The generation and utilization of a cancer-oriented representation of the human transcriptome by using expressed sequence tags (original) (raw)
Abstract
Whereas genome sequencing defines the genetic potential of an organism, transcript sequencing defines the utilization of this potential and links the genome with most areas of biology. To exploit the information within the human genome in the fight against cancer, we have deposited some two million expressed sequence tags (ESTs) from human tumors and their corresponding normal tissues in the public databases. The data currently define ≈23,500 genes, of which only ≈1,250 are still represented only by ESTs. Examination of the EST coverage of known cancer-related (CR) genes reveals that <1% do not have corresponding ESTs, indicating that the representation of genes associated with commonly studied tumors is high. The careful recording of the origin of all ESTs we have produced has enabled detailed definition of where the genes they represent are expressed in the human body. More than 100,000 ESTs are available for seven tissues, indicating a surprising variability of gene usage that has led to the discovery of a significant number of genes with restricted expression, and that may thus be therapeutically useful. The ESTs also reveal novel nonsynonymous germline variants (although the one-pass nature of the data necessitates careful validation) and many alternatively spliced transcripts. Although widely exploited by the scientific community, vindicating our totally open source policy, the EST data generated still provide extensive information that remains to be systematically explored, and that may further facilitate progress toward both the understanding and treatment of human cancers.
Human cancer results from the accrual of genetic mutations or epigenetic changes in the genomes of individual somatic cells. These exert their effect via alterations in the structure and abundance of individual mRNA molecules that, in turn, alter crucial protein-mediated cellular functions. One step in the path toward building a comprehensive molecular portrait of human cancer is the definition of the genes actively expressed in specific tumors and corresponding normal tissues. It is within these sets of genes that we must search for cancer-defining mutations and epigenetic changes and delineate the extent of the molecular milieu within which we will deepen our understanding of cancer. Very short sequence tags of transcripts and hybridization techniques such as microarrays identify many of the genes expressed in tumors and are widely used for measuring the relative levels of gene expression (1,2). However, longer transcript sequences such as ESTs permit genes expressed in individual cells and tissues to be identified in a completely unambiguous manner, provide additional data on transcript and gene variants, and represent a key source for the search for as yet incompletely characterized genes.
In two large projects, extensive EST sequencing of human tumor tissues has been undertaken: the Cancer Genome Anatomy Project (CGAP) (3) and the Fundaçao de Amparo à Pesquisa do Estado de Sao Paulo/Ludwig Institute for Cancer Research-Human Cancer Genome Project (HCGP) (4,5). CGAP, launched in 1997 by the National Cancer Institute, has used single-pass sequencing from the 5′ and/or 3′ extremities of cDNA clones for sequence generation (6). The HCGP project adopted an alternative EST-based strategy, termed ORESTES, which generates sequences biased toward the central coding regions of transcripts (7). The data gathered by these two projects are thus complementary and have been combined into an International Database of Cancer Gene Expression (5), available athttp://cgap.nci.nih.gov. They also constitute the basis of the Human Cancer Index at TIGR (www.tigr.org/tdb/tgi/hcgi).
These sequencing initiatives are unique in providing a very large disease-oriented transcriptional database that contains an unprecedented amount of information on expressed human genes. To maximize the benefit of these data, we have made them all publicly available immediately on generation. We here provide a description of these data, as well as their utility for the identification of human genes, the tissue specificity of their expression, and the structure of some of their transcript variants. We find that, although the data have been widely used by the scientific community, much additional information remains untapped.
Materials and Methods
Transcript Sequencing. ORESTES sequences were generated as previously described (7). Tumors and corresponding normal tissues were mostly obtained from the Hospital de Câncer A.C. Camargo, São Paulo, with the exception of purified breast tissue samples that were obtained from University College London (7). In addition, extensive use was made of both breast and colon cell lines. Identification of the source of mRNA for all libraries is available at the CGAP homepage. CGAP sequences were generated from the 3′ and 5′ extremities of both standard and normalized cDNA libraries from a variety of sources as described elsewhere (5).
Transcript to Genome Mapping. A comprehensive reconstruction of human transcripts based on genome data were undertaken based on two datasets: (i) a set of alignments between transcripts and genome regions, thoroughly filtered to eliminate the effects of pseudogenes, highly conserved gene families, repetitive elements and EST sequencing errors; (ii) a mapping onto the genome of all polyadenylation sites that could be extracted from the chromatograms of the EST sequencing projects, thus marking sites where polyadenylation has been experimentally documented (8). To be included as a spliced cluster, we stipulated that canonical splice sites must be present on at least one of the transcript sequences. ORF identification required that for the longest ORF predicted by ESTScan (www.ch.embnet.org/software/ESTScan.html) for each transcript cluster, the ESTScan score divided by the ORF length had to be >1 (9), an empirical measure that covers >99% of human SwissProt entries. In addition, the ESTScan-predicted ORFs had to have at least three nucleotides 5′ and 3′, as a measure of having at least some 5′ and 3′ UTR.
Generation of a Representative Set of CR Genes. This manually curated compilation was drawn up during a week-long meeting of the Annotation Consortium in August of 2001.‡ It is a nonredundant list comprising 1,127 human cancer-associated genes based on initial querying of GenBank (www.ncbi.nlm.nih.gov/GenBank/index.html), GenCard (http://bioinfo.weizmann.ac.il/cards/index.html), and Harvard University (http://sbweb.med.harvard.edu/research/breast_cancer/currentlistofgenes.htm) with the words “cancer” and “tumor.” The list is available athttp://bit.fmrp.usp.br/jamborestes.
Detection of Single-Nucleotide Polymorphisms (SNPs). The transcript sequences were aligned against one another by using FASTA and base quality values files generated by using PHRED (10,11). A SNP was considered for further analysis only if indicated by reads from at least two different ORESTES libraries, at least two different CGAP reads, or one read from each source. All selected SNPs were compared with SNPs already deposited in dbSNP (www.ncbi.nlm.nih.gov/SNP) by BLASTN. Only newly identified human SNPs were included in experimental validation, by PCR with DNA from a panel of 150 Brazilian individuals from three different ethnic backgrounds: 50 whites (mostly of Western and Southern European ancestry), 50 blacks (mulattos excluded), and 50 Japanese. All groups reported no racial admixture in their four grandparents.
Alternative Splicing. Two approaches were used. In the first, exon skipping was detected by using j_explorer (available for download fromwww.sanbi.ac.za/exon_skipping), which reduces the complexity of the gene sequences to a set of possible splice junctions used to search for ESTs spanning the annotated exon-exon junctions (12). The second approach listed all variant exons as revealed by transcript to genome alignments by using only those cDNAs that span at least two exons. Variants are represented by a binary matrix where each row corresponds to a sequence, and each column corresponds to an exon (33).
Results and Discussion
Gene Identification. Collectively, the Fundaçao de Amparo à Pesquisa do Estado de Sao Paulo/Ludwig Institute for Cancer Research-Human Cancer Genome Project and the Cancer Genome Anatomy Project have deposited over two million sequences from tumors and normal tissues in GenBank (Table 1). To give perspective, this number of sequences is greater than that required to complete the high-quality shotgun sequencing of ≈40 bacterial genomes. The two projects are the largest individual contributors to the public human EST database and are responsible for >40% of all publicly available human EST data available (dbEST release 122002,www.ncbi.nlm.nih.gov/dbEST).
Table 1. HCGP and CGAP transcript sequence generation and clustering.
| Form of gene representation | Number of sequences, clusters, or genes |
|---|---|
| ORESTES submitted to GenBank | 823,121 sequences |
| CGAP EST submitted to GenBank | 1,214,358 sequences |
| TOTAL EST submitted to GenBank | 2,037,479 sequences |
| Total clusters | 32,129 clusters |
| Total clusters with known genes | 22,152 clusters |
| Clusters without known genes | 9,977 clusters |
| Clusters without known genes but with coding potential | 1,285 clusters |
| Estimated total genes based on HCGP and CGAP data | 23,437 genes |
The first, and most obvious, use of ESTs is to define genes within the human genome. The precise experimental definition of these structures is crucial not only for CR research but also for research undertaken in all aspects of human biology. To achieve this, we have organized our EST data by aligning them, together with all other publicly available transcript sequences, to the human genome assembly. This allows an accurate organization of sequences into clusters corresponding to individual genes even in cases of minimal overlap or high similarity between paralogs. The transcript to genome alignment also provides an arbiter of quality, in that ESTs and EST clusters that span splice sites (referred to as spliced ESTs and spliced clusters, respectively) but correspond to still incompletely defined genes can be unambiguously distinguished from EST sequences derived from contaminating DNA or immature mRNA molecules.
We identified transcript clusters that contain at least one EST derived from one of the two cancer transcript sequencing projects, and which we judge to have a high probability of being derived from authentic expressed genes due to the presence of a reportedly full-length cDNA sequence, evidence of splicing or the presence of a predicted ORF. A total of 22,152 of the clusters contain full-length cDNAs. We use the presence of such full-length cDNA-containing transcript clusters as the gold standard for identification of human genes and refer to the genes they define as known genes. The known genes mapped by our ESTs comprise a subset of the total of the 29,332 known genes currently contained within the human genome, indicating that ≈75.5% of known human genes are expressed in the tumors and normal tissues we have studied. The 25% of genes that were not represented by our ESTs are defined by ESTs from other projects or full length cDNAs from various sources. We assume that these genes are either not expressed or are expressed at low levels in the cells and tissues that we have studied. Some 4,000 of the known genes to which our ESTs map have been defined since the publication of the draft human genome in 2001 (13,14). In the original published draft, these genes were represented only by the EST clusters. Interestingly, this number is 75% of all novel genes defined in this period, demonstrating that our ESTs equally well represent previously known genes and those still being defined. The preexistence of the EST clusters led directly to the generation of full-length cDNAs of a number of CR genes. The ESTs served either to indicate the existence of genes within defined regions of the human genome or as evidence of previously unknown members of paralogous families (Table 2 and cited in detail in Table 6, which is published as supporting information on the PNAS web site).
Table 2. Novel genes and definition of genes with restricted expression patterns utilizing EST clusters containing sequences from the HCGP and CGAP databases.
| Gene type | No. of genes |
|---|---|
| Positionally cloned genes | 5 |
| Genes cloned on the basis of homology | 16 |
| Tissue restricted expression | |
| Brain | 9 |
| Breast | 14 |
| Colon | 11 |
| Prostate | 15 |
| Cancer/testis | 4 |
| Various | 13 |
| Total | 87 |
To investigate the extent to which our ESTs represent known CR genes, we compiled a list of 1,127 human genes known or presumed to play a role in the process of transformation to malignancy, which we refer to as CR (cancer-related) genes (seehttp://bit.fmrp.usp.br). The CR set contains extensively studied CR genes such as TP53, RB1, BRCA1, CDKN2, and ERBB2, as well as members of paralogous gene families that function in critical signal-transducing pathways, such as cadherins, integrins, and mitogen-activated protein kinases. Although incomplete, we believe the CR set is representative of well characterized genes relevant to the development of human cancer. Most importantly, many of these have been cloned on the basis of strategies, such as positional cloning, that do not depend on transcript abundance. Of the genes in the list, we found that 1,009 (89%) have at least one corresponding ORESTES sequence; 1,099 genes (97%) have at least one CGAP sequence; and 1,102 genes (97%) have EST sequences derived from at least one of the two projects. Of the 25 genes for which we have not generated ESTs, 18 have no EST coverage at all, indicating that their overall expression is at very low levels in the human body.
The 9,977 clusters composed only of ESTs represent an unknown number of additional genes for which a full-length cDNA is not yet available (Table 1). A particular difficulty in relating these clusters to genes lies in the very high level of heterogeneity at their 3′ ends (8). Thus, noncoding, spliced clusters may be extensions of known genes; those that are not might represent multiple different forms of as-yet-undefined genes. To avoid the complications of the 3′ heterogeneity of transcripts, and also to exclude clusters that represent noncoding transcripts, one approach is to use only clusters that contain an identifiable ORF (9). Although we identified 9,977 EST clusters with splicing, only 1,285 are apparently coding sequences. We take these as each representing an individual human gene in the present discussion, although this one-to-one correlation remains to be demonstrated by full-length cDNA sequencing. Nevertheless, on the basis of these criteria, the two million or so EST sequences that we have generated translate into 23,437 genes, of which 1,285 are currently not defined by full-length cDNAs. Taking our data to include 75.5% of all genes, on the basis of the coverage of known genes described above, this value would predict a total of 31,042 genes in the human genome, in line with other recent estimates (13-16). It should be noted, however, that this value is likely to steadily increase as more small, nonspliced, and nonprotein-coding genes are discovered (16-18). These analyses indicate, however, that about the same number of genes confirmed by full-length cDNA sequencing since the publication of the draft genome sequence could be added to the genome immediately on the basis of the EST clusters with predicted ORFs to which we have contributed. These data are likely to contain a rich variety of novel genes relevant to cancer. For example, we found in September 2001 that among clusters of this kind, there were 19 that apparently encode novel paralogs of genes in the CR list. Of these, five have already been deposited by others in the GenBank database: CAMK1 (accession no. BC032726), MBD2 (accession no. AY038022), TLN2 (accession no. NM_015059), BCHE (accession no. BC028713), and CNK (accession no.AK054808). The remainder will be described in detail elsewhere as the determination of the full-length sequences is completed.
We have previously shown that the number of ESTs corresponding to a gene is an indication of its level of expression (19). Thus, genes more frequently expressed in tumors than in normal tissues, for example, can be identified from the EST data. Several examples of genes with differential gene expression identified from our ESTs are included inTable 2 and are listed in detail in Table 7, which is published as supporting information on the PNAS web site. It should be pointed out, however, that in general we have rather less data from normal than tumor-derived tissues, which limits the power of this approach at present. Moreover, both normal and tumor samples can be highly heterogeneous, with normal tissues in particular being mixtures of different cell types. Thus, all conclusions drawn from analysis of our sequencing data require confirmation both in subsequent RT-PCR experiments and ultimately by histochemical analysis at the protein level. Nevertheless, EST cluster size appears to be a useful general indicator of at least overall gene expression and comparison of the number of CGAP and ORESTES sequences with serial analysis of gene expression (SAGE) tags for the same genes are positively correlated (r = 0.6), despite the very different sets of tissues that have been accessed by the two databases. The average cluster size for all of the known human genes for which we have generated ESTs to date is 606, whereas the spliced EST clusters with predicted ORFs contain an average of only 19 ESTs. Thus, the latter represent genes much less frequently expressed than the average in the tissues we have studied. The average cluster size of the novel known genes added to the databases since the publication of the draft genome is 55, indicating that with time genes with lower levels of expression are being defined.
Tissue Specificity of Gene Expression. The genomes of all cells in the human body are copied from that in the fertilized egg, although they are continually modified over the lifetime of the individual due to the relentless acquisition of somatic mutations. Although it is these somatic alterations that are ultimately responsible for human cancer, their relative importance will depend on the transcriptional programming of the cell in which they occur. Thus, the careful documentation of the tissue specificity of gene expression is vital to understanding the genetic basis of cancer. For this purpose, the availability of details of the tissue origin of our ESTs represents a powerful resource. All information on the tissue origins of our ESTs is available on the CGAP web site (http://cgap.nci.nih.gov). For each of seven human tissues (brain, head and neck, colon, lung, breast, uterus, and kidney), we have generated >100,000 EST sequences providing, in each case, a deep survey of gene expression. This value is comparable to the number of serial analysis of gene expression tags typically generated in individual experiments for the detection of differential gene expression and far in excess of any other EST compilation for individual tissues (20).
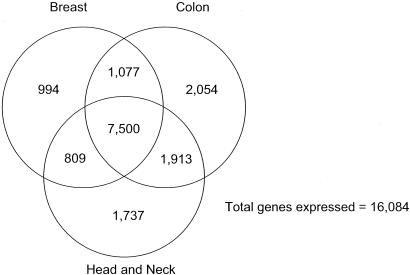
On the basis of ESTs that correspond to known genes and spliced EST clusters with predicted ORFs, we have evidence for the expression of between 10,000 and 13,500 genes for the seven tissue types that we have explored in depth, with lung having the highest number of expressed genes so far (Table 3). This indicates that not >57% of all of the genes defined by our ESTs genes are expressed in any one tissue type. It would thus appear that gene utility is quite variable between tissues. For example, if we take the 16,084 genes collectively expressed in breast, colon, and head and neck, there is evidence for expression for only 7,500 (47%) in all three tissues, whereas 4,785 (30%) are expressed in only one of the three tissues (Fig. 1). These numbers are arrived at despite the undoubted impurity of the tissue samples, in particular those derived from bulk tumors. Indeed, such impurity is readily detected by comparing ESTs present in libraries derived from bulk tumors but not corresponding cell lines, as can be easily accomplished by using the tools at the CGAP home page (http://cgap.nci.nih.gov). In the case of colon, for example, genes represented by ESTs from tumors but not cell lines include many examples (see Table 8, which is published as supporting information on the PNAS web site) that would not be expected to be present in epithelial cells. Given this tissue contamination, we might thus presume tissue specificity to be even higher than that measured, because the same contaminating lymphoid, fibroblast, endothelial, and smooth muscle cells are likely to be contaminants common to tumors from most locations.
Table 3. Documentation of gene expression in individual human tissues and estimation of tissue transcriptome complexity.
| Tissue | ESTs | Total clusters with known genes | Clusters with splicing and coding potential | Estimated total tissue transcriptome |
|---|---|---|---|---|
| Brain | 185,193 | 12,746 | 282 | 13,028 |
| Breast | 137,867 | 10,153 | 227 | 10,380 |
| Colon | 186,870 | 12,218 | 326 | 12,544 |
| Head/neck | 186,298 | 11,698 | 261 | 11,959 |
| Kidney | 115,096 | 12,368 | 341 | 12,709 |
| Lung | 166,469 | 13,076 | 314 | 13,390 |
| Ovary | 53,070 | 7,472 | 137 | 7,609 |
| Prostate | 85,910 | 10,004 | 224 | 10,228 |
| Uterus | 125,079 | 11,614 | 244 | 11,858 |
| Others | 795,627 | 19,818 | 1,079 | 20,897 |
Fig. 1.
Coexpression of genes among breast, colon, and head/neck.
On the other hand, we wondered whether the apparent high level of differential gene expression between tissues and between bulk tumors and cell lines might be exaggerated due to a still-insufficient level of sequence coverage. To address this possibility, we used data from the recently completed generation of several million transcript tags by Massively Parallel Signature Sequencing (MPSS) (21) from two human cell lines, one from breast and one from colon. In the MPSS experiment, the depth of sequencing guarantees the identification of all transcripts present at the level of at least one copy per cell (22). We found that in the MPSS experiment, ≈10,000 and 15,000 genes were expressed in the breast and colon cells, respectively, of which only 8,500 were expressed in both cell lines (21). In addition, inspection of the MPSS data also revealed that the great majority of the ESTs that we hypothesized were not derived from tumor epithelial cells and were also not detected in the colon cell line sequenced by MPSS, and those that were, were at very low levels (Table 8). Thus on both counts, the levels of EST coverage that we have achieved appear to provide a view of gene expression not significantly altered by very much deeper levels of transcript tagging.
A pairwise analysis of the tissues for which we have generated >100,000 ESTs indicates a consistency of shared and specific gene expression, with ≈70% of genes being expressed in common by any given pair (Table 4). These findings are consistent with the structure and function of human tissues being defined by the usage of highly variable permutations of genes, with almost all being expressed in more than one tissue. This would contrast, for example, with a model that requires a fixed set of ubiquitously expressed housekeeping genes with a substantial number of entirely tissue specific genes for individual tissues.
Table 4. Pairwise comparison of gene expression in human tissues.
| Tissue | Brain | Breast | Colon | Head/neck | Kidney | Lung | Uterus |
|---|---|---|---|---|---|---|---|
| Total genes | 13,028 | 10,380 | 12,544 | 11,959 | 12,709 | 13,390 | 11,858 |
| Brain | × | 8,484 | 9,745 | 9,370 | 9,797 | 10,337 | 9,502 |
| Breast | × | 8,577 | 8,309 | 8,434 | 8,715 | 8,354 | |
| Colon | × | 9,413 | 9,806 | 10,176 | 9,610 | ||
| Head/neck | × | 9,316 | 9,771 | 9,155 | |||
| Kidney | × | 10,299 | 9,621 | ||||
| Lung | × | 9,922 | |||||
| Uterus | × |
The EST data have proved extremely robust when used for the identification of genes with defined patterns of tissue specificity.Table 2 indicates examples of published studies that have identified CR genes with particular expression profiles and where the clusters representing the genes contained ESTs from our projects. The details of these studies are listed in Table 7. Of particular interest has been the identification of genes that are restricted to organs such as the breast and prostate that could serve as therapeutic targets for cancers in these organs. In addition, growing interest is being focused on genes restricted in expression to normal testis and tumors (CT-antigens) that are important potential targets of therapeutic vaccines. The relatively sparse use of the data for tissue types other than breast and prostate indicates there are likely to be many more genes with restricted expression still to be identified by using our data.
Identification of Gene and Transcript Variants. In addition to gene definition and identification of the tissues in which a gene is expressed, ESTs also provide information concerning the existence of germline variants and alternatively spliced transcript forms. The former can have a significant impact on an individual's predisposition to cancer. Nevertheless, because such variants take the form of SNPs for the most part, the relatively high sequencing error rate (≈1%) of the single-pass transcript sequences we have generated requires that potential variants have to be carefully evaluated. Nevertheless, many studies have reported large-scale analyses of SNPs that have incorporated the ESTs we have generated (23,24).
By using the original chromatograms, we identified a total of 237 SNPs for genes in the CR list, which are already listed in dbSNP. Of these, 47 were identified only with ORESTES data, 72 only with CGAP data, and 118 with both. In addition, a total of 210 further putative novel SNPs were identified by using the ORESTES data, and 295 were identified by using the CGAP data. Other studies have indicated that a high percentage of SNPs based on conventional ESTs, such as those generated in the CGAP project, can be subsequently verified (24). We suspected that, because ORESTES is a PCR-based methodology, a lower percentage of potential variants may be real. To test this, we examined experimentally 20 of the nonsynonymous SNPs identified with the ORESTES data alone and found that three (15%) were present in a bank of 100 normal human DNA sequences (Table 5). Although this percentage is (as expected) low, the ORESTES-based variants tend to be of particular interest, because they most frequently lie within the coding regions, thus potentially generating altered protein molecules. On the basis of our validation rate, we predict there are likely to be >30 true novel nonsynonymous SNPs identifiable by the ORESTES data alone within the CR list.
Table 5. Validated new human SNPs based on ORESTES clusters.
| Gene name | Accession no. | Frequency of the variant allele in 150 Brazilian individuals | |||
|---|---|---|---|---|---|
| Variation | Whites | Blacks | Japanese | ||
| TM4SF3 | NM_004616 | G/C G73A | 38% C | 8% C | 27% C |
| CGM2 | X98311 | T/A F1201 | 39% A | 63% A | 62% A |
| KISS1 | U43527 | A/G Q36R | 8% G | — | — |
Alternative splicing generates variability at the transcriptome level and is conceivably of direct relevance to the generation of the malignant phenotype (25-27). Extensive utilization has been made of the ESTs we have generated for the identification of alternatively spliced genes within the human genome (12,28-32). We explored the degree of variability due to alternative exon usage in the set of 1,124 CR genes by using two different approaches. The first approach, which detects transcripts displaying exon skipping through the use of a software called j-explorer (12), predicts possible splice junctions and then searches the EST databases to identify transcripts spanning the annotated exon-exon junction. The second approach lists all variants based on transcript to genome mapping and is thus less stringent (33). The former found evidence of alternative splicing for 21.3% of all CR genes, with an average of 1.4 variants per gene, and the latter found that 47.5% of all genes in the list undergo alternative splicing with a total of 3,179 variants, and an average of 3.17 variants per gene. A total of 210 genes were found by both approaches as having more than one splicing variant. We sought to validate exon-skipping events identified in three of the genes: the skipping of exon 7 in CD 53 (NM_00560), the skipping of exon 26 in PTPN13 (NM_006264), and the insertion of an additional exon between denominated exons 1 and 2 in NM23A (NM_000269). We had detected each of these only in ESTs derived from tumors, thus suggesting that the truncated transcripts might be tumor specific. However, by using a strategy that enhanced the amplification of the transcript with the skipped exon with an RT-PCR primer that spanned the novel splice site formed, we were able, in each case, to detect the alternative transcript in both normal and tumor derived samples. Thus, although carefully documented alternative splicing events can almost always be readily validated, tumor-specific alternative splicing may be rare.
Conclusion
The data we have generated have contributed to the identification of genes within the human genome, the definition of the tissue specificity of their expression, and the discovery of SNPs and alternatively spliced transcripts. It has been our intent that this should speed progress in the diagnosis, treatment, and understanding of human cancer. The number and variety of published studies in which the data we have generated appear to have played a significant role attest to the value of the open-source approach. Undoubtedly, vastly more progress has been made than if we had opted to concentrate on in-house utilization of our findings. Additionally, the thousands of high-quality EST clusters that are both spliced and contain predicted ORFs, the hundreds of genes that appear to have strongly tissue-restricted expression, as well as the thousands of potential SNPs and alternative splice forms that our data define, suggest that much more remains to be discovered within the existing publicly available data. We aim to continue generating sequence-based CR data with the aim of further improving the accurate transcriptional description of human tumors and the normal tissues from which they arise. We believe this will not only provide an important underlying support for broad-based cancer research but will also serve as the source for novel discoveries and hypotheses that will ultimately lead to improved cancer care.
Supplementary Material
Supporting Tables
Acknowledgments
We are indebted Lloyd J. Old, Scientific Director of the Ludwig Institute for Cancer Research, for support and stimulation; to J. Fernando Perez, Scientific Director of the Fundaçao de Amparo à Pesquisa do Estado de Sao Paulo (FAPESP); and to Richard Klausner, Director of the National Cancer Institute, without whom these studies would not have been possible. We are also greatly indebted to Dr. Jucara Parra for dedication to the successful execution of these studies through her role as Project Manager for the FAPESP/Ludwig Institute for Cancer Research-Human Cancer Genome Project.
Abbreviations: EST, expressed sequence tag; CGAP, Cancer Genome Anatomy Project; HCGP, Human Cancer Genome Project; SNP, single-nucleotide polymorphism; CR, cancer-related.
Footnotes
‡
The Human Cancer Genome Project/Cancer Genome Anatomy Project Annotation Consortium, Jamborestes, Aug. 20-25, 2001, São Paulo, Brazil.
References
- 1.Lal, A., Lash, A. E., Altschul, S. F., Velculescu, V., Zhang, L., McLendon, R. E., Marra, M. A., Prange, C., Morin, P. J., Polyak, K., et al. (1999) Cancer Res. 59**,** 5403-5407. [PubMed] [Google Scholar]
- 2.DeRisi, J., Penland, L., Brown, P. O., Bittner, M. L., Meltzer, P. S., Ray, M., Chen, Y., Su, Y. A. & Trent, J. M. (1996) Nat. Genet. 14**,** 457-460. [DOI] [PubMed] [Google Scholar]
- 3.Strausberg, R. L., Dahl, C. A. & Klausner, R. D. (1997) Nat. Genet. 15**,** Spec. No., 415-416. [DOI] [PubMed] [Google Scholar]
- 4.Bonalume, N. R. (1999) Nature 398**,** 450. [DOI] [PubMed] [Google Scholar]
- 5.Strausberg, R. L., Camargo, A. A., Riggins, G. J., Schaefer, C. F., De Souza, S. J., Grouse, L. H., Lal, A., Buetow, K. H., Boon, K., Greenhut, S. F., et al. (2002) Pharmacogenomics. J. 2**,** 156-164. [DOI] [PubMed] [Google Scholar]
- 6.Strausberg, R. L., Buetow, K. H., Emmert-Buck, M. R. & Klausner, R. D. (2000) Trends Genet. 16**,** 103-106. [DOI] [PubMed] [Google Scholar]
- 7.Dias, N. E., Garcia, C. R., Verjovski-Almeida, S., Briones, M. R., Nagai, M. A., da Silva, W., Jr., Zago, M. A., Bordin, S., Costa, F. F., Goldman, G. H., et al. (2000) Proc. Natl. Acad. Sci. USA 97**,** 3491-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iseli, C., Stevenson, B. J., De Souza, S. J., Samaia, H. B., Camargo, A. A., Buetow, K. H., Strausberg, R. L., Simpson, A. J., Bucher, P. & Jongeneel, C. V. (2002) Genome Res. 12**,** 1068-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iseli, C., Jongeneel, C. V. & Bucher, P. (1999) Proc. Int. Conf. Intell. Syst. Mol. Biol., 138-148. [PubMed]
- 10.Ewing, B. & Green, P. (1998) Genome Res. 8**,** 186-194. [PubMed] [Google Scholar]
- 11.Ewing, B., Hillier, L., Wendl, M. C. & Green, P. (1998) Genome Res. 8**,** 175-185. [DOI] [PubMed] [Google Scholar]
- 12.Hide, W. A., Babenko, V. N., van Heusden, P. A., Seoighe, C. & Kelso, J. F. (2001) Genome Res. 11**,** 1848-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409**,** 860-921. [DOI] [PubMed] [Google Scholar]
- 14.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291**,** 1304-1351. [DOI] [PubMed] [Google Scholar]
- 15.Roest, C. H., Jaillon, O., Bernot, A., Dasilva, C., Bouneau, L., Fischer, C., Fizames, C., Wincker, P., Brottier, P., Quetier, F., et al. (2000) Nat. Genet. 25**,** 235-238. [DOI] [PubMed] [Google Scholar]
- 16.Ewing, B. & Green, P. (2000) Nat. Genet. 25**,** 232-234. [DOI] [PubMed] [Google Scholar]
- 17.Reymond, A., Camargo, A. A., Deutsch, S., Stevenson, B. J., Parmigiani, R. B., Ucla, C., Bettoni, F., Rossier, C., Lyle, R., Guipponi, M.,et al. (2002) Genomics 79**,** 824-832. [DOI] [PubMed] [Google Scholar]
- 18.Eddy, S. R. (2002) Cell 109**,** 137-140. [DOI] [PubMed] [Google Scholar]
- 19.Camargo, A. A., Samaia, H. P., Dias-Neto, E., Simao, D. F., Migotto, I. A., Briones, M. R., Costa, F. F., Nagai, M. A., Verjovski-Almeida, S., Zago, M. A., et al. (2001) Proc. Natl. Acad. Sci. USA 98**,** 12103-12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boon, K., Osorio, E. C., Greenhut, S. F., Schaefer, C. F., Shoemaker, J., Polyak, K., Morin, P. J., Buetow, K. H., Strausberg, R. L., De Souza, S. J. et al. (2002) Proc. Natl. Acad. Sci. USA 99**,** 11287-11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jongeneel, C. V., Iseli, C., Stevenson, B. J., Riggins, G. J., Lal, A., Mackay, A., Harris, R. A., O'Hare, M. J., Neville, A. M., Simpson, A. J.,et al. (2003) Proc. Natl. Acad. Sci. USA 100**,** 4702-4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenner, S., Johnson, M., Bridgham, J., Golda, G., Lloyd, D. H., Johnson, D., Luo, S., McCurdy, S., Foy, M., Ewan, M., et al. (2000) Nat. Biotechnol. 18**,** 630-634. [DOI] [PubMed] [Google Scholar]
- 23.Clifford, R., Edmonson, M., Hu, Y., Nguyen, C., Scherpbier, T. & Buetow, K. H. (2000) Genome Res. 10**,** 1259-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irizarry, K., Kustanovich, V., Li, C., Brown, N., Nelson, S., Wong, W. & Lee, C. J. (2000) Nat. Genet. 26**,** 233-236. [DOI] [PubMed] [Google Scholar]
- 25.Caballero, O. L., De Souza, S. J., Brentani, R. R. & Simpson, A. J. (2001) Dis. Markers 17**,** 67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chun, S. Y., Bae, O. S. & Kim, J. B. (2000) J. Korean Med. Sci. 15**,** 696-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez, L. M., Hajos, S. E., Basilio, F. M., Mongini, C. & Alvarez, E. (2001) Oncol. Rep. 8**,** 145-151. [PubMed] [Google Scholar]
- 28.Brett, D., Pospisil, H., Valcarcel, J., Reich, J. & Bork, P. (2002) Nat. Genet. 30**,** 29-30. [DOI] [PubMed] [Google Scholar]
- 29.Kan, Z., States, D. & Gish, W. (2002) Genome Res. 12**,** 1837-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Modrek, B., Resch, A., Grasso, C. & Lee, C. (2001) Nucleic Acids Res. 29**,** 2850-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie, H., Zhu, W. Y., Wasserman, A., Grebinskiy, V., Olson, A. & Mintz, L. (2002) Genomics 80**,** 326-330. [DOI] [PubMed] [Google Scholar]
- 32.Xu, Q., Modrek, B. & Lee, C. (2002) Nucleic Acids Res. 30**,** 3754-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakabe, N. J., Souza, J. E. S., Galante, P. F. A., Oliveira, P. S. L., Passetti, F., Brentani, H., Osorio, E. C., Zaiats, A., Leerkes, M. R., Kitajima, J. P., et al. (2003) Proc. Fr. Acad. Sci., in press.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Tables