Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns (original) (raw)
Abstract
CHD7 is a member of the chromodomain helicase DNA binding domain family of ATP-dependent chromatin remodeling enzymes. De novo mutation of the CHD7 gene is a major cause of CHARGE syndrome, a genetic disease characterized by a complex constellation of birth defects (Coloboma of the eye, Heart defects, Atresia of the choanae, severe Retardation of growth and development, Genital abnormalities, and Ear abnormalities). To gain insight into the function of CHD7, we mapped the distribution of the CHD7 protein on chromatin using the approach of chromatin immunoprecipitation on tiled microarrays (ChIP-chip). These studies were performed in human colorectal carcinoma cells, human neuroblastoma cells, and mouse embryonic stem (ES) cells before and after differentiation into neural precursor cells. The results indicate that CHD7 localizes to discrete locations along chromatin that are specific to each cell type, and that the cell-specific binding of CHD7 correlates with a subset of histone H3 methylated at lysine 4 (H3K4me). The CHD7 sites change concomitantly with H3K4me patterns during ES cell differentiation, suggesting that H3K4me is part of the epigenetic signature that defines lineage-specific association of CHD7 with specific sites on chromatin. Furthermore, the CHD7 sites are predominantly located distal to transcription start sites, most often contained within DNase hypersensitive sites, frequently conserved, and near genes expressed at relatively high levels. These features are similar to those of gene enhancer elements, raising the possibility that CHD7 functions in enhancer mediated transcription, and that the congenital anomalies in CHARGE syndrome are due to alterations in transcription of tissue-specific genes normally regulated by CHD7 during development.
CHARGE syndrome is a complex genetic disorder that occurs in approximately one in every 10,000 births worldwide (Kallen et al. 1999). CHARGE is an acronym for Coloboma of the eye, Heart defects, Atresia of the choanae, severe Retardation of growth and development, Genital abnormalities, and Ear abnormalities (Pagon et al. 1981). The clinical presentation of CHARGE syndrome is highly variable, and many additional features have been observed as part of the clinical spectrum. Some of these features include cranial nerve abnormalities, cleft lip and/or palate, hypotonia, kidney abnormalities, and limb or other skeletal abnormalities (Blake et al. 1998).
In 2004, the major gene for CHARGE syndrome was discovered, CHD7 (Vissers et al. 2004). CHD7 encodes Chromodomain Helicase DNA-binding protein 7, and mutations in CHD7 have been found in 58%–71% of affected individuals (Aramaki et al. 2006; Lalani et al. 2006). Although some missense mutations are reported, the majority are nonsense and frameshift mutations that arise de novo. Haploinsufficiency is the most likely mechanism, because most mutations are predicted to lead to premature truncation of the protein. Studies in mice support the haploinsufficiency model. Specifically, mice that are homozygous for null mutations in Chd7 die at E10.5–E11.5 (Bosman et al. 2005; Hurd et al. 2007). Heterozygous Chd7 mice are born with many malformations that resemble those found in humans with CHARGE syndrome, including defects of the inner ear, choanae, heart, eye, genitals, and palate (Bosman et al. 2005). The expression of CHD7 is widespread in early fetal development, with high levels in epithelial cell types, ganglia, and several areas of the brain. CHD7 expression remains ubiquitous in later stages of fetal development, with relatively high levels in organs affected in CHARGE syndrome, including the eye, olfactory epithelium, ear, kidney, and the vascular system (Vissers et al. 2004; Bosman et al. 2005; Lalani et al. 2006; Sanlaville et al. 2006).
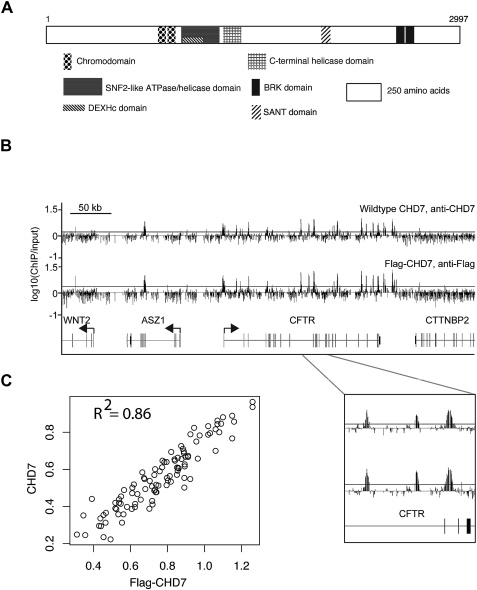
The function of the CHD7 protein is unknown. A role in transcription through ATP-dependent chromatin remodeling is hypothesized based on the presence of several highly conserved domains as well as homology with other proteins within the nine member CHD superfamily. Specifically, CHD7 contains two chromodomains suspected to mediate binding to methylated histones, two helicase domains, a SANT domain that may mediate binding to either DNA or modified histones, and two BRK domains of unknown function (Fig. 1A). In mammalian cells, CHD1, which also contains chromo and helicase domains, was shown to facilitate recruitment of factors involved in both transcription and pre-mRNA splicing (Sims III et al. 2007). In Drosophila, CHD1 is essential for incorporation of histone H3.3 into chromatin, a histone variant that serves as a key substrate for replication-independent chromatin assembly (Konev et al. 2007). CHD3 and CHD4, distinguished by the presence of two plant homeodomains (PHD), are components of the NuRD (Nucleosome Remodeling and histone Deacteylation) complex associated with transcriptional repression (Bowen et al. 2004). CHD8, which contains all of the same domains as CHD7 except for the SANT domain, blocks gene expression through interactions with the insulator binding protein CTCF (Ishihara et al. 2006). CHD8 also interacts with beta-catenin and can negatively regulate expression of beta-catenin target genes (Thompson et al. 2008). CHD7, CHD8, and CHD9 are orthologous to Drosophila kismet, a positive regulator of transcriptional elongation (Daubresse et al. 1999; Srinivasan et al. 2005).
Figure 1.
CHD7 binds to specific sites on chromatin. (A) Structure of CHD7. (B) ChIP-chip analysis of wild-type and Flag-tagged CHD7 in DLD1 cells. CHD7 binding profiles from a 500-kb region on chromosome 7. Median raw ratio data of three biological replicates from the indicated ChIP-Chip experiments are plotted, along with the relative locations of known genes and their transcriptional orientation (arrows). The top 5% is displayed as a horizontal gray line. An expanded view of three positive signals along the CFTR gene is shown below. (C) The maximum signal intensity ratios for each CHD7 occupied site in CHD7 and Flag ChIP-chip experiments are plotted on the _y_- and _x_-axes, respectively (n = 96).
Here we report the first detailed analysis of the distribution of CHD7 protein on chromatin using the technique of chromatin immunoprecipitation on tiled microarrays. Our results indicate that CHD7 is recruited to specific sites on chromatin in a manner that is highly dependent on cell context. The cell-type specific pattern of occupancy of CHD7 is highly correlated with the location of mono- and dimethylated lysine 4 of histone H3, and direct interactions between CHD7 and methylated H3K4 are likely mediated by CHD7 chromodomains. Correlations between the location of the CHD7 binding sites, species conservation, DNase hypersensitivity, gene expression, and known enhancer elements, as well as the results of luciferase reporter assays, suggest a role for CHD7 in enhancer-mediated transcription.
Results
CHD7 is a nuclear protein that localizes to discrete regions on chromatin
CHD7 lacks an apparent DNA binding domain, and there is no evidence that CHD7 binds directly to DNA. However, CHD7 is hypothesized to associate with chromatin, an interaction mediated by either the SANT domain or the tandem chromodomains (Fig. 1A; Boyer et al. 2002; Flanagan et al. 2005). To investigate this hypothesis, we mapped interactions between CHD7 and chromatin using the technique of chromatin immunoprecipitation on microarrays (ChIP-chip) using an antibody to CHD7. The results from human colorectal carcinoma cells (DLD1) using tiled microarrays spanning six ENCODE regions indicate that CHD7 localizes to discrete regions on chromatin. In contrast to many other factors that localize predominantly to transcription start sites (TSSs), including E2F4 (Xu et al. 2007), menin (Scacheri et al. 2006b), and RNA Polymerase II (Guenther et al. 2007), CHD7 is highly enriched both at TSSs and distal regions. To confirm that the CHD7 sites are not due to nonspecific interactions related to antibody specificity, we introduced DNA encoding a triple Flag epitope tag (3 × FLAG) into the CHD7 locus using a rAAV-mediated somatic cell knock-in approach (Zhang et al. 2008), and then performed ChIP-chip analysis of Flag-tagged CHD7 using anti-Flag antibody. ChIP-chip results using antibodies to Flag-CHD7 and wild-type CHD7 ChIP-chip experiments appear strikingly similar for the ENCODE region shown in Figure 1B. To systematically determine the overlap of CHD7 binding sites for all six ENCODE regions, we selected all sites that were identified by ChIP-chip with antibodies to Flag (n = 85) or wild-type CHD7 (n = 84), and plotted the maximum mean signal intensity value for each site in a scatter plot (Fig. 1C). The plot reveals excellent correlation between sites identified using Flag antibodies and those identified using CHD7 antibodies, suggesting that the vast majority of binding sites identified between experiments overlap. These results indicate that the CHD7 antibodies are specific, and that the extensive binding pattern of CHD7 is likely genuine.
CHD7 sites are hypersensitive to DNase I digestion
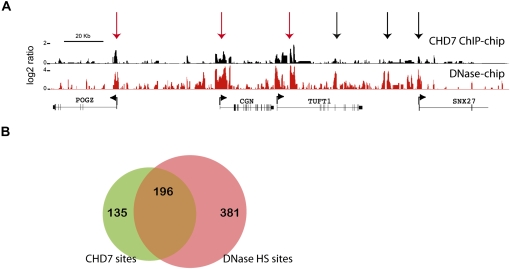
Regulatory elements including promoters, enhancers, silencers, insulators, and locus control regions are contained within regions of open chromatin that are hypersensitive to DNase I digestion (DNase HS) (Keene et al. 1981; McGhee et al. 1981). To test whether CHD7 sites correlate with the location of regulatory elements, we mapped DNase HS sites in DLD1 cells using the method of DNase-chip (Crawford et al. 2006), and compared these sites to CHD7 sites identified by ChIP-chip across all 44 ENCODE regions. Of 331 CHD7 sites detected at high confidence (1 × 10−12), the majority (59%–66%) overlap with a DNase HS site, but not vice versa (Fig. 2B), suggesting that CHD7 binds a subset of regulatory elements. Moreover, the majority (61%) of overlapping CHD7-DNase HS sites were located >2 kb from GENCODE TSSs, which is consistent with the location of regulatory elements that operate at large distances from the basal transcriptional machinery, such as enhancers and insulators.
Figure 2.
CHD7 overlaps with a subset of regions known to harbor regulatory elements. (A) Regulatory elements were mapped using the approach of DNase-chip (red), and compared to CHD7 sites mapped by ChIP-chip (black). Median smoothed raw ratio data are displayed. Note that most of the CHD7 binding sites are contained within DNase I hypersensitive sites (red arrows), but not vice-versa (black arrows). (B) Venn diagram depicting the overlap between high confidence (P ≤ 1 × 10−12) CHD7 (green) and DNase-HS sites (pink) across the ENCODE regions.
Recent studies have shown that a large percentage of distal DNase HS sites that are constitutively or ubiquitously open between different cell types overlap with CTCF (Xi et al. 2007), a protein with known enhancer-blocking insulator activity and reported to physically associate with CHD8 (Ishihara et al. 2006). Of the 331 CHD7 sites identified by ChIP-chip, 99 are located within 2 kb of a TSS and 232 are distal to a TSS. Of the distal CHD7 sites, less than 3% overlap a known ubiquitous DNase HS site predicted to be targeted by CTCF. Moreover, none of the CHD7 sites, proximal or distal, are significantly enriched for a CTCF insulator binding motif (data not shown). These observations suggest that CHD7 probably binds a class of functional elements that differs from those bound by CTCF and CHD8.
Characterization of CHD7 binding sites in multiple cell types
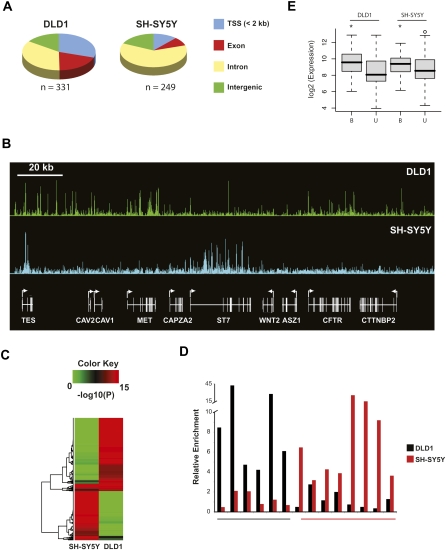
Next, we mapped 249 CHD7 binding sites in SH-SY5Y cells, a neural-like cell line derived from a metastatic neuroblastoma, and with growth and morphological characteristics that are very different from DLD1 cells. Approximately 65% of the CHD7 binding sites in DLD1 and SH-SY5Y cells overlap regions that are conserved between multiple species. We also determined that 70% of CHD7 sites in DLD1 cells and 88% of sites in SH-SY5Y map >2 kb from a GENCODE TSS (Fig. 3A; Harrow et al. 2006; Washietl et al. 2007). Further characterization of the CHD7 sites reveals that many are located within intra- and intergenic regions, with most of the intragenic CHD7 sites located within introns. We also detected striking differences in the pattern of CHD7 occupancy between the two cell types (Fig. 3B). To depict these differences more quantitatively, we selected sites occupied by CHD7 in either DLD1 or SH-SY5Y cells at high confidence (P < 1 × 10−12) across all ENCODE regions, and then plotted the corresponding _P_-values for each binding site as a heat map (Fig. 3C). Aside from a small proportion of CHD7 sites that overlap between the two cell types (∼10%), the heat map shows that most CHD7 sites differ, indicating that the binding specificity of CHD7 is largely dependent on cell lineage. The cell-type specific occupancy of CHD7 was validated by conventional ChIP-PCR for 6/6 loci in DLD1 cells and 7/8 loci in SH-SY5Y cells (Fig. 3D).
Figure 3.
Characterization of CHD7 sites in DLD1 and SH-SY5Y cells. (A) Pie charts showing the distribution of CHD7 sites relative to GENCODE genes. (B) CHD7 ChIP-chip profiles from DLD1 and SH-SY5Y cells along ENCODE region ENm001. Raw ratio data are plotted. (C) Plotted in the heat map are _P_-values corresponding to signal intensities of all CHD7 sites identified by ChIP-chip in DLD1 and SH-SY5Y cells. (D) Confirmatory ChIP-PCR of CHD7 sites unique to DLD1 cells (underlined in black), and 8 CHD7 sites unique to SH-SY5Y cells (underlined in red). (E) Average expression of genes within 10 kb of a CHD7 site (B) and greater than 10 kb from a CHD7 site (U) in DLD1 and SH-SY5Y cells. The asterisk denotes significant differences as determined by Wilcoxon test (DLD1 P = 0.00005, SH-SY5Y P = 0.03).
As an additional means to assess the regulatory potential of CHD7 sites, we compared the location of CHD7 sites to an independent set of _cis_-regulatory modules (predicted regulatory modulators [PReMods]) that were computationally predicted based on clustering of conserved transcription factor binding motifs (Blanchette et al. 2006). Consistent with the CHD7 sites possessing regulatory potential, a significant number of CHD7 distal sites overlapped PReMods in both DLD1 and SH-SY5Y (24.1% and 28.3%, respectively: P < 10−16).
CHD7 binding correlates with transcriptional activity
CHD7 sites are prominently located distal to TSSs, often susceptible to DNase digestion, conserved, and largely cell-type specific. These features are consistent with proteins known to function as gene enhancers. These results prompted us to assess whether CHD7 occupancy correlates with high levels of gene expression. A systematic comparison between ChIP-chip and expression data revealed that the level of expression of genes located within 10 kb of a CHD7 site is significantly higher than nontarget genes in both DLD1 and SH-SY5Y cells (Fig. 3E). These data suggest that CHD7 is generally associated with transcriptionally active genes.
CHD7 binding correlates with the location of methylated lysine 4 of histone H3
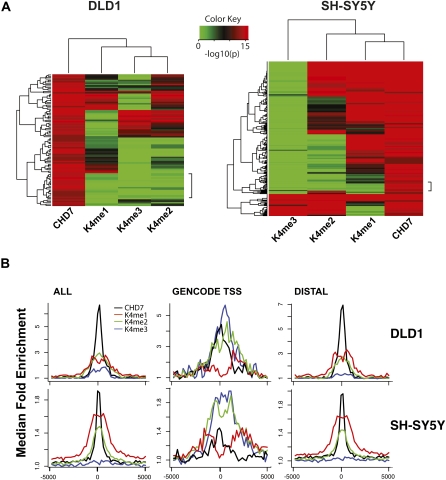
Recent studies have shown that enhancer elements harbor a distinct chromatin “signature” that is primarily defined by the presence of mono-methylated H3K4 at sites distal to TSSs, while 50%–60% of these sites also contain dimethylated H3K4 (Heintzman et al. 2007). Other studies have reported that many enhancers are also associated with trimethylated H3K4 (Barski et al. 2007; Wang et al. 2008). To investigate whether methylated H3K4 could account for the distribution CHD7 on chromatin, we mapped the distribution of mono-, di-, and trimethylated H3K4 (H3K4me1,2,3) in both DLD1 and SH-SY5Y along ENCODE regions. Similar to previously described results, trimethylated H3K4 was detected primarily at the TSS, and mono-methyl H3K4 was detected at distal regions (Schneider et al. 2004; The ENCODE Project Consortium 2007; Heintzman et al. 2007; Mikkelsen et al. 2007). Di-methyl H3K4 was observed at both proximal and distal regions, and almost always overlapped either mono- or trimethyl H3K4. To compare the location of the H3K4 methyl marks to CHD7 sites, we selected high confidence CHD7 sites (P > 1 × 10−12), and plotted the corresponding _P_-values for each K4 methyl site as a heat map (Fig. 4A,B). In both cell types, significant overlap between CHD7 and all three K4 methyl marks was apparent, although the strongest correlations were observed between CHD7, H3K4me1, and H3K4me2. Specifically, at high (P < 1 × 10−12) to medium (P < 1 × 10−8) confidence, 18%–30% of the CHD7 sites in DLD1 cells overlap with H3K4me1, 24%–42% with H3K4me2, and 17%–24% with H3K4me3, and the heat map illustrates instances where the marks are colocalized. In SH-SY5Y cells, 67%–80% of the CHD7 sites are marked with H3K4me1; 43%–57% contain H3K4me2, and 14% contain H3K4me3. Although the heat maps also reveal some examples where CHD7 binds in the absence of H3K4me, these regions harbor low levels of H3K4me1 that are apparent upon visual inspection of the raw data, and this category virtually disappears when the heat maps are plotted at lower significance thresholds (data not shown). Also of interest, although most CHD7 sites overlap with methylated H3K4, the minority of methylated H3K4 sites (25%–27%) contain CHD7 (data not shown).
Figure 4.
Comparison of CHD7 and H3K4 methylation patterns. (A) Heat maps illustrating the overlap of CHD7 and K4 mono-, di-, and trimethylation across 6 ENCODE regions (left) in DLD1 cells and all 44 ENCODE regions in SH-SY5Y cells (right), respectively. The regions denoted by the side brackets harbor low levels of H3K4me1 that are apparent at lower significance thresholds. (B) Aggregate plots of ChIP-chip signals. Raw ChIP-chip data from each cell line were quantile normalized. The median signal intensity of CHD7 peaks (from −5 to +5 kb with respect to the center of the peak) was then calculated and plotted, along with signal intensities for mono-, di-, and trimethyl K4 at the equivalent regions in both DLD1 (top left) and SH-SY5Y cells (bottom left). This was repeated for CHD7 sites located at GENCODE transcription start sites (center) and distal CHD7 sites (right).
The correlations between CHD7 and H3K4 methylation are also evident when ChIP-chip signals from CHD7 and the histone modifications are aggregated and plotted (Fig. 4B). Furthermore, as expected, given the previously reported locations of the K4 methyl marks, the colocalization of CHD7 and H3K4me3 occurs predominantly at TSSs (Fig. 4B, center), whereas the CHD7 and H3K4me1 occurs predominantly at distal sites (Fig. 4B, right). H3K4me2, which is typically found at both TSS and distal sites, correlates with CHD7 at both TSSs and distal sites (The ENCODE Project Consortium 2007; Heintzman et al. 2007). It is also apparent from the aggregate plots that, when the CHD7 signals at TSS and distal sites are separated, the signal intensity of CHD7 at TSSs is lower than at distal CHD7 binding sites. The differences in binding affinity may reflect a scenario whereby CHD7 is binding directly to distal enhancer elements and indirectly looping to TSSs, although further studies are required to test this model.
CHD7 chromodomains interact with all methylated forms of H3K4 in vitro
CHD1 chromodomains have been shown to bind directly and selectively to methylated H3K4 in peptide pulldown assays (Sims III et al. 2005). Although CHD1 chromodomains can interact with all three forms of methylated H3K4, di- and trimethyl K4 bind with threefold higher affinity than monomethyl H3K4. The specificity of CHD1 for methylated H3K4 is determined by the amino acid sequence of the chromodomains, which form cages around the methyl ammonium groups of the methylated lysine residues (Flanagan et al. 2005). The critical amino acid residues in the CHD1 chromodomains important for methyl-lysine binding are conserved in CHD7, i.e., both are aromatic and hydrophobic, and thus, CHD7 would be predicted to bind to methylated H3K4. None of the CHD proteins are thought to directly methylate chromatin at H3K4. Consistent with this notion, knockdown of CHD7 by RNAi in HEK293 cells does not alter bulk levels of mono-, di-, or trimethylated H3K4 (data not shown).
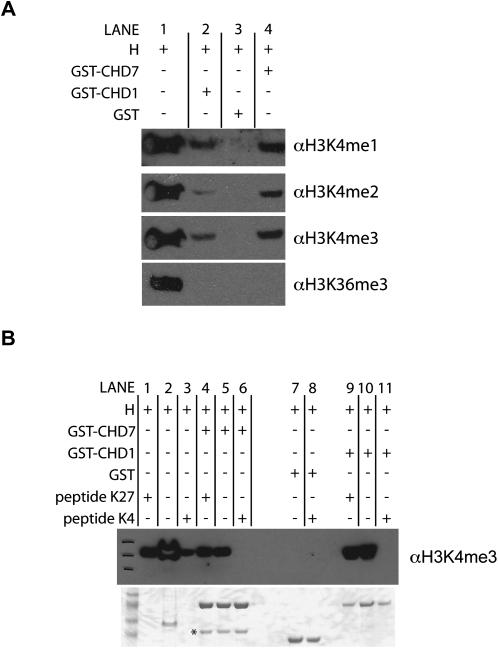
To test if CHD7 can directly bind to methylated histones, we performed in vitro binding assays using purified histones and GST-fusion proteins containing both CHD7 chromodomains. GST-fusion protein containing the CHD1 chromodomains was used as a positive control. Compared to GST protein alone, GST-CHD1 and GST-CHD7 demonstrated physical interactions with mono-, di-, and trimethylated H3K4 (Fig. 5A, cf. lanes 2 and 4 to lane 3). However, interactions with trimethylated H3K36 or H3K27 were not detected (Fig. 5; data not shown). As an additional measure of specificity, the interactions between CHD7 and CHD1 chromodomains to mono-, di-, and trimethyl H3K4 were tested by peptide competition. The interactions were competed away with methylated H3K4 peptides, but not H3K27me3 peptide (Fig. 5B, cf. lanes 6 and 11 to lanes 4 and 9). Together, the results of the in vivo ChIP assays and the in vitro binding assays suggest that CHD7, like CHD1, directly binds to methylated H3K4, and this interaction is mediated by the chromodomains. However, given that only a subset of methylated H3K4 is bound by CHD7, specificity for particular sites on chromatin cannot be determined by binding to methylated H3K4 alone.
Figure 5.
In vitro binding of CHD7 chromodomains to methylated H3K4. (A) Histone pulldown assays were performed using purified GST, GST-CHD7, and GST-CHD1 fusion proteins and purified histones (H) containing all possible covalent modifications. Bound proteins were analyzed by SDS-PAGE and Western blotting with the indicated antibodies. Lane 1 corresponds to input. (B, top) Peptide competition assay in which purified histones (H) were incubated with either H3K4me3 peptide (peptide K4) or H3K27me3 peptide (peptide K27) prior to incubating with indicated GST proteins. Pulldowns were performed with GST beads, and bound proteins were analyzed by SDS-PAGE and Western blotting with H3K4me3 antibody. Similar results were obtained with H3K4me1 and H3K4me2 peptides and antibodies (data not shown). Lanes 1–3 are input. A Coomassie-stained gel showing the purified GST proteins is shown on the bottom. The asterisk denotes a nonspecific band.
Analysis of CHD7 sites in ES cells differentiating along a neural path
Accumulating evidence has established that the patterns of histone modifications change during cell-lineage decisions (Mikkelsen et al. 2007, 2008; Meissner et al. 2008). We reasoned that if H3K4me is part of the epigenetic signature that specifies association of CHD7 with chromatin, then the location of CHD7 sites should change concomitantly with H3K4me patterns during cell differentiation. To test this hypothesis, and to evaluate the genomic occupancy of CHD7 in primary cell types that are not subject to the limitations of transformed cell lines, we differentiated mouse embryonic stem (ES) cells into neural progenitor (NP) cells for CHD7 ChIP-chip analysis. The NP cells were a pure population, as determined by immunopositive nestin staining, and were morphologically similar to those previously described (Conti et al. 2005). In addition, ES and NP cells express high levels of CHD7 as measured by real-time RT-PCR and Western blot (Supplemental Fig. 1A,B).
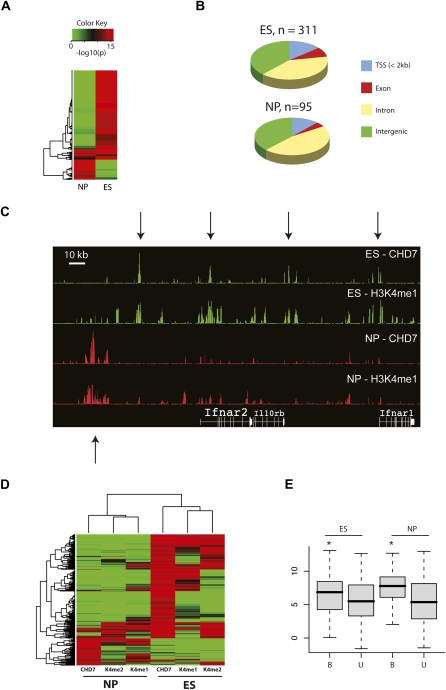
Through ChIP-chip analyses using microarrays containing oligos tiled across the orthologous mouse ENCODE regions, we detected 311 CHD7 sites in ES cells, and 95 CHD7 sites in NP cells. The reason for the difference in the number of CHD7 sites between the two cell types is unknown, but could be due to varying levels of CHD7-ChIP enrichment. Despite these differences, the majority of CHD7 sites differ between ES and NP cells (Fig. 6A,C,D). Specifically, 19 of 95 (20%) CHD7 sites in NP cells were detected in ES cells. Not surprising given that ES and NP cells are of similar lineage, the overlap of CHD7 sites in ES and NP cells is higher than in DLD1 and SH-SY5Y cells (20% versus 10%). Comparing the location of CHD7 sites to mouse RefSeq genes, we find that 14% of CHD7 sites in ES cells map to TSSs, 48% are intragenic, and 38% are intergenic (Fig. 6B). Similar percentages were determined in the NP cells, indicating that despite the cell-type specific differences in CHD7 occupancy, by and large the location of sites relative to known genes is similar before and after differentiation. 54.6% and 87.3% of the CHD7 sites are highly conserved across multiple species in ES and NP cells, respectively (data not shown). Also, a significant number of distal CHD7 sites in both ES and NP cells (17.0% and 31.0%, respectively: P < 10−16) contain independently PreMods identified based on clustering of conserved transcription factor binding motifs (Blanchette et al. 2006). Moreover, as in the DLD1 and SH-SYSY cells, genes located within 10 kb of a CHD7 binding site tend to show higher expression than genes lacking a CHD7 site (Fig. 6E). The findings suggest that the function of CHD7 is not only conserved between mice and humans, but also between the earliest stages of lineage development.
Figure 6.
Characterization of CHD7 sites in ES and NP cells. (A) Heat map of _P_-values corresponding to CHD7 ChIP-chip signal intensities in ES and NP cells. (B) Pie charts showing distribution of CHD7 sites with respect to RefSeq genes in regions of the mouse genome that are orthologous to the human ENCODE regions. (C) CHD7 and H3K4me1 ChIP-chip profiles in ES and NP cells. (D) Heat map of _P_-values corresponding to CHD7, H3K4me1, and H3K4me2 ChIP-chip signals in ES and NP cells. (E) Average expression of genes within 10 kb of a CHD7 site (B) and greater than 10 kb from a CHD7 site (U) in ES and NP cells. The asterisk denotes significant differences as determined by Wilcoxon test (ES P = 0.03, NP P = 0.02).
Next, we compared the location of CHD7 sites to mono-, di-, and trimethyl H3K4 in both ES and NP cells. Trimethylated H3K9, a mark of repressive chromatin, was also investigated in ES cells. In ES cells, 36%–52% of the CHD7 sites are monomethylated at H3K4; 42%–50% are di-methylated at H3K4, and 21%–28% are trimethylated at H3K4 (Supplemental Fig. 2A). In contrast to the significant overlap between CHD7 and all three H3K4 methyl marks, an insignificant proportion of CHD7 sites (<3%, _P_ > 0.5) overlapped trimethylated H3K9. In NP cells, 35%–61% of the CHD7 sites are monomethylated at H3K4; 23%–44% are dimethylated at H3K4, and 12%–16% are trimethylated at H3K4 (Supplemental Fig. 2B). In both ES and NP cells, the percentage of overlap between CHD7 and H3K4me sites increases when the comparisons are performed at slightly lower stringencies (data not shown). Moreover, as in the terminally differentiated cell types, a relatively small fraction (<25%) of the methylated H3K4 sites are bound by CHD7 in the ES and NP cells. Similar to previous studies, we observed that the locations of many mono- and dimethyl K4 marks change upon ES cell differentiation, while the trimethyl K4 mark is relatively static (Mikkelsen et al. 2007). Comparing the location of these marks to CHD7 sites, we find that most of the CHD7 sites change concomitantly with mono- and dimethyl H3K4 patterns before and after differentiation into NP cells (Fig. 6C,D), consistent with the notion that H3K4 methylation patterns partly determine the distribution of CHD7 on chromatin.
CHD7 binds the beta-globin HS2 enhancer element
We next determined whether any of the CHD7 sites overlap known enhancer elements within the ENCODE regions. The locus control/enhancer region (LCR) of the human and mouse beta-globin clusters is defined by five DNase I hypersensitive sites, HS1, HS2, HS3, HS4, and HS5, located far upstream of the multiple genes that they regulate. HS1, HS3, and HS4 are erythroid specific, whereas HS2 and HS5 have been observed in non-erythroid lineages (Tuan et al. 1985; Forrester et al. 1986; Dhar et al. 1990; Jimenez et al. 1992). We detected significant CHD7 ChIP-chip signal directly over the location of the HS2 enhancer in DLD1 cells (Supplemental Fig. 3). As expected, this site was also determined by DNase-ChIP to be hypersensitive to DNase digestion. Moderate CHD7 signals at the orthologous HS2 enhancer in the mouse genome was observed in mouse ES cells, indicating evolutionary conservation of CHD7 binding at this site. CHD7 sites in the beta-globin LCR were undetectable in SH-SY5Y and NP cells. Although additional studies are required to determine the functional relevance of CHD7 at the HS2 site in DLD1 and ES cells, the data provide additional evidence that a subset of the CHD7 sites are indeed enhancer elements.
A subset of CHD7 sites can enhance transcription
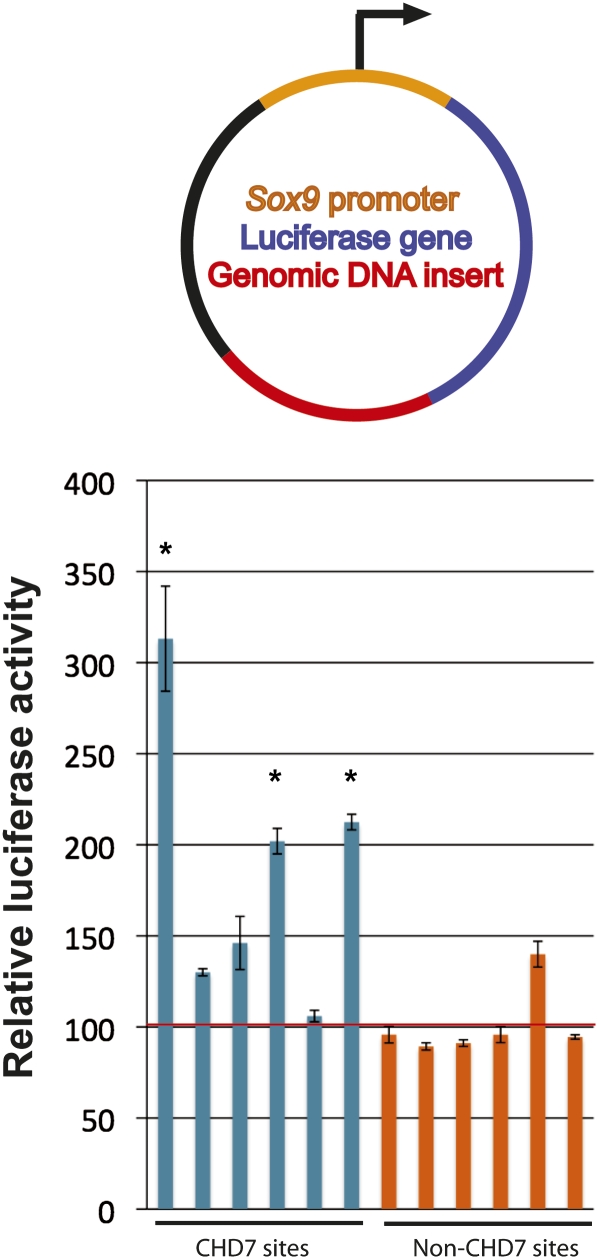
CHD7 sites identified by ChIP-chip in DLD1 cells were assayed for enhancer activity. Specifically, six CHD7 sites were cloned downstream from a luciferase reporter driven by a minimal Sox9 promoter (Fig. 7). These constructs were transfected into DLD1 cells, and luciferase activity was measured. Three of the six constructs containing CHD7 sites showed significant enhancer activity ranging from two- to threefold more than that of constructs containing genomic regions that were devoid of CHD7 binding in DLD1 cells (Fig. 7). The findings are consistent with the notion that a subset of the CHD7 sites function as enhancers, although further studies are necessary to determine whether enhancer activity is dependent on CHD7.
Figure 7.
Analysis of CHD7 sites in reporter assays. (Top) Genomic fragments (red) were cloned downstream from the luciferase gene (blue) driven by the minimal Sox9 promoter (orange). (Bottom) Compared to cells transfected with constructs containing genomic inserts devoid of CHD7 binding in DLD1 cells (non-CHD7 sites, orange bars), 3/6 constructs containing CHD7 sites (blue bars) show significant increases in luciferase activity when transfected into DLD1 cells. The asterisks denote significance as determined by two-tailed _t_-tests. The red horizontal line denotes the average of the non-CHD7 sites.
Discussion
Prior to this study, CHD7 was predicted to interact with chromatin based on homology with other CHD proteins, but it was not known whether specific histone modifications mediate the interactions, and no detailed information was available about the location of CHD7 sites relative to known genes and functional elements. Using a combination of chromatin immunoprecipitation and microarray (ChIP-chip) experiments, we generated high-resolution maps of CHD7 distribution along 30 Mb of the human and mouse genomes in multiple cell types. This is the first ChIP-chip analysis of a member of the CHD protein family in mammalian cells, and the results provide important descriptive insights about the CHD7 protein. First, CHD7 occupancy virtually always coincides with at least one H3K4 methyl mark, a modification that is generally associated with active gene transcription. The correlations between CHD7 and H3K4 methylation are observed in cell types of diverse lineage, and persist as H3K4 methylation patterns change during ES cell differentiation. The correlations occur in both human and mouse cells, indicating that the interactions between CHD7 and H3K4 methylation are functionally conserved between species. The results of histone pulldown assays indicate that the interactions between CHD7 and methylated histones are mediated by tandem chromodomains within CHD7, similarly to CHD1.
Many of the distal CHD7 sites demonstrate several features of enhancers. First, as previously demonstrated for a set of enhancers validated in luciferase reporter assays, the distal CHD7 sites are generally marked with high levels of H3K4me1 and lack H3K4me3 (Heintzman et al. 2007). Additionally, most enhancers (>80%) are located >2.5 kb from a TSS, approximately half are evolutionarily conserved, and a third overlap computationally predicted transcriptional regulatory modules (Heintzman et al. 2007). The CHD7 sites we identified here have nearly identical characteristics. Moreover, consistent with tissue-specific enhancer elements, and in stark contrast to insulator elements, the vast majority of CHD7 sites are cell-type specific and often correlate with expressed genes. More than half of the CHD7 sites overlapped with DNase HS sites, including the well-characterized HS2 enhancer within the LCR of the beta-globin locus (Li et al. 2002). Last, three out of six CHD7 sites act as enhancers when placed downstream from a luciferase reporter. These multiple lines of evidence provide support that the distal CHD7 sites represent a subset of enhancers.
We propose two hypothetical models for CHD7 function. First, similar to other coactivators, like p300/CBP, CHD7 could itself function to increase transcriptional rate by mediating long-range communication between enhancer elements and promoters through mechanisms involving chromosomal looping (Hatzis and Talianidis 2002). On average, CHD7 signals are stronger at distal sites than at TSSs, and the weaker signals could be indicative of indirect, transient interactions occurring as a result of chromatin looping. Rather than directly functioning as enhancer, a second possibility is that CHD7 participates in locally opening chromatin structure to provide access for transcriptional coactivators. Consistent with this model, CHD7 contains two ATP-dependent helicase domains postulated to remodel chromatin, and the majority of CHD7 sites are contained within open regions of chromatin. Our data currently do not distinguish between these models and functional studies are required. Specifically, experiments to test whether expression of CHD7 target genes is influenced by CHD7 levels could shed light on whether CHD7 directly functions as a transcriptional enhancer. Testing the second model could be achieved by determining whether sites that are normally bound by CHD7 are resistant to DNase digestion in CHD7-null cells.
How might mutations in CHD7 lead to the developmental abnormalities in CHARGE syndrome? Given the potential role in enhancer-mediated transcription, one attractive hypothesis is that the various birth defects that arise as a result of CHD7 mutation are due to dysregulated transcription of key target genes that are normally regulated by CHD7 during development. These genes could affect cell proliferation, migration, and/or differentiation. The number of CHD7 sites we detected in 1% of the human and mouse genomes ranged from 95 in neural precursor cells to 311 in colon cancer cells. Extrapolating to the whole genome, the total number of CHD7 sites could be as few as 9000, or as many as 35,000 or more, depending on cell type. Given these estimates and current gene estimates of 20,000–25,000, the total number of genes that may be regulated by CHD7 could be quite large. However, measurement of chromatin binding does not necessarily imply regulation, and target gene activation is likely to be cofactor dependent. Indeed, recent ChIP studies of various transcriptional factors in ES cells showed that activation of genes associated with pluripotency requires colocalization of as many as five transcription factors on selective sites (Chen et al. 2008). Moreover, enhancers are scattered throughout the genome, and gene activation often involves more than one enhancer. Examples include the parentally imprinted insulin growth factor 2 gene and H19, and genes located within the beta globin cluster (Tuan et al. 1985; Dhar et al. 1990; Leighton et al. 1995; Webber et al. 1998). Regardless of whether the total number of CHD7-regulated genes is large or small, given the cell-specific binding of CHD7, our findings suggest that the critical CHD7 targets are likely to be cell-context dependent, and pinpointing these genes could prove to be particularly challenging as multiple, seemingly disparate tissues are affected in CHARGE syndrome.
The CHD7 ChIP-chip data presented here should serve as a public resource for (1) functional analyses of putative target genes, and (2) comparison to other publicly available ENCODE or genome-wide ChIP data sets. The latter could be particularly useful for identifying factors that may colocalize with CHD7 sites, and thus might directly interact with CHD7 or participate in coregulation of target genes. Last, CHD1 is the sole homolog in Saccharomyces cerevisiae, and it is not clear why nine CHD proteins exist in mammalian cells. All nine CHD proteins contain structural motifs that are predicted to bind methylated histones and many are coexpressed in a wide variety of tissues. These features hint at some degree of functional redundancy among CHD family members, yet mounting evidence from the study of CHD1, CHD3, CHD4, and CHD8 suggests that the functions of the CHD proteins are quite diverse (Nishioka et al. 2002; Zegerman et al. 2002; Bowen et al. 2004; Flanagan et al. 2005; Pray-Grant et al. 2005; Sims III et al. 2005; Yuan et al. 2007; Thompson et al. 2008). Our study raises the possibility that functional diversity is dictated by specific histone modifications that determine binding specificity to different classes of functional elements. Comparing the CHD7 ChIP-chip data set with the eight remaining CHD proteins could lend support to this hypothesis, once their interactions with chromatin are delineated. Comparing all nine CHD proteins could also provide important clues to how and why such gene families have evolved.
Methods
Antibodies used for ChIP
Affinity purified rabbit polyclonal CHD7 antibodies (ab31824) were obtained from Abcam. The CHD7 antibody recognizes an epitope at the C terminus of human CHD7. Flag monoclonal antibody (M5 F4042; Sigma-Aldrich) was used for chromatin immunoprecipitation of CHD7 in DLD1 cells expressing 3 × Flag-tagged CHD7. The following Abcam polyclonal antibodies were used in histone modification ChIP reactions: H3K4 monomethyl (ab8895), H3K4 dimethyl (ab7766), H3K4 trimethyl (ab8580), and H3K9 trimethyl (ab8898).
Cell culture
The method used to epitope tag endogenous CHD7 in DLD1 cells is previously described (Zhang et al. 2008). All cell lines were maintained at 37°C with 5% CO2. DLD1 cells were grown in 10% fetal bovine serum (SH30070, HyClone) and 90% McCoy's 5a medium (16600, Invitrogen). SH-SY5Y cells were grown in 10% fetal bovine serum, 90% 1:1 mix of F12 (11765, Invitrogen) and MEM media (11095, Invitrogen) supplemented with L-glutamine (2 mM final). Mouse embryonic cells lines were maintained in Iscove's MDM with 20% FBS, 0.1 mM 2-mercaptoethanol, 0.1 mM MEM nonessential amino acids, ESGRO LIF at 1.0 U/mL, and penicillin/streptomycin (1000 U/mL and 1000 μg/mL, respectively). Neural pan-precursor cells were generated and maintained as previously described (Conti et al. 2005). Briefly, ES cells were grown without serum in neurobasal medium plus B27 plus modified N2 for five days, then replated in euromed media plus N2 with EGF and FGF2 growth factors. Floating cell aggregates were observed within 3–5 d and were harvested and replated in fresh media. Neural pan-precursors forming from cellular aggregates were validated by positive nestin immunoblot and immunohistochemistry. The relative level of CHD7 expression in each of the four cell lines is shown in Supplemental Figure 1A,B.
ChIP-chip experiments
ChIP-chip experiments were performed as previously described (Scacheri et al. 2006b). Briefly, 1–2 × 108 cells were cross-linked with 1% formaldehyde solution for 20 min at room temperature. Cells were harvested and nuclei were extracted, lysed, and sonicated. DNA fragments were then enriched by immunoprecipitation with factor-specific antibodies. Twenty micrograms of CHD7 antibody were used for each CHD7-ChIP reaction. Eight micrograms of antibodies were used for each histone modification. After heat reversal of cross links, ChIP enriched fragments were amplified by ligation-mediated PCR (LM-PCR) and labeled with Cy5-labeled dUTP (Enzo Lifesciences). Genomic DNA not subjected to ChIP was amplified by LM-PCR and labeled with Cy3-labeled dUTP. Labeled samples were cohybridized to either Agilent or NimbleGen arrays according to manufacturer's instructions and scanned using an Agilent Technologies microarray scanner. The Agilent arrays contained ∼44,000 oligonucleotides tiled across the following six ENCODE regions: ENr231, ENr112, ENm001, ENm010, ENr232, and ENm005. NimbleGen arrays contained oligos tiled along all 44 human ENCODE regions. For ChIP-chip studies in ES and NP cells, a “Mouse ENCODE” array containing oligos corresponding to the orthologous ENCODE regions from mouse was utilized. CHD7 and Flag-CHD7 ChIP-chip experiments in DLD1 cells using Agilent arrays containing the six ENCODE regions were each performed in biological triplicate. CHD7 ChIP-chip in DLD1 cells using ENCODE NimbleGen arrays were performed once. CHD7 ChIP-chip in mouse ES cells was performed in biological triplicate. CHD7 ChIP-chip in SH-SY5Y and NP cells were performed in duplicate. ChIP-chip experiments involving each of the various histone marks were performed once. To account for potential artifacts due to variations in copy number, ChIP'd DNA was always cohybridized with total genomic DNA from corresponding cell lines. Raw array data from NimbleGen arrays were normalized using bi-weight mean using the NimbleGen Version 2.3 software. Log 2 ratios corresponding to ChIP DNA/total genomic DNA were calculated. Log 2 ratios from biological replicates were medianed. The software package ACME (Algorithm for Capturing Microarray Enrichment) was used to identify potential sites of enrichment (Scacheri et al. 2006a,b). This program works by sliding a 500 base pair (bp) window stepwise along tiled regions. Hybridization signals of probes within each window were tested by χ2 analysis to determine if the window contained a higher than expected number of probes above the 95th percentile. These calculations result in a single _P_-value associated with each median data point. Signals at a threshold of P ≤ 1 × 10−12 were considered significant at high confidence. Medium confidence signals were considered those in which P ≤ 1 × 10−8. Peaks separated by <1000 bp were consolidated. The P ≤ 1 × 10−12 threshold has been previously determined to accurately predict genuine binding sites for a given factor (Crawford et al. 2006; Scacheri et al. 2006a,b; Zhang et al. 2008). All raw and processed ChIP-chip data are publicly available in GEO. All CHD7 sites detected by ChIP-chip at P ≤ 1 × 10−12 are listed in Supplemental Tables I–IV.
DNase-ChIP
DNase-ChIP was performed as previously described (Crawford et al. 2006). To summarize, nuclei from ∼5 × 107 DLD1 cells were treated with six different DNase I (Roche) concentrations (0–12 units) for 10 min at 37°C. The DNase digestion reaction was stopped by adding 50 mM EDTA and subsequently DNA was embedded in 1% InCert low-melt gel agarose plugs (Bio-Rad). Plugs were incubated overnight at 37°C in LIDS buffer (1% lithium dodecyl sulfate, 10 mM Tris-HCl [pH 7.5], 100 mM EDTA), washed five times in 50 mM EDTA and stored at 4°C. Pulsed field gel electrophoresis (PFGE) was performed to determine the optimal concentrations of DNase I to be used for DNase-ChIP. Based on PFGE results, two DNA plugs from each of the two DNase I concentrations of 4 and 12 units were combined, blunt ended in-gel using T4 DNA polymerase and purified by phenol-chloroform extraction and ethanol precipitation. For the reference sample, genomic DNA from the DLD1 cells was randomly sheared, and vigorously vortexed to generate similar sized fragments as the DNase I-treated samples. Biotinylated primers were ligated to the DNase I-treated and randomly sheared samples and DNA fragments were captured on streptavidin-coated beads. The DNase I-treated and reference samples were further blunt-ended using T4 DNA polymerase, ligated to non-biotinylated linkers, and amplified by ligation-mediated PCR (LM-PCR). Similar to DNA fragments enriched by ChIP, LM-PCR products from DNase treated and randomly sheared samples were fluorescently labeled, purified, and co-hybridized to NimbleGen ENCODE tiling arrays. Analysis of DNase-chip data was performed using ACME as previously described (Scacheri et al. 2006a,b).
Real-time PCR validation
Standard ChIP with CHD7 antibodies was performed on DLD1 cells and SH-SY5Y cells. PCR primers were designed to amplify 175–200 bp fragments from genomic regions determined to be enriched by ChIP-chip at a threshold of P ≤ 1 × 10−10. Eight CHD7 enriched regions specific for DLD1 and SH-SY5Y cells were selected. Three nonenriched regions devoid of CHD7-binding were amplified for comparison. Real-time PCR was performed with ChIP samples and input genomic DNA samples in duplicate using SYBR green PCR mix (Applied Biosystems) in an Applied Biosystems 7300 Real-Time PCR system. To determine the difference between ChIP and input samples, the C t of the ChIP samples was subtracted from the C t of the input samples and 2 was raised to this Δ_C_ t value. Enrichment values were calculated by comparing the relative 2Δ_C_t values between target and nontarget regions. Standard ChIP-PCR of a CHD7 target region in DLD1 cells is shown in Supplemental Figure 1C.
Comparison of ChIP-chip and expression data
Publicly available expression data from DLD1, SH-SY5Y, ES, and NP cell were downloaded from the Gene Expression Omnibus (GEO). The accession numbers are as follows: DLD1: GSM206467_1, GSM206467_2; SHSY5Y: GSM102825, GSM102842, GSM102870; ES: GSM198062, GSM198063, GSM198064; and NP: GSM198065, GSM198066, GSM198067. The DLD1 and SH-SY5Y data sets were generated using Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays (Peddada et al. 2006; Wagner et al. 2007). ES and NP data sets were generated using Affymetrix GeneChip Mouse Genome 430 2.0 Array (Mikkelsen et al. 2007). Data from the DLD1 and SH-SY5Y cells were normalized to one another using dChip software (Li and Wong 2001). Replicate data sets were averaged and the averaged raw intensity values were converted to log base 2. The expression values corresponding to genes located within the human and mouse ENCODE regions were then selected. Genes with TSSs located within 10 kb of a CHD7 binding site were considered “bound” and merged to expression data sets by gene symbol. The expression of “unbound” genes, defined as those contained within the ENCODE regions but located >10 kb from a CHD7 site, was then determined and compared to the bound genes. For genes represented by multiple probe sets, the highest value was retained in the analysis. Wilcoxon tests were used for statistical comparison of bound and unbound genes.
Comparison of CHD7 peaks to the annotated genome, evolutionary constrained sequences, and predicted regulatory modules
Using the Table Browser function in the UCSC Genome Browser, CHD7 peaks identified by ChIP-chip analyses in DLD1 and SH-SY5Y cells were compared to GENCODE genes based on the HG17 genome assemblies (Washietl et al. 2007). For comparisons between CHD7 sites and conserved sequences, the CHD7 sites were compared to consensus element tracks at both moderate and strict stringencies. The consensus elements in this track were generated by the ENCODE Multi-Species Analysis group from the nine different combinations of three conservation algorithms (phastCons, binCons, and GERP) and three sequence alignment methods (TBA, MLAGAN, and MAVID) applied to the ENCODE region sequences of 28 vertebrate species as defined in the September 2005 ENCODE MSA (Multi-Species Sequence Analysis) sequence freeze and the MSA species guide tree. CHD7 peaks identified in ES and NP cells were compared to RefSeq gene annotations based on the Mm7 genome assembly. CHD7 sites were compared to the Most Conserved (phastConsElements) track in the UCSC Genome Browser. This track is a measure of evolutionary conservation in 17 vertebrates, including mammalian, amphibian, bird, and fish species, based on a phylogenetic hidden Markov model. For comparison of CHD7 binding sites to computationally PReMods, data were downloaded from http://genomequebec.mcgill.ca/PReMod/ and loaded onto the UCSC browser. Modules that overlapped a CHD7 site were then determined using the Tables Browser function. Significance was determined using a binomial test comparing the overlap between CHD7 sites and PReMods to that of a randomly generated set of 311 chromosome coordinates, or “random peaks” located within the ENCODE regions. Randomly selected peaks were size matched to CHD7 peaks identified in DLD1 cells by ChIP-chip.
Histone pulldown assays
The histone pulldown assays are previously described (Tie et al. 2003). Constructs were made containing GST alone, GST, and CHD7 chromodomains, and GST and CHD1 chromodomains. Constructs were separately expressed in Escherichia coli, and fusion proteins were purified on Glutathione Sepharose Transferase beads. Beads coupled to purified fusion proteins were then incubated with acid-extracted histones from DLD1 cells at 4°C overnight in buffer containing 15 mM HEPES, pH 7.5, 0.4 M NaCl, 0.11 M KCl, 5% glycerol, and 0.5% NP 40. Complexes were washed 5 times in the same buffer, and 1 time in buffer containing 15 mM HEPES, pH 7.5, 5% glycerol, and 0.5% NP 40. The complexes were then separated by SDS-PAGE and Western blots were performed with Abcam antibodies directed against H3K4me1 (ab8895), H3K4me2 (ab7766), H3K4me3 (ab8580), and H3K36me3 (ab9050). H3K27me3 antibodies were purchased from Upstate/Millipore (07449). Peptide competition assays were performed similarly, except that 1 μg of H3K4me1, H4K4me2, H3K4me3, or H3K27me3 biotinylated peptides (Upstate) were incubated with histones prior to incubation with GST proteins.
Luciferase assays
Six CHD7 binding sites and six sites absent for CHD7 binding (850–2400 bp) were amplified from genomic DNA and cloned into Invitrogen's TA cloning vector, pCR2.1. Inserts were partially sequenced to confirm identity, then were moved into the BamHI site, downstream from the firefly luciferase gene, of pGL4.10[luc2] (Promega) that had been modified to contain a Sox9 promoter (∼530 bp) at the HindIII site. DLD1 cells were transfected with reporter constructs using Lipofectamine 2000 (Invitrogen). The Renilla luciferase plasmid pGL4.74-hRluc (Promega) was cotransfected as an internal control. Cells were harvested ∼40 h after transfection, and the relative luciferase activities of cell lysates were measured using the Dual-luciferase reporter assay system (Promega). All assays were performed in triplicate. The chromosome coordinates of inserts in the order shown in Figure 7 are as follows: chr5:55968880-55970127, chr6:132257491-132258201, chr6:41606801-41608296, chr8:118928834-118929701, chr11:5598396-5599386, chr18:23755258-23756628, chr4:118655000-118655936, chr6:108456927-108458572, chr7:125932546-125934653, chr8:119018143-119019789, chr14:53108186-53109124, chr16:61300365-61302500.
Acknowledgments
We thank Peter J. Harte for helpful discussions and critically reading this manuscript, and Feng Tie for technical assistance. This work was supported by grants from the National Institute of Child Health and Development (R01HD056369) and the National Cancer Institute (KCA103843) to P.C.S., as well as the National Human Genome Research Institute (R01HG004722) to both P.C.S and Z.W. M.P.S. is supported in part by a training grant from the National Institute of Child Health and Development (T32 HD007104).
Footnotes
[Supplemental material is available online at www.genome.org. ChIP-chip array data from this study have been submitted to GEO under accession no. GSE14460.]
References
- Aramaki M., Udaka T., Kosaki R., Makita Y., OkamAto N., Yoshihashi H., Oki H., Nanao K., Moriyama N., Oku S., et al. Phenotypic spectrum of CHARGE syndrome with CHD7 mutations. J. Pediatr. 2006;148:410–414. doi: 10.1016/j.jpeds.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Blake K.D., Davenport S.L., Hall B.D., Hefner M.A., Pagon R.A., Williams M.S., Lin A.E., Graham J.M., Jr CHARGE association: An update and review for the primary pediatrician. Clin. Pediatr. (Phila.) 1998;37:159–173. doi: 10.1177/000992289803700302. [DOI] [PubMed] [Google Scholar]
- Blanchette M., Bataille A.R., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosman E.A., Penn A.C., Ambrose J.C., Kettleborough R., Stemple D.L., Steel K.P. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum. Mol. Genet. 2005;14:3463–3476. doi: 10.1093/hmg/ddi375. [DOI] [PubMed] [Google Scholar]
- Bowen N.J., Fujita N., Kajita M., Wade P.A. Mi-2/NuRD: Multiple complexes for many purposes. Biochim. Biophys. Acta. 2004;1677:52–57. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Langer M.R., Crowley K.A., Tan S., Denu J.M., Peterson C.L. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell. 2002;10:935–942. doi: 10.1016/s1097-2765(02)00634-2. [DOI] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J., et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Conti L., Pollard S.M., Gorba T., Reitano E., Toselli M., Biella G., Sun Y., Sanzone S., Ying Q.L., Cattaneo E., et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford G.E., Davis S., Scacheri P.C., Renaud G., Halawi M.J., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S. DNase-chip: A high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods. 2006;3:503–509. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubresse G., Deuring R., Moore L., Papoulas O., Zakrajsek I., Waldrip W.R., Scott M.P., Kennison J.A., Tamkun J.W. The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development. 1999;126:1175–1187. doi: 10.1242/dev.126.6.1175. [DOI] [PubMed] [Google Scholar]
- Dhar V., Nandi A., Schildkraut C.L., Skoultchi A.I. Erythroid-specific nuclease-hypersensitive sites flanking the human β-globin domain. Mol. Cell. Biol. 1990;10:4324–4333. doi: 10.1128/mcb.10.8.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan J.F., Mi L.Z., Chruszcz M., Cymborowski M., Clines K.L., Kim Y., Minor W., Rastinejad F., Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- Forrester W.C., Thompson C., Elder J.T., Groudine M. A developmentally stable chromatin structure in the human beta-globin gene cluster. Proc. Natl. Acad. Sci. 1986;83:1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., et al. GENCODE: Producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl)(S4):1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzis P., Talianidis I. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell. 2002;10:1467–1477. doi: 10.1016/s1097-2765(02)00786-4. [DOI] [PubMed] [Google Scholar]
- Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hurd E.A., Capers P.L., Blauwkamp M.N., Adams M.E., Raphael Y., Poucher H.K., Martin D.M. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm. Genome. 2007;18:94–104. doi: 10.1007/s00335-006-0107-6. [DOI] [PubMed] [Google Scholar]
- Ishihara K., Oshimura M., Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol. Cell. 2006;23:733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Jimenez G., Griffiths S.D., Ford A.M., Greaves M.F., Enver T. Activation of the beta-globin locus control region precedes commitment to the erythroid lineage. Proc. Natl. Acad. Sci. 1992;89:10618–10622. doi: 10.1073/pnas.89.22.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen K., Robert E., Mastroiacovo P., Castilla E.E., Kallen B. CHARGE association in newborns: A registry-based study. Teratology. 1999;60:334–343. doi: 10.1002/(SICI)1096-9926(199912)60:6<334::AID-TERA5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Keene M.A., Corces V., Lowenhaupt K., Elgin S.C. DNase I hypersensitive sites in Drosophila chromatin occur at the 5′ ends of regions of transcription. Proc. Natl. Acad. Sci. 1981;78:143–146. doi: 10.1073/pnas.78.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konev A.Y., Tribus M., Park S.Y., Podhraski V., Lim C.Y., Emelyanov A.V., Vershilova E., Pirrotta V., Kadonaga J.T., Lusser A., et al. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317:1087–1090. doi: 10.1126/science.1145339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani S.R., Safiullah A.M., Fernbach S.D., Harutyunyan K.G., Thaller C., Peterson L.E., McPherson J.D., Gibbs R.A., White L.D., Hefner M., et al. Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype–phenotype correlation. Am. J. Hum. Genet. 2006;78:303–314. doi: 10.1086/500273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton P.A., Saam J.R., Ingram R.S., Stewart C.L., Tilghman S.M. An enhancer deletion affects both H19 and Igf2 expression. Genes & Dev. 1995;9:2079–2089. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- Li C., Wong W.H. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Peterson K.R., Fang X., Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J.D., Wood W.I., Dolan M., Engel J.D., Felsenfeld G. A 200 base pair region at the 5′ end of the chicken adult β-globin gene is accessible to nuclease digestion. Cell. 1981;27:45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B., et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B.E., Jaenisch R., Lander E.S., Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:794. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K., Chuikov S., Sarma K., Erdjument-Bromage H., Allis C.D., Tempst P., Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes & Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagon R.A., Graham J.M., Jr, Zonana J., Yong S.L. Coloboma, congenital heart disease, and choanal atresia with multiple anomalies: CHARGE association. J. Pediatr. 1981;99:223–227. doi: 10.1016/s0022-3476(81)80454-4. [DOI] [PubMed] [Google Scholar]
- Peddada S., Yasui D.H., LaSalle J.M. Inhibitors of differentiation (ID1, ID2, ID3 and ID4) genes are neuronal targets of MeCP2 that are elevated in Rett syndrome. Hum. Mol. Genet. 2006;15:2003–2014. doi: 10.1093/hmg/ddl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant M.G., Daniel J.A., Schieltz D., Yates J.R., III, Grant P.A. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Sanlaville D., Etchevers H.C., Gonzales M., Martinovic J., Clement-Ziza M., Delezoide A.L., Aubry M.C., Pelet A., Chemouny S., Cruaud C., et al. Phenotypic spectrum of CHARGE syndrome in fetuses with CHD7 truncating mutations correlates with expression during human development. J. Med. Genet. 2006;43:211–317. doi: 10.1136/jmg.2005.036160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacheri P.C., Crawford G.E., Davis S. [14] Statistics for ChIP-chip and DNase hypersensitivity experiments on NimbleGen arrays. Methods Enzymol. 2006a;411:270–282. doi: 10.1016/S0076-6879(06)11014-9. [DOI] [PubMed] [Google Scholar]
- Scacheri P.C., Davis S., Odom D.T., Crawford G.E., Perkins S., Halawi M.J., Agarwal S.K., Marx S.J., Spiegel A.M., Meltzer P.S., et al. Genome-wide analysis of menin binding provides insights into MEN1 tumorigenesis. PLoS Genet. 2006b;2:e51. doi: 10.1371/journal.pgen.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R., Bannister A.J., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Sims R.J., III, Chen C.F., Santos-Rosa H., Kouzarides T., Patel S.S., Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R.J., III, Millhouse S., Chen C.F., Lewis B.A., Erdjument-Bromage H., Tempst P., Manley J.L., Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription post-initiation factors and pre-mRNA splicing. Mol. Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Armstrong J.A., Deuring R., Dahlsveen I.K., McNeill H., Tamkun J.W. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA polymerase II. Development. 2005;132:1623–1635. doi: 10.1242/dev.01713. [DOI] [PubMed] [Google Scholar]
- Thompson B.A., Tremblay V., Lin G., Bochar D.A. CHD8 is an ATP-dependent chromatin remodeling factor that regulates β-catenin target genes. Mol. Cell. Biol. 2008;28:3894–3904. doi: 10.1128/MCB.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F., Prasad-Sinha J., Birve A., Rasmuson-Lestander A., Harte P.J. A 1-megadalton ESC/E(Z) complex from Drosophila that contains polycomblike and RPD3. Mol. Cell. Biol. 2003;23:3352–3362. doi: 10.1128/MCB.23.9.3352-3362.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan D., Solomon W., Li Q., London I.M. The “beta-like-globin” gene domain in human erythroid cells. Proc. Natl. Acad. Sci. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers L.E., van Ravenswaaij C.M., Admiraal R., Hurst J.A., de Vries B.B., Janssen I.M., van der Vliet W.A., Huys E.H., de Jong P.J., Hamel B.C., et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat. Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- Wagner K.W., Punnoose E.A., Januario T., Lawrence D.A., Pitti R.M., Lancaster K., Lee D., von Goetz M., Yee S.F., Totpal K., et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat. Med. 2007;13:1070–1077. doi: 10.1038/nm1627. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zang C., Rosenfeld J.A., Schones D.E., Barski A., Cuddapah S., Cui K., Roh T.Y., Peng W., Zhang M.Q., et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S., Pedersen J.S., Korbel J.O., Stocsits C., Gruber A.R., Hackermuller J., Hertel J., Lindemeyer M., Reiche K., Tanzer A., et al. Structured RNAs in the ENCODE selected regions of the human genome. Genome Res. 2007;17:852–864. doi: 10.1101/gr.5650707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber A.L., Ingram R.S., Levorse J.M., Tilghman S.M. Location of enhancers is essential for the imprinting of H19 and Igf2 genes. Nature. 1998;391:711–715. doi: 10.1038/35655. [DOI] [PubMed] [Google Scholar]
- Xi H., Shulha H.P., Lin J.M., Vales T.R., Fu Y., Bodine D.M., McKay R.D., Chenoweth J.G., Tesar P.J., Furey T.S., et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Bieda M., Jin V.X., Rabinovich A., Oberley M.J., Green R., Farnham P.J. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17:1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C.C., Zhao X., Florens L., Swanson S.K., Washburn M.P., Hernandez N. CHD8 associates with human Staf and contributes to efficient U6 RNA polymerase III transcription. Mol. Cell. Biol. 2007;27:8729–8738. doi: 10.1128/MCB.00846-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P., Canas B., Pappin D., Kouzarides T. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J. Biol. Chem. 2002;277:11621–11624. doi: 10.1074/jbc.C200045200. [DOI] [PubMed] [Google Scholar]
- Zhang X., Guo C., Chen Y., Shulha H.P., Schnetz M.P., LaFramboise T., Bartels C.F., Markowitz S., Weng Z., Scacheri P.C., et al. Epitope tagging of endogenous proteins for genome-wide ChIP-chip studies. Nat. Methods. 2008;5:163–165. doi: 10.1038/nmeth1170. [DOI] [PMC free article] [PubMed] [Google Scholar]