Induction of TLR4-target genes entails calcium/calmodulin-dependent regulation of chromatin remodeling (original) (raw)
Abstract
Upon toll-like receptor 4 (TLR4) signaling in macrophages, the mammalian Swi/Snf-like BAF chromatin remodeling complex is recruited to many TLR4 target genes where it remodels their chromatin to promote transcription. Here, we show that, surprisingly, recruitment is not sufficient for chromatin remodeling; a second event, dependent on calcium/calmodulin (CaM), is additionally required. Calcium/CaM directly binds the HMG domain of the BAF57 subunit within the BAF complex. Calcium/CaM antagonists, including a CaM-binding peptide derived from BAF57, abolish BAF-dependent remodeling and gene expression without compromising BAF recruitment. BAF57 RNAi and BAF57 dominant negative mutants defective in CaM binding similarly impair the induction of BAF target genes. Our data implicate calcium/CaM in TLR4 signaling, and reveal a previously undescribed, recruitment-independent mode of regulation of the BAF complex that is probably achieved through a direct CaM-BAF interaction.
Keywords: innate immunity, Brg1, macrophages
The BAF complex is the prototypical mammalian chromatin-remodeling complex and has important roles in gene regulation in diverse systems, including the immune system (1). Cells use sophisticated mechanisms, involving sequence-specific transcription factors, phosphorylation of BAF subunits, inositol lipids, and/or histone acetylation, to tightly regulate recruitment of the BAF complex to target genes, and BAF recruitment appears to invariably coincide with remodeling (see, for example, ref. 2). Thus, the BAF complex appears to be regulated primarily at the level of recruitment; little is known about other modes of BAF regulation. The BAF complex consists of over 10 subunits, but the remodeling activity in vitro can be fully reconstituted with only four subunits, including the catalytic subunit Brg (3). The functions of the remaining subunits are largely obscure, except that some of these subunits, including the HMG-box protein BAF57 (4), help recruit the BAF complex (5–8).
Calmodulin (CaM) is the major calcium sensor in our body. On binding calcium, CaM undergoes a conformational change, which allows it to bind and regulate diverse target proteins by means of multiple mechanisms (9). CaM is present in both the cytoplasm and the nucleus, but its nuclear functions are poorly understood, as is calcium in the nucleus (10). CaM binds certain basic helix–loop–helix transcriptional factors to block their DNA binding (11, 12), and in vitro interactions between CaM and transcription factors have also been documented in plants (13–15), indicating CaM can directly regulate transcription factors.
Toll-like receptors (TLRs) sense invading pathogens and trigger innate and ultimately adaptive immune responses (16). The founding member of mammalian TLRs is TLR4 (17), which is highly expressed on macrophages and senses lipopolysacchride (LPS), a component of Gram-negative bacteria. On LPS stimulation, macrophages express many proinflammatory genes. These genes can be divided into 3 categories based on induction characteristics (18). The “early primary response genes” are rapidly induced (within minutes), whereas the “delayed primary response genes” are induced with slower kinetics (within hours). The primary response genes do not require de novo protein synthesis for induction, and stand in contrast to the “secondary response genes,” the third class of LPS target genes. The promoter chromatin of the early primary response genes is already “open” in the resting state before TLR4 signaling, which presumably poises the genes for rapid induction. In contrast, the promoters of the delayed primary and secondary response genes must undergo extensive chromatin remodeling on LPS stimulation, which presumably contributes to their slower induction kinetics. The remodeling of both the delayed primary and secondary response genes is catalyzed by the BAF complex, which is recruited to the promoters of these genes on LPS stimulation, and BAF recruitment coincides with remodeling of the target genes (18). The roles of CaM in TLR4-dependent gene expression are essentially unknown.
Here, we show that CaM binds BAF57 and stimulates BAF-catalyzed chromatin remodeling in macrophages independently of the recruitment of the BAF complex, thus revealing a previously undescribed mode of regulation of the BAF complex that is essential for TLR4 signaling.
Results
Calcium/CaM Binds the BAF Complex.
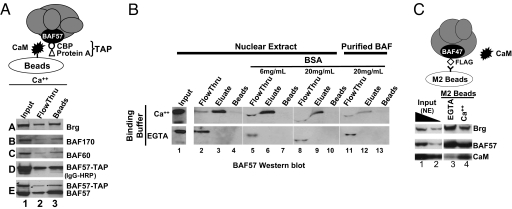
In an effort to study the role of BAF57 in chromatin remodeling, we made a HeLa cell line expressing BAF57 fused to a “tandem-affinity purification tag” (TAP) consisting of protein A and a CaM-binding peptide (CBP); this system would enable us to purify the BAF complex bearing WT or mutant BAF57. To test whether the CBP tag is active, we used CaM beads to pull down the tagged protein by means of CaM-CBP interaction as described (Fig. 1A) (19). The tagged protein was expressed at a low level relative to the endogenous protein (Fig. 1A, blot E, lane 1). Surprisingly, CaM beads captured not only the tagged protein but also endogenous BAF57 (Fig. 1A, blot E), together with other BAF subunits (Fig. 1A, blots A–C), suggesting that CaM beads bind endogenous BAF complex. This binding is calcium-dependent, because ethylene glycol tetraacetic acid (EGTA) could elute the BAF complex from CaM beads (Fig. 1B Upper, lanes 3 and 4), and prevent CaM beads from capturing the BAF complex if added to the binding buffer (Fig. 1B Lower, lanes 3 and 4). In contrast, BSA failed to abolish CaM-BAF interaction even at high concentrations (Fig. 1B, lanes 5–10). The CaM-BAF interaction was also observed between purified BAF complex and CaM beads (Fig. 1B, lanes 11–13), indicating that CaM beads directly bind the BAF complex.
Fig. 1.
CaM binds the BAF complex. (A) CaM beads pulled down endogenous BAF complex in addition to affinity-tagged BAF complex from crude HeLa NEs. The integrity of the tag, fused to BAF57, was confirmed by Western blotting using an HRP-conjugated IgG that recognizes the protein A moiety of the tag (blot D). (B) CaM-BAF interactions are calcium-dependent and direct. CaM beads were mixed with 50 μg of HeLa NEs (lanes 2–10) or 0.5 μg of purified BAF complex (lanes 11–13) in the presence of CaCl2 (Upper), EGTA (Lower), or high concentrations of BSA that distorted the gels (lanes 5–13); the BAF complex was affinity-purified by using M2 beads as described (20). CaM beads were washed under the binding condition and eluted with EGTA, and the samples probed with anti-BAF57. (C) M2-beads, which recognize Flag-tagged BAF complex, pulled down endogenous CaM. 1% and 0.3% of NE in the immunoprecipitation (IP) reaction (lanes 1–2) and 50% of precipitated material (lanes 3–4) were loaded on the gel. The IP reaction contained either 5 mM EGTA (lane 3) or 2 mM CaCl2 (lane 4), the latter appearing to impair the IP efficiency.
To determine whether endogenous CaM also binds the BAF complex, we took advantage of a HeLa cell line widely used for affinity-purification of the BAF complex. This line stably expresses Flag-tagged BAF47, which is quantitatively incorporated into the BAF complex (20). Thus, Flag-tagged BAF47 is not overexpressed in this line. Importantly, the buffer condition for BAF purification using this system is compatible with CaM-BAF interaction (see Materials and Methods). We found that CaM was present in the HeLa nuclear extract (NE) (Fig. 1C, lanes 1 and 2), a fraction of which was indeed copurified with the BAF complex in a calcium-dependent manner (Fig. 1C, lanes 3 and 4).
CaM Binds the HMG Domain of BAF57.
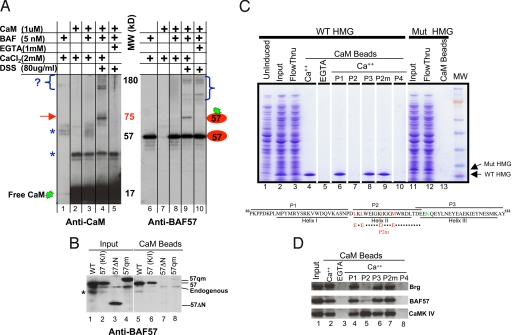
To identify CaM-binding subunit(s) in the BAF complex, we mixed purified BAF complex with excessive amounts of recombinant CaM, which drove the binding to completion. We then irreversibly cross-linked interacting partners with Disuccinimidyl suberate (DSS) and used Western blotting to identify cross-linking products. The gel was first probed with an anti-CaM antibody. There was a prominent, calcium-dependent cross-linker product migrating at ≈75 kDa, as well as several larger (≈180 kDa) products (Fig. 2A, lanes 4 and 5). Because the molecular masses of CaM and BAF57 are 17 and 57 kDa, respectively, the 75-kDa band might represent a CaM-BAF57 heterodimer, which was confirmed using a BAF57 antibody (Fig. 2A, lane 9). The identities of the large (≈180 kDa) CaM-binding proteins are unknown, but all of the candidate BAF subunits have been excluded (data not shown), suggesting that these proteins are contaminants instead of genuine BAF subunits. The BAF57 antibody also detected multiple calcium-independent cross-linking products, which presumably reflects cross-linking between BAF57 and its neighboring subunits (Fig. 2A, lanes 9 and 10). Together, these data suggest that CaM directly binds BAF57. To confirm this hypothesis, we overexpressed Flag-tagged BAF57 in 293T cells. CaM beads pulled down not only the endogenous but also recombinant BAF57 from 293T NE (Fig. 2B, lanes 1 and 5), demonstrating that CaM can bind BAF57 independently of other BAF subunits. BAF57 contains an HMG-box domain (4). The HMG-box domains in 2 other proteins (SRY and HMG1) appear to bind CaM (21), which prompted us to examine whether CaM binds the BAF57 HMG domain. Indeed, an N-terminal deletion mutant of BAF57 lacking amino acid 1–133 that encompasses the HMG domain (amino acid 63–133) is defective in CaM binding (Fig. 2B, lanes 3 and 7). To confirm that the BAF57 HMG domain directly binds CaM, we expressed the BAF57 N terminus (amino acid 1–169) in Escherichia coli, where this fragment is known to be stable (4). CaM beads indeed specifically pulled down the recombinant protein from crude bacterial lysate (Fig. 2C, lanes 2–5). CaM typically binds a target protein by means of interactions with a short (≈12–30 residues) amphipathic α-helix on the target, with the 2 globular domains of CaM engulfing the α-helix. The BAF57 HMG box consists of 3 α-helices (Fig. 2C Lower). To identify which helix binds CaM, we derived peptides from each of the helices and tested their ability to block CaM-HMG interaction. The peptide derived from helix 2 (P2), but not P1 or P3, blocked this interaction (Fig. 2C, lanes 6–8), as did a peptide (P4) derived from the CaM binding site in CaMK II (Fig. 2C, lane 10).
Fig. 2.
CaM binds the HMG domain of BAF57. (A) In vitro cross-linking assay. CaM was cross-linked to the BAF complex and the reaction products resolved by SDS/PAGE before being probed sequentially with antibodies against CaM (Left) and BAF57 (Right). Brackets denote crosslinking between CaM and unknown proteins (Left), or between BAF57 and its neighboring subunits (Right). Arrow, crosslinking between CaM and BAF57. Asterisks, nonspecific immunoreactivity. (B) Anti-BAF57 Western blotting showing that CaM beads captured BAF57 (WT) and the BAF57 point mutant defective in DNA binding [57(K/I)], but not the deletion mutant (57 ΔN) lacking the N-terminal 133 residues or the quadruple point mutant (57qm) bearing substitutions in hydrophobic residues as depicted in C. WT and mutant BAF57, Flag-tagged to distinguish them from endogenous BAF57, were transiently overexpressed in 293T cells; 293T cells were used because they are highly transfectable. Asterisk, nonspecific reactivity or BAF57 degradation product. (C) Coomassie blue-stained gel showing that CaM beads pulled down WT BAF57 HMG domain (lanes 1–5), but not the point mutant (lanes 11–13) from bacterial lysates, and the effects of various peptides (100 μM) on CaM-BAF57 HMG interaction (lanes 6–10). Illustrated at the bottom are the WT and mutant BAF57 HMG boxes together with the corresponding peptides. The hydrophobic residues involved in CaM binding and the lysine residue essential for DNA binding are highlighted in red and green, respectively. P2m, mutant P2 bearing the quadruple point mutations. (D) Western blots showing the effects of various peptides (100 μM) on CaM-BAF and CaM-CaMK IV interactions in thymocyte NE where CaMK IV is readily detectable. The peptides were added to the NE together with CaM beads.
We next sought to identify the residues in Helix 2 important for CaM binding. Helix 2 is a 17-residue peptide predicted to be amphipathic, but does not fit into any known CaM-binding sequence-motif (22). Because most of the binding energy of CaM-target interactions results from the interactions between the hydrophobic patches of CaM and hydrophobic residues on the target peptide, we asked whether mutating hydrophobic residues on Helix 2 could disrupt CaM-Helix 2 interaction. There are 9 hydrophobic residues on Helix 2, mutating 2 of which (I99D and M103E) failed to significantly affect CaM binding (data not shown), but mutating 4 (L91E, L93E, I99D, and M103E) abolished the ability of P2 to block CaM-BAF57 HMG interaction (Fig. 2C, lane 9). In agreement with this observation, bacterially expressed recombinant BAF57 (1–169) protein bearing the quadruple point mutations failed to bind CaM (Fig. 2C, lane 13), and neither could the full-length BAF57 protein bearing the same mutations expressed in 293T cells (Fig. 2B, lanes 4 and 8; note the slower mobility of the point mutants in Fig. 2 B and C). As a control, we show that a point mutation in the BAF57 HMG domain (K112I), which is known to impair the ability of the HMG domain to bind 4-way junction DNA (4) and regulate CD4/CD8 expression (23), had no effect on BAF57-CaM interaction (Fig. 2B, lanes 2 and 6), indicating that DNA binding by the HMG domain can be dissociated from its binding to CaM.
We also tested the specificity of P2 and found that P2 does not generally block CaM-target interactions: CaM beads pulled down both the BAF complex and CaMK IV from thymocyte NEs (Fig. 2D, lanes 2 and 3), but P2 only blocked CaM-BAF interaction (Fig. 2D, lane 5). As expected, P1, P3 or P2m failed to block this interaction (Fig. 2D, lanes 4, 6, 7). The failure of P2 to block CaM-CaMK IV interaction is not an artifact, because P4 disrupted CaM-CaMK IV interaction in addition to CaM-BAF interaction (Fig. 2D, lane 8). Of note, CaM beads could not pull down HMG1 from the NEs, suggesting that CaM binds BAF57 more tightly than it binds HMG1 (data not shown).
Calcium/CaM Antagonists Block BAF-Dependent Chromatin Remodeling Without Compromising BAF Recruitment in Macrophages.
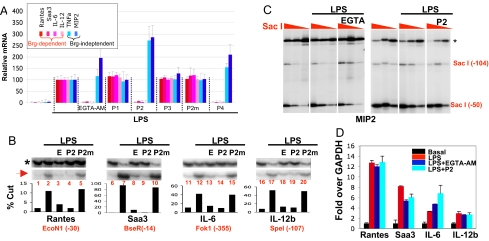
We next explored the functional relevance of CaM-BAF interaction in macrophages where many BAF target genes have been identified. On LPS stimulation, macrophages express 3 categories of genes: the early primary response genes, the delayed primary response genes, and the secondary response genes, the expression of the latter 2 classes depending on Brg (18). We examined the effects of calcium/CaM antagonists on a total of 6 TLR4 target genes, 2 from each class: Saa3 and Rantes (delayed primary response genes), IL-6 and IL-12b (secondary response genes), and MIP2 and TNFa (early primary response genes). The use of calcium/CaM antagonists is the standard approach for probing the functions of CaM; CaM is encoded by 5 nonallelic genes, making RNAi or gene knockout unfeasible. Macrophages were stimulated with LPS for 3 h in the presence or absence of calcium/CaM antagonists before we analyzed gene expression. The calcium/CaM antagonists include EGTA, EGTA-tetraacetoxymethyl ester [EGTA-AM, a cell-permeable version of EGTA], BAPTA-AM, an intracellular calcium chelator structurally different from EGTA-AM, and the aforementioned peptides made cell-permeable by fusion to a string of 9 arginines (24). We first determined the role of calcium in LPS signaling. We found that EGTA could not prevent LPS from inducing its target genes, and ionomycin could not enhance the effect of LPS, indicating that extracellular calcium is irrelevant to LPS signaling (data not shown). In contrast, EGTA-AM, BAPTA-AM, P2, and P4 each selectively repressed Brg-dependent genes, whereas P2 as well as P4 somewhat stimulated Brg-independent genes (Fig. 3A and data not shown). As expected, P2m, P1, or P3 did not affect TLR4 target genes (Fig. 3A). Thus, Calcium/CaM appears selectively required for the expression of Brg-dependent genes.
Fig. 3.
Effects of calcium/CaM antagonists in macrophages. We stimulated bone marrow-derived primary macrophages with LPS (0.1 μg/mL) for 3 h in the presence or absence of EGTA-AM (100 μM) and peptides (30 μM) before analyzing gene expression, chromatin accessibility, and Brg recruitment. (A) RT-qPCR analysis of 6 TLR4 target genes. The expression levels in the cells treated with LPS alone were set as 100. The data were averaged from 3 independent experiments. (B) Accessibility of Brg-dependent genes. The restriction enzymes (first enzymes) used for digesting chromatin in macrophages were indicated at the bottom, together with the positions of their cleavage sites relative to the transcription start site (+1). DNA was then purified and completely digested with a second enzyme, and both cleavages were detected by LM-PCR (Upper), with the second cleavage (asterisk) serving as a normalization control for the first (red arrow). The bands were quantified by phosphorImager to yield the percentages of cleavages (Lower). E, EGTA-AM. (C) Accessibility of the Brg-independent MIP2 gene was not affected by EGTA-AM (Left) or P2 (Right). Cells were digested with 3-fold dilutions of SacI (red triangles) before DNA purification. The experiments presented in Left and Right were done independently. EGTA in the Left denotes EGTA-AM. (D) Anti-Brg ChIP. Precipitated DNA was quantified by qPCR and plotted relative to basal levels of Brg binding in resting cells. Shown is a representative experiment out of >3 independent experiments; error bars are SDs from triplicate PCR. Independent experiments are shown in Fig. S2. Although there is variability, the trend is clearly upheld.
To determine whether the defects in transcription result from defects in remodeling, we used restriction enzyme accessibility assays. Macrophages were stimulated with LPS in the presence of various drugs. The cells were then permeabilized and digested with restriction enzymes recognizing the promoters of the Brg-target genes. DNA was then purified and digested to completion with a second enzyme that cuts in the vicinity of the first enzyme to generate normalization controls. Both cleavages were detected by LM-PCR, which produced 2 bands, the lower and upper bands representing the cleavage generated by the first and second enzymes, respectively; the relative abundance of the 2 bands is taken as a measure of accessibility and hence of remodeling. As expected, LPS significantly stimulated remodeling of the Brg-dependent genes (Fig. 3B, lanes 2, 7, 12, and 17). Importantly, EGTA-AM and P2 abolished LPS-induced remodeling (Fig. 3B, lanes 3, 4, 8, 9, 13, 14, 18, and 19), whereas P2m, P1, or P3 could not (Fig. 3B, lanes 5, 10, 15, and 20; data not shown), consistent with the effects of these reagents on mRNA expression. In addition, neither EGTA nor ionomycin influenced remodeling, reinforcing the notion that calcium influx is irrelevant to TLR4 signaling [supporting information (SI) Fig. S1; data not shown]. As a control, we show neither EGTA-AM nor P2 could decrease the accessibility of the MIP2 promoter, which is constitutively open in resting macrophages before LPS stimulation (Fig. 3C).
Signaling pathways are well known to regulate the recruitment of the BAF complex. To determine whether calcium/CaM antagonists block BAF recruitment, macrophages were stimulated with LPS in the presence of P2 or EGTA-AM before Brg recruitment was assessed with the ChIP assays. LPS stimulation induced Brg recruitment to the target genes, as reported (Fig. 3D) (18). Surprisingly, neither EGTA-AM nor P2 impaired this recruitment, indicating that calcium/CaM controls BAF activity but not recruitment (Fig. 3D; Fig. S2).
We also examined the effects of W7, a small-molecule inhibitor of CaM. As in the case of other calcium/CaM antagonists, W7 inhibited remodeling without impairing Brg recruitment, although interpretation of the data are confounded by the toxicity of the drug (Fig. S3).
We conclude that in the presence of calcium/CaM antagonists, LPS can still induce recruitment of the BAF complex to its target genes, but the promoter-tethered complex is unable to remodel chromatin.
Roles of BAF57 in the Expression of Brg-Target Genes.
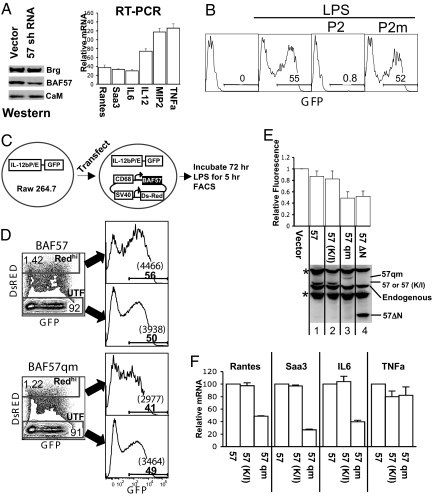
The pharmacological data described above predict that BAF57 is required for the expression of Brg-dependent genes in macrophages. To test this idea, we infected murine macrophage J774 cells with a retrovirus vector expressing an shRNA targeting BAF57. The shRNA reduced BAF57 protein level and indeed impaired induction of Brg-dependent genes without affecting Brg-independent genes (Fig. 4A). To corroborate these findings, we attempted to use BAF57 dominant negative mutants defective in CaM binding to interfere with the induction of Brg-dependent genes. Despite much effort, we were unable to make macrophage lines highly expressing BAF57 mutants. Therefore, we sought to transiently express BAF57 mutants and assay their activities on a stably integrated reporter plasmid bearing the IL-12b promoter/enhancer upstream of GFP; the GFP expression cassette is flanked by insulators to minimize interference from neighboring chromatin (25). It is known that macrophage J774 cells stably transfected with this plasmid express GFP on LPS stimulation, accompanied by remodeling at the integrated IL-12b promoter (25). Thus, GFP expression provides readout for IL-12b promoter activity. Because J774 lines are difficult to transfect, we integrated the reporter plasmid into the Raw 264.7 macrophage line known to support robust induction of reporter genes driven by the IL-12b promoter, even though endogenous IL-12b is hardly inducible in Raw 264.7 cells (26). As expected, GFP was induced by LPS, and the induction was blocked by P2 but not P2m (Fig. 4B). We then transfected plasmids expressing BAF57 or its mutants together with the marker protein DsRED, followed by LPS stimulation (Fig. 4C). GFP induction in cells expressing highest levels of DsRed (REDhi) was reduced (≈2-fold) by BAF57qm and BAF57ΔN, but not by WT BAF57 or BAF57 (K112I) (Fig. 4D and Fig. 4E Upper), even though the expression levels of these proteins were comparable, except that BAF57ΔN was more abundant (Fig. 4E Lower). Given that the K112I mutation in BAF57 can impair the classic DNA binding function of the BAF57 HMG domain, the failure of BAF57 (K112I) to inhibit GFP induction reinforces the notion that a previously undescribed activity of the BAF57 HMG domain is used to regulate the BAF complex in macrophages. To determine whether BAF57qm also inhibited the expression of endogenous Brg target genes, we sorted out cells highly expressing DsRed, and analyzed mRNA expression. As expected, BAF57qm, but not BAF57 (K112I), impaired the induction of Brg target genes without affecting the expression of TNFa (Fig. 4F).
Fig. 4.
Roles of BAF57 in the expression of Brg-dependent genes. (A) Effects of BAF57 knockdown. Macrophage J774 cells were infected with a retrovirus expressing BAF57 shRNA, which knocked down BAF57 protein (but not Brg or CaM; Left). Cells were stimulated with LPS for 3 h, and mRNA levels of the TLR4 target genes quantified and plotted relative to that in cells infected with the empty vector (Right); the basal mRNA levels before LPS stimulation are <1% of the activated levels (data not shown). The RT-PCR data are from a representative experiment, the error bars being SDs of triplicate values. An independent experiment is shown in Fig. S4. (_B–F_) Effects of dominant negative BAF57 mutants. (_B_) A Raw 264.7 line harboring IL-12b reporter plasmid was stimulated with LPS (0.1 μg/mL) for 1 h in the presence or absence of P2 (30 μM) before the cells were washed. Incubations were continued for 4 h before FACS analysis. (_C_) The experimental strategy. The transfected plasmid expresses BAF57 (or its mutants) and DsRed from the CD68 and SV40 promoters, respectively. (_D_) Representative FACS plots. Transiently transfected cells were stimulated with LPS and analyzed for DsRed/GFP expression on LSR II-Green. The majority of the cells were untransfected (UTF), whereas ≈5% of the cells expressed detectable levels of DsRed, but only the subset highly expressing DsRed (Redhi) had a consistent defect in GFP induction when BAF57qm was co-expressed (lower plots), manifested as decreases in both the percentages and the mean fluorescent intensity (MFI) of GFP+ cells (histograms at the right, where the numbers in bold font and those within parentheses denote the percentages of GFP+ cells and their MFI, respectively). (_E_) The effects of various BAF57 mutants on GFP induction. The percentages and MFI of GFP+ cells in the Redhi subset were multiplied, and their products normalized to that in UTF population and plotted relative to that in the samples transfected with the empty vector, the latter arbitrarily set as 1. The values were averaged from >3 independent experiments. The bottom gel is a Western blot detecting BAF57 and its mutants. Asterisks, nonspecific bands. It is unknown why BAF57ΔN is no more effective than BAF57qm in impairing GFP induction, given its higher expression; perhaps the deletion impaired incorporation of the mutant into the BAF complex. (F) DsRedhi cells were sorted, and the expression of endogenous genes quantified and plotted relative to the mRNA level of cells transfected with the vector expressing BAF57. Error bars are SDs from duplicate PCR values. IL12b mRNA could not be accurately quantified due to its extremely low induction level (data not shown).
We conclude that the BAF57 HMG domain is necessary for efficient expression of Brg target genes in macrophages, and the function of the HMG domain appears to depend on its interaction with CaM instead of DNA.
Discussion
In this work, we demonstrated that (i) calcium/CaM is essential for the induction of certain subsets of TLR4 target genes; (ii) calcium/CaM regulates these genes by stimulating BAF-dependent chromatin remodeling; and (iii) this stimulatory effect is independent of BAF recruitment, and appears to be mediated by a direct interaction between calcium/CaM and BAF57. Our data reveal unexpected roles of calcium/CaM in TLR4 signaling and a previously undescribed mode of regulation of chromatin remodeling.
Roles of Calcium in LPS Signaling.
Although calcium is a ubiquitous signaling molecule, its role in LPS signaling remains largely unexplored. We showed that calcium/CaM is essential for TLR4 to activate many of its target genes, which is consistent with the fact that TMB-8, which prevents calcium release from intracellular stores, compromises LPS-induced tumoricidal activity of macrophages (27, 28). Paradoxically, LPS cannot induce any increase in intracellular calcium concentrations (27–30), or at best induces very weak calcium signals (31, 32). However, it has been consistently reported that basal calcium concentrations in macrophages (100–200 nM) are higher than T cells (33–35), which is minimally sufficient for activating the CaM-Calcineurin pathway, leading to NFATc nuclear translocation and repression of basal-level NFkb activation in resting macrophages (33, 36). The basal level calcium might also be (minimally) sufficient for CaM to regulate the BAF complex, especially in conjunction with the putative weak calcium signal induced on LPS stimulation. In any case, it is clear that LPS-stimulated gene expression does not require dramatic, global increases in intracellular calcium concentrations. Of note, phorbol ester is able to activate CaM-CaMK II pathway in T cells without invoking a detectable increase in intracellular calcium (37, 38), reinforcing the notion that strong calcium signals are not needed for activating some CaM-dependent events.
CaM-Dependent Regulation of Remodeling: Beyond BAF Recruitment.
On LPS signaling, BAF complex is recruited to target genes but cannot remodel them without a second, CaM-dependent event, which is unexpected because BAF recruitment appears to invariably coincide with remodeling. The mechanism underlying such a counterintuitive phenomenon is unclear. Purified BAF complex can efficiently disrupt nucleosomes even in the absence the BAF57 HMG domain (4), and CaM cannot further stimulate remodeling in vitro (data not shown). Neither is CaM generally needed for BAF complex to remodel endogenous targets, because overexpression of a dominant negative mutant of BAF57 lacking the HMG domain produces little defects in T cell development beyond misregulation of the CD4/CD8 genes, whereas deletion of Brg blocks thymocyte survival, proliferation, and differentiation (23, 39). Collectively, these data reveal a specific role of BAF57 in LPS-induced gene expression in macrophages. We speculate that some repressors of remodeling are present in macrophages, and CaM is needed to overcome the repression. This idea is inspired by the fact that on LPS stimulation, Mi-2b, a component of the chromatin remodeling NuRD complex, is corecruited with the BAF complex to BAF target genes where it antagonizes the BAF complex and limits the expression levels of BAF target genes (18). However, we have excluded Mi-2b as a relevant repressor, because Mi-2b RNAi failed to bypass the need of CaM for the expression of BAF target genes (data not shown). A genetic screen using shRNA library might uncover the putative repressors.
How exactly does CaM stimulate remodeling? We found that CaM binds the BAF complex in vitro and that pharmacological and genetic disruption of CaM-BAF interactions blocks BAF function in macrophages. These data suggest that CaM stimulates remodeling in macrophages, at least in part, by means of physical interactions with the BAF complex, which is consistent with the presence of CaM in the macrophage nucleus (Fig. S5). However, we are unable to detect CaM at the Brg target genes by ChIP assays (data not shown), which is perhaps not surprising. The BAF57 HMG domain binds not only CaM but also DNA. DNA can disrupt CaM binding to the SRY HMG domain (21), suggesting that it can also disrupt CaM-BAF57 interaction. We imagine that CaM is associated with the BAF complex and corecruited to BAF target genes, but subsequently released from the BAF complex once BAF57 has engaged DNA, thus escaping detection in the ChIP assay requiring relatively stable binding at the target genes. In this scenario, CaM stimulates remodeling in a “hit and run” manner. CaM might use BAF57 to hitchhike to the promoters, where it quickly acts on chromatin to help initiate remodeling. Alternatively, CaM may be required to maintain some conformational changes on BAF57 till BAF57 binds DNA. Identification of the putative repressors is critical for addressing these issues.
Last, we note that, although most of the BAF subunits have several (tissue-specific) isoforms that are combinatorially assembled into perhaps several hundred functionally divergent BAF complexes (6), BAF57 appears to be monomorphic, ubiquitously expressed, and present in distinct BAF complexes (4), suggesting that CaM can regulate various BAF complexes in diverse tissues and presumably also the homologous remodeling complexes in lower organisms, given that CaM is one of the most conserved eukaryotic proteins and BAF57 is conserved in Drosophila (40).
Materials and Methods
In Vitro CaM-BAF Cross-Linking.
We mixed 5 μL (≈0.5 μg) of the BAF complex affinity-purified from the HeLa line expressing Flag-tagged BAF47 with 1 μL (1 μg) of recombinant CaM (Millipore) in 40 μL of reaction buffer (20 mM Hepes pH7.6/1 mM MgAc/1 mM Imidazole/0.1% Nonidet P-40/1% Tween-20/0.1 M NaCl) containing either 2 mM CaCl2 or 5 mM EGTA. The reaction was incubated for 3 min on ice before addition of DSS to 80 μg/mL. The cross-liking reaction was stopped 30 min thereafter with Laemmlie sample buffer, the sample boiled to dissociate noncross-linked subunits, and analyzed by Western blotting using anti-CaM monoclonal antibody (Millipore no. 05–173) and BAF57 antibody. Under this condition, the majority of the BAF subunits remained uncross-linked, which ensured that only directly interacting partners were subject to cross-linking. Other experimental procedures are relatively standard, and are described in SI Materials and Methods.
Supplementary Material
Supporting Information
Acknowledgments.
We thank P. Cresswell, M. Ikura, R. Kingston (Harvard University, Boston, MA), and D. Greaves (University of Oxford, Oxford OX1 3RE, UK) for advice/reagents, and R. Medzhitov for critical reading of the manuscript. This work is supported in part by National Institutes of Health Grants R01AI063554–02 (to T.C.) and GM56244 (to A.N.I.). T.G. is supported by grants from the Swedish Research Council and the Swedish Cancer Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Chi T. A BAF-centred view of the immune system. Nat Rev Immunol. 2004;4:965–977. doi: 10.1038/nri1501. [DOI] [PubMed] [Google Scholar]
- 2.Agalioti T, et al. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 3.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 4.Wang W, et al. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc Natl Acad Sci USA. 1998;95:492–498. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nie Z, et al. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol. 2000;20:8879–8888. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu JI, et al. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 7.Lickert H, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 8.Belandia B, Orford RL, Hurst HC, Parker MG. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 2002;21:4094–4103. doi: 10.1093/emboj/cdf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoeflich KP, Ikura M. Calmodulin in action: Diversity in target recognition and activation mechanisms. Cell. 2002;108:739–742. doi: 10.1016/s0092-8674(02)00682-7. [DOI] [PubMed] [Google Scholar]
- 10.Bachs O. Calcium and Calmodulin in the Cell Nucleus. Austin, TX: RG Landes Company; 1995. [Google Scholar]
- 11.Hauser J, Saarikettu J, Grundstrom T. Calcium regulation of myogenesis by differential calmodulin inhibition of basic helix-loop-helix transcription factors. Mol Biol Cell. 2008;19:2509–2519. doi: 10.1091/mbc.E07-09-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauser J, et al. B-cell receptor activation inhibits AID expression through calmodulin inhibition of E-proteins. Proc Natl Acad Sci USA. 2008;105:1267–1272. doi: 10.1073/pnas.0708220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szymanski DB, Liao B, Zielinski RE. Calmodulin isoforms differentially enhance the binding of cauliflower nuclear proteins and recombinant TGA3 to a region derived from the Arabidopsis Cam-3 promoter. Plant Cell. 1996;8:1069–1077. doi: 10.1105/tpc.8.6.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoo JH, et al. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in arabidopsis. J Biol Chem. 2005;280:3697–3706. doi: 10.1074/jbc.M408237200. [DOI] [PubMed] [Google Scholar]
- 15.Bouche N, et al. A novel family of calmodulin-binding transcription activators in multicellular organisms. J Biol Chem. 2002;277:21851–21861. doi: 10.1074/jbc.M200268200. [DOI] [PubMed] [Google Scholar]
- 16.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 17.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 18.Ramirez-Carrozzi VR, et al. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 20.Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harley VR, Lovell-Badge R, Goodfellow PN, Hextall PJ. The HMG box of SRY is a calmodulin binding domain. FEBS Lett. 1996;391:24–28. doi: 10.1016/0014-5793(96)00694-1. [DOI] [PubMed] [Google Scholar]
- 22.Yap KL, et al. Calmodulin target database. J Struct Funct Genom. 2000;1:8–14. doi: 10.1023/a:1011320027914. [DOI] [PubMed] [Google Scholar]
- 23.Chi TH, et al. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 2002;418:195–199. doi: 10.1038/nature00876. [DOI] [PubMed] [Google Scholar]
- 24.Futaki S, Goto S, Sugiura Y. Membrane permeability commonly shared among arginine-rich peptides. J Mol Recognit. 2003;16:260–264. doi: 10.1002/jmr.635. [DOI] [PubMed] [Google Scholar]
- 25.Zhou L, et al. An inducible enhancer required for Il12b promoter activity in an insulated chromatin environment. Mol Cell Biol. 2007;27:2698–2712. doi: 10.1128/MCB.00788-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinmann AS, et al. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nat Immunol. 2001;2:51–57. doi: 10.1038/83168. [DOI] [PubMed] [Google Scholar]
- 27.Gorecka-Tisera AM, Snowdowne KW, Borle AB. Implications of a rise in cytosolic free calcium in the activation of RAW-264 macrophages for tumor cell killing. Cell Immunol. 1986;100:411–421. doi: 10.1016/0008-8749(86)90040-7. [DOI] [PubMed] [Google Scholar]
- 28.Drysdale BE, Yapundich RA, Shin ML, Shin HS. Lipopolysaccharide-mediated macrophage activation: The role of calcium in the generation of tumoricidal activity. J Immunol. 1987;138:951–956. [PubMed] [Google Scholar]
- 29.Prpic V, et al. Effects of bacterial lipopolysaccharide on the hydrolysis of phosphatidylinositol-4,5-bisphosphate in murine peritoneal macrophages. J Immunol. 1987;139:526–533. [PubMed] [Google Scholar]
- 30.Watanabe N, Suzuki J, Kobayashi Y. Role of calcium in tumor necrosis factor-alpha production by activated macrophages. J Biochem. 1996;120:1190–1195. doi: 10.1093/oxfordjournals.jbchem.a021540. [DOI] [PubMed] [Google Scholar]
- 31.Letari O, et al. Activation by bacterial lipopolysaccharide causes changes in the cytosolic free calcium concentration in single peritoneal macrophages. J Immunol. 1991;147:980–983. [PubMed] [Google Scholar]
- 32.Kim Y, et al. Ca2+/calmodulin-dependent protein phosphatase calcineurin mediates the expression of iNOS through IKK and NF-kappaB activity in LPS-stimulated mouse peritoneal macrophages and RAW 264.7 cells. Biochem Biophys Res Commun. 2004;314:695–703. doi: 10.1016/j.bbrc.2003.12.153. [DOI] [PubMed] [Google Scholar]
- 33.Kang YJ, et al. Calcineurin negatively regulates TLR-mediated activation pathways. J Immunol. 2007;179:4598–4607. doi: 10.4049/jimmunol.179.7.4598. [DOI] [PubMed] [Google Scholar]
- 34.Beppu M, et al. Substrate-bound fibronectin enhances scavenger receptor activity of macrophages by calcium signaling. Arch Biochem Biophys. 2001;390:243–252. doi: 10.1006/abbi.2001.2381. [DOI] [PubMed] [Google Scholar]
- 35.Korhonen R, et al. Bi-directional effects of the elevation of intracellular calcium on the expression of inducible nitric oxide synthase in J774 macrophages exposed to low and to high concentrations of endotoxin. Biochem J. 2001;354:351–358. doi: 10.1042/0264-6021:3540351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conboy IM, Manoli D, Mhaiskar V, Jones PP. Calcineurin and vacuolar-type H+-ATPase modulate macrophage effector functions. Proc Natl Acad Sci USA. 1999;96:6324–6329. doi: 10.1073/pnas.96.11.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes K, Edin S, Antonsson A, Grundstrom T. Calmodulin-dependent kinase II mediates T cell receptor/CD3- and phorbol ester-induced activation of IkappaB kinase. J Biol Chem. 2001;276:36008–36013. doi: 10.1074/jbc.M106125200. [DOI] [PubMed] [Google Scholar]
- 38.Hughes K, Antonsson A, Grundstrom T. Calmodulin dependence of NFkappaB activation. FEBS Lett. 1998;441:132–136. doi: 10.1016/s0014-5793(98)01537-3. [DOI] [PubMed] [Google Scholar]
- 39.Chi TH, et al. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19:169–182. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- 40.Papoulas O, et al. The HMG-domain protein BAP111 is important for the function of the BRM chromatin-remodeling complex in vivo. Proc Natl Acad Sci USA. 2001;98:5728–5733. doi: 10.1073/pnas.091533398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Onions J, Hermann S, Grundstrom T. A novel type of calmodulin interaction in the inhibition of basic helix-loop-helix transcription factors. Biochemistry. 2000;39:4366–4374. doi: 10.1021/bi992533u. [DOI] [PubMed] [Google Scholar]
- 42.Rikihisa Y, Zhang Y, Park J. Role of Ca2+ and calmodulin in ehrlichial infection in macrophages. Infect Immun. 1995;63:2310–2316. doi: 10.1128/iai.63.6.2310-2316.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W, et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 44.Wang W, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 45.Weinmann AS, Plevy SE, Smale ST. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 1999;11:665–675. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
- 46.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 47.Chi T, Yan Z, Xue Y, Wang W. Purification and functional analysis of the mammalian SWI/SNF-family of chromatin-remodeling complexes. Methods Enzymol. 2004;377:299–316. doi: 10.1016/S0076-6879(03)77018-9. [DOI] [PubMed] [Google Scholar]
- 48.Yu M, et al. Nucleoprotein structure of the CD4 locus: Implications for the mechanisms underlying CD4 regulation during T cell development. Proc Natl Acad Sci USA. 2008;105:3873–3878. doi: 10.1073/pnas.0800810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gough PJ, Gordon S, Greaves DR. The use of human CD68 transcriptional regulatory sequences to direct high-level expression of class A scavenger receptor in macrophages in vitro and in vivo. Immunology. 2001;103:351–361. doi: 10.1046/j.1365-2567.2001.01256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information