowledge Database Clusters) or GO/IPR over-representation analysis of primary screening data. All measurements represent the mean of at least four assays, and activities not tested or unconfirmed are depicted in grey. To identify those factors most likely to influence the intracellular steps of HIV-1 replication, a subgroup of genes were further assayed and confirmed to inhibit infection by an HIV-1 vector pseudotyped with the 10A1 MuLV envelope protein, a viral envelope protein that directs pH-independent viral entry at the plasma membrane. These genes are indicated with an asterisk (*)(See also Table S3).
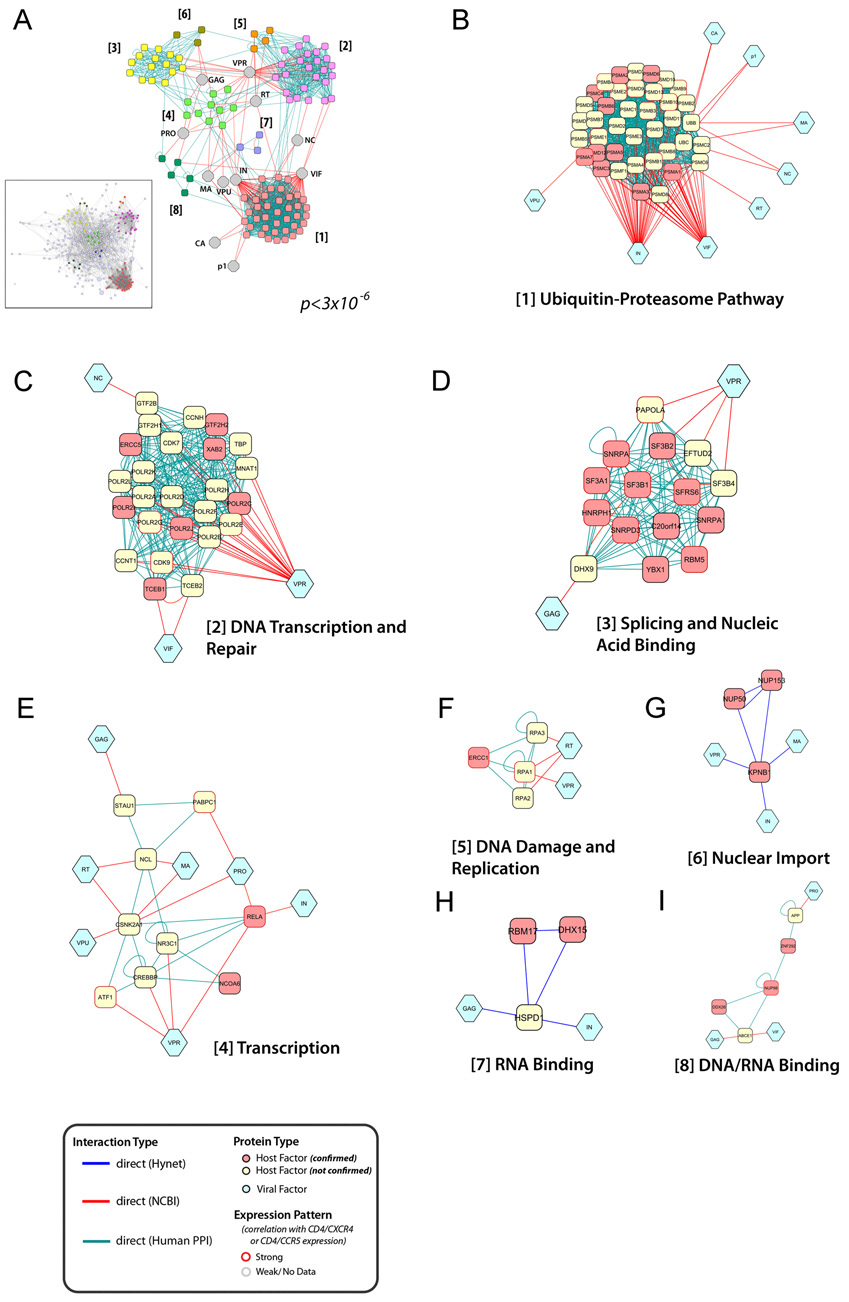
Figure 3. Network topology of HIV-host protein interactions.
The interaction network was elucidated based upon protein-protein binding data derived from the Hynet yeast two-hybrid database (blue connectors) and additional various human protein interaction databases (green connectors). Furthermore, connections to HIV encoded proteins (light blue) was completed by incorporation of data from the HIV-1 Human interaction database (NIAID) (red connectors). (a, inset) The resulting network contained 2291 direct protein interactions, connecting 213 confirmed host cellular factors, 11 HIV proteins, and 169 additional human proteins, which interact directly to at least two confirmed HIV host factors and one HIV-encoded protein (Figure S6). Using permutation testing, the density of protein interactions in this network was found to be significantly enriched (p<3×10−6). (a) sTo identify potential molecular complexes, this network was analyzed for highly-connected local network modules (MCODE). (b–i) Importantly, a number of these densely-connected areas formed distinct functional subgroups, suggesting that they represent multi-protein complexes, which directly interact with viral factors to facilitate HIV replication. HIV encoded proteins were abbreviated as follows: CA: Capsid, GAG: gag polyprotein, MA: Matrix, VPR: Vpr, NC: Nucleocapsid, IN: Integrase, VIF: Vif, VPU: Vpu, RT: Reverse Transcriptase, PRO: Protease, P1: p1
Next, we re-analyzed these confirmed factors to identify over-represented biological annotations based on gene-ontology, or protein families based on Interpro (IPR), databases, respectively (p<0.01; Figure S5, Table S4). This analysis elucidated 176 statistically enriched biological terms, including over 25 functional processes or protein domains that influence HIV (VSV-G) infection (Figure 2b). Since functional over-representation was one of the criteria used to select genes for further study, many of these classifications reflected functional categories identified in the OPI analysis conducted across the four genome-wide screens (Figure S4, Table S2). Among others, a collection of genes involved in DNA repair, Nucleoside Diphosphate (NUDIX) hydrolase activity, as well as members of the Tripartite Motif (TRIM) family of proteins were implicated as factors likely to be important for early HIV (VSV-G) infection (Figure 2c).
To visualize the global topography of HIV-host interactions, we reconstructed a network based upon the 295 HIV host cellular factors confirmed in secondary assays. Using the Hynet, as well as additional protein-protein interaction databases (see Experimental Procedures), we assessed the network connectivity of these confirmed factors and additional host and HIV encoded proteins (Figure 3a, inset, Figure S6, Table S5). This host-pathogen interaction network contained 213 functionally validated and 169 predicted HIV host cellular nodes, which were internally connected via 2291 binary protein interactions, and 318 interactions to HIV encoded proteins. To identify densely-connected local network neighborhoods, we executed a Molecular Complex Detection (MCODE) analysis, which revealed several sub-networks with high local network connectivity (Figure 3a). Strikingly, the functional activities of these protein clusters reflect classifications identified using the GO and OPI analysis, including Ubiquitin-Proteasome Pathway, DNA Transcription and Repair, and Nucleic Acid Binding, further underscoring their importance in the direct regulation of early stage HIV replication (Figure 3b–3i).
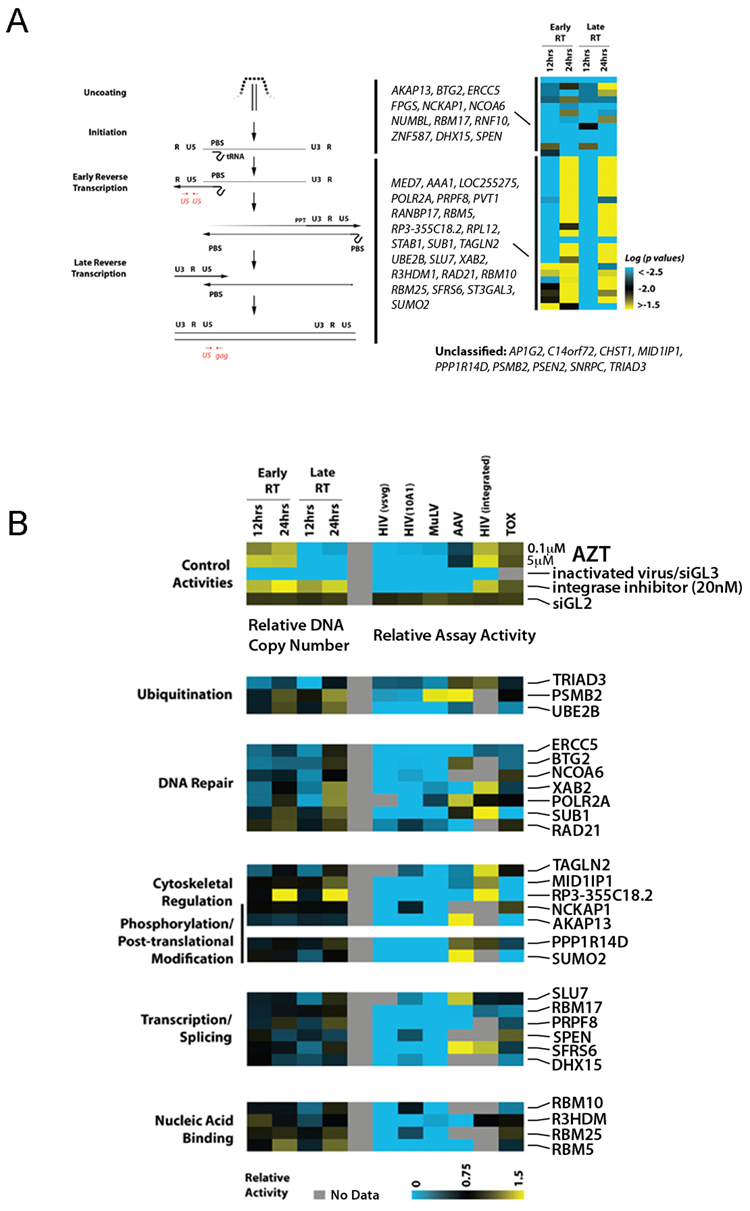
Quantitative PCR analysis of viral DNA products was performed to determine the effects of siRNAs on specific steps of early HIV-1 replication (Figure 4a, left panel). To analyze the accumulation of reverse transcription products, cells transfected with siRNAs targeting confirmed host factors were infected with the VSV-G pseudotyped HIV-1 vector, and total DNA was extracted for analysis at either 12 hours or 24 hours post-infection. A reduction in viral DNA indicates that the targeted genes were acting either to promote synthesis or inhibit degradation. Control tests performed with inactivated virus or with an inhibitor of reverse transcription (AZT) showed strong inhibition at the expected steps of virus replication (Figure 4b, top panel).
Figure 4. Host factors important for HIV-1 Reverse Transcription.
(a) 293T cells were transfected with siRNAs targeting indicated genes and, after 48h, infected with pNL43R+E-luc(VSV-G). Real-time quantitative PCR amplification analysis on DNA extracts was performed at the indicated timepoints to assess for the amount of early and late viral RT products. The left panel depicts a schematic of the reverse transcription process and the corresponding primer/probe sets utilized for the quantitative assay (red). Multiple experiments for one of more active siRNAs targeting the same gene were considered to determine statistical significance compared to negative controls (Wilcoxon signed-rank test) (right panel). A pair-wise comparison across the assays (data not shown) was also performed, and then the factors were categorized into two functional groups (indicated by the black bars). The black bars in the left panel indicate the potential steps during reverse transcription where the two separate groups of factors are proposed to act. Additional factors that influenced reverse transcription could not be statistically segregated into either of these two groups are indicated (unclassified). (b) As in panel (a), reverse transcription was monitored with 293T cells that had been transfected with siRNAs targeting indicated host mRNAs (lower panel) or with a negative control siRNA GL2 (upper panel). For control purposes, assays were also performed with the HIV-1 RT inhibitor AZT, an HIV-1 integrase inhibitor, or with heat-inactivated virus. (Top Panel). Quantitative PCR values were normalized to an internal control gene and then to the control siRNA GL2 values. The right section of the heatmap depicts the effects of the gene inhibition on various cellular assays, including infection by VSV-G or 10A1 pseudotyped HIV-1 vector, VSV-G pseudotyped MuLV vector, or by AAV, as indicated, as well on HIV-1 LTR-mediated transcription (HIV integrated) and on cellular viability (TOX) as described in Figure 1. Luciferase signal values were also normalized to control siRNA GL2. The median relative activity is depicted in a continuum of low (blue) to high (yellow). Only the genes which, upon depletion, significantly alter the initiation or rate of reverse transcription are shown (p<0.001, student t-test)
Analysis of the siRNA activities indicates that factors affecting reverse transcription can be segregated into at least two distinct functional classes, those that are absolutely required for viral DNA synthesis (AKAP13 to SPEN), and those that appear to affect the kinetics of viral DNA synthesis (MED7 to SUMO2) (Figure 4a, right panel, Table S6). A third set of proteins were found to affect HIV-1 uncoating or reverse transcription (AP1G2 to TRIAD3), but based upon statistical criteria, could not be placed into either functional class (Figure 4a, right panel). We hypothesize that factors which fall within the first group influence a step that is important for capsid uncoating or initiation of reverse transcription, or else inhibit degradation of the viral DNA after synthesis. Conversely, we predict that the siRNAs, which alter the kinetics of reverse transcription target cellular co-factors which facilitate, but are not essential to the process (Figure 4A). Further studies will be required to determine if delayed viral DNA synthesis in the absence of these genes produce PICs that are competent for integration. However, we anticipate that these kinetic factors are likely to play critical roles in maintaining viral fitness in vivo.
Additional PCR-based assays were used to identify those genes that play roles in either nuclear import of the viral PIC or viral DNA integration. The amount of 2-LTR circular forms of viral DNA were quantified and served as markers of blocks either to nuclear import (reduced 2-LTR circles) or to integration (accumulation of 2-LTR circles). In addition, viral DNA integration was quantified by taking advantage of amplification of HIV DNA sequences adjacent to cellular Alu repeats (Butler et al., 2001)(Figure 5a). For control purposes, parallel infections were performed in the presence of an HIV-1 integrase inhibitor (Hazuda et al., 2000).
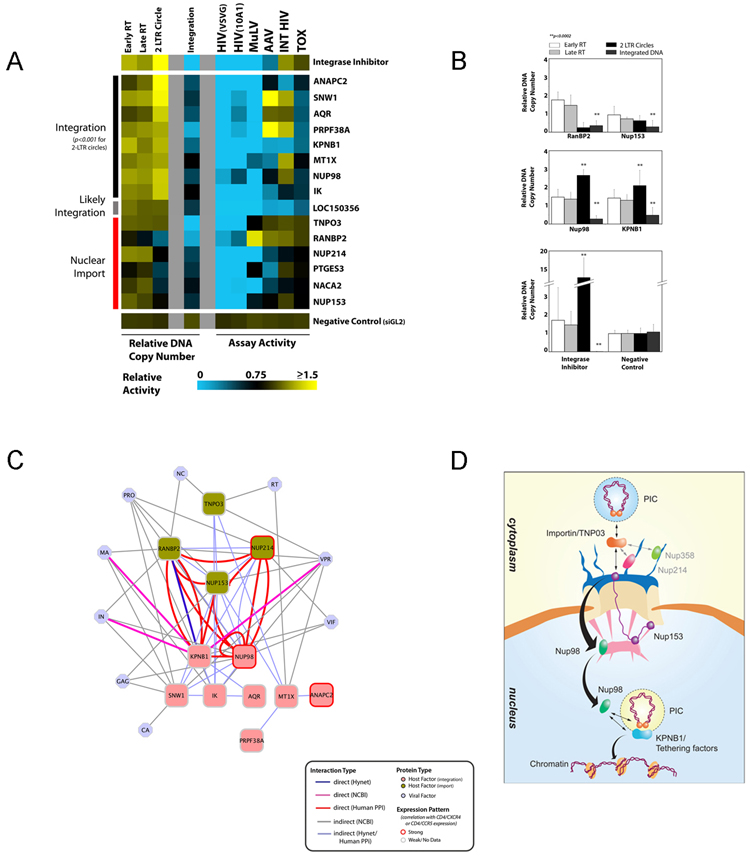
Figure 5. Host factors important for Nuclear Import of HIV-1 PICs and Viral DNA Integration.
(a) 293T cells were transfected with siRNAs targeting indicated host factors or control siRNA (lower panel) or treated with an HIV-1 integrase inhibitor and, subsequently challenged with pNL43R+E-luc(VSV-G). Real-time quantitative PCR amplification analysis was then used to measure early and late RT products at 24 h post-infection, as well as levels of 2-LTR circles and of integrated viral DNA at 24 and 48 h post-infection, respectively. siRNA-transfected 293 cells were also subjected to the same infection, proviral expression and toxicity screens as described in figure 4b (right panel of heatmap). The median of at least three replicate experiments for one or more active siRNAs targeting the same gene is shown in a blue to yellow continuum. Genes which, when inhibited, significantly impede viral integration are shown (p<0.01, student t-test). Segregation of factors into Nuclear Import, Likely Integration, and Integration classes is primarily based on the level of 2LTR circles, and is also supported by additional statistical significance calculations based upon the collective activity across all PCR assays in comparison to positive (integrase inhibitor) and negative (siGL2) controls (Figure S7). (b) Representative single siRNA activities of nuclear envelope/import-associated genes in quantitative PCR assays reflect the dual categorization of this class of proteins in both Nuclear Import (upper panel), and integration (middle panel). Relative DNA copy numbers of early RT, late RT, 2LTR circle formation and Integrated DNA as well statistical significance for 2LTR circles and integrated DNA are shown (p<0.0001, student t-test). (c) Biochemical relationships, based upon the network analysis shown in Figure 2, between proteins involved in integration (red) and nuclear import (green) and direct or indirect interactions amongst those proteins and with proteins encoded by HIV (blue) are depicted. (d) A model for the molecular coupling nuclear import of the viral PIC and proviral DNA integration processes. Proteins were organized as predicted from the protein interaction data in 5c and oriented on the basis of the quantitative PCR data (5a and 5b).
Fifteen genes were identified which, when targeted by cognate siRNAs, significantly reduced viral DNA integration (p<0.001; Figure 5a, Table S7). The activities of all functional siRNAs targeting the same gene were combined to classify these factors into two categories based on the quantitative PCR assays. In the first, RNAi knockdowns resulted in a reduction in integration and an increase in 2 LTR-circles, indicating effects on viral DNA integration itself (ANAPC2 to IK) (Figure 5a). In the second, little or no reduction in 2 LTR circles was seen, indicating a block to nuclear import (TNPO3 to NUP153) (Figure 5a). One gene, CHADL/LOC150356, could initially not be definitely placed, but further analysis suggests that it is likely involved in regulating viral DNA integration (Figure S7).
Several of the identified factors were members of the nucleocytoplasmic transport machinery. Unexpectedly, inhibition of these factors affected HIV-1 infection at two different steps. NUP153 and RANBP2 seemed to be involved in nuclear import of the PIC (Figure 5b, upper panel). In contrast, the nuclear import protein, KPNB1 (importin β-1), as well as Nup98 seemed to be required for viral integration. (Figure 5b, middle panel). These results suggest that karyopherin and nucleoporin can regulate viral DNA integration at a post nuclear entry step, though additional studies will be helpful to determine whether they act directly on viral nucleoprotein complexes, or regulate a host cellular factor required for integration.
Four out of six of the genes that inhibited nuclear import were selective for HIV, including NUP358/RANBP2, NUP153 and the importin-β-family protein Transportin-SR2/TNPO3 (Figure 5a). TNPO3 was identified by Brass et al. (Brass et al., 2008) as being important for HIV-1 infection, and here we demonstrate its involvement in nuclear import of the PIC (Figure 5a). In contrast, all genes which influence integration also affected infection by MuLV (Figure 5a). Thus the ability of the lentivirus HIV-1 to replicate in non-dividing cells (Lewis and Emerman, 1994), as compared with MuLV, is reflected in the differences of host factors needed for nuclear import (De Rijck et al., 2007). Interestingly, the HIV PIC was unable to integrate into the host genome independently of nuclear translocation in cycling cells (post-nuclear envelope breakdown), suggesting that HIV nuclear import and integration may be functionally coupled processes (see discussion).
Discussion
Here, we describe a genome-wide assay using an arrayed siRNA library to identify genes required for early stages of HIV infection. Inhibition of gene function of putative host factors may induce cellular responses that destabilize the viral cDNA, accelerate turnover, or otherwise indirectly interfere with viral replication. To identify those proteins which are more likely to be direct regulators of the viral life cycle, and thus critical for HIV-1 pathogenesis, we developed further selection criteria, which took into consideration a number of additional, and statistically significant lines of evidence, including protein interactions, mRNA expression, and gene ontology (Figure 1d; Supplemental Data). Importantly, since this approach does not only rely on identifying host factors based solely on ranking siRNA activities, we were able to more effectively mine the genetic datasets to identify those factors more likely to be immediate regulators of HIV-1 infection, and establish a network of host-pathogen interactions which coordinate HIV-1 infection. Retrospective analysis showed that evidence scores of confirmed genes are, on average, 40% higher than those that did not confirm (p=9.7×10−15), indicating that our approach enriched for biologically relevant activities (Table S3, data not shown).
Further validation studies have identified over 40 host factors that regulate capsid uncoating and reverse transcription steps of early HIV-1 replication (Figure 4). Additionally, we have elucidated 15 cellular factors that facilitate nuclear entry of the HIV-1 pre-integration complex and integration of proviral DNA (Figure 5). Importantly, only genes which regulated infection with HIV-1 virus pseudotyped with both VSV-G and 10A1 envelopes were considered for further analysis, thus excluding factors that may regulate endosomal function associated with VSV-G-mediated entry. Taken together, these studies indicate that host cellular factors are involved in a variety of different cellular processes that influence HIV-1 reverse transcription, nuclear import, and integration.
Comparison with Reported HIV Host Factors Identified Through RNAi-based Functional Screening (Brass et al.)
A recent genome-wide RNAi analysis by Brass et al. has identified approximately 284 genes as host cellular factors required for HIV replication (Brass et al., 2008). Comparison with the 295 confirmed genes presented here reveals a modest, but statistically significant overlap of 13 genes (p=0.00021). We speculate that this moderate concordance is largely due to differences in the analysis and experiment methodologies used. For example, the criteria Brass et al. employ to report host cellular factors are genes targeted by one or more siRNAs with activities >2 standard deviations from the mean. Since approximately 154 of the genes in the Brass et al. study were supported by the activity of only a single siRNA, it is likely that a fraction of these reported host factors represent false positive readouts due to off-target RNAi activity(Echeverri et al., 2006).
If we apply the criteria used in Brass et al. to the data presented here, we can identify 60 genes that are in common between the two RNAi studies (p=0.024, Table S8). Further reinforcing the parallels, we find that, based upon protein network analysis, an additional 64 genes reported by Brass et al. directly interact with a confirmed gene in our study (p=0.019, Table S8). In our study, we have also prioritized genes activities not only based upon siRNA activity, but also considered comparative activities in additional screens, HIV-host protein interaction data, as well as gene expression and ontology analysis. We anticipate that this approach enabled us to enrich for the most relevant host cellular factors that promote HIV infection, but this also likely contributed to the observed differences between the two host factor datasets.
Several experimental differences must also be considered when comparing these studies (see also Supplemental Table S8). Two independent RNAi libraries, constructed using separate design criteria and arrayed in different formats, were used to conduct these screens. Also, we have employed a single cycle replication-defective HIV vector pseudotyped with a VSV-G envelope in HEK293 cells and measured viral infection at a 24-hour time point. Brass et al. monitored the replication of a wild-type X4 strain of HIV in CD4/CXCR4-expressing Hela cells over 48 hours. Thus, we would not be able to identify factors involved in CD-4-mediated viral entry, as well as host molecules that regulate late-stage HIV replication. In contrast, however, we monitored single-cycle virus infectivity at an early (24hr) timepoint, which likely enabled us to elucidate a more comprehensive set of host factors specifically involved in early stages of replication, including uncoating, reverse transcription, and integration. These also encompassed proteins that regulate the kinetics of these processes (Figure 4a). Thus, while false positive activities are an inherent part of large-scale analyses, it is likely that variations in both experimental and data analysis techniques can largely account for the differences between our results and those reported by Brass et al.
The Role of Cytoskeletal Proteins in Early HIV Replication
The actin cytoskeleton was previously implicated in regulating the initiation of HIV-1 reverse transcription (Bukrinskaya et al., 1998) as well as in the movement of intracellular viral nucleoprotein complexes (NPCs) (Arhel et al., 2006). Consistently, the present study has revealed important roles in the earliest steps of HIV-1 infection for AKAP13, a RhoA-specific guanine nucleotide exchange factor (GEF) that regulates actin stress fiber formation, for NCKAP1 which associates with WAVE proteins that regulate actin nucleation/organization, and for TAGLN-2, a putative actin crosslinking/gelling protein (Table 1). Microtubules, previously shown to be involved in the intracellular movement of HIV-1 RTCs (Arhel et al., 2006; Naghavi et al., 2007) may also play a regulatory role in reverse transcription, since viral DNA levels were influenced by RP3-355C18.2, a predicted tubulin tyrosine-ligase, and MID1IP1, involved in bundling and stabilizing microtubules (Table 1). MID1IP1 was also identified recently by Elledge and colleagues (Brass et al., 2008), but its role in HIV-1 replication was not defined.
Table 1.
Selected siRNA phenotypes in early steps of HIV-1 replication
| Gene Names | Functional Categorya | Proposed Role in HIV Replication | Replication Block |
|---|---|---|---|
| ERCC5, BTG2, NCOA6 | DNA damage repair | DNA repair during RT | RT initiation |
| DHX15 | Nucleic Acid Binding | putative RNA helicase | |
| RBM17 | Nucleic Acid Binding (RNA splicing) | RNA Binding | |
| AKAP13 | Regulation of Cytoskeleton | PKA scaffold/actin organization | |
| NCKAP1 | actin organization | ||
| SPEN | Signal Transduction | Wnt-signaling regulation | |
| NUMBL | Notch-signaling regulation | ||
| POLR2A, SUB1, XAB2, RAD21 | DNA damage repair | DNA repair during RT | RT kinetics or viral DNA stability |
| R3HDM1 | Nucleic Acid Binding | Single-stranded RNA/DNA binding | |
| RBM5, RBM10, RBM25 | Nucleic Acid Binding (RNA splicing) | RNA Binding | |
| SFRS6, PRPF8, SLU7 | |||
| RANBP17 | Nuclear Import | ||
| SUMO2 | Post-translational Modification | SUMOylation | |
| RP3-355C18.2 | Regulation of Cytoskeleton | putative tubulin tyrosine ligase | |
| TAGLN2 | actin organization | ||
| UBE2B | Ubiquitin/Proteasome | ubiquitin ligase | |
| SNRPC | Nucleic Acid Binding (RNA splicing) | RNA Binding | |
| PPP1R14D | Post-translational Modification | Phosphatase Activity | RT (unclassified) |
| MID1IP1 | Regulation of Cytoskeleton | Microtubule organization | |
| PSEN2 | Signal Transduction | Notch signaling | |
| TRIAD3 | Ubiquitin/Proteasome | ubiquitin ligase | |
| PSMB2 | Proteasome component | ||
| TNPO3 | Nuclear Import | Translocation of PIC | Nuclear Import |
| RANBP2 | Nuclear Import/Post-Translation Modification | ||
| NUP153 | Nuclear pore | ||
| NUP214 | component | ||
| PTGES3 | prostaglandin E synthesis | ||
| KPNB1 | Nuclear import | Integration Coupling Factors | Integration |
| NUP98 | Nuclear pore component | ||
| ANAPC2 | anaphase promotion/ubiquitin ligase activity | Putative Tethering Factors | |
| AQR, PRPF38A | Nucleic Acid Binding | ||
| SNW1 | Transcriptional regulation |
Involvement of the DNA Damage Repair Pathway in HIV Uncoating/Reverse Transcription
Cellular DNA repair machinery has been implicated in playing roles in viral DNA integration and in completion of viral DNA synthesis following integration (Goff, 2007). An unanticipated finding here was that proteins involved in DNA damage response and repair also influence the initiation of reverse transcription and the accumulation of HIV-1 DNA products prior to integration (Table 1). Moreover, we have found two locally dense networks of proteins containing host factors that participate in DNA repair (Figure 3c: DNA transcription/repair and Figure 3f: DNA damage/replication). Both clusters contain multiple confirmed factors, including POLR2A, XAB2 and ERCC5, which have been mapped to early steps in the viral life cycle (Table 1). The viral interface for this host-pathogen interaction is mediated by Vpr, a component of the RTC/PIC that was recently being linked to the DNA damage response pathway (Schrofelbauer et al., 2007). Several other DNA repair proteins including MUS81, ERCC1, MRE11, involved in nucleotide excision repair, were also implicated in the early events of HIV-1 infection (Figure 2c).
Nucleic Acid Binding Proteins Participate in Early Stages of HIV Replication
Retroviral reverse transcription presumably involves the unwinding of RNA-RNA, RNA-DNA, and DNA-DNA strands, suggesting, that one or more cellular helicases may participate in viral DNA synthesis. Our results indicate that DHX15, a putative ATP-dependent RNA helicase that plays a role in pre-mRNA splicing, is important for early viral DNA synthesis in target cells (Table 1). Other nucleic acid-binding proteins, including RBM17 were also implicated in regulating HIV-1 DNA synthesis (Table 1). DHX15 and RBM17 are contained in a densely connected network (Figure 3h), supporting their status as proximal regulators of HIV-1 replication. In addition, several factors involved in transcription or mRNA splicing were also identified that regulate viral uncoating or reverse transcription (Table 1). Correspondingly, two MCODE protein complexes, which regulate transcription, splicing and nucleic acid binding, were identified through our network analysis (Figure 3d and 3e). The latter cluster consists of 11 confirmed factors, of which two were required for reverse transcription, suggesting that these proteins collectively coordinate either uncoating or DNA synthesis steps of HIV replication. It remains to be determined whether these host proteins act by interacting directly with the viral nucleic acids or whether they regulate other host factors required for HIV infection.
The Ubiquitin-Proteasome Pathway is Required for Capsid Uncoating and Viral DNA Synthesis
The ubiquitin-proteasome pathway has previously been associated with early steps of HIV replication, where it acts negatively to destroy incoming viral replication complexes (Butler et al., 2002; Schwartz et al., 1998). Our studies have revealed a positive role also for this pathway. The ubiquitin ligases UBE2B (RAD6) and TRIAD3, as well as the proteasome component PSMB2, were each important for HIV-1 reverse transcription (Table 1). Network analysis also revealed that the viral integrase and Vif proteins have multiple interactions with a cluster of proteins that function in the ubiquitin-proteasome pathway (Figure 3b), indicating that these viral factors may play a structural role in the HIV RTC to recruit the proteosomal machinery and facilitate uncoating or reverse transcription.
Post-Translational Modifications in Early HIV Replication
Previously, the cAMP-dependent protein kinase (PKA) was implicated in regulating early steps of HIV-1 replication (Cartier et al., 2003). AKAP13 is a PKA scaffold protein (Table 1) and as such, may be involved in mediating PKA-dependent regulation of HIV-1 reverse transcription. Protein dephosphorylation events may also act to regulate HIV-1 reverse transcription as indicated by the importance of protein phosphatase 1 regulatory subunit 14D (PPP1R14D), a negative regulator of the catalytic subunit of the serine/threonine protein phosphatase 1 (Table 1).
SUMOylation events have been proposed to be important in early steps of MuLV infection (Yueh et al., 2006). We have found that SUMO-2, one of three small ubiquitin-related modifier proteins, is important during the late stage of HIV-1 (and MuLV) reverse transcription (Table 1). In addition, RANBP2, a SUMO-1 E3 ligase that is a component of the cytoplasmic filaments of the nuclear pore complex was required for nuclear import of the HIV-1 DNA (Table 1) perhaps through the sumoylation of viral proteins in the PIC or host factors required in the PIC. Since RANBP2 influenced HIV but not MuLV infection (Figure 4b), different SUMO conjugating systems may be important for each of these two viruses.
Host Factors Required for Nuclear Import of the Viral Pre-Integration Complex
The mechanism of HIV nuclear import is controversial with multiple proteins and nucleic acids proposed to play a role (Suzuki and Craigie, 2007). Our studies, combined with those of Brass et al. (Brass et al., 2008), indicate the involvement of Nup153, RANBP2, and TNPO3 as factors involved in HIV-1 PIC import (Table 1). We have also uncovered roles for Nup214, the nascent polypeptide-associated complex alpha subunit 2, NACA2, and prostaglandin E synthase, PTGES3 (Table 1). The potential role of prostaglandins in HIV-1 nuclear import is particularly intriguing, because these factors are already known to regulate the import of other types of nuclear cargo (Gomez et al., 2005) (Malki et al., 2005) and they may represent a new therapeutic target for HIV-1 infection.
Host Proteins and Viral DNA Integration
Our studies have also revealed several cellular factors important for HIV-1 DNA integration. The first is ANAPC2, a component of the anaphase-promoting complex which promotes the metaphase-anaphase transition. ANAPC2 is a cullin protein with a role in ubiquitin-ligase activity (Table 1) suggesting that ubiquitin-modification and/or proteolysis of virus PIC components may play a regulatory role during the integration stage of viral DNA replication. Recent reports have suggested that passage through mitosis may promote HIV as well as MLV DNA integration (Mannioui et al., 2004; Roe et al., 1993), providing a possible role for ANAPC2. The second factor SNW1, is a transcriptional coactivator which associates with a cyclophilin-like protein, peptidyl-prolyl isomerase-like 1 (PPIL1) (Table 1). The structure of PPIL1 resembles other members of the cyclophilin family, in particular Cyclophilin A, (Xu et al., 2006) suggesting that cyclophilin proteins might be involved in regulating both late (integration) as well as early (uncoating) (Luban, 2007), replication steps. A third factor, aquarius or AQR, also regulates HIV-1 DNA integration (Figure 5a). This factor associates with XAB2, a protein involved in transcription-coupled repair (Kuraoka et al., 2008) suggesting that this DNA repair pathway might play a role during viral DNA integration.
A Model for Import-coupled Integration
Protein networks that are implicated in nuclear import and integration of the HIV PIC are shown in Figure 5c. This network was constructed based upon known or experimentally determined interactions between host factors and HIV-encoded proteins, and proteins are organized and oriented on the basis of the quantitative PCR mapping data (Figure 5a). Based upon these data, we propose a model wherein nuclear import of the PIC and proviral DNA integration are molecularly coupled events mediated by nucleoporins, karyopherin, and putative tethering factors (Figure 5d). Specifically, we hypothesize that nuclear transport of the viral PIC through the nuclear pore complex is mediated by soluble transport receptors, such as TNPO3, and the nuclear pore complex (Stewart, 2007). Next, the PIC cargo is transferred consecutively to phenylalanine-glycine (FG) repeat domains of variant nucleoporins, for example Nup358/RANBP2. Subsequently, in this model, the cargo is delivered to the FG repeats of Nup214 (anchored to the cytoplasmic ring) and Nup153 (distal ring). The FG-repeats of both Nup214 and Nup153 are highly flexible and can either reach down to the nuclear basket (Nup214) or towards the cytoplasmic periphery of the central pore (Nup153) (Paulillo et al., 2005), thereby possibly acting to accelerate PIC translocation. As an alternative, we cannot rule out that RNAi-silencing of nuclear import machinery components may be acting indirectly by altering the localization of protein(s) required for HIV nuclear import.
Upon traversing the nuclear pore, we hypothesize that the PIC is then released into the nucleus through interaction with the FG-containing Nup98 located near the nuclear basket, which dynamically associates with and dissociates from the nuclear pore (Griffis et al., 2004). Our data indicates that Nup98 is essential for viral integration, suggesting that Nup98 likely directs the viral PIC from the nuclear pore to the proximity of the chromatin. The intranuclear mobility of Nup98 has previously been linked to active transcription sites, possibly through direct interactions with the transcriptional machinery or with newly produced transcripts and RNP complexes (Griffis et al., 2004). Additionally, the PIC is anchored to chromatin through potential “tethering” factors. Targeted integration into active transcription units is mediated by PSIP1/LEDGF/p75 (Ciuffi and Bushman, 2006)(and references therein). Additional factors may help direct integration to transcriptionally active regions, as suggested by the finding that histone post-translational modifications associated with expression positively correlate with HIV integration frequency (Wang et al., 2007). We have identified several proteins that are required for proviral integration and are linked to transcription, signaling and splicing (ANAPC2, MT1X, SNW1, IK, PRPF38A, and AQR). We hypothesize that these proteins might act to target the PIC to the chromatin and/or act as enzymatic co-factors for DNA integration. Our data indicates that karyopherin KPNB1 is also required for viral integration, suggesting that it may either co-operate with NUP98 to direct the PIC to transcriptionally active chromatin, or, alternatively, could serve as an intermediate to couple the viral PIC to the tethering factors. These data support a model in which components of nuclear import machinery facilitate both HIV nuclear entry and integration, and suggest that the coupling of these steps are required for the establishment of the HIV provirus.
Taken together, this integrative approach towards genome-wide host pathogen interaction analysis has revealed cellular factors that coordinately regulate the early steps of HIV viral replication, including those required for reverse transcription and proviral DNA integration. Altogether new pathways are suggested for the first time, such as possible involvement of prostaglandins in nuclear import. An understanding of their role in cell types targeted by HIV during infection, such as helper T-cells, macrophages, dendritic and microglial cells, will be critical to characterizing the contribution of these factors towards disease progression. Development of small molecules that modulate the activity of these proteins may provide novel strategies for treatment of HIV/AIDS, particularly since the inhibition of these stages in the viral life cycle has already proven to be therapeutically effective.
Experimental Procedures
For detailed Experimental Procedures see the Supplemental Data.
High-throughput and Reconfirmation siRNA screens
Genome-wide siRNA libraries targeting 19,628 human genes (2 siRNAs/well, approx. 3 wells/gene) were screened in duplicate or triplicate runs using a high-throughput robotic system to identify siRNAs that influence infection of human 293T cells by either a VSV-G pseudotyped HIV-1 reporter virus encoding luciferase, by a VSV-G pseudotyped MLV vector encoding luciferase or by an AAV vector encoding luciferase. In addition, the library was counter-screened to identify siRNAs that influence cell viability. siRNAs for reconfirmation were individually rearrayed in 384 wells in duplicate and screened at least in four independent experiments and analyzed as described in Supplemental Material.
Expression vectors and reporter viruses
For the HIV and MLV screens, pNL43-Luc-E−R+ (HIV-1 wild-type Δenv, encoding firefly luciferase GL3) vector was used to generate VSV-G pseudotyped lentiviral supernatant and the MLV-based retroviral vector pVGIP3, a derivative of pBabe, to generate a Moloney-based virus. For the AAV screen, GL3 luciferase was introduced into an ITR-flanked AAV expression vector to generate AAV particles encoding luciferase. For secondary confirmation assays, an amphotropic 10A1 envelope pseudotyped HIV virus was generated.
Secondary confirmation analysis (life cycle staging)
Genes subject to staging of their effects in the HIV life cycle were also screened in parallel for their activities on HIV-1 (VSV-G), HIV-1 (10A1), MLV (VSV-G) and AAV reporter viruses as well on HIV-1 LTR-mediated transcription and cell viability in 96-well-based assays.
Viral DNA quantitation by real-time PCR
HEK-293T cells were transfected with siRNAs and two days later infected with HIV-1 (VSV-G). Total DNA was isolated at 12 hours post infection (hpi), 24 hpi and 48 hpi. For control purposes, a reverse transcription inhibitor or integrase inhibitor was added at the time of infection. DNA was subject to real-time PCR to quantify specifically HIV Early RT products, Late RT products, 2-LTR-circle forms and Proviral DNA using Alu-based nested PCR. To normalize the amount of DNA in each PCR assay, the copy number of the cellular gene porphobilinogen deaminase (PBGD) was quantified. With each experiment, a standard curve was generated.
Yeast Two-Hybrid screen with HIV proteins
A yeast two-hybrid screen was performed using the entire HIV HXB2 proteome as bait and screening against a human leukocyte cDNA library cloned into the GAL4 expression vector.
Datasets and Network analysis
Multiple human protein-protein interaction databases were incorporated in the Network analysis: Yeast-two-Hybrid databases and other curated literature-based protein-protein interaction databases (Bind, HPRD, MINT, Reactome). In addition, two human-HIV interaction databases, the NCBI Human-HIV Interaction database and the Y2H in-house database, and the GNF Tissue Atlas Gene Expression database were used in this study. Detailed methodology of the multi-scale strategy for hit selection and network analysis can be found in Supplemental Data.
Supplementary Material
01. Supplemental Data.
Experimental Procedure
Figures. S1 to S8
Tables S1 to S19
10
02
03
04
05
06
07
08
09
Acknowledgements
Tracy L. Diamond is a Leukemia and Lymphoma Society Fellow (Grant # 5217-06). We thank U. O’Doherty and J.J. Yu for technical advice with the Alu-PCR assay and Zhang J. for excellent technical assistance, Walter Tian and Eric Lader (Qiagen GMBGH) for facilitating the acquisition of reagents used for this study, and John Hogenesch for critical feedback on the manuscript. This work was supported by a grant from the US National Institutes of Health (1 RO1 AI072645-01 to JAY and RO1 AI052845 to FDB) and the University of Pennsylvania Center for AIDS Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arhel N, Genovesio A, Kim KA, Miko S, Perret E, Olivo-Marin JC, Shorte S, Charneau P. Quantitative four-dimensional tracking of cytoplasmic and nuclear HIV-1 complexes. Nat Methods. 2006;3:817–824. doi: 10.1038/nmeth928. [DOI] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of Host Proteins Required for HIV Infection Through a Functional Genomic Screen. Science. 2008 doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Bukrinskaya A, Brichacek B, Mann A, Stevenson M. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J Exp Med. 1998;188:2113–2125. doi: 10.1084/jem.188.11.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- Butler SL, Johnson EP, Bushman FD. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. Journal of virology. 2002;76:3739–3747. doi: 10.1128/JVI.76.8.3739-3747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier C, Hemonnot B, Gay B, Bardy M, Sanchiz C, Devaux C, Briant L. Active cAMP-dependent protein kinase incorporated within highly purified HIV-1 particles is required for viral infectivity and interacts with viral capsid protein. The Journal of biological chemistry. 2003;278:35211–35219. doi: 10.1074/jbc.M301257200. [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Bushman FD. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006;22:388–395. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- De Rijck J, Vandekerckhove L, Christ F, Debyser Z. Lentiviral nuclear import: a complex interplay between virus and host. Bioessays. 2007;29:441–451. doi: 10.1002/bies.20561. [DOI] [PubMed] [Google Scholar]
- Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- Goff SP. Host factors exploited by retroviruses. Nat Rev Microbiol. 2007;5:253–263. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- Gomez PF, Pillinger MH, Attur M, Marjanovic N, Dave M, Park J, Bingham CO, 3rd, Al-Mussawir H, Abramson SB. Resolution of inflammation: prostaglandin E2 dissociates nuclear trafficking of individual NF-kappaB subunits (p65, p50) in stimulated rheumatoid synovial fibroblasts. J Immunol. 2005;175:6924–6930. doi: 10.4049/jimmunol.175.10.6924. [DOI] [PubMed] [Google Scholar]
- Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Molecular biology of the cell. 2004;15:1991–2002. doi: 10.1091/mbc.E03-10-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, et al. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- Konig R, Chiang CY, Tu BP, Yan SF, DeJesus PD, Romero A, Bergauer T, Orth A, Krueger U, Zhou Y, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat Methods. 2007;4:847–849. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- Kuraoka I, Ito S, Wada T, Hayashida M, Lee L, Saijo M, Nakatsu Y, Matsumoto M, Matsunaga T, Handa H, et al. Isolation of XAB2 complex involved in pre-mRNA splicing, transcription, and transcription-coupled repair. The Journal of biological chemistry. 2008;283:940–950. doi: 10.1074/jbc.M706647200. [DOI] [PubMed] [Google Scholar]
- Lewis PF, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. Journal of virology. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luban J. Cyclophilin A, TRIM5, and resistance to human immunodeficiency virus type 1 infection. Journal of virology. 2007;81:1054–1061. doi: 10.1128/JVI.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malki S, Nef S, Notarnicola C, Thevenet L, Gasca S, Mejean C, Berta P, Poulat F, Boizet-Bonhoure B. Prostaglandin D2 induces nuclear import of the sex-determining factor SOX9 via its cAMP-PKA phosphorylation. Embo J. 2005;24:1798–1809. doi: 10.1038/sj.emboj.7600660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannioui A, Schiffer C, Felix N, Nelson E, Brussel A, Sonigo P, Gluckman JC, Canque B. Cell cycle regulation of human immunodeficiency virus type 1 integration in T cells: antagonistic effects of nuclear envelope breakdown and chromatin condensation. Virology. 2004;329:77–88. doi: 10.1016/j.virol.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Mukherji M, Bell R, Supekova L, Wang Y, Orth AP, Batalov S, Miraglia L, Huesken D, Lange J, Martin C, et al. Genome-wide functional analysis of human cell-cycle regulators. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:14819–14824. doi: 10.1073/pnas.0604320103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naghavi MH, Valente S, Hatziioannou T, de Los Santos K, Wen Y, Mott C, Gundersen GG, Goff SP. Moesin regulates stable microtubule formation and limits retroviral infection in cultured cells. Embo J. 2007;26:41–52. doi: 10.1038/sj.emboj.7601475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Saib A. Early steps of retrovirus replicative cycle. Retrovirology. 2004;1:9. doi: 10.1186/1742-4690-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulillo SM, Phillips EM, Koser J, Sauder U, Ullman KS, Powers MA, Fahrenkrog B. Nucleoporin domain topology is linked to the transport status of the nuclear pore complex. Journal of molecular biology. 2005;351:784–798. doi: 10.1016/j.jmb.2005.06.034. [DOI] [PubMed] [Google Scholar]
- Roe T, Reynolds TC, Yu G, Brown PO. Integration of murine leukemia virus DNA depends on mitosis. Embo J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrofelbauer B, Hakata Y, Landau NR. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4130–4135. doi: 10.1073/pnas.0610167104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O, Marechal V, Friguet B, Arenzana-Seisdedos F, Heard JM. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. Journal of virology. 1998;72:3845–3850. doi: 10.1128/jvi.72.5.3845-3850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nature reviews. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Craigie R. The road to chromatin - nuclear entry of retroviruses. Nat Rev Microbiol. 2007;5:187–196. doi: 10.1038/nrmicro1579. [DOI] [PubMed] [Google Scholar]
- Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome research. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Zhang J, Huang X, Sun J, Xu Y, Tang Y, Wu J, Shi Y, Huang Q, Zhang Q. Solution structure of human peptidyl prolyl isomerase-like protein 1 and insights into its interaction with SKIP. The Journal of biological chemistry. 2006;281:15900–15908. doi: 10.1074/jbc.M511155200. [DOI] [PubMed] [Google Scholar]
- Yueh A, Leung J, Bhattacharyya S, Perrone LA, de los Santos K, Pu SY, Goff SP. Interaction of moloney murine leukemia virus capsid with Ubc9 and PIASy mediates SUMO-1 addition required early in infection. Journal of virology. 2006;80:342–352. doi: 10.1128/JVI.80.1.342-352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar J. Biostatistical Analysis. Prentice Hall, NJ: 1999. p. 523. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01. Supplemental Data.
Experimental Procedure
Figures. S1 to S8
Tables S1 to S19
10
02
03
04
05
06
07
08
09