MERIT40 facilitates BRCA1 localization and DNA damage repair (original) (raw)
Abstract
The product of breast cancer susceptibility gene 1, BRCA1, plays pivotal roles in the maintenance of genomic integrity. Mounting evidence indicates that BRCA1 associates with many proteins or protein complexes to regulate diverse processes important for the cellular response to DNA damage. One of these complexes, which mediates the accumulation of BRCA1 at sites of DNA breaks, involves the ubiquitin-binding motif (UIM)-containing protein RAP80, a coiled-coil domain protein CCDC98/Abraxas, and a deubiquitinating enzyme BRCC36. Here we describe the characterization of a novel component of this complex, MERIT40 (Mediator of Rap80 Interactions and Targeting 40 kd), which together with an adaptor protein BRE/BRCC45, enforces the BRCA1-dependent DNA damage response. MERIT40 is assembled into this RAP80/CCDC98-containing complex via its direct interaction with BRE/BRCC45. Importantly, MERIT40 regulates BRCA1 retention at DNA breaks and checkpoint function primarily via a role in maintaining the stability of BRE and this five-subunit protein complex at sites of DNA damage. Together, our study reveals that a stable complex containing MERIT40 acts early in DNA damage response and regulates damage-dependent BRCA1 localization.
Keywords: MERIT40, RAP80, CCDC98/Abraxas, BRE/BRCC45, BRCC36, BRCA1
The breast cancer susceptible gene 1 (BRCA1) is a tumor suppressor that plays critical roles in diverse cellular processes that include DNA damage repair, ubiquitination, and transcriptional regulation, and dysregulation of which predisposes women to early onset familial breast and ovarian cancers (Boulton 2006; Mullan et al. 2006; Greenberg 2008; Venkitaraman 2009). Emerging evidence indicates that BRCA1 participates in various tumor-suppressing functions through its ability to form distinct protein complexes in the cell, supporting a multifactorial model in which the concerted actions of these BRCA1 complexes play instrumental roles in the maintenance of genomic stability through regulation of checkpoint activation, DNA repair, and/or other cellular functions.
The BRCA1 protein contains two well-characterized functional domains, namely, its N-terminal RING finger domain, which encodes for its ubiquitin ligase activity (Joazeiro and Weissman 2000; Hashizume et al. 2001; Baer and Ludwig 2002), and a C-terminal tandem BRCT repeat. Importantly, patient mutations have been found within these domains, implicating their importance for BRCA1 function. Notably, its BRCT domain, which constitutes a phosphopeptide recognition domain that binds specifically to phospho-SXXF motifs (Manke et al. 2003; Yu et al. 2003), has been documented to associate with a list of interacting partners. Through this BRCT domain, BRCA1 was recently reported to exist in at least three distinct protein complexes. Our previous work demonstrated that BRCA1 associates with BACH1 (BRCA1-associated C-terminal helicase) (Cantor et al. 2001) in a phosphorylation-dependent manner to mediate S-phase checkpoint control (Manke et al. 2003; Yu et al. 2003). Subsequent studies revealed that the association of BRCA1 with CtIP (CtBP-interacting protein) (Yu et al. 1998; Li et al. 1999) is also phosphorylation-dependent and is important for the BRCA1-dependent G2/M checkpoint control (Yu and Chen 2004). More recently, a coiled-coil domain-containing protein CCDC98/Abraxas was also found to interact with BRCA1, which again is mediated by the binding of BRCA1 BRCT domains with a phosphorylated SXXF motif on CCDC98/Abraxas (Kim et al. 2007b; Liu et al. 2007; Wang et al. 2007). The current working hypothesis is that BACH1 and CtIP are likely to act downstream from or in parallel with BRCA1, and thus they do not contribute to proper BRCA1 localization at DNA damage sites (Greenberg et al. 2006). On the other hand, CCDC98 and its associated proteins RAP80 and BRCC36 are all involved in the recruitment of BRCA1 to DNA damage sites (Chen et al. 2006; Kim et al. 2007a; Sobhian et al. 2007; Wang et al. 2007). This recruitment is controlled by protein ubiquitination at sites of DNA breaks (Polanowska et al. 2006; Zhao et al. 2007) and relies on a recently identified signaling pathway from pH2AX/MDC1 to an E3 ubiquitin ligase RNF8 (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007; Bennett and Harper 2008).
Since RAP80 contains tandem ubiquitin-binding motifs (UIMs), which is required for its localization at DNA damage sites, it is now generally accepted that RAP80 recognizes the ubiquitinated proteins at sites of DNA breaks, which in turn facilitates the recruitment of BRCA1 via a direct interaction between BRCA1 and CCDC98 (Kim et al. 2007a; Sobhian et al. 2007; Wang et al. 2007). In this study, we isolated a multiprotein complex containing RAP80, CCDC98, BRCC36, and BRCA1. We described the identification and characterization of a novel component in this complex, Hspc142 (C19orf62), which participates in RAP80 complex formation and is required for BRCA1 localization, cell cycle checkpoint control, and cell survival following DNA damage. To better reflect its cellular function, we named Hspc142 as MERIT40 (Mediator of Rap80 Interactions and Targeting 40 kd) in the text.
Results
MERIT40 is a novel component of RAP80/CCDC98 complex
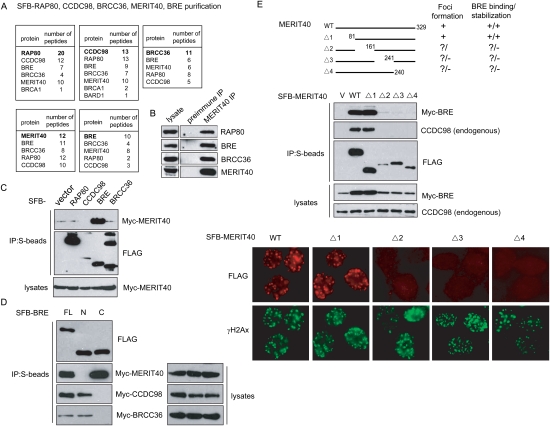
Previous studies have reported association between RAP80, CCDC98/Abraxas, BRCC36, and BRCA1 (Kim et al. 2007a; Sobhian et al. 2007; Wang and Elledge 2007; Wang et al. 2007). However, exactly how this complex is assembled in vivo remains elusive. To better understand how the RAP80/CCDC98/BRCC36 protein complex is assembled in the cell, we generated 293T derivative cell lines stably expressing triple-tagged (S-Flag-Streptavidin-binding peptide) RAP80, CCDC98, or BRCC36 with an aim to identify additional components important for the complex formation. Following a tandem affinity purification (TAP) scheme, proteins associated with RAP80, CCDC98, or BRCC36 were identified by mass spectrometry analyses (Fig. 1A, top panel). Interestingly, beside RAP80, CCDC98, and BRCC36, we also identified BRE (brain and reproductive organ-expressed protein) (Li et al. 1995), also known as BRCC45 (Dong et al. 2003)] and an uncharacterized protein, Hspc142 (C19orf62), among the three purified protein complexes. As indicated above, we renamed this protein Hspc142/C19orf62 as MERIT40. To further confirm that MERIT40 and BRE exist in the same complex with RAP80, CCDC98, and BRCC36, we generated stable cells expressing triple-tagged MERIT40 and BRE, respectively. Notably, mass spectrometry analyses of MERIT40 or BRE-associated proteins revealed peptides that corresponded to RAP80, CCDC98, and BRCC36 (Fig. 1A; bottom panel), suggesting that these five proteins likely form a stable complex in vivo.
Figure 1.
Identification of MERIT40 as a new component in the RAP80/CCDC98 protein complex. (A) 293T cells stably expressing SFB-tagged (S-tag, Flag epitope tag, and streptavidin-binding peptide tag)-RAP80, CCDC98, BRCC36, MERIT40, or BRE were used for TAP of protein complexes specifically from chromatin fractions isolated from irradiated cells. Tables are summaries of proteins identified by Mass spectrometry analysis. Letters in bold indicate the bait proteins. (B) Immunoprecipitations were performed using preimmune serum or anti-MERIT40 antibodies. Interactions between endogenous MERIT40 and RAP80, BRE, or BRCC36 were assessed by immunoblotting using antibodies as indicated. (C) Myc-tagged MERIT40 interacted strongly with SFB-tagged BRE. 293T cells were transiently transfected with plasmids encoding SFB-tagged RAP80, CCDC98, BRE, BRCC36, or empty vector together with plasmids encoding myc-tagged MERIT40. Cell lysates were precipitated with S-protein beads and immunoblotted with indicated antibodies. (D) CCDC98, BRCC36, or MERIT40 binds to different regions on BRE. 293T cells were transfected with plasmids encoding SFB-tagged full-length (FL), N-terminal part (N; residues 1–200), or C-terminal part (C; residues 201–383) of BRE, together with plasmids encoding Myc-tagged CCDC98, BRCC36, or BRE. Cell lysates were subjected to precipitation (IP) using S-protein beads, and immunoblotting was conducted using antibodies as indicated. (E) MERIT40 associates with BRE and forms IRIF. (Top panel) 293T cells were transfected with plasmids encoding SFB-tagged wild-type MERIT40 or its deletion mutants (Δ1–Δ4) together with plasmids encoding Myc-tagged BRE. Co-IP experiments were performed as described in C. (Bottom panel) 293T cells stably expressing SFB-tagged wild-type (WT) or deletion mutants (Δ1–Δ4) of MERIT40 were irradiated with 10 Gy, allowed to recove for 4 h before fixation and immunostaining with anti-Flag (red) and anti-γH2AX (green) antibodies.
BRE was previously identified as a subunit in the BRCA1/BRCA2-containing protein complex (Dong et al. 2003), whereas MERIT40 is a 329-amino-acid protein with unknown function. Neither BRE nor MERIT40 has any known functional motif. To verify the association among MERIT40, BRE, BRCC36, and RAP80, we performed coimmunoprecipitation (co-IP) experiments and found that endogenous MERIT40 immunoprecipitated RAP80, BRE, and BRCC36 (Fig. 1B), suggesting that these proteins indeed form a complex in vivo. Because the molecular weight of CCDC98 (∼50 KDa) is similar to the immunoglobulin heavy chain, we did not include it in our co-IP experiments. Taken together, our data reveal the presence of a stable RAP80-containing protein complex in the cell and suggest that MERIT40 is a novel component that resides in this complex.
Next, we sought to determine how this complex is assembled in vivo. Ectopically expressed MERIT40 interacted strongly with BRE, but only weakly with other components in co-overexpression experiments (Fig. 1C), indicating that MERIT40 may interact directly with BRE. This observation is consistent with the data obtained from several independent large-scale studies of protein–protein interactions, which suggest that BRE binds to MERIT40 (Rual et al. 2005; Ewing et al. 2007). To determine the region on BRE responsible for its interaction with MERIT40, we generated two BRE truncation mutants. Similar to that between full-length BRE and MERIT40, we observed a robust interaction between the C terminus of BRE (residues 201–383) with MERIT40 (Fig. 1D). Interestingly, while the N terminus (residues 1–200) of BRE is not required for its binding to MERIT40, it is essential for the association of BRE with CCDC98 and BRCC36 (Fig. 1D), suggesting that BRE interacts with MERIT40 and CCDC98/BRCC36 via different regions.
Next, we generated a series of MERIT40 internal deletion mutants to map the BRE-binding domain on MERIT40. Although the very N terminus of MERIT40 is dispensable for the MERIT40/BRE interaction, we found that the C-terminal three-quarters of MERIT40 all seem to be involved in its interaction with BRE (Fig. 1E, top panel), albeit the expression of these C-terminal deletion mutants of MERIT40 is lower than that of wild-type MERIT40 or the N-terminal deletion mutant (Fig. 1E, middle panel). Surprisingly, coexpression of MERIT40 with BRE also stabilized BRE in the cell and this stabilization effect of MERIT40 on BRE correlates with the interaction between the two proteins (Fig. 1E, middle panel). Conversely, MERIT40 stability does not depend on BRE (see also Fig. 3A, below; data not shown).
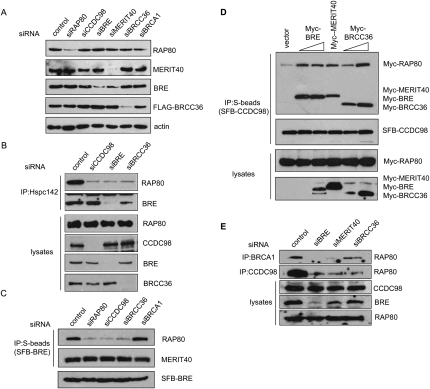
Figure 3.
MERIT40 is critical for maintaining the integrity of RAP80/CCDC98-containing protein complex. (A) U2OS cells were transfected with a scrambled control siRNA or siRNAs specific for the indicated genes. Cell lysates were prepared and immunoblotted with indicated antibodies. (B) CCDC98, BRE, and BRCC36 are required for the association of MERIT40 with RAP80. HeLa cells were transfected with control siRNA or siRNAs targeting CCDC98, BRE, or BRCC36. Cell lysates were immunoprecipitated with antibody against MERIT40, and immunoblotting experiments were carried out using antibodies as indicated. (C) CCDC98 and BRCC36 are required for the binding of BRE to RAP80, but are dispensable for BRE/MERIT40 interaction. 293T cells stably expressing SFB-BRE were transfected with control siRNA or siRNAs targeting RAP80, CCDC98, BRCC36, or BRCA1. Cell lysates were immunoprecipitated using anti-MERIT40 antibodies. Immunoblots were performed using antibodies as indicated. (D) Overexpression of BRE, MERIT40, or BRCC36 promotes RAP80/CCDC98 interaction. 293T cells were transfected with plasmids encoding SFB-tagged CCDC98 and Myc-tagged RAP80, together with empty vector or variables amount of plasmids encoding Myc-tagged BRE, MERIT40, or BRCC36 as indicated. Cell lysates were prepared, immunoprecipitated with S-protein beads, and immnoblotted with indicated antibodies. (E) Depletion of BRE, MERIT40, or BRCC36 impaired endogenous RAP80/CCDC98 or RAP80/BRCA1 interaction. HeLa cells were transfected with control siRNA or siRNAs targeting BRE, MERIT40, or BRCC36. Cell lysates were immunoprecipitated using anti-BRCA1 or anti-CCDC98 antibodies, and immunoblotting was carried out using indicated antibodies.
Interestingly, MERIT40 mutations that disrupted its binding with BRE also abolished the MERIT40–CCDC98 interaction (Fig. 1E, middle panel), which coincided with the regions required for MERIT40 foci formation (Fig. 1E, bottom panel). Together, these data suggest that MERIT40 may be assembled into the RAP80/CCDC98 complex through its association with BRE.
BRE bridges the interaction between MERIT40 and RAP80/CCDC98/BRCC36
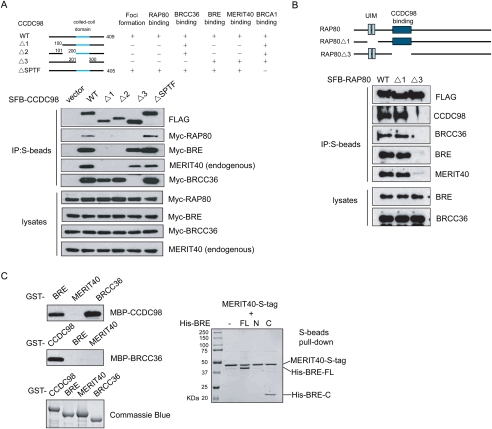
Earlier reports have shown that BRCC36 interacts with CCDC98 through the coiled-coil domain of the latter (Wang and Elledge 2007). RAP80 associates with a large region containing N-terminal three-fourths of CCDC98 (Kim et al. 2007b; Wang and Elledge 2007), while BRCA1 binds directly to the C-terminal SPTF motif of CCDC98 in a phosphorylation-dependent manner (Kim et al. 2007b; Liu et al. 2007; Wang et al. 2007). Thus, it appears that CCDC98 is at the center of this protein complex. Indeed, the interaction between MERIT40 and CCDC98 seems to correlate with its interaction with BRE (Fig. 1E, middle panel), implying that BRE may bind directly to CCDC98. Therefore, we used a series of CCDC98 mutants to examine how CCDC98 mediates this complex formation (Fig. 2A).
Figure 2.
BRE mediates the interaction between MERIT40 and CCDC98. (A) RAP80, BRE, and BRCC36 bind to different regions on CCDC98. 293T cells were transfected with plasmids encoding SFB-tagged wild-type or internal deletion mutants of CCDC98 together with plasmids encoding Myc-tagged RAP80, BRE, or BRCC36. Cell lysates were prepared, precipitated with S-protein beads, and immnoblotted with indicated antibodies. (B) The CCDC98-interacting region, but not the UIM domains of RAP80 is required for the interaction between RAP80 and BRE, BRCC36, or MERIT40. 293T cells were transiently transfected with plasmids encoding SFB-tagged wild-type RAP80 (WT), UIM deletion mutant (Δ1), or CCDC98-interacting region deletion mutant (Δ3) of RAP80. Cell lysates were subjected to precipitation (IP) with S-protein beads and immunoblotting with indicated antibodies. (C, left panels) BRE and BRCC36 bind directly to CCDC98 in vitro. Bacterially expressed MBP-tagged CCDC98, BRE, MERIT40, or BRCC36 were incubated with GST fusion proteins as indicated. Proteins bound to the beads were eluted by boiling in SDS sample buffer, separated by SDS-PAGE, and immunoblotted with anti-MBP antibody. GST fusion proteins used in these pull-down assays were analyzed by Commassie blue staining. (Right panels) BRE associates with MERIT40 in vitro. S-tagged MERIT40 was coexpressed with His-tagged wild-type or deletion mutants of BRE using bicistronic vectors in BL21 cells. Proteins bound to S-beads were eluted and visualized by Commassie blue staining.
While wild-type CCDC98 binds strongly to BRE, mutations in CCDC98, Δ1, and Δ2 resulted in a loss of the interaction between CCDC98 and BRE/MERIT40 (Fig. 2A). Importantly, although BRE and BRCC36 bound to different regions on CCDC98, both of these regions overlapped with the RAP80-binding domain on CCDC98 (Fig. 2A). We thus investigated whether RAP80 could exist in a single protein complex containing not only CCDC98, but also BRCC36, BRE, and MERIT40. As shown in Figure 2B, all four of these proteins coimmunoprecipited with wild-type RAP80 and RAP80 UIM deletion mutant (Δ1). However, they failed to interact with RAP80Δ3 mutant, which lacks the CCDC98-interacting region (Fig. 2B). These data support the hypothesis that all five of these proteins exist in one protein complex. While RAP80, BRE, and BRCC36 seem to share overlapping binding regions on CCDC98, they associate with each other in a nonexclusive manner to form a single entity.
Next, we used pull-down assays to further confirm the direct interactions among these proteins. Bacterially expressed and purified MBP-CCDC98 bound directly to GST-BRE and GST-BRCC36, but not to GST-MERIT40 (Fig. 2C). Similarly, GST-CCDC98 was also able to pull down MBP-BRE or MBP-BRCC36 (Fig. 2C), but not MBP-MERIT40 (data not shown), which agrees with our in vivo interaction data. In addition, when BRE and MERIT40 were coexpressed in Escherichia coli, they formed a stable complex (Fig. 2C). The association between BRE and MERIT40 in bacteria requires the C terminus of BRE (Fig. 2C), which again confirms our in vivo study (Fig. 1D). These data indicate that MERIT40 binds directly to BRE.
MERIT40 and BRE are required for maintaining the integrity of RAP80/CCDC98 complex
A multicomponent protein complex sometimes relies on one or several of its components for the stability of the protein complex. Therefore, we depleted each one of these subunits in the RAP80/CCDC98 protein complex and examined the stability of other components and also the complex formation under these situations. As shown in Figure 3A, depletion of RAP80, CCDC98, BRE, or BRCC36 did not affect the stability of the remaining four components in this complex. However, while depletion of MERIT40 by siRNA did not affect the steady levels of RAP80, CCDC98, or BRCC36, it led to a dramatic decrease in BRE protein level (Fig. 3A). This suggests that BRE is normally stabilized in vivo by MERIT40, which is entirely consistent with our overexpression results shown in Figure 1E.
To further investigate the nature of this complex formation, we immunoprecipitated endogenous MERIT40 from control cells or cells depleted of CCDC98, BRE, or BRCC36. Knockdown of CCDC98 dramatically decreased endogenous RAP80 and MERIT40 interaction (Fig. 3B). In addition, when BRE was depleted, the binding of MERIT40 to RAP80 was also greatly diminished (Fig. 3B), further supporting the hypothesis that MERIT40 is assembled into the RAP80/CCDC98 complex through its interaction with BRE. Strikingly, depletion of BRCC36 also reduced the interaction between MERIT40 and RAP80 (Fig. 3B). Likewise, knockdown of CCDC98 or BRCC36 by siRNAs in 293T cells stably expressing SFB-BRE also led to decreased binding between RAP80 and SFB-BRE, without any detectable change in BRE/MERIT40 association (Fig. 3C). In addition, knockdown of BRE or BRCC36 also led to decreased interaction between SFB-MERIT40 with RAP80 or CCDC98 (Supplemental Fig. 1A). These data imply that the interactions of CCDC98 with BRE/MERIT40 or with BRCC36 influence the whole complex formation.
This notion is supported by the observations that the regions on CCDC98 responsible for BRE and BRCC36 binding overlap with the region required for RAP80 binding (Fig. 2A). It is possible that all of these proteins bind to CCDC98 in a cooperative manner. To test this idea, we determined the complex formation between Myc-RAP80 and SFB-CCDC98 in the absence or the presence of overexpressed BRE, Hpsc142, or BRCC36. Interestingly, overexpression of any of these three proteins (BRE, MERIT40, and BRCC36) resulted in an increased RAP80/CCDC98 interaction (Fig. 3D). Furthermore, knockdown of endogenous BRE, MERIT40, or BRCC36 weakened the interaction between endogenous RAP80 and endogenous CCDC98 or BRCA1 (Fig. 3E), thereby demonstrating that these proteins bind cooperatively to maintain the assembly of a stable protein complex in vivo.
RAP80/CCDC98/BRCC36/BRE/MERIT40 depend on each other for their focus localization following DNA damage
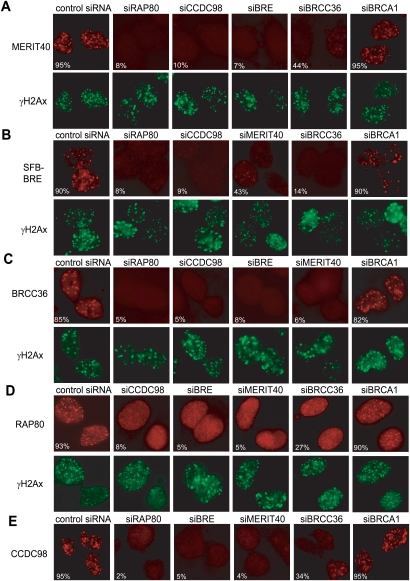
RAP80, CCDC98, BRCC36, and BRCA1 all localize to sites of DNA breaks in cells exposed to ionizing radiation (IR) (Scully et al. 1997; Kim et al. 2007a,b; Liu et al. 2007; Sobhian et al. 2007; Wang et al. 2007). Given that MERIT40 and BRE exist in the RAP80/CCDC98 complex and that MERIT40 also localizes to damage-induced foci (Fig. 1E), we would like to examine how they influence each other's IR-induced foci (IRIF) formation. As shown in Figure 4, A and B, both endogenous MERIT40 and ectopically expressed BRE formed IRIF, which colocialized with those of the DNA damage marker γH2AX (Fig. 4A,B; Paull et al. 2000), indicating that MERIT40 and BRE indeed localize to DSB sites. As shown in Figure 4, IRIF of BRCC36, RAP80, CCDC98 were significantly decreased in BRE or MERIT40-depleted cells and vice versa, suggesting RAP80/CCDC98/BRCC36/BRE/MERIT40 depend on each other for their IRIF formation. Interestingly, depletion of BRCA1 did not affect the focus formation of any of the four components in the protein complex (Fig. 4), which agrees with previous reports that BRCA1 acts downstream from the RAP80/CCDC98 complex (Kim et al. 2007a,b; Liu et al. 2007; Sobhian et al. 2007; Wang et al. 2007) and that BRCA1 is not a stable component of this protein complex (Fig. 1A).
Figure 4.
IRIF formation of RAP80, CCDC98, BRCC36, BRE, and MERIT40 requires all four other components in this protein complex. (A,C–E) U2OS cells or 293T cells stably expressing SFB-tagged BRE (B) were transfected with control siRNAs or siRNAs specifically targeting indicated genes. Cells were irradiated, fixed, and immunostained with anti-MERIT40 (A), anti-Flag (BRE) (B), anti-BRCC36 (C), anti-RAP80 (D), or anti-CCDC98 (E) antibodies with or without costaining with anti-γH2AX antibodies. More than 200 cells were counted to determine the percentages of foci-forming cells in each sample.
A stable protein complex consisting of RAP80/CCDC98/BRCC36/BRE/MERIT40 is required for efficient BRCA1 localization and function following DNA damage
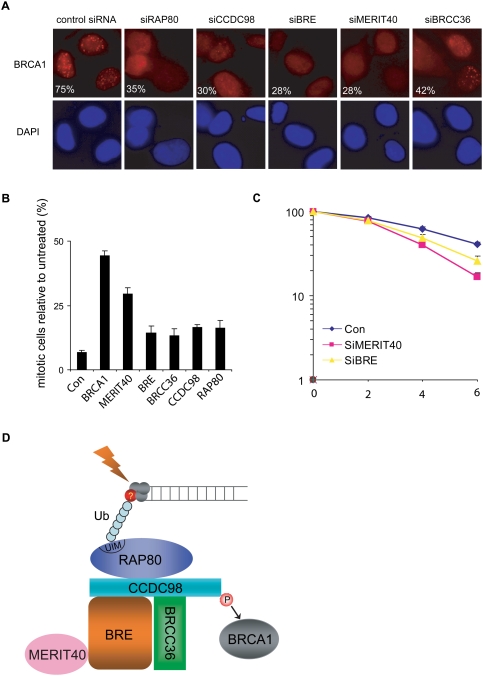
RAP80/CCDC98 and BRCC36 are all known to act upstream of BRCA1 and are required for targeting BRCA1 to DNA damage foci (Chen et al. 2006; Kim et al. 2007a,b; Liu et al. 2007; Sobhian et al. 2007; Wang and Elledge 2007; Wang et al. 2007). Since MERIT40 and BRE are components of this five-subunit protein complex and are required for the overall stability and the focus formation of this protein complex following DNA damage, we speculated that MERIT40 and BRE might also be required for BRCA1 foci formation. As shown in Figure 5A, similar to RAP80-, CCDC98-, or BRCC36-depleted cells, BRCA1 IRIF formation was compromised in cells transfected with MERIT40 or BRE siRNAs, suggesting that this protein complex containing MERIT40 and BRE acts upstream of BRCA1 in the DNA damage response pathway. We used a second MERIT40 siRNA and observed similar reduction of RAP80 and BRCA1 foci formation following DNA damage (Supplemental Fig. 1B). In addition, MERIT40 or BRE depletion led to a defective G2/M checkpoint control (Fig. 5B) and reduced cell survival following IR (Fig. 5C; also see Supplemental Fig. 1C), highlighting the importance of these subunits in regulating BRCA1 and cellular response to DNA damage.
Figure 5.
MERIT40 and BRE participate in IR-induced DNA damage response. (A) Defective BRCA1 foci formation was observed in RAP80-, CCDC98-, BRE-, MERIT40-, or BRCC36-depleted cells. U2OS cells were transfected with different siRNAs as indicated. Cells were irradiated, fixed, and immunostained with anti-BRCA1 antibody. More than 200 cells were counted to determine the percentages of cells containing BRCA1 foci. Two independent siRNAs were used for the depletion of CCDC98, BRE, BRCC36, or MERIT40, and similar results were obtained (data not shown). (B) IR-induced G2/M checkpoint is defective in cells depleted of MERIT40, BRE, or other components in the RAP80/CCDC98 complex. HeLa cells were transfected with indicated siRNAs and percentages of mitotic cells before and after radiation were determined by FACS analysis as described in the Materials and Methods. The figure represents the value obtained from three separate experiments. Error bars indicate standard deviations. (C) Cells depleted of MERIT40 or BRE display increased radiation sensitivity. HeLa cells were transfected with control siRNAs or siRNAs specifically targeting indicated genes. Cells were irradiated with various doses of IR. Percentages of surviving colonies were determined 11 d later. Experiments were done in triplicate. Results shown are averages of two or three independent experiments at each dose, and error bars indicate standard deviation. (D) A model describing the assembly of the RAP80/CCDC98/BRCC36/BRE/MERIT40 complex at sites of DNA breaks. See the text for details.
Discussion
In this study, we adopted the TAP approach to isolate RAP80/CCDC98-containing protein complex and identified a novel component, MERIT40, in this complex. Similar to RAP80 and CCDC98, MERIT40 localizes to sites of DNA break, participates in G2/M checkpoint control, and promotes cell survival following DNA damage, suggesting that MERIT40 is an important factor within the RAP80 complex that plays crucial roles in the DNA damage response. More importantly, we analyzed the RAP80/CCDC98 protein complex in great detail. Our data suggest that this RAP80/CCDC98 complex consists of five core subunits (Fig. 5D), with CCDC98 as the central component that facilitates the assembly of this protein complex. CCDC98 binds to RAP80 via a large N-terminal region (Kim et al. 2007b; Wang and Elledge 2007) and interacts with the DUB BRCC36 through its C-terminal coiled-coil domain (Wang and Elledge 2007). CCDC98 also directly interacts with BRE/BRCC45 via its N terminus. While MERIT40 does not associate directly with CCDC98, it is brought into this complex through its interaction with BRE. We were able to reconstitute this five-protein complex in vitro using baculovirus expression system (Supplemental Fig. 1D). Interestingly, we showed that a stable complex formation among these five subunits is not only required for their own IRIF formation, but is also involved in the recruitment of BRCA1 to the sites of DNA breaks. Previous studies indicated that the very C terminus of CCDC98 is phosphorylated and directly recruits BRCA1 (Kim et al. 2007b; Liu et al. 2007; Wang et al. 2007). Here, we showed that in the absence of MERIT40, BRE, or BRCC36, we also observed a significant reduction in BRCA1 foci formation. This is mediated by the destabilization of this protein complex, especially by the reduced interaction between RAP80 and CCDC98 in cells missing any one of these components (Fig. 3).
Early studies have already suggested that the localization of this RAP80/CCDC98-containing protein complex to DNA damage sites requires the interaction between the RAP80 UIM domain and ubiquitin chains formed at the sites of DNA breaks (Kim et al. 2007a,b; Liu et al. 2007; Sobhian et al. 2007; Wang and Elledge 2007; Wang et al. 2007). Intriguingly, we observed that depletion of any one of the four other components beside RAP80 also greatly reduced RAP80 focus formation (Fig. 4), a phenomenon that was also noticed in an early study in cells with CCDC98 depletion (Wang and Elledge 2007). Exactly how these proteins, of which their IRIF formation all depend on RAP80, can also feedback and influence RAP80 DSB localization is still elusive. One plausible explanation is that the stable complex formation among these proteins may lead to important conformational changes that enhance the interaction between RAP80 and the ubiquitination chains formed at the sites of DNA breaks. An alternative but not exclusive explanation could be that there is a second chromatin-binding protein that exists in this complex that may facilitate the stable accumulation of the entire complex at or near the sites of DNA breaks. Another interesting phenomenon is that while RAP80 IRIF can be easily detected, we have to use a pre-extraction method to visualize BRE/MERIT40 and BRCC36 foci formation. This implies that only a fraction of cellular BRE/MERIT40 and BRCC36 localizes to the sites of DNA breaks, raising the possibility that BRE/MERIT40 and BRCC36 may also have other functions in the cell.
A prominent feature of the recently advanced DNA damage signaling pathway that involves BRCA1 is the presence of many ubiquitination and presumably deubiquitination events. Several groups, including ourselves, have shown that an E3 ligase RNF8 localizes to DNA damage sites and probably initiates K63-linked ubiquitin chains at the vicinity of DNA breaks (Bennett and Harper 2008; Yan and Jetten 2008). This initiation ubiquitination event is believed to lead to the accumulation of a RAP80-containing complex to the sites of DNA damage, at least in part, via a direct interaction between protein ubiquitination chains and the RAP80 UIM domains. There are several additional ubiquitination-related events after that. First, the BRCA1/BARD1 heterodimer is a ubiquitin ligase that promotes K6-linked protein ubiquitination and has a number of substrates including CtIP involved in DNA damage response (Morris and Solomon 2004; Yu et al. 2006). Second, BRCC36 is a deubiquitinating enzyme that specifically hydrolyzes K63-linked polyubiquitination chains, but not K48-linked polyubiquitin chains (Sobhian et al. 2007). Third, an early study has also suggested that BRCC36 may facilitate BRCA1 ubiquitin ligase activity (Dong et al. 2003). Putting these together, an attractive model is that there may be a ubiquitin-editing event that occurs at or near the sites of DNA breaks, which could be similar to the case of A20 in the NF-κB pathway (Wertz et al. 2004; Balakirev and Wilkinson 2008). Such editing activity may be mediated by BRCC36 and thus convert the initial K63-linked polyubiquitination chains to BRCA1-dependent, K6-linked polyubiquitination chains for subsequent accumulation of yet-to-be-identified DNA damage checkpoint and repair proteins. Future studies will be conducted to test this hypothesis.
It is also noteworthy to mention that checkpoint defect or IR sensitivity observed in cells that are deficient of RAP80 or any other component in this five-subunit protein complex are usually not as severe as those observed in BRCA1-deficient cells. In addition, incomplete BRCA1 localization at laser-induced DSBs still occurs in the absence of RAP80 (Sobhian et al. 2007). This agrees with an early study suggesting that transient BRCA1 localization at DNA damage sites can occur in the absence of H2AX (Celeste et al. 2003). All of these point out that the possibility of a parallel pathway that acts cooperatively with the H2AX/MDC1/RNF8/RAP80 pathway to regulate the localization and function of BRCA1 in response to DNA damage.
Materials and methods
Cell culture and IR
U2OS, 293T, and HeLa cells were maintained in RMPI 1640 supplemented with 10% fetal bovine serum. Cells were irradiated using JL Spepherd 137Cs radiation source at indicated doses. Irradiated cells were then returned to culture conditions and maintained for the indicated periods of time specified in the figure legends.
TAP
293T cells were transfected with plasmids encoding SFB-tagged RAP80, CCDC98, MERIT40, BRE, or BRCC36, respectively, to establish stable cell lines expressing their corresponding tagged proteins. Cells were irradiated (10 Gy) and harvested 4 h later. Cells were lysed in NETN buffer (20 mM Tris-Hcl at pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40) containing 50 mM β-glycerophosphate, 10 mM NaF, and 1 μg/mL each of pepstatin A and aprotinin. Following centrifugation, the pellet was sonicated for 40 sec in high-salt solution (20 mM HEPES at pH 7.8, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, protease inhibitor) to extract chromatin-bound proteins fractions. The supernatants were cleared at 14,000 rpm to remove debris and then incubated with streptavidin-conjugated beads (Amersham) for 1 h at 4°C. The immunocomplexes were washed three times with NETN buffer and then bead-bound proteins were eluted with NETN buffer containing 2 mg/mL biotin (Sigma). The elutes were incubated with S protein beads (Novagen). After three washes, the immunocomplexes were analyzed by SDS-PAGE, and Mass spectrometry analyses were performed by the Harvard Medical School Taplin Biological Mass Spectrometry facility.
SiRNA, transfection and immunoprecipitation
All siRNAs were synthesized by Dharmacon, Inc. The siRNAs were 21 base pairs and sequences are as follows: MERIT40 siRNA, CAGAGAACGUGCAGACGAUdTdT; BRCC45 siRNA, GAGGAUAACUGACUUAAAAdTdT; BRCC36 siRNA, CAUAAUGGCUCAGUGUUUAdTdT; RAP80 siRNA, GAAGGAUGUGGAAACUACCdTdT; CCDC98 siRNA, ACACAAGACAAACGAUCUAUU; BRCA1 siRNA, UCACAGUGUCCUUUAUGUAdTdT; and control siRNA, UUCAAUAAAUUCUUGAGGUdTdT.
Plasmid transfection was performed using Lipofectamine2000 (Invitrogen). Oligofectamine (Invitrogen) was used for siRNA transfection. U2OS or HeLa cells were transfected with siRNA duplexes twice at 24-h intervals, while 293T cells were transfected with siRNAs three times at 24-h intervals. Twenty-four hours after the last transfection, cells were collected or irradiated.
Cell lysates and immunoprecipitation were performed similar to that described previously (Kim et al. 2007b), except that cell extracts were prepared using NETN buffer containing higher salt concentration (400 mM NaCl).
Antibodies
Anti-Flag and anti-actin antibodies were purchased from Sigma. Anti-phospho-H3 antibody was purchased from Upstate Biotechnologies. Anti-Myc monoclonal antibody, anti-MBP antibody, and anti-BRCC36 polyclonal antibody were obtained from Santa Cruz Biotechnologies, New England Biolabs, and ProSci, Inc., respectively. RAP80, CCDC98, and BRCA1 polyclonal antibodies have been described previously (Kim et al. 2007b). Rabbit anti-MERIT40 and anti-BRE antisera were raised by immunizing rabbits, respectively, with GST-MERIT40 (residues 130–329) and GST-BRE (residues 1–200) fusion proteins expressed and purified from E. coli. Antisera were affinity-purified using AminoLink Plus Immobilization and Purification Kit (Pierce).
Pull-down assay
GST- or MBP-fused CCDC98, BRE, MERIT40, and BRCC36 were expressed in BL21 cells (Novagen). GST fusion proteins were purified with glutathione Sepharose beads (Amersham Biosciences) according to the manufacturer's instructions. MBP fusion proteins were purified with Amylose beads (New England Biolabs) according to the manufacturer's instructions. For pull-down assay, 1 μg of maltose-eluted MBP fusion proteins were incubated with GST fusion proteins in NETN buffer for 2 h at 4°C. Beads were washed five times with NETN buffer. Proteins bound to the beads were separated by SDS-PAGE and analyzed by Western blotting or Coomassie Blue staining as indicated in the figure legends. For coexpression of MERIT40 with wild-type or deletion mutants of BRE, the corresponding coding sequences were cloned into pETduet-1 vector (Novagen). Bacterial expression and purification were performed according to the manufacturer's instructions (Novagen).
Immunofluorescent staining
Cells grown on coverslips were fixed with 3% paraformaldehyde at room temperature for 20 min and then permeabilized with PBS containing 0.5% Triton X-100 for 5 min at room temperature. The coverslips were blocked with PBS containing 5% goat serum for 30 min at room temperature before incubation with indicated primary antibodies for 1 h at room temperature. After extensive washing at least three times with PBS, cells were incubated with secondary antibodies (FITC-conjugated goat anti-mouse IgG, rhodamine-conjugated goat anti-rabbit IgG, or rhodamine-conjugated goat anti-mouse IgG; Jackson Immuno-Reserach Laboratories, Inc.) for 1 h at room temperature. DAPI (4, 6-diamidino-2-phenylindole) was used to counterstain the nuclei. After a final wash with PBS, coverslips were mounted with glycerin containing p-phenylenediamine. All images were obtained with a Nikon ECLIPSE E800 fluorescence microscope.
G2/M checkpoint assay
HeLa cells in a 35-mm plate were transfected twice with control siRNA or siRNAs specifically targeting RAP80, CCDC98, BRCC45, BRCC36, MERIT40, or BRCA1 at 24-h intervals. Twenty-four hours after the second siRNA transfection, cells were transferred into 100-mm dishes. Cells were incubated for another 24 h before they were mock treated or irradiated using a JL Spepherd 137Cs radiation source at indicated doses. One hour after irradiation, cells were fixed with 70% ethanol for 24 h at −20°C. Cells were then stained with rabbit anti-phospho-histone H3 antibody (pH3), followed by incubation with FITC-conjugated goat anti-rabbit immunoglobulin secondary antibody. The stained cells were treated with RNaseA, incubated with propidium iodide, and then analyzed by flow cytometry to determine the percentages of mitotic cells.
Colony-forming assay
HeLa cells in a 35-mm plate were transfected twice with control siRNA or siRNAs specifically targeting RAP80, CCDC98, BRCC45, BRCC36, MERIT40, or BRCA1, similar to that mentioned above. Twenty-four hours after the second transfection, cells were split and transferred into 60-mm dishes. Cells were incubated for 24 h before they were irradiated using a JL Spepherd 137Cs radiation source at various doses. Eleven days after irradiation, cells were washed with PBS, fixed, and stained with 2% methylene blue, and the numbers of colonies were determined.
Acknowledgments
We thank all colleagues in J.C.'s laboratory, especially Dr. Michael S.Y. Huen, for insightful discussion, technical assistance, and proofreading of this manuscript. We also thank Dr. Roger Greenberg at the University of Pennsylvania for sharing unpublished observations. This work was supported in part by grants from the National Institutes of Health (CA089239 and CA092312 to J.C.). J.C is a recipient of an Era of Hope Scholar award from the Department of Defense and is a member of the Mayo Clinic Breast SPORE program.
Footnotes
References
- Baer R., Ludwig T. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr. Opin. Genet. Dev. 2002;12:86–91. doi: 10.1016/s0959-437x(01)00269-6. [DOI] [PubMed] [Google Scholar]
- Balakirev M.Y., Wilkinson K.D. OTU takes the chains OUT. Nat. Chem. Biol. 2008;4:227–228. doi: 10.1038/nchembio0408-227. [DOI] [PubMed] [Google Scholar]
- Bennett E.J., Harper J.W. DNA damage: Ubiquitin marks the spot. Nat. Struct. Mol. Biol. 2008;15:20–22. doi: 10.1038/nsmb0108-20. [DOI] [PubMed] [Google Scholar]
- Boulton S.J. BRCA1-mediated ubiquitylation. Cell Cycle. 2006;5:1481–1486. doi: 10.4161/cc.5.14.2930. [DOI] [PubMed] [Google Scholar]
- Cantor S.B., Bell D.W., Ganesan S., Kass E.M., Drapkin R., Grossman S., Wahrer D.C., Sgroi D.C., Lane W.S., Haber D.A., et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- Celeste A., Fernandez-Capetillo O., Kruhlak M.J., Pilch D.R., Staudt D.W., Lee A., Bonner R.F., Bonner W.M., Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- Chen X., Arciero C.A., Wang C., Broccoli D., Godwin A.K. BRCC36 is essential for ionizing radiation-induced BRCA1 phosphorylation and nuclear foci formation. Cancer Res. 2006;66:5039–5046. doi: 10.1158/0008-5472.CAN-05-4194. [DOI] [PubMed] [Google Scholar]
- Dong Y., Hakimi M.A., Chen X., Kumaraswamy E., Cooch N.S., Godwin A.K., Shiekhattar R. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol. Cell. 2003;12:1087–1099. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- Ewing R.M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M.D., O'Connor L., Li M., et al. Large-scale mapping of human protein–protein interactions by mass spectrometry. Mol. Syst. Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg R.A. Recognition of DNA double strand breaks by the BRCA1 tumor suppressor network. Chromosoma. 2008;117:305–317. doi: 10.1007/s00412-008-0154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg R.A., Sobhian B., Pathania S., Cantor S.B., Nakatani Y., Livingston D.M. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes & Dev. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume R., Fukuda M., Maeda I., Nishikawa H., Oyake D., Yabuki Y., Ogata H., Ohta T. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro C.A., Weissman A.M. RING finger proteins: Mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- Kim H., Chen J., Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007a;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- Kim H., Huang J., Chen J. CCDC98 is a BRCA1–BRCT domain-binding protein involved in the DNA damage response. Nat. Struct. Mol. Biol. 2007b;14:710–715. doi: 10.1038/nsmb1277. [DOI] [PubMed] [Google Scholar]
- Kolas N.K., Chapman J.R., Nakada S., Ylanko J., Chahwan R., Sweeney F.D., Panier S., Mendez M., Wildenhain J., Thomson T.M., et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Yoo H., Becker F.F., Ali-Osman F., Chan J.Y. Identification of a brain- and reproductive-organs-specific gene responsive to DNA damage and retinoic acid. Biochem. Biophys. Res. Commun. 1995;206:764–774. doi: 10.1006/bbrc.1995.1108. [DOI] [PubMed] [Google Scholar]
- Li S., Chen P.L., Subramanian T., Chinnadurai G., Tomlinson G., Osborne C.K., Sharp Z.D., Lee W.H. Binding of CtIP to the BRCT repeats of BRCA1 involved in the transcription regulation of p21 is disrupted upon DNA damage. J. Biol. Chem. 1999;274:11334–11338. doi: 10.1074/jbc.274.16.11334. [DOI] [PubMed] [Google Scholar]
- Liu Z., Wu J., Yu X. CCDC98 targets BRCA1 to DNA damage sites. Nat. Struct. Mol. Biol. 2007;14:716–720. doi: 10.1038/nsmb1279. [DOI] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Manke I.A., Lowery D.M., Nguyen A., Yaffe M.B. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- Morris J.R., Solomon E. BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum. Mol. Genet. 2004;13:807–817. doi: 10.1093/hmg/ddh095. [DOI] [PubMed] [Google Scholar]
- Mullan P.B., Quinn J.E., Harkin D.P. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854–5863. doi: 10.1038/sj.onc.1209872. [DOI] [PubMed] [Google Scholar]
- Paull T.T., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- Polanowska J., Martin J.S., Garcia-Muse T., Petalcorin M.I., Boulton S.J. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. EMBO J. 2006;25:2178–2188. doi: 10.1038/sj.emboj.7601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Scully R., Chen J., Ochs R.L., Keegan K., Hoekstra M., Feunteun J., Livingston D.M. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- Sobhian B., Shao G., Lilli D.R., Culhane A.C., Moreau L.A., Xia B., Livingston D.M., Greenberg R.A. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman A.R. Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Annu. Rev. Pathol. 2009;4:461–487. doi: 10.1146/annurev.pathol.3.121806.151422. [DOI] [PubMed] [Google Scholar]
- Wang B., Elledge S.J. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Matsuoka S., Ballif B.A., Zhang D., Smogorzewska A., Gygi S.P., Elledge S.J. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz I.E., O'Rourke K.M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D.L., et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Yan J., Jetten A.M. RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites. Cancer Lett. 2008;271:179–190. doi: 10.1016/j.canlet.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Wu L.C., Bowcock A.M., Aronheim A., Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- Yu X., Chini C.C., He M., Mer G., Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- Yu X., Fu S., Lai M., Baer R., Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes & Dev. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G.Y., Sonoda E., Barber L.J., Oka H., Murakawa Y., Yamada K., Ikura T., Wang X., Kobayashi M., Yamamoto K., et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol. Cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]