The Phosphotyrosine Interactome of the Insulin Receptor Family and Its Substrates IRS-1 and IRS-2 (original) (raw)
Abstract
The insulin signaling pathway is critical in regulating glucose levels and is associated with diabetes, obesity, and longevity. A tyrosine phosphorylation cascade creates docking sites for protein interactions, initiating subsequent propagation of the signal throughout the cell. The phosphotyrosine interactome of this medically important pathway has not yet been studied comprehensively. We therefore applied quantitative interaction proteomics to exhaustively profile all potential phosphotyrosine-dependent interaction sites in its key players. We targeted and compared insulin receptor substrates 1 and 2 (IRS-1 and IRS-2) as central distributors of the insulin signal, the insulin receptor, the insulin-like growth factor 1 receptor, and the insulin receptor-related receptor. Using the stable isotope labeling by amino acids in cell culture (SILAC) approach with phosphorylated versus non-phosphorylated bait peptides, we found phosphorylation-specific interaction partners for 52 out of 109 investigated sites. In addition, doubly and triply phosphorylated motifs provided insight into the combinatorial effects of phosphorylation events in close proximity to each other. Our results retrieve known interactions and substantially broaden the spectrum of potential interaction partners of IRS-1 and IRS-2. A large number of common interactors rationalize their extensive functional redundancy. However, several proteins involved in signaling and metabolism interact differentially with IRS-1 and IRS-2 and thus provide leads into their different physiological roles. Differences in interactions at the receptor level are reflected in multisite recruitment of SHP2 by the insulin-like growth factor 1 receptor and limited but exclusive interactions with the IRR. In common with other recent reports, our data furthermore hint at non-SH2 or phosphotyrosine-binding domain-mediated phosphotyrosine binding.
Regulated protein-protein interactions form the basis of cellular signal transduction, and frequently posttranslational modifications constitute the molecular switch to facilitate the association or dissociation of proteins. Phosphorylation is a prominent instrument in the toolbox of signaling, and tyrosine phosphorylation in particular is important in the upstream events following ligand binding to receptor tyrosine kinases. The mere binding to a phosphorylated motif can modulate enzymatic activity in some cases. However, the primary effect of the interaction is usually to increase the local concentration of the recruited protein and scaffold it together with its upstream or downstream effectors. Common recognition modules for tyrosine phosphorylated sequences are the Src homology 2 (SH2)1 domain and the phosphotyrosine-binding (PTB) domain (1, 2). A large part of the free binding energy to an SH2 domain is provided by the phospho-moiety itself. Another part, and most importantly the binding specificity, is contributed by interactions with the residues C-terminal to the phosphotyrosine (Tyr(P)). As determined by degenerate peptide library screening, motifs for SH2 domains typically encompass residues +1 to +4 relative to the Tyr(P) (3). Some SH2 domains also exploit amino acids at the N-terminal side for binding (3, 4). In few cases even more extended contacts from −6 to +6 are formed (5, 6). The SH2 domain of SLAM-associated protein (SAP), for example, engages so many residues that already the non-phosphorylated form shows considerable binding, and phosphorylation only enhances this binding 5-fold (7). The general interaction mode of SH2 domains directs the sequence of the partner protein perpendicular to the central β-sheet of the SH2 domain in an extended conformation. Therefore the interaction is largely independent of the structural context in the native protein. This allows studying SH2 binding using short synthetic peptides (3). The genomes of humans and mice contain 120 different SH2 domains in 110 different proteins.
The other canonical interaction domain for Tyr(P), the PTB domain, occurs 56 times in the genome; its binding mode can vary considerably and is less conserved than is the case for SH2 domains (8, 9). PTB domains are divided into three classes: IRS-1/DOK-like, Shc-like, and Dab-like. Only members of the first two classes, which account for just 25% of all PTB domains, actually bind in a phosphorylation-dependent manner (1, 8). Recently several reports have described Tyr(P)-dependent protein interactions that do not involve SH2 or PTB domains. In one case binding is attributed to a C2 domain (10), but in other cases the responsible modules are still elusive (11). Even though it is still unclear whether those cases represent exceptions or instead more general principles, they demonstrate that Tyr(P)-mediated binding is not limited to the classical interaction domains.
A particularly important and clinically relevant pathway that involves tyrosine phosphorylation is the insulin-signaling network. Malfunction of insulin signaling can lead to type II diabetes if insulin resistance cannot be compensated by pancreatic β-cells any longer. Initially, insulin resistance is counterbalanced by increased insulin secretion by β-cells, but chronic hyperinsulinemia results in increased insulin resistance and finally leads to a decline of β-cell mass and function, facilitating hyperglycemia and diabetes (12). The main target tissues of insulin are skeletal muscle, liver, and adipose tissue, with muscle accounting for more than 70% of glucose disposal (13). One of the most important metabolic effects of insulin is the translocation of the glucose transporter GLUT4 from intracellular storage vesicles to the plasma membrane (14).
A complex and still incompletely understood network of signaling proteins connects the insulin receptor (InsR) with its downstream effectors. Insulin receptor substrate proteins (IRSs) play a pivotal role as interaction platforms that become phosphorylated on multiple tyrosines by the InsR and subsequently attract various signaling proteins to spread the stimulus within the cell. All six members of the IRS family contain a PH domain and a PTB domain at the N terminus, but they vary in the length of their C-terminal part. IRS-1 and IRS-2 are large proteins with many tyrosine phosphorylation sites and are ubiquitously expressed. IRS-3 is short and only present in rodents. IRS-4 has similar dimensions as IRS-1 and IRS-2, but is only expressed in very few and specialized tissues. IRS-5 and IRS-6 (also termed DOK-4 and DOK-5) possess very short C-terminal parts and consequently very few potential phosphotyrosine motifs (15, 16).
By far the most important players in insulin signaling are IRS-1 and IRS-2. Apart from their role in metabolic signaling, they propagate proliferative and anti-apoptotic signals and are consequently overexpressed or activated more strongly in most cancers (17). Furthermore many of the triggers of insulin resistance like excess lipids (13, 18, 19), inflammatory cytokines (18), or reactive oxygen species (20), exert their undesired effect through activation of multiple kinases that phosphorylate IRSs on serine residues (13). IRS-1 and IRS-2 share 75% amino acid sequence identity in their N-terminal domains and 35% in their C-terminal part. Despite their homology and many similar tyrosine-phosphorylation motifs, studies in knockout mice and knockout cell lines indicate that these two IRS proteins also serve complementary, rather than completely redundant, roles in insulin and IGF-1 signaling (21). In general, IRS-1 plays a prominent role in growth, while the main functions of IRS-2 are in glucose homeostasis and proper function of pancreatic β-cells (22). Knockout of either of them results in insulin resistance, but only IRS-2-knockout leads to diabetes. Tissue-specific differences between IRS-1 and IRS-2 add a further level of complexity (23). In muscle, IRS-1 is more closely associated with glucose uptake, whereas IRS-2 stimulates the MAP kinase pathway (24, 25). In liver, both are involved in metabolic regulation, but IRS-2 has a more pronounced role in lipid metabolism (26, 27). Adipose tissue engages IRS-1 mainly for differentiation whereas IRS-2 serves insulin-stimulated glucose uptake (28).
Similarly, among the three different receptors, InsR, insulin-like growth factor 1 receptor (IGF1R), and insulin receptor-related receptor (IRR), relatively small differences in the C-terminal, cytosolic sequence lead to considerable diversity in the signaling output. The insulin receptor and its ligand maintain metabolic homeostasis, whereas IGF-1 and its receptor preferentially control developmental and growth processes (29). Despite a similar degree of homology, the IRR is still an orphan receptor with obscure function and is expressed in few tissues (30, 31). Its greatest differences compared with the other receptors are in the C-terminal part, where it lacks the last 50 amino acids compared with InsR and IGF1R.
Here we undertook a systematic and exhaustive profiling of all potential phosphotyrosine-dependent interaction sites in IRS-1, IRS-2, InsR, IGF1R, and IRR. Because this kind of interaction is mediated by short, unstructured sequence motifs (2, 32, 33), and based on our previous results with ErbB receptors (34), bacterial proteins and the histone code (35), we employed peptide bait fishing from cell lysates combined with state-of-the-art mass spectrometry as readout. Our peptide pulldown approach, combined with the SILAC technique (36), allows straightforward discrimination between specific interaction partners and background binders (37). In contrast to in vitro experiments using purified components (38), the interactions take place within the environment of a whole cell lysate. In addition to the site-specific information obtained, the unbiased nature of the approach facilitates the discovery of unexpected interactions. This makes it possible, in principle, to uncover novel kinds of interactions mediated by modules other than the currently known SH2 and PTB domains in Tyr(P)-dependent signaling.
Our large scale experiment resulted in a global overview of specificity, redundancy, and distribution of protein interaction sites in the insulin signaling platform. It furthermore allows comparing the principle signaling capabilities between IRS-1 and IRS-2 as well as between InsR, IGF1R, and IRR. The Tyr(P)-interactomes of the IRS proteins were largely similar, accounting for the large functional redundancy. However, we did observe specific differences in the binding of SH2/PTB domain containing proteins and intriguingly in some proteins with other functions such as fatty acid degrading enzymes.
A main difference between InsR and IGF1R turned out to be the recruitment of the tyrosine phosphatase SHP2 to more sites in the IGF1R. The cryptic function of the IRR could be attributed to lack of interactions in its C-terminal part, perhaps combined with the unique recruitment of a membrane-associated guanylate kinase discovered here. Finally, in some instances combinatorial tyrosine phosphorylation either abolished or enabled certain protein interactions.
EXPERIMENTAL PROCEDURES
SILAC Cell Culture and Lysis—
Murine C2C12 muscle cells were cultured in Dulbecco's modified Eagle's medium containing 4.5% glucose and deficient in arginine (Arg) and lysine (Lys), supplemented with 10% dialyzed fetal calf serum and antibiotics. One cell population was supplied with normal Arg and Lys, and the other one with the stable isotope-labeled heavy analogues 13C615N4-Arg (or 13C6-Arg) and 13C615N2-Lys from Sigma Isotec. For triple labeling experiments 13C6-Arg and D4-Lys as well as 13C615N4-Arg and 13C615N2-Lys were employed. Cells were expanded as myoblasts for at least five doublings, and differentiation into myotubes was initiated by lowering serum content to 2% in confluent dishes. After 8 days myotube cultures typically contained less than 15% mononucleated cells. Harvesting was carried out by washing dishes with phosphate-buffered saline and adding ice-cold lysis buffer to the dishes for 15 min. Lysis buffer consisted of 1% Igepal (Nonidet P-40; v/v), 150 mm NaCl, 50 mm Tris-HCl, pH 7.5, 1 mm dithiothreitol, protease inhibitor mixture (Roche complete tablets), and 1 mm sodium orthovanadate as tyrosine phosphatase inhibitor. Cells were scraped off the dishes and vortexed vigorously. Following centrifugation at 16,000 × g for 15 min the supernatant was used for peptide affinity pulldown experiments.
Peptide Synthesis—
Peptides were synthesized as pairs in phosphorylated and non-phosphorylated form on a solid-phase peptide synthesizer using amide resin (Intavis, Cologne, Germany). To enhance accessibility, a short flexible linker of one serine and one glycine preceded the actual sequence. To further account for steric limitations and to build on current knowledge of binding modes of SH2 and PTB domains, we chose the sequence stretch as a 15-mer with 8 residues N-terminal of the Tyr(P) and 6 residues C-terminal. The peptides were synthesized with an N-terminal desthiobiotin for coupling to streptavidin-coated beads and efficient elution via biotin. Identity and purity of the synthetic peptides were confirmed by mass spectrometric analysis.
Peptide Pulldown Procedure—
Peptides were bound to streptavidin-coated magnetic beads (Dynal MyOne, Invitrogen), and cell lysate typically corresponding to 1.5 mg of protein (∼5 mg/ml protein) was added to 75 μl of beads containing an estimated amount of 2 nmol peptide. Heavy SILAC-labeled lysate was incubated with the phosphorylated version of the peptide, whereas light SILAC-labeled lysate was added to the non-phosphorylated counterpart. In parallel a crossover experiment was conducted, where the incubation was inverted, that is heavy lysate was incubated with the non-phosphorylated peptide and light lysate with the phosphorylated version. After rotation at 4 °C for six hours or overnight, the beads were washed for at least 3 times by vortexing with lysis buffer. Beads from each peptide pair were combined, and bound proteins were eluted using 20 mm biotin. Eluted proteins were then precipitated by adding 5 volumes of ethanol together with sodium acetate and 20-μg glycoblue (Ambion).
In-solution Digestion of Proteins—
Proteins were resuspended in 20 μl of 6 m urea, 2 m thiourea, 20 mm Tris-HCl, pH 8.0 and reduced by adding 1 μg of dithiothreitol for 30 min, followed by alkylation of cysteines by incubating with 5-μg iodoacetamide for 20 min. Digestion was started by adding endoproteinase Lys-C (Wako). After three hours samples were diluted with four volumes of 50 mm NH4HCO3, and trypsin (Promega) was added for overnight incubation. Proteases were applied in a ratio of 1:50 to protein material, and all steps were carried out at room temperature. Digestion was stopped by acidifying with trifluoroacetic acid, and the samples were loaded onto StageTips (39, 40) packed with reversed-phase-C18 Empore disks, (3M, St. Paul, MN) for desalting, and concentration prior to LC-MS-analysis.
NanoLC-MS/MS—
Digested peptide mixtures were separated by online reversed phase nanoscale capillary liquid chromatography and analyzed by electrospray tandem mass spectrometry. Experiments were performed with an Agilent 1100 nanoflow system connected to an LTQ-Orbitrap or LTQ-FT mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a nanoelectrospray ion source (Proxeon Biosystems, Odense, Denmark). Binding and chromatographic separation of the peptides took place in a 15 cm fused silica emitter (75-μm inner diameter) in-house packed (41) with reversed-phase ReproSil-Pur C18-AQ 3-μm resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany).
Peptide mixtures were injected onto the column with a flow of 500 nL/min and subsequently eluted with a flow of 250 nL/min from 2% to 40% MeCN in 0.5% acetic acid, in a 100-min gradient. The mass spectrometer was operated in data-dependent mode to automatically switch between MS and MS/MS (MS2) acquisition. Survey full scan MS spectra with m/z 300–1600 were acquired in the Orbitrap with a resolution of 60,000 at m/z 400 after accumulation to a target value of 1 million charges in the linear ion trap using the lock mass option for internal calibration of each spectrum (42). The five most intense ions were sequentially isolated for fragmentation in the linear ion trap using collisionally induced dissociation with normalized collision energy of 30% at a target value of 5000. The resulting fragment ions were recorded in the linear ion trap with unit resolution. Ions already selected for MS/MS were dynamically excluded for 60 s. For LTQ-FT measurement resolution was set to 25,000 for full scan; 5 million ions were accumulated; and the top three ions were selected for sequencing. In FT-ICR, selected ion monitoring scans were acquired before fragmentation for enhanced mass accuracy and signal-to-noise of the parent ion (resolution 50,000 at m/z 400, target value 50,000). MS3 was performed on the most abundant fragment ion for increased certainty of identification (43).
Peptide Identification and Quantitation—
Peak lists for database searching were generated from the raw data using in-house developed software called Raw2msm. From each fragment spectrum the six most intense peaks per 100 Th were extracted. Proteins were identified by automated database searching (Mascot version 2.1, Matrix Science) against an in-house curated version of the mouse IPI database (versions ranged from 3.00 to 3.37 and contained between 40,613 and 68,655 entries complemented with frequently observed contaminants like porcine trypsin and human keratins). Carbamidomethyl-cysteine was used as a fixed modification; variable modifications were oxidation of methionine, protein _N_-acetylation, deamidation of Asn and Gln, _N_-pyroglutamate, and heavy versions of Arg and Lys. We required full tryptic specificity (cleavage at Arg-Pro and Lys-Pro as well as Asp-Pro was included), a maximum of two missed cleavages, and maximal mass deviation of 5 ppm for the parent ion and 0.5 Da for fragment ions. As initial identification threshold for peptides we chose a false positive rate of maximum 5% as judged by searching a concatenated database consisting of normal and reverse sequences. However, we required peptides with twice that score for final protein identification for phosphorylation-specific interaction partners (for example, MASCOT score 50 if score 25 corresponded to 5% false positive rate). Identification confidence of interaction partners was further enhanced by requirement of a SILAC ratio different from 1:1, and the identification of the binder in an independent, crossover experiment with inversed ratio. For the relative quantitation of SILAC peptide pairs our in-house developed software MSQuant was used. All peptides from proteins appearing with an elevated SILAC-ratio were verified and re-analyzed manually.
Determination of Significant Binding Partners—
A typical pulldown experiment yielded hundreds of identified proteins. The vast majority had a SILAC ratio close to 1:1, indicating that they represent background binders of the beads irrespective of the phosphorylation state of the peptide. All protein ratios were normalized against the median. Proteins with a ratio more than 2–3 standard deviations above the median were considered as phosphorylation-specific binders, given that they had a correspondingly inverted ratio in the crossover experiment. It was not appropriate to decide on a fixed cut-off value, since the number of interactors with ratio different from 1:1 influences the standard deviation. In borderline cases (referring to the ratio or the amount of identified peptides), or whenever an unexpected binding partner was discovered (e.g. a protein without known Tyr(P)-binding domain), the experiment was repeated at least once to verify the significance of binding.
Note that the value of the ratio itself does not directly reflect the affinity or the stoichiometry of the interaction. In fact, repetitions of an experiment can yield fluctuations in the ratios, due to minor variations in the stringency of washing steps without, however, changing the significance of the interaction. Furthermore, in many cases the differences between binding to the bait and the control are so great that the unlabeled peptides (representing binding to the control bait) become undetectable and hence unquantifiable (44). Even though this clearly indicates phosphorylation-specific interaction, it leads to imprecise ratios. For this reason, we considered the outlier significance (distance from mean) in addition to the absolute ratio.
RESULTS
Quantitative Proteomics for Unbiased Identification of Interactions—
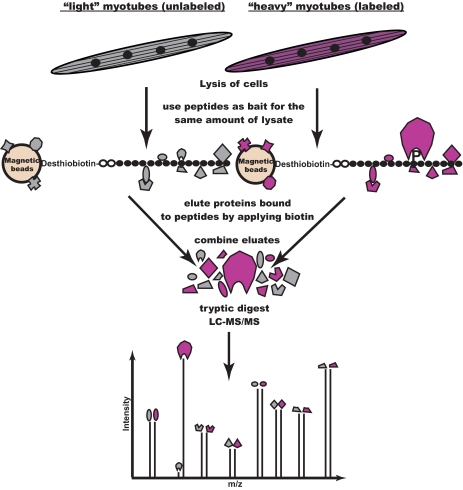
For the identification of site-specific, Tyr(P)-dependent interaction partners, we applied our previously established peptide pulldown approach, which is based on quantitative proteomics (45). Due to the central role of skeletal muscle in insulin signaling, we employed whole cell lysate from the murine muscle cell line C2C12, which we differentiated to myotubes before harvest. The SILAC technique was applied by incubating the phosphorylated peptide with lysate from cells grown in medium containing stable isotope labeled arginine and lysine, whereas unlabeled lysate was added to the control peptides (Fig. 1). Phosphorylation-specific binders were uncovered through their high ratios between heavy and light labeled peptides in the MS spectra (Fig. 2), which distinguished them from hundreds of background proteins and which was much more specific and sensitive than the classical approach of differential staining (supplemental Fig. 1) (46).
Fig. 1.
Proteomic screening for interaction partners of tyrosine phosphorylated sequences. Peptides corresponding to potential tyrosine phosphorylation sites are synthesized in phosphorylated and non-phosphorylated form. Cell populations are metabolically labeled using the SILAC technique to allow discrimination based on different peptide masses. Cell lysate from the population labeled with heavy Arg and Lys is incubated with the phosphorylated version of the peptide, whereas the control cell lysate is incubated with the non-phosphorylated peptide. Eluted proteins from those parallel pulldown experiments are combined and digested with trypsin. Peptides from unspecific background binders appear as pairs with abundance ratios close to 1:1 in the mass spectra. Phosphorylation-specific binders are identified as such through their high abundance ratio between heavy and light labeling states. In a crossover experiment, the incubation scheme is swapped, resulting in inverted ratios.
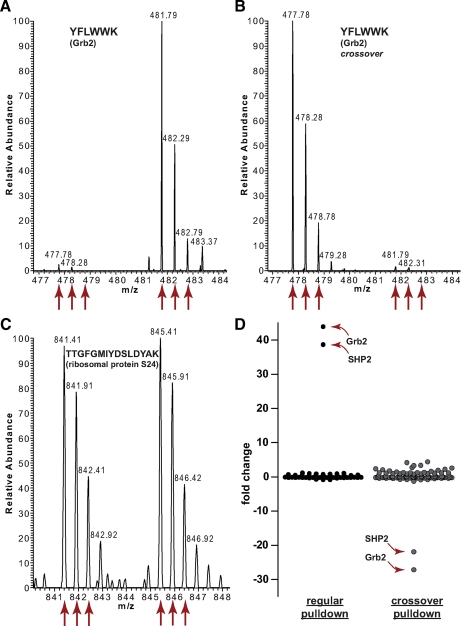
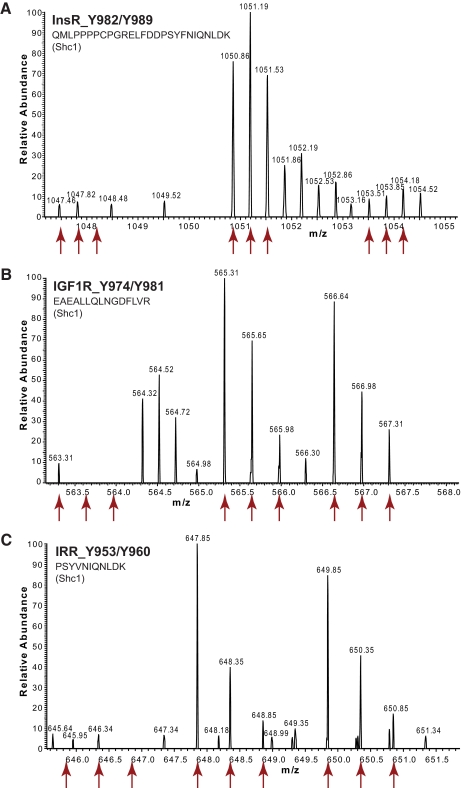
Fig. 2.
Typical result of a SILAC peptide pulldown experiment, exemplified by the bait peptide IRS-1 Tyr-0891. A, a peptide derived from the interaction partner Grb2 is about 40 times more abundant in the heavy form than in the light form, indicating that Grb2 binds specifically to IRS-1 phosphorylated on Tyr-0891. B, in a crossover experiment (swapped SILAC labels) the abundance ratio of the same peptide is inverted. C, most other proteins have a 1:1 ratio, indicating that they are unspecific binders to the peptides or the magnetic beads that are bound irrespective of phosphorylation. D, a plot of the protein abundance ratios shows that Grb2 and SHP2 are significant outliers in both the Tyr-0891 pulldown and the crossover experiment. Every dot represents one protein.
In total, we performed pulldowns for 109 individual sites in forward and crossover experiments, where interactors were required to have inverted ratios. The majority of pulldowns were additionally repeated in separate experiments to validate novel interactors. Fourteen sites were furthermore targeted in combinatorial binding experiments (see below). The interaction data is provided in supplemental Tables 1–3 and summarized in Figs. 3 and 4.
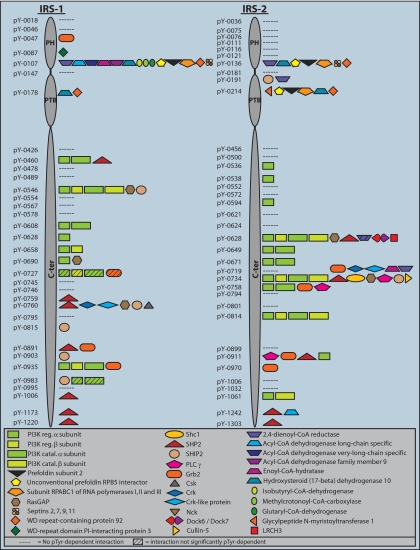
Fig. 3.
The phosphotyrosine interactome of IRS-1 and IRS-2. Tyr(P)-specific interaction partners obtained in peptide pulldown experiments are depicted as symbols along the primary structure of IRS-1 and IRS-2. Detailed data for every pulldown experiment is provided in supplemental Table 1.
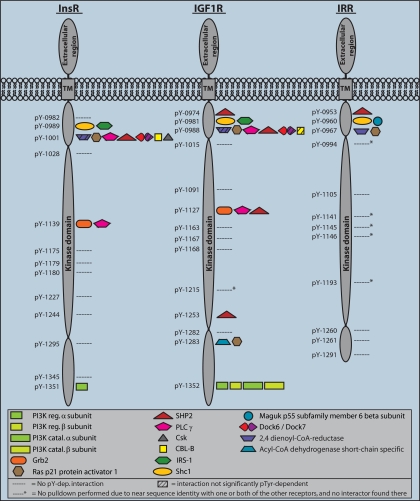
Fig. 4.
The phosphotyrosine interactome of InsR, IGF1R, and IRR. Tyr(P)-specific interaction partners obtained in peptide pulldown experiments are depicted as symbols along the primary structure of the intracellular regions of the receptors. The cytoplasmic part starts at position 968 for the InsR, 961 for the IGF1R, and 944 for the IRR, according to the Swiss-Prot database. Detailed data for every pulldown experiment is provided in supplemental Table 1.
The Tyr(P)-interactome of IRS-1 and IRS-2—
IRS-1 and IRS-2 have been recognized as interaction platforms soon after their discovery, and literature reviews report ∼10 known Tyr(P)-dependent interaction partners, albeit sometimes with little supporting evidence (see under “Discussion”). Our study, for the first time, provides a systematic study of the Tyr(P) interactome of these proteins. However, the interaction partners depicted in Fig. 3, although specific to the phosphorylated baits, are not validated as biological interactions of the full-length endogenous proteins in the cell, nor do we know for all Tyr(P) sites if they are indeed phosphorylated upon insulin stimulation. Therefore we refer to them as potential interaction partners.
We first checked if our large scale screen yielded known interaction partners. We indeed found these proteins, including the phosphatidylinositol-3-kinase (PI3K) subunits, Grb2, SHP2, Nck, C-terminal Src-kinase (Csk), and Crk. Additionally, the majority of interaction partners in supplemental Table 1 have SH2 or PTB domains, suggesting that these interactions are direct and specific. Thus we conclude that the screen performed well in retrieving interaction partners known from literature. By extension, most of the novel interaction partners are likely to have functional roles in insulin signaling as well.
IRS proteins contain multiple interaction motifs for PI3K. For each of these motifs, we found the alpha isoform of the SH2 domain containing regulatory subunit, suggesting that each of the 9 (IRS-1) or 11 (IRS-2) is indeed capable of binding to PI3K. This is not self-evident because some of these motifs are imperfect, i.e. they lack methionine in the +3 position. The beta isoform of the regulatory PI3K was found less often, perhaps due to its low expression levels. Likewise, the catalytic subunits of PI3K were frequently identified due to their tight association with the regulatory subunits.
The protein tyrosine phosphatase SHP2 and the adaptor protein Grb2 are also well-established IRS interactors. Here we mapped the specific sites on IRS-1 and IRS-2 that these effector proteins could dock to (7 and 4 sites for SHP2, respectively and 2 and 3 for Grb2; Fig. 3 and supplemental Table 1). In addition, the inositol-5-phosphatase SHIP-2 and the Ras GTPase-activating protein RasGAP can bind to various sites on IRS-1 and IRS-2. In contrast, the adaptor molecules Crk and Crk-like protein were pulled down by only one specific tyrosine in each IRS protein. Interestingly, this tyrosine occurs at a similar position (Y0760 in IRS-1 and Y0734 in IRS-2 in the mouse sequence) but the binding sequences are very different. Apart from Crk and Crk-like protein, the site in IRS-2, Tyr-0734, attracts many different proteins simultaneously. Even though our washing steps were optimized to detect direct binders to the phosphorylated bait peptide, it is possible that some of the detected interactions occur via indirect binding such as noted above for the catalytic subunits of PI3K.
Most interactors were recruited by both IRS proteins. However, cases of exclusive interactions also occurred. Phospholipase C gamma (PLCγ), for example, docks to several sites in IRS-2 but none in IRS-1 in our experiment. Similarly, Shc, Cullin-5, LRCH3, and DOCK-6 and -7 as well as glycylpeptide _N_-myristoyltransferase 1 were exclusively recruited by IRS-2-derived peptides. IRS-1-specific binders include Csk, WD-repeat domain phosphoinositide-interacting protein 3, and several enzymes involved in fatty and amino acid degradations. Detailed results of all pulldown experiments are listed in supplemental Table 1.
The ubichinol-cytochrome c reductase complex chaperone CBP3 homolog (Swiss-Prot Q9CWU6) was exclusively and repeatedly found in pulldowns with the two homologous sites IRS-1 Tyr-0107 and IRS-2 Tyr-0136 with a slightly elevated ratio, but in most cases it was not significantly phosphorylation-dependent (and is hence not listed in supplemental Table 1). It could therefore be an indirect binder recruited via one of the fatty acid metabolizing enzymes. The protein tyrosine kinase Fyn has been reported as an IRS-2 interactor previously (47), and we similarly found consistent but relatively weak ratios when binding to Tyr-0734 of IRS-2.
The Phosphotyrosine Interactome of InsR, IGF1R, and IRR—
As expected, the receptors yielded fewer interactors than the IRS proteins (Fig. 4). InsR and IGF1R share a very similar Tyr(P)-interaction profile. However, the IGF1R contains multiple sites for recruitment of SHP2 and a site that binds RasGAP as well as short chain-specific acyl-CoA dehydrogenase. The physiological importance of the latter site is questionable, however, because it is not conserved in the human IGF1R sequence. Strikingly, the IRR lacks interactions in its C-terminal part. InsR Tyr-1001 and IGF1R Tyr-0988 can recruit many different proteins, and apparently minor differences in the sequence N-terminal of the Tyr(P) in the corresponding site IRR Tyr-0967 impede some of these binding events. Interestingly, 2,4-dienoyl-CoA reductase shows phosphorylation-dependent binding only for the latter site but is strongly bound by the other sites irrespective of their phosphorylation state. This behavior was observed for a few sites in IRS proteins as well and is indicated by skew stripes in the protein symbols in Figs. 3 and 4. Binding of IRS-1 and Shc to the NPEY sites in the receptors was only detectable when the bait sequence was enlarged by six amino acids toward the N-terminal side. This observation can be rationalized on structural grounds: crystal structures of the IRS-1 PTB domain bound to the InsR reveal that a relatively long sequence at the N-terminal side of the Tyr(P) is involved. Residues −8 to −3 form a β-strand that establishes hydrogen bonds with a β-strand of the PTB domain. The leucine at position −8 is particularly important and hydrophobic residues from −6 to −8 are favored by Shc and IRS-1 (48, 49). These findings suggest the desirability of providing longer and freely accessible N-terminal regions for sequences that are involved in PTB domain recruitment. However, IRS-1 was still only present with few peptides, and IRS-2 was not detected. The PTB domains of Shc and IRS-1 are electrostatically polarized similar to PH domains and can therefore associate with phospho-inositides in the membrane through their positively charged surface (1). Since these additional, stabilizing interactions are not present in our experiment, reduced affinity (but not reduced specificity) is to be expected. We conclude that PTB domain interactions are more challenging to detect in our assay and that they might need further optimization. In contrast to its binding to InsR and IGF1R, IRS-1 was not detected in the experiment with the bait sequence of the IRR, likely due to the challenges of detection. However, the membrane-associated guanylate kinase (MAGUK) p55 subfamily member 6 was identified as an interactor with IRR. We did not find interactors for a fourth receptor, IGF2R (also called mannose-6-phosphate receptor), that had two cytosolic tyrosines.
One of the advantages of conducting a large number of peptide pulldown experiments in the same system is facilitated discrimination of biochemical noise. We were initially puzzled by the identification of certain unexpected proteins with elevated ratios that passed the significance threshold in many pulldown experiments with completely unrelated bait sequences. We soon realized that the combination of frequent observation across multiple experiments and irreproducibility in repetitions with the same bait sequence classified this kind of candidates as unlikely to be biologically relevant. Indeed, 7 out of those 8 “uninvited guests” are RNA-binding proteins, providing a ready explanation for their specific binding to phosphorylated peptides: the negative charges on the phosphorylated bait peptides introduce an ion exchange effect with the positively charged RNA-binding proteins, somewhat mimicking their binding to RNA. We list those proteins here and advise to take care in categorizing such candidates in similar studies: 60 S ribosomal protein L11, RNA-binding protein SiahBP homolog, activated RNA polymerase II transcriptional coactivator p15 precursor, eukaryotic translation initiation factor 5, splicing factor U2AF 35 and 65 kDa subunit, peptidylprolyl isomerase B, ATP-dependent RNA helicase DDX3X.
In principle, our setup allows the simultaneous detection of proteins whose interaction with the bait sequence is abolished by the phosphorylation. Those candidates would then appear with a ratio much lower than one in the pulldown experiments. However, we did not reproducibly observe such proteins and they are therefore not listed in the supplementary tables.
Doubly and Triply Phosphorylated Motifs—
To assess the effect of phosphorylation events in close proximity to each other, we performed experiments with doubly or triply phosphorylated bait peptides for some of the sequence stretches that contained neighboring tyrosines (supplemental Table 2). We reasoned that combinatorial phosphorylation might either prevent binding events compared with the mono-phosphorylated counterpart or even generate new binding interfaces. We indeed encountered some sites whose interaction capability was modified by additional phosphorylations. For example, PI3K was not displaced by phosphorylations N-terminal of its binding sequences around IRS-2 Tyr-0538 and InsR Tyr-1351. However, it failed to interact with the IRS-2 Tyr-0628 peptide sequence, which matched to a weaker consensus motif, if positions Tyr-0621 and Tyr-0624 were phosphorylated at the same time. Most strikingly, the triply phosphorylated peptide resembling the kinase activation loop of the InsR recruited several interactors (dedicators of cytokinesis 6 and 7, RasGAP, LRCH3), whereas a doubly and the singly phosphorylated versions did not recruit any proteins (see under “Discussion”).
For precise mapping of differences between mono- and doubly phosphorylated versions, we performed triple SILAC labeling experiments for selected cases (supplemental Table 3). In this type of pulldown experiments, non-phosphorylated peptide was incubated with unlabeled cell lysate (Arg-0 + Lys-0), mono-phosphorylated peptide was incubated with a medium labeled state (Arg-6 + Lys-4) and doubly phosphorylated peptide was incubated with heavily labeled lysate (Arg-10 + Lys-8). Peptides appear as triplets in mass spectra, enabling accurate comparison between the three conditions. Using this assay we encountered differential effects of a phosphorylation located seven residues upstream of the NPEY motif in the receptors. The recruitment of Shc was not affected by this additional phosphorylation within the binding motif of the PTB domain in case of IGF1R and IRR. However, binding of Shc to the respective sequence in the InsR proved to be sensitive to this change, and the interaction was abolished (Fig. 5). In case of IRS-1 Tyr-0546/Tyr-0554, the second phosphorylation neither increased nor diminished the binding of interactors. On the other hand, for IRS-1 Tyr-0759/Tyr-0760 the double phosphorylation induced novel interactions. Although SHP2, Csk, Crk, and Crk-like protein were equally attracted by mono- and doubly phosphorylated version, the binding of RasGAP and SHIP-2 was enhanced by the additional phosphorylation. Some proteins were even exclusively recruited by the doubly phosphorylated sequence, namely Nck2, PLCγ, Grb2, PI3K, and Cbl-b.
Fig. 5.
Triple labeling pulldown reveals distinct combinatorial effects of double phosphorylation within the NPEY motifs in the insulin receptor family. A, when phosphorylated at the tyrosine within the NPEY motif, the InsR recruits Shc, as demonstrated by the high ratio between the medium-label state and the unlabeled state. If the tyrosine located seven amino acids further N-terminal also carries a phosphorylation, the interaction with Shc is abolished. B and C, when IGF1R or IRR exhibits this phosphorylation pattern, the interaction between Shc and the receptor is not influenced by the second phosphorylation event (supplemental Table 3).
DISCUSSION
Capabilities and Limitations of Quantitative Proteomics Combined with Peptide Bait Fishing—
MS-based quantitative proteomics has become a powerful and versatile tool for comparing complex proteomes and their modifications (50–52). In particular, metabolic labeling by the SILAC technique has proven to be a very accurate method for interaction proteomics (53) (37). Here we uncovered Tyr(P)-dependent binding events on proteins that serve as interaction platforms in the insulin signaling pathway using synthetic phosphorylated peptides as baits in pulldown experiments from whole cell lysates. In comparison to purely in vitro experiments such as chip-based assays, the unbiased nature of the mass spectrometric approach enables discovery of novel interactors, unconstrained by prior hypotheses, as well as the specific binding site. Furthermore, in contrast to in vitro binding assays, our experiment involves the original expression level of the interacting proteins in the context of thousands of other proteins. This is made possible by the ability to discriminate between specific phosphorylation-dependent interactors and background binders by quantitative proteomics.
However, even though all significant interactors found in our experiments are per definition specific for the phosphorylated form of the peptide, they can only take place in vivo if the site is accessible, becomes phosphorylated by a kinase, and if the binding partner is available for the interaction. For example, an interaction will not take place if the potential interactors are present in different subcellular locations or expressed in different tissues. Affinity, relative expression levels, effects of the neighboring sequence, and the involvement of additional interaction domains furthermore co-determine the actual occurrence of the Tyr(P)-mediated interaction in vivo. For most interacting proteins additional information, such as known subcellular localization and the presence of known interaction domains can help in determining the likelihood that the interaction is biologically relevant. Thus our results represent a catalogue of interactions that are proven to be biophysically possible and for interactions that fulfill the above criteria, likely to occur in vivo. In this regard, the present study is complemented by our recent, systematic investigation of the tyrosine phosphoproteome induced by insulin and IGF-1 stimulation (54). Below we first discuss the common and distinct interactomes of the IRS proteins and the three receptors that involve classical SH2 or PTB domain-mediated interactions. We then discuss interactions with proteins not containing these binding modules.
Common Interactors of IRS-1 and IRS-2—
Most of the interaction partners identified in this work contain an SH2 domain that mediates binding to the Tyr(P) sequence. Recognition motifs for many SH2 domains have been determined by peptide library screening (3, 55). SH2 domains do not bind to arbitrary sequences surrounding the Tyr(P) but instead bind only to a subset of consensus motifs. Conversely, the same peptide sequence can bind to different SH2 domains (3), which is reflected here by the recruitment of a range of binding partners in many of our peptide pulldowns.
Among the proteins recruited to both IRS proteins, PI3K bound to the largest number of different sites. PI3K is a critical node in insulin signaling (28), which activates the Akt kinase through generation of phosphatidylinositol-3,4,5-triphosphate at the plasma membrane. Akt in turn has several downstream pathway branches important for growth and metabolism. Our prevailing observation of the alpha isoform of the catalytic subunit of PI3K agrees with its predominant role in insulin signaling (56). The adaptor protein Grb2, which interacts with multiple phosphorylated peptides from IRS-1 and IRS-2, triggers signaling via the activation of the Ras to MAP-kinase axis. Intriguingly, the suppressor of Ras signaling RasGAP binds to multiple sites in IRS proteins. This novel finding suggests that the activity of RasGAP is regulated by recruitment to the IRS platform.
The tyrosine phosphatase SHP2 plays a dynamic role in the activity of the pathway by both providing negative feedback for PI3K activation via dephosphorylating Tyr(P) sites on IRS proteins that bind PI3K (57) and stimulating mitogenic signaling via insulin (58–60). Here we systematically map binding sites of SHP2 on the IRS signaling platforms.
The lipid phosphatase SHIP-2 dephosphorylates phosphatidylinositol phosphates at the 5′-position. Even though SHIP-2 action does not abolish the ability of PIPs to activate Akt, it clearly antagonizes insulin signaling (61). Here we mapped five interaction sites of SHIP-2 to IRS-1 and two to IRS-2.
Crk family adaptors mediate protein complex formation in various signaling pathways and have been shown to influence mitogenesis, cytoskeletal rearrangements, and insulin-stimulated glucose uptake (62, 63). While binding to IRS-1 and IRS-2 has been observed before (47), our data provides the specific docking sites.
Differential Interactions between IRS-1 and IRS-2—
The physiological effects of insulin can vary greatly in different target tissues due to different modes of signal transmission and modulation inside cells. For example in liver synthesis of glycogen, proteins and lipids are triggered along with inhibition of hepatic glucose production and very low density lipoprotein secretion. In muscle, the response mainly involves glucose uptake and glycogen synthesis (28, 64). Previously many of these differences were attributed to stronger activation of IRS-1 or IRS-2. However, differences in binding partners between IRS-1 and IRS-2 are not only determined at the level of their primary sequence motifs, but also by differing time courses of phosphorylation, dose-response curves toward hormones, and intracellular compartmentalization (24). The expression level of IRSs and their interactors also play a role, and those factors can even result in opposite functions of an IRS protein depending on the cell type investigated (65). Illustrating this complexity, a recent study found that IRS-1 and IRS-2 trigger the same downstream signals, but IRS-1 was more active in the postprandial state, whereas IRS-2 was employed during fasting (66).
Given the extensive and often contradictory investigations into the differential roles of IRS-1 and IRS-2, our experiment at the least provides a large-scale data set delineating potential common and differential Tyr(P)-mediated interactors. In general, our results reinforce the notion of a large overlap in the signaling capabilities between IRS-1 and IRS-2. Most Tyr(P)-dependent interaction partners involved in growth and metabolic signaling were recruited by both proteins. However, we also detected clear differences in the interactomes. For example, as noted above SHIP-2 binds to substantially more sites in IRS-1. Exclusive binding to IRS-1 was observed for Csk, which phosphorylates members of the Src family of kinases at C-terminal tyrosines, inhibiting their activity. Csk has been shown to bind to IRS-1 via its SH2 domain (67), whereas interaction or lack thereof with IRS-2 has not previously been noted. The Csk-mediated inactivation of Src-family kinases leads to an insulin-dependent decrease of Tyr(P) on focal adhesion kinase (FAK) and Paxillin, which reduces actin stress fibers and allows insulin to influence the reorganization of the cytoskeleton. Our observation that Csk is recruited only to IRS-1-derived peptides and not to IRS-2-derived peptides is in line with the observation that IRS-1, but IRS-2, mediates actin remodeling in myotubes (24).
Shc and PLCγ were only found as interactors of IRS-2. Shc is an adaptor protein best known for its ability to bind Grb2 and accordingly promote Ras activation. PLCγ is the only PLC that contains SH2 domains. Binding via its N-terminal SH2 domain to a tyrosine kinase leads to phosphorylation of PLCγ on several tyrosines, thereby activating it and ultimately stimulating PKC activation (68). PLCγ has previously been shown to bind to IRS proteins in an insulin-dependent manner (47). Our finding that PLCγ is recruited to three sites in IRS-2 and none in IRS-1 suggests one mechanism for specific functions of the two signaling platforms. With the positive effect on GLUT4-translocation mediated by PKC in mind, the exclusive recruitment of PLCγ to IRS-2 might in part explain the stronger metabolic role of IRS-2 versus IRS-1 observed in other studies.
The Tyr(P)-interactome of InsR, IGF1R, and IRR—
The insulin signaling pathway is activated by the InsR as well as the IGF1R. Signaling through InsR has metabolic functions whereas IGF1R, as its name implies, has mainly growth and mitogenic effects. However, there is also a significant level of cross-talk between these functions. Experiments with chimeric receptors have shown that it is mainly the intracellular part that determines specificity, not the extracellular ligand binding domains (69). The most apparent difference between the interactomes of IGF1R and InsR as determined here is the larger number of SHP2 binding sites to IGF1R (four versus one). This observation helps to explain the stimulatory role of this phosphatase in growth signaling. Both are able to recruit Cbl, even though the interaction was only weakly Tyr(P)-dependent in the case of the IGF1R. Cbl is an E3 ligase and allosterically activates an E2 enzyme for ubiquitinylation, which leads to the internalization and subsequent degradation of the receptor (70).
IRR is thought to have a less prominent role in the insulin/IGF pathway, and few functions have been described. We found that IRR is less prone to participate in Tyr(P)-dependent binding events and detected just three interactors with SH2 domains. Only the membrane proximal sites recruit interaction partners. Our failure to detect IRS-1 binding to IRR might be due to very weak binding of the IRS PTB domain under the conditions of this assay (see under “Results”).
The NPEY motif containing phosphopeptide derived from IRR did bind to MAGUK p55 subfamily member 6, also termed PALS2, one of the non-SH2 domain containing proteins otherwise discussed below. Little functional information is known about MAGUK p55 but it appears to be involved in proper targeting of receptors in polarized cells, as well as in stabilization of receptors and acting as signaling scaffold in non-polarized cells (71). This potential interaction would be unique to the IRR as compared with its other family members.
Doubly and Triply Phosphorylated Motifs—
Another interesting difference between the receptors became apparent when studying the combinatorial effects of an additional phosphorylation at the tyrosine located seven residues N-terminal of the NPEY motif. In the doubly phosphorylated peptide, this site bound to SHIP-2 and SHP2 (in InsR and IGF1R + IRR, respectively) just as in the case of the mono-phosphorylated peptide (supplemental Table 3). Likewise the doubly phosphorylated peptide still bound IRS-1. However, binding of Shc to the NPEY motif was abrogated specifically for the InsR sequence-derived peptide. Since Shc activates MAP kinase signaling when associated with tyrosine kinases this is a possible mechanism of differential control, toward metabolic signaling, between the receptors.
In most other cases of combinatorial phosphorylation, we did not observe any changes. The kinase activation loop, interestingly, yielded several binding partners in its triply phosphorylated state. This may ensure that the interactions can only take place after full activation of the kinase. Selective binding to the triply phosphorylated sequence of the activation loop has previously been reported for adaptor protein with a pleckstrin homology and Src homology 2 domain (APS) and IRS-2 (72, 73). The sequence in this region is identical between InsR and IGF1R and has only one Ile/Val substitution in IRR, therefore this result likely applies to all of the receptors.
IRS-1 has two tyrosines adjacent to each other (Tyr-0759/Tyr-0760). In pulldowns with singly phosphorylated peptides Tyr(P)-0760 interacts with SHP2 as well as five other proteins containing SH2 domains; however, Tyr(P)-0759 only interacts with SHP2. The doubly phosphorylated peptide still binds to all interactors and strikingly to several additional ones. These proteins, Grb2, PLCγ, Nck2, and Cbl, all contain SH2 domains and for two of them binding to doubly phosphorylated peptides has previously been described. PLCγ, for example, usually binds hydrophobic sequences with its C-terminal SH2 domain, but following a conformational change it creates a second Tyr(P)-binding pocket and can then also bind doubly phosphorylated motifs, such as pYESPpYAD in the activated Syk tyrosine kinase (74). For Grb2, which binds selectively to the pYpY sequence here, a similar observation has been made for a pYpY motif in Shc (75). This doubly phosphorylated peptide is the only one binding Nck2 in our study. Nck has previously been observed to bind to IRS-1 in an insulin-dependent manner and is engaged in cytoskeletal rearrangements and mitogenic signaling (76, 77), but the mode of its binding has not been described. The doubly phosphorylated version clearly bound the Src-kinase Fyn, albeit not in a significantly Tyr(P)-dependent manner. Fyn has been reported to bind to IRS-1 and IRS-2 (47) and to tyrosine phosphorylated Cbl after insulin stimulation (78).
Tyr(P) Binding Independent of SH2 and PTB Domains—
The interactions described above are all readily explained by Tyr(P)-SH2 or -PTB domain-mediated binding. Most of them recapitulate and extend known interactions or can be readily understood in terms of the biology of this well studied pathway. A further goal of our experiments was to possibly detect novel interaction partners and interaction modes, not mediated by known interaction domains or interactions with known members of the insulin signaling pathway. Our results indeed contain a number of such interaction partners. Since indirect binding is possible in our experimental setup, they might have been recruited as secondary interactors. For some of the investigated sites, however, we exclusively found non-SH2 domain containing proteins. Furthermore, even for those sites where SH2-containing proteins might indirectly recruit other proteins, this may be unlikely since it should happen at all sites with which they interact. We therefore consider it unlikely that indirect binding is the only explanation for these binders.
As demonstrated by the relatively late discovery of the Cbl SH2 domain, genome annotation algorithms can sometimes miss an SH2 domain if it has an atypical sequence (79). More importantly, alternative Tyr(P)-binding domains have recently been described. The C2 domain of PKCδ has been shown to bind Tyr(P) (80). A very recent report by the Cantley group (also using the SILAC technology) showed that a yet uncharacterized region in pyruvate kinase M2 binds specifically to Tyr(P) containing peptides (11). This was interpreted to provide a direct link to mitochondria-based metabolic functions. In total we found 21 “non-classical” interaction partners and based on co-occurrence estimate that at least ten of these bind directly to Tyr(P) containing bait peptides.
Note that some of the interactions might not necessarily occur in insulin or IGF1 signaling because IRS-1 and IRS-2 can also be engaged by other receptors. Those include the prolactin (81), androgen (82), growth hormone (81, 83), and vascular endothelial growth factor receptors (84), as well as members of the integrin receptor family (85, 86) and several cytokine receptors (87).
Cytosolic interactors of the IRS proteins included two WD repeat proteins. The function of WDR92 (WD repeat-containing protein 92) is still elusive, but it has been suggested to act as a modulator of apoptosis (88). The function of WD-repeat domain phosphoinositide-interacting protein 3, which bound to a phosphopeptide derived from IRS-1, is also not yet known. It potentially binds to membrane phosphoinositides in addition to the interaction with Tyr-0087 in the PH domain of IRS-1, which is membrane-associated, and thus could enhance membrane anchoring of IRS-1 after stimulation. Several septins were found as interactors of both IRS proteins. Septins behave like filaments or scaffold proteins and play a role in cytokinesis and in cytoskeleton and membrane organization (89).
Among the proteins exclusively interacting with peptides derived from IRS-2, Cullin-5 is a scaffold protein and part of an E3 ubiquitin ligase complex. As such it might be involved in proteasomal degradation of IRS proteins. Another cullin, Cullin-7, triggers proteasomal degradation of IRSs (90). LRCH3 (leucine-rich repeat and calponin homology domain-containing protein 3) is a protein with unknown function, and DOCK-6 and -7 (dedicator of cytokinesis) act as guanine nucleotide exchange factor for small G-proteins of the Rho family according to their UniProt annotation. This connects them to regulation of cytoskeletal changes, one of the known effects of insulin and IGF-1 signaling. Glycylpeptide _N_-myristoyltransferase 1 attaches myristoyl groups to proteins with a glycine at their N terminus. Myristoylation of proteins equips them with a membrane anchor, targeting them to cellular membranes.
Intriguingly, we detected several effectors of metabolic regulation in insulin signaling, which bound to homologous sites in IRS-1 and IRS-2. We encountered a number of enzymes associated with fatty acid catabolism in mitochondria. Acyl-CoA dehydrogenases catalyze the first step in beta-oxidation, and enoyl-CoA hydratase is responsible for the second step. 2,4-dienoyl-CoA reductase participates in beta-oxidation by feeding unsaturated fatty acids into the pathway. Hydroxysteroid (17-beta) dehydrogenase 10 is crucial in the degradation of branched chain fatty acids and isoleucine as well as in the metabolism of steroid hormones (91). The activity of these enzymes is known to exert an effect back on IRS signaling. Long-chain acyl-CoA can be metabolized to diacylglycerol, which activates PKCθ (92). PKCθ directly and indirectly (via c-Jun NH2-terminal kinase (JNK) and inhibitor of kappaB kinase (IKK)) leads to phosphorylation of IRS-1 on inhibitory serine residues (93). A recent bioinformatic analysis even reports that two related acyl-CoA dehydrogenases (family members 10 and 11) contain kinase domains, pointing to a direct link between fatty acid catabolism and cell signaling (94).
Furthermore, several enzymes involved in amino acid catabolism were found to be able to interact with IRS-1-derived phosphopeptides. However, their mitochondrial localization makes a physiological interaction with IRS-1 questionable. Methylcrotonoyl-CoA-carboxylase degrades leucine, isobutyryl-CoA-dehydrogenase degrades valine, and glutaryl-CoA-dehydrogenase degrades lysine and tryptophane. Branched chain amino acids play an important role in the regulation of translation via mammalian target of rapamycin (mTOR) signaling and in insulin signaling (95, 96).
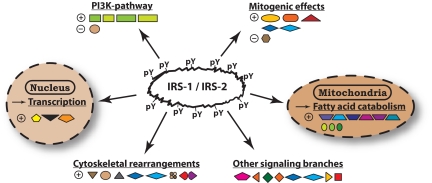
Finally, a complex consisting of prefoldin, RPABC1/RPB5, and “unconventional prefoldin RPB5 interactor” binds to homologous sites in the IRS-1 and IRS-2 PH and PTB domains. This complex shuttles between cytosol and nucleus, where it is believed to mediate transcriptional effects of nutrient signaling via mammalian target of rapamycin (mTOR) (97). Fig. 6 summarizes the various pathways that are potentially directly linked to IRS-1 and IRS-2 via their Tyr(P)-dependent interaction partners as measured in this study.
Fig. 6.
Molecular functions potentially influenced by IRS-1 and IRS-2 through direct binding. The Tyr(P)-dependent interaction partners of IRS-1 and IRS-2 identified in this study are grouped according to their function and subcellular location. Stimulatory and inhibitory effects on the pathways are indicated by [⊕] and [⊖], respectively.
CONCLUSIONS
Here we have used quantitative interaction proteomics based on the SILAC technology in a systematic study of phosphotyrosine binding in the insulin signaling pathway. With this work we hope to have contributed new knowledge to the insulin signaling pathway, the malfunction of which underlies diabetes, a disease that will soon afflict 300 million patients worldwide and that threatens the very functioning of national health systems (98). High accuracy mass spectrometry and relative quantitation between phosphopeptide pulldowns and control pulldowns from cell extracts ensured that our data represent specific phosphopeptide-protein interactions. Further studies are needed to validate these interactions in the context of endogenous, full-length proteins. However, for most of our interactors, previous knowledge of binding modes and involvement in the pathway makes this extremely likely. For the potential interaction partners with non-traditional binding modes, our data raise interesting hypotheses that can be followed up in a directed way by researchers in the field. Recent work from a number of laboratories encourage us to believe that at least some of these interactions may point to novel and as yet unstudied mechanisms in insulin signaling.
Acknowledgments
We thank Dr. Waltraud Schulze for her skilled advice on the peptide pulldown procedure and Dr. Jesper V. Olsen and Dr. Mara Monetti for critically reading the manuscript. We are grateful to other members of our department for fruitful discussions.
Footnotes
Published, MCP Papers in Press, November 11, 2008, DOI 10.1074/mcp.M800407-MCP200
1
The abbreviations used are: SH2, Src homology 2; PH, pleckstrin homology; MS, mass spectrometry; Csk, C-terminal Src-kinase; Tyr(P), phosphotyrosine; LC, liquid chromatography; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; MS/MS, tandem mass spectrometry; PLCγ, phospholipase C gamma; LTQ, linear quadrupole ion trap; SILAC, stable isotope labeling by amino acids in cell culture; PI3K, phosphatidylinositol-3-kinase; Arg, arginine; Lys, lysine; PTB, phosphotyrosine binding; IRS, insulin receptor substrate; InsR, insulin receptor; IGF1R, insulin-like growth factor 1 receptor; IRR, insulin receptor-related receptor; IGF, insulin-like growth factor; MAP, mitogen-activated protein; ErbB, epidermal growth factor family of receptor tyrosine kinases; Crk, chicken tumor virus no. 10 regulator of kinase; NPEY, Asn-Pro-Glu-Tyr.
*
This work was supported, in whole or in part, by National Institutes of Health Grant DK 60837 through the Diabetes Genome Anatomy Project consortium. This work was also supported by Interaction Proteome, a 6th Framework program of the European Commission.
S
The on-line version of this article (available at http://www.mcp.org) contains supplemental material.
REFERENCES
- 1.Schlessinger, J., and Lemmon, M. A. ( 2003) SH2 and PTB domains in tyrosine kinase signaling. Sci. STKE 2003, RE12 [DOI] [PubMed]
- 2.Pawson, T. ( 2004) Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 116, 191–203 [DOI] [PubMed] [Google Scholar]
- 3.Huang, H., Li, L., Wu, C., Schibli, D., Colwill, K., Ma, S., Li, C., Roy, P., Ho, K., Songyang, Z., Pawson, T., Gao, Y., and Li, S. S. ( 2008) Defining the specificity space of the human SRC homology 2 domain. Mol. Cell. Proteomics 7, 768–784 [DOI] [PubMed] [Google Scholar]
- 4.Miller, M. L., Hanke, S., Hinsby, A. M., Friis, C., Brunak, S., Mann, M., and Blom, N. ( 2008) Motif decomposition of the phosphotyrosine proteome reveals a new N-terminal binding motif for SHIP2. Mol. Cell. Proteomics 7, 181–192 [DOI] [PubMed] [Google Scholar]
- 5.Hu, J., and Hubbard, S. R. ( 2005) Structural characterization of a novel Cbl phosphotyrosine recognition motif in the APS family of adapter proteins. J. Biol. Chem. 280, 18943–18949 [DOI] [PubMed] [Google Scholar]
- 6.Pascal, S. M., Singer, A. U., Gish, G., Yamazaki, T., Shoelson, S. E., Pawson, T., Kay, L. E., and Forman-Kay, J. D. ( 1994) Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-gamma 1 complexed with a high affinity binding peptide. Cell 77, 461–472 [DOI] [PubMed] [Google Scholar]
- 7.Li, S. C., Gish, G., Yang, D., Coffey, A. J., Forman-Kay, J. D., Ernberg, I., Kay, L. E., and Pawson, T. ( 1999) Novel mode of ligand binding by the SH2 domain of the human XLP disease gene product SAP/SH2D1A. Curr. Biol. 9, 1355–1362 [DOI] [PubMed] [Google Scholar]
- 8.Uhlik, M. T., Temple, B., Bencharit, S., Kimple, A. J., Siderovski, D. P., and Johnson, G. L. ( 2005) Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J. Mol. Biol. 345, 1–20 [DOI] [PubMed] [Google Scholar]
- 9.Smith, M. J., Hardy, W. R., Murphy, J. M., Jones, N., and Pawson, T. ( 2006) Screening for PTB domain binding partners and ligand specificity using proteome-derived NPXY peptide arrays. Mol. Cell. Biol. 26, 8461–8474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sondermann, H., and Kuriyan, J. ( 2005) C2 can do it, too. Cell 121, 158–160 [DOI] [PubMed] [Google Scholar]
- 11.Christofk, H. R., Vander Heiden, M. G., Wu, N., Asara, J. M., and Cantley, L. C. ( 2008) Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 452, 181–186 [DOI] [PubMed] [Google Scholar]
- 12.Saltiel, A. R. ( 2001) New perspectives into the molecular pathogenesis and treatment of type 2 diabetes. Cell 104, 517–529 [DOI] [PubMed] [Google Scholar]
- 13.Sesti, G. ( 2006) Pathophysiology of insulin resistance. Best Prac. Res. Clin. Endocrinol. Metab. 20, 665–679 [DOI] [PubMed] [Google Scholar]
- 14.Bryant, N. J., Govers, R., and James, D. E. ( 2002) Regulated transport of the glucose transporter GLUT4. Nat. Rev. Mol. Cell Biol. 3, 267–277 [DOI] [PubMed] [Google Scholar]
- 15.White, M. F. ( 2002) IRS proteins and the common path to diabetes. Amer. J. Physiol. 283, E413–E422 [DOI] [PubMed] [Google Scholar]
- 16.Cai, D., Dhe-Paganon, S., Melendez, P. A., Lee, J., and Shoelson, S. E. ( 2003) Two new substrates in insulin signaling, IRS5/DOK4 and IRS6/DOK5. J. Biol. Chem. 278, 25323–25330 [DOI] [PubMed] [Google Scholar]
- 17.Dearth, R. K., Cui, X., Kim, H. J., Hadsell, D. L., and Lee, A. V. ( 2007) Oncogenic transformation by the signaling adaptor proteins insulin receptor substrate (IRS)-1 and IRS-2. Cell Cycle 6, 705–713 [DOI] [PubMed] [Google Scholar]
- 18.Summers, S. A. ( 2006) Ceramides in insulin resistance and lipotoxicity. Prog. Lipid Res. 45, 42–72 [DOI] [PubMed] [Google Scholar]
- 19.Petersen, K. F., and Shulman, G. I. ( 2006) Etiology of insulin resistance. Amer. J. Med. 119, S10–S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houstis, N., Rosen, E. D., and Lander, E. S. ( 2006) Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature 440, 944–948 [DOI] [PubMed] [Google Scholar]
- 21.Sesti, G., Federici, M., Hribal, M. L., Lauro, D., Sbraccia, P., and Lauro, R. ( 2001) Defects of the insulin receptor substrate (IRS) system in human metabolic disorders. FASEB J. 15, 2099–2111 [DOI] [PubMed] [Google Scholar]
- 22.Withers, D. J., Burks, D. J., Towery, H. H., Altamuro, S. L., Flint, C. L., and White, M. F. ( 1999) Irs-2 coordinates Igf-1 receptor-mediated beta-cell development and peripheral insulin signaling. Nat. Genet. 23, 32–40 [DOI] [PubMed] [Google Scholar]
- 23.Kido, Y., Burks, D. J., Withers, D., Bruning, J. C., Kahn, C. R., White, M. F., and Accili, D. ( 2000) Tissue-specific insulin resistance in mice with mutations in the insulin receptor, IRS-1, and IRS-2. J. Clin. Investig. 105, 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, C., Thirone, A. C., Huang, X., and Klip, A. ( 2005) Differential contribution of insulin receptor substrates 1 versus 2 to insulin signaling and glucose uptake in l6 myotubes. J. Biol. Chem. 280, 19426–19435 [DOI] [PubMed] [Google Scholar]
- 25.Araki, E., Lipes, M. A., Patti, M. E., Bruning, J. C., Haag, B., 3rd, Johnson, R. S., and Kahn, C. R. ( 1994) Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature 372, 186–190 [DOI] [PubMed] [Google Scholar]
- 26.Taniguchi, C. M., Ueki, K., and Kahn, R. ( 2005) Complementary roles of IRS-1 and IRS-2 in the hepatic regulation of metabolism. J. Clin. Investig. 115, 718–727 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Withers, D. J., Gutierrez, J. S., Towery, H., Burks, D. J., Ren, J. M., Previs, S., Zhang, Y., Bernal, D., Pons, S., Shulman, G. I., Bonner-Weir, S., and White, M. F. ( 1998) Disruption of IRS-2 causes type 2 diabetes in mice. Nature 391, 900–904 [DOI] [PubMed] [Google Scholar]
- 28.Taniguchi, C. M., Emanuelli, B., and Kahn, C. R. ( 2006) Critical nodes in signalling pathways: insights into insulin action. Nat. Rev. Mol. Cell Biol. 7, 85–96 [DOI] [PubMed] [Google Scholar]
- 29.Nakae, J., Kido, Y., and Accili, D. ( 2001) Distinct and overlapping functions of insulin and IGF-I receptors. Endocr. Rev. 22, 818–835 [DOI] [PubMed] [Google Scholar]
- 30.Hirayama, I., Tamemoto, H., Yokota, H., Kubo, S. K., Wang, J., Kuwano, H., Nagamachi, Y., Takeuchi, T., and Izumi, T. ( 1999) Insulin receptor-related receptor is expressed in pancreatic beta-cells and stimulates tyrosine phosphorylation of insulin receptor substrate-1 and -2. Diabetes 48, 1237–1244 [DOI] [PubMed] [Google Scholar]
- 31.Klammt, J., Garten, A., Barnikol-Oettler, A., Beck-Sickinger, A. G., and Kiess, W. ( 2005) Comparative analysis of the signaling capabilities of the insulin receptor-related receptor. Biochem. Biophys. Res. Commun. 327, 557–564 [DOI] [PubMed] [Google Scholar]
- 32.Neduva, V., and Russell, R. B. ( 2006) Peptides mediating interaction networks: new leads at last. Curr. Opin. Biotechnol. 17, 465–471 [DOI] [PubMed] [Google Scholar]
- 33.Diella, F., Haslam, N., Chica, C., Budd, A., Michael, S., Brown, N. P., Trave, G., and Gibson, T. J. ( 2008) Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci. 13, 6580–6603 [DOI] [PubMed] [Google Scholar]
- 34.Schulze, W. X., Deng, L., and Mann, M. ( 2005) Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol. Syst. Biol. 1: 2005.2008 [DOI] [PMC free article] [PubMed]
- 35.Vermeulen, M., Mulder, K. W., Denissov, S., Pijnappel, W. W., van Schaik, F. M., Varier, R. A., Baltissen, M. P., Stunnenberg, H. G., Mann, M., and Timmers, H. T. ( 2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 [DOI] [PubMed] [Google Scholar]
- 36.Ong, S. E., Blagoev, B., Kratchmarova, I., Kristensen, D. B., Steen, H., Pandey, A., and Mann, M. ( 2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 37.Vermeulen, M., Hubner, N. C., and Mann, M. ( 2008) High confidence determination of specific protein-protein interactions using quantitative mass spectrometry. Curr. Opin. Biotechnol. 19, 331–337 [DOI] [PubMed] [Google Scholar]
- 38.Jones, R. B., Gordus, A., Krall, J. A., and MacBeath, G. ( 2006) A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature 439, 168–174 [DOI] [PubMed] [Google Scholar]
- 39.Rappsilber, J., Ishihama, Y., and Mann, M. ( 2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 40.Rappsilber, J., Mann, M., and Ishihama, Y. ( 2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 41.Ishihama, Y., Rappsilber, J., Andersen, J. S., and Mann, M. ( 2002) Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. 979, 233–239 [DOI] [PubMed] [Google Scholar]
- 42.Olsen, J. V., de Godoy, L. M., Li, G., Macek, B., Mortensen, P., Pesch, R., Makarov, A., Lange, O., Horning, S., and Mann, M. ( 2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 43.Olsen, J. V., and Mann, M. ( 2004) Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc. Natl. Acad. Sci. U. S. A. 101, 13417–13422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanke, S., Besir, H., Oesterhelt, D., and Mann, M. ( 2008) Absolute SILAC for accurate quantitation of proteins in complex mixtures down to the attomole level. J. Proteome Res. 7, 1118–1130 [DOI] [PubMed] [Google Scholar]
- 45.Schulze, W. X., and Mann, M. ( 2004) A novel proteomic screen for peptide-protein interactions. J. Biol. Chem. 279, 10756–10764 [DOI] [PubMed] [Google Scholar]
- 46.Mann, M. ( 2008) Can proteomics retire the Western blot? J. Proteome Res. 7, 3065. [DOI] [PubMed] [Google Scholar]
- 47.Sun, X. J., Pons, S., Wang, L. M., Zhang, Y., Yenush, L., Burks, D., Myers, M. G., Jr., Glasheen, E., Copeland, N. G., Jenkins, N. A., Pierce, J. H., and White, M. F. ( 1997) The IRS-2 gene on murine chromosome 8 encodes a unique signaling adapter for insulin and cytokine action. Mol. Endocrinol. 11, 251–262 [DOI] [PubMed] [Google Scholar]
- 48.Eck, M. J., Dhe-Paganon, S., Trub, T., Nolte, R. T., and Shoelson, S. E. ( 1996) Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell 85, 695–705 [DOI] [PubMed] [Google Scholar]
- 49.Wolf, G., Trub, T., Ottinger, E., Groninga, L., Lynch, A., White, M. F., Miyazaki, M., Lee, J., and Shoelson, S. E. ( 1995) PTB domains of IRS-1 and Shc have distinct but overlapping binding specificities. J. Biol. Chem. 270, 27407–27410 [DOI] [PubMed] [Google Scholar]
- 50.Ong, S. E., and Mann, M. ( 2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 51.Witze, E. S., Old, W. M., Resing, K. A., and Ahn, N. G. ( 2007) Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 4, 798–806 [DOI] [PubMed] [Google Scholar]
- 52.Jensen, O. N. ( 2006) Interpreting the protein language using proteomics. Nat. Rev. Mol. Cell Biol. 7, 391–403 [DOI] [PubMed] [Google Scholar]
- 53.Mann, M. ( 2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 7, 952–958 [DOI] [PubMed] [Google Scholar]
- 54.Kruger, M., Kratchmarova, I., Blagoev, B., Tseng, Y. H., Kahn, C. R., and Mann, M. ( 2008) Dissection of the insulin signaling pathway via quantitative phosphoproteomics. Proc. Natl. Acad. Sci. U. S. A. 105, 2451–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Songyang, Z., Shoelson, S. E., Chaudhuri, M., Gish, G., Pawson, T., Haser, W. G., King, F., Roberts, T., Ratnofsky, S., Lechleider, R. J., Neel, B. G., Birge, R. B., Fajardo, J. E., Chou, M. M., Hanafusa, H., Schaffhausen, B., and Cantley, L. C. ( 1993) SH2 domains recognize specific phosphopeptide sequences. Cell 72, 767–778 [DOI] [PubMed] [Google Scholar]
- 56.Knight, Z. A., Gonzalez, B., Feldman, M. E., Zunder, E. R., Goldenberg, D. D., Williams, O., Loewith, R., Stokoe, D., Balla, A., Toth, B., Balla, T., Weiss, W. A., Williams, R. L., and Shokat, K. M. ( 2006) A pharmacological map of the PI3-K family defines a role for p110 alpha in insulin signaling. Cell 125, 733–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Myers, M. G., Jr., Mendez, R., Shi, P., Pierce, J. H., Rhoads, R., and White, M. F. ( 1998) The COOH-terminal tyrosine phosphorylation sites on IRS-1 bind SHP-2 and negatively regulate insulin signaling. J. Biol. Chem. 273, 26908–26914 [DOI] [PubMed] [Google Scholar]
- 58.Li, W., Nishimura, R., Kashishian, A., Batzer, A. G., Kim, W. J., Cooper, J. A., and Schlessinger, J. ( 1994) A new function for a phosphotyrosine phosphatase: linking GRB2-Sos to a receptor tyrosine kinase. Mol. Cell. Biol. 14, 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Milarski, K. L., and Saltiel, A. R. ( 1994) Expression of catalytically inactive Syp phosphatase in 3T3 cells blocks stimulation of mitogen-activated protein kinase by insulin. J. Biol. Chem. 269, 21239–21243 [PubMed] [Google Scholar]
- 60.Noguchi, T., Matozaki, T., Horita, K., Fujioka, Y., and Kasuga, M. ( 1994) Role of SH-PTP2, a protein-tyrosine phosphatase with Src homology 2 domains, in insulin-stimulated Ras activation. Mol. Cell. Biol. 14, 6674–6682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dyson, J. M., Kong, A. M., Wiradjaja, F., Astle, M. V., Gurung, R., and Mitchell, C. A. ( 2005) The SH2 domain containing inositol polyphosphate 5-phosphatase-2: SHIP2. Int. J. Biochem. Cell Biol. 37, 2260–2265 [DOI] [PubMed] [Google Scholar]
- 62.Feller, S. M. ( 2001) Crk family adaptors-signalling complex formation and biological roles. Oncogene 20, 6348–6371 [DOI] [PubMed] [Google Scholar]
- 63.Chang, L., Chiang, S. H., and Saltiel, A. R. ( 2004) Insulin signaling and the regulation of glucose transport. Mol. Med. 10, 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saltiel, A. R., and Kahn, C. R. ( 2001) Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799–806 [DOI] [PubMed] [Google Scholar]
- 65.Sadagurski, M., Weingarten, G., Rhodes, C. J., White, M. F., and Wertheimer, E. ( 2005) Insulin receptor substrate 2 plays diverse cell-specific roles in the regulation of glucose transport. J. Biol. Chem. 280, 14536–14544 [DOI] [PubMed] [Google Scholar]
- 66.Haeusler, R. A., and Accili, D. ( 2008) The double life of Irs. Cell Metab. 8, 7–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tobe, K., Sabe, H., Yamamoto, T., Yamauchi, T., Asai, S., Kaburagi, Y., Tamemoto, H., Ueki, K., Kimura, H., Akanuma, Y., Yazaki, Y., Hanafusa, H., and Kadowaki, T. ( 1996) Csk enhances insulin-stimulated dephosphorylation of focal adhesion proteins. Mol. Cell. Biol. 16, 4765–4772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berridge, M. J., and Irvine, R. F. ( 1989) Inositol phosphates and cell signaling. Nature 341, 197–205 [DOI] [PubMed] [Google Scholar]
- 69.Kim, J. J., and Accili, D. ( 2002) Signaling through IGF-I and insulin receptors: where is the specificity? Growth Horm. IGF Res. 12, 84–90 [DOI] [PubMed] [Google Scholar]
- 70.Yaffe, M. B. ( 2002) Phosphotyrosine-binding domains in signal transduction. Nat. Rev. Mol. Cell Biol. 3, 177–186 [DOI] [PubMed] [Google Scholar]
- 71.Kamberov, E., Makarova, O., Roh, M., Liu, A., Karnak, D., Straight, S., and Margolis, B. ( 2000) Molecular cloning and characterization of Pals, proteins associated with mLin-7. J. Biol. Chem. 275, 11425–11431 [DOI] [PubMed] [Google Scholar]
- 72.Hu, J., Liu, J., Ghirlando, R., Saltiel, A. R., and Hubbard, S. R. ( 2003) Structural basis for recruitment of the adaptor protein APS to the activated insulin receptor. Mol. Cell 12, 1379–1389 [DOI] [PubMed] [Google Scholar]
- 73.Sawka-Verhelle, D., Tartare-Deckert, S., White, M. F., and Van Obberghen, E. ( 1996) Insulin receptor substrate-2 binds to the insulin receptor through its phosphotyrosine-binding domain and through a newly identified domain comprising amino acids 591–786. J. Biol. Chem. 271, 5980–5983 [DOI] [PubMed] [Google Scholar]
- 74.Groesch, T. D., Zhou, F., Mattila, S., Geahlen, R. L., and Post, C. B. ( 2006) Structural basis for the requirement of two phosphotyrosine residues in signaling mediated by Syk tyrosine kinase. J. Mol. Biol. 356, 1222–1236 [DOI] [PubMed] [Google Scholar]
- 75.Velazquez, L., Gish, G. D., van Der Geer, P., Taylor, L., Shulman, J., and Pawson, T. ( 2000) The shc adaptor protein forms interdependent phosphotyrosine-mediated protein complexes in mast cells stimulated with interleukin 3. Blood 96, 132–138 [PubMed] [Google Scholar]
- 76.Lee, C. H., Li, W., Nishimura, R., Zhou, M., Batzer, A. G., Myers, M. G., Jr., White, M. F., Schlessinger, J., and Skolnik, E. Y. ( 1993) Nck associates with the SH2 domain-docking protein IRS-1 in insulin-stimulated cells. Proc. Natl. Acad. Sci. U. S. A. 90, 11713–11717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buday, L., Wunderlich, L., and Tamas, P. ( 2002) The Nck family of adapter proteins: regulators of actin cytoskeleton. Cell. Signal. 14, 723–731 [DOI] [PubMed] [Google Scholar]
- 78.Virkamaki, A., Ueki, K., and Kahn, C. R. ( 1999) Protein-protein interaction in insulin signaling and the molecular mechanisms of insulin resistance. J. Clin. Investig. 103, 931–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meng, W., Sawasdikosol, S., Burakoff, S. J., and Eck, M. J. ( 1999) Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature 398, 84–90 [DOI] [PubMed] [Google Scholar]
- 80.Benes, C. H., Wu, N., Elia, A. E., Dharia, T., Cantley, L. C., and Soltoff, S. P. ( 2005) The C2 domain of PKCdelta is a phosphotyrosine binding domain. Cell 121, 271–280 [DOI] [PubMed] [Google Scholar]
- 81.Yamauchi, T., Kaburagi, Y., Ueki, K., Tsuji, Y., Stark, G. R., Kerr, I. M., Tsushima, T., Akanuma, Y., Komuro, I., Tobe, K., Yazaki, Y., and Kadowaki, T. ( 1998) Growth hormone and prolactin stimulate tyrosine phosphorylation of insulin receptor substrate-1, -2, and -3, their association with p85 phosphatidylinositol 3-kinase (PI3-kinase), and concomitantly PI3-kinase activation via JAK2 kinase. J. Biol. Chem. 273, 15719–15726 [DOI] [PubMed] [Google Scholar]
- 82.Lanzino, M., Garofalo, C., Morelli, C., Le Pera, M., Casaburi, I., McPhaul, M. J., Surmacz, E., Ando, S., and Sisci, D. ( 2008) Insulin receptor substrate 1 modulates the transcriptional activity and the stability of androgen receptor in breast cancer cells. Breast Can. Res. Treat. 10.1007/s10549-008-0079-1 [DOI] [PubMed]
- 83.Liang, L., Zhou, T., Jiang, J., Pierce, J. H., Gustafson, T. A., and Frank, S. J. ( 1999) Insulin receptor substrate-1 enhances growth hormone-induced proliferation. Endocrinology 140, 1972–1983 [DOI] [PubMed] [Google Scholar]
- 84.Senthil, D., Ghosh Choudhury, G., Bhandari, B. K., and Kasinath, B. S. ( 2002) The type 2 vascular endothelial growth factor receptor recruits insulin receptor substrate-1 in its signalling pathway. Biochem. J. 368, 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vuori, K., and Ruoslahti, E. ( 1994) Association of insulin receptor substrate-1 with integrins. Science 266, 1576–1578 [DOI] [PubMed] [Google Scholar]
- 86.Shaw, L. M. ( 2001) Identification of insulin receptor substrate 1 (IRS-1) and IRS-2 as signaling intermediates in the α6β4 integrin-dependent activation of phosphoinositide 3-OH kinase and promotion of invasion. Mol. Cell. Biol. 21, 5082–5093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yenush, L., and White, M. F. ( 1997) The IRS-signalling system during insulin and cytokine action. Bioessays 19, 491–500 [DOI] [PubMed] [Google Scholar]
- 88.Saeki, M., Irie, Y., Ni, L., Yoshida, M., Itsuki, Y., and Kamisaki, Y. ( 2006) Monad, a WD40 repeat protein, promotes apoptosis induced by TNF-alpha. Biochem. Biophy. Res. Comm. 342, 568–572 [DOI] [PubMed] [Google Scholar]
- 89.Weirich, C. S., Erzberger, J. P., and Barral, Y. ( 2008) The septin family of GTPases: architecture and dynamics. Nat. Rev. Mol. Cell Biol. 9, 478–489 [DOI] [PubMed] [Google Scholar]
- 90.Xu, X., Sarikas, A., Dias-Santagata, D. C., Dolios, G., Lafontant, P. J., Tsai, S. C., Zhu, W., Nakajima, H., Nakajima, H. O., Field, L. J., Wang, R., and Pan, Z. Q. ( 2008) The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Mol. Cell 30, 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.He, X. Y., and Yang, S. Y. ( 2006) Roles of type 10 17beta-hydroxysteroid dehydrogenase in intracrinology and metabolism of isoleucine and fatty acids. Endoc. Metab. Immune Disord. Drug Targets 6, 95–102 [DOI] [PubMed] [Google Scholar]
- 92.Yu, C., Chen, Y., Cline, G. W., Zhang, D., Zong, H., Wang, Y., Bergeron, R., Kim, J. K., Cushman, S. W., Cooney, G. J., Atcheson, B., White, M. F., Kraegen, E. W., and Shulman, G. I. ( 2002) Mechanism by which fatty acids inhibit insulin activation of insulin receptor substrate-1 (IRS-1)-associated phosphatidylinositol 3-kinase activity in muscle. J. Biol. Chem. 277, 50230–50236 [DOI] [PubMed] [Google Scholar]
- 93.Gual, P., Le Marchand-Brustel, Y., and Tanti, J. F. ( 2005) Positive and negative regulation of insulin signaling through IRS-1 phosphorylation. Biochimie 87, 99–109 [DOI] [PubMed] [Google Scholar]
- 94.Briedis, K. M., Starr, A., and Bourne, P. E. ( 2008) Analysis of the human kinome using methods including fold recognition reveals two novel kinases. PLoS ONE 3, e1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Proud, C. G. ( 2002) Regulation of mammalian translation factors by nutrients. Eur. J. Biochem. 269, 5338–5349 [DOI] [PubMed] [Google Scholar]
- 96.Patti, M. E., Brambilla, E., Luzi, L., Landaker, E. J., and Kahn, C. R. ( 1998) Bidirectional modulation of insulin action by amino acids. J. Clin. Investig. 101, 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gstaiger, M., Luke, B., Hess, D., Oakeley, E. J., Wirbelauer, C., Blondel, M., Vigneron, M., Peter, M., and Krek, W. ( 2003) Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science (New York, N. Y.) 302, 1208–1212 [DOI] [PubMed] [Google Scholar]
- 98.Adeghate, E., Schattner, P., and Dunn, E. ( 2006) An update on the etiology and epidemiology of diabetes mellitus. Ann. N. Y. Acad. Sci. 1084, 1–29 [DOI] [PubMed] [Google Scholar]