Mouse models of human AML accurately predict chemotherapy response (original) (raw)
Abstract
The genetic heterogeneity of cancer influences the trajectory of tumor progression and may underlie clinical variation in therapy response. To model such heterogeneity, we produced genetically and pathologically accurate mouse models of common forms of human acute myeloid leukemia (AML) and developed methods to mimic standard induction chemotherapy and efficiently monitor therapy response. We see that murine AMLs harboring two common human AML genotypes show remarkably diverse responses to conventional therapy that mirror clinical experience. Specifically, murine leukemias expressing the AML1/ETO fusion oncoprotein, associated with a favorable prognosis in patients, show a dramatic response to induction chemotherapy owing to robust activation of the p53 tumor suppressor network. Conversely, murine leukemias expressing MLL fusion proteins, associated with a dismal prognosis in patients, are drug-resistant due to an attenuated p53 response. Our studies highlight the importance of genetic information in guiding the treatment of human AML, functionally establish the p53 network as a central determinant of chemotherapy response in AML, and demonstrate that genetically engineered mouse models of human cancer can accurately predict therapy response in patients.
Keywords: AML, AML1/ETO, MLL, chemotherapy, mouse models
Cancer arises as a consequence of accumulating genetic lesions in proto-oncogenes and tumor suppressor genes, which impact primary pathology and trajectory of individual tumors and ultimately dictate their response to therapy. Cancer genomes are complex and heterogeneous, and it is this genetic heterogeneity that underlies the variable responses of individual patients to therapy, even within the same pathological cancer type. It follows that a better understanding of these genotype–response relationships will enable a more effective use of established regimens and guide the development and evaluation of new therapeutic agents.
Acute myeloid leukemia (AML) is a malignant disorder of progenitor cells in myeloid hematopoiesis and exemplifies a genetically heterogeneous cancer. To date, >100 genetic aberrations have been implicated in this disease. The emergence of AML is thought to require cooperation between proproliferative mutations typically involving Ras signaling pathways and defects in myeloid differentiation, which often result from chromosomal translocations (Gilliland et al. 2004). Numerous clinical studies have assessed frequency and co-occurrence as well as pathological and clinical associations of common mutations in AML (for review see, Estey and Dohner 2006). While most of these data sets are limited to selected candidate genes, recent advances in genome analysis now allow for a comprehensive survey of leukemia oncogenomes (Mullighan et al. 2007; Ley et al. 2008).
Patients with AML show a heterogeneous response to therapy. The current “standard of care” for AML patients involves an initial phase of intense chemotherapy (induction) followed by post-remission treatment including additional chemotherapy cycles and/or allogeneic stem cell transplantation. The combination of cytarabine (cytosine arabinoside, Ara-C) with an anthracycline is the backbone of induction therapy; nevertheless, treatment responses and outcome to this regimen vary from primary refractory disease to cure (Estey and Dohner 2006). In addition to patient age, disease etiology, and initial response to therapy, certain mutations have been associated with favorable, average, or adverse treatment outcome (Grimwade et al. 1998; Byrd et al. 2002; Schlenk et al. 2008). For example, deletions of chromosomes 5q and 7q and certain 11q23 rearrangements resulting in MLL fusion proteins are associated with adverse therapy response (for review, see Estey and Dohner 2006). On the contrary, translocations t(8;21) and t(16;16)/inv(16), resulting in expression of the anti-differentiation fusion proteins AML1/ETO and CBFβ/MYH11, respectively, are associated with a favorable outcome (Grimwade et al. 1998). Despite these associations, the use of genotypic information to guide treatment decisions has only recently entered into clinical practice, and not in any standardized way.
A better understanding of how cancer genotypes influence treatment outcome in AML and other cancers will undoubtedly improve the use of existing drugs and facilitate the development of more effective therapies. Genetic analysis of human AML has provided valuable correlative data, yet the complexity of AML genomes complicates its use in predicting treatment response (Dohner 2007). In contrast, functional studies enable a cause-and-effect relationship between particular genes and treatment response, but can be confounded by the lack of predictive experimental systems. Human-derived cancer cell lines, while useful in high-throughput drug screening, are often poorly defined at the genotypic level and fail to engage the microenvironment, which has decisive roles in both tumorigenesis (Wei et al. 2008) and therapy response (Schmitt et al. 2000). As a consequence, it has been debated whether such systems are accurate predictors of drug action in patients. On the other hand, genetically engineered mouse models enable the study of drug action in a physiological environment and thereby provide a means to dissect the relationship between complex disease genetics and therapy response in a way that is not possible in humans. However, potential differences in drug behavior, as well as the technical challenges associated with animal studies, have made it difficult to determine whether these models accurately mirror treatment behavior in the clinic.
In this study, we use AML as a model cancer type to study the genotype–response relationship of genetically defined murine cancers to frontline chemotherapy. We exploited information on genetic lesions occurring together in human leukemias to generate new models of common AML genotypes, established an induction chemotherapy protocol for mice, and integrated sensitive imaging methods to monitor leukemia response in vivo. We then used these tools to characterize genotype–response relationships in AML and to explore the underlying molecular basis for these effects. Our studies provide insights into the differential response patterns of human leukemia, suggest strategies for improving the use of conventional chemotherapy, and produce tractable preclinical systems for testing new therapeutic approaches.
Results
Oncogenic Ras cooperates with anti-differentiation fusion proteins to produce AML
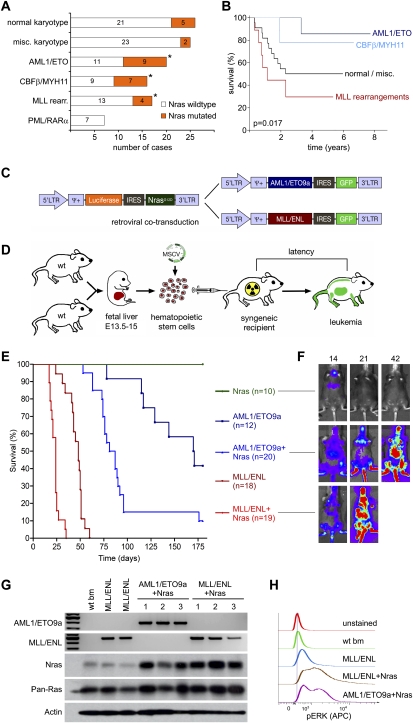
To generate mouse models that reflect common genetic themes in human AML, we examined a new data set of 111 pediatric AML cases that integrates conventional karyotyping, genome-wide SNP array analysis, and detailed sequencing data for 21 oncogenes and tumor suppressors (I. Radtke and J.R. Downing, unpubl.). Most cases harbored, on average, mutations in two known oncogenes or tumor suppressor genes, which occurred in various combinations. Among the most common karyotype aberrations were translocations producing AML1/ETO (n = 20), CBFB/MYH11 (n = 16), and MLL rearrangements (n = 17). Remarkably, all three translocations were most commonly associated with activating mutations in NRAS that were detected in 45%, 44%, and 24% of the cases, respectively (Fig. 1A). Consistent with previous reports on adult AML, patients harboring leukemias containing AML1/ETO or CBFB/MYH11 fusion genes showed an improved response to therapy and better clinical course than those harboring MLL fusions (Fig. 1B). Based on these findings, together with studies indicating that anti-differentiation and proproliferative genes cooperate to promote AML in mice (Kelly et al. 2002; Ono et al. 2005), we reasoned that mouse leukemias coexpressing either AML1/ETO or MLL/ENL together with oncogenic Nras would provide accurate yet distinct models of human AML associated with dramatically different clinical outcomes.
Figure 1.
Generation of genetically defined mosaic mouse models based on common genetic associations in human AML. (A) Frequency of Nras mutations in 111 cases of pediatric AML in major karyotype groups. Asterisks indicate significant association in multifactor dimensionality reduction analysis (Ritchie et al. 2001). (B) Kaplan-Meier plot showing the overall survival of pediatric AML patients treated after 1998 depending on major karyotype group, including t(8;21) = AML1/ETO (n = 10), inv(16)/t(16;16) = CBFB/MYH11 (n = 9), and 11q23/MLL rearrangements (n = 9) compared with other subtypes (n = 22, excluding patients with PML/RARα positive AML). The presence of AML1/ETO and MLL fusion proteins has opposite effects on long-term therapy outcome. In an independent analysis using the same data set, Nras mutations were associated with a slightly better 5-yr survival with low-statistic significance (5-yr survival 79.4 vs. 50.4%, P = 0.09). (C) MSCV-based retroviral constructs used to coexpress AML oncogenes with fluorescent and bioluminescent markers. (D) Schematic overview of mosaic AML mouse models. Wild-type C57BL/6 FLCs isolated at E13.5–E15 were (co)transduced with oncogenic retroviruses and used to reconstitute the hematopoietic system of lethally irradiated recipient mice. (E) Mice reconstituted with FLCs transduced with the indicated transgenes were monitored for illness for 180 d and died or were euthanized at a terminal disease stage. The data are presented in a Kaplan–Meier format showing the percentage of mouse survival at various time points post-transplantation. Five recipients of FLCs transduced with Nras only were followed up further; three succumbed to leukemias after 212, 287, and 292 d, likely after accumulating additional lesions. (F) Luciferase imaging of recipient mice of FLCs transduced with the indicated genes at 14, 21, and 42 d following transplantation. Transduction of Luciferase-IRES-Nras rapidly induces onset of Luciferase-positive disease only in concert with AML1/ETO9a or MLL/ENL. (G) Expression analysis of retroviral oncogenes in wild-type bone marrow (wt bm) and independent primary AMLs (1–3) with indicated genotypes. Expression of human AML1/ETO9a and MLL/ENL transcripts was verified by RT–PCR using fusion site-specific primers. Reverse transcriptase free control reactions were negative in all samples (data not shown). Western blot analysis using pan-Ras and Nras-specific antibodies demonstrating Nras overexpression in leukemia lysates derived from Nras-cotransduced FLCs. Overall Ras levels (pan-Ras) do not show significant elevation. (H) Baseline phospho-Erk levels are strongly elevated in leukemias deriving from Nras-cotransduced FLCs. Levels of phosphorylated Erk were measured using phospho-specific flow cytometry in wild-type whole bone marrow and GFP-positive MLL/ENL, MLL/ENL + Nras, and AML1/ETO9a + Nras leukemias. Representative histograms are shown.
To rapidly generate mice harboring leukemias with various genetic alterations, we applied a “mosaic” approach involving retroviral transduction of oncogenes into hematopoietic stem and progenitor cells, followed by retransplantation of the genetically altered cells into syngeneic recipients (Fig. 1D). This strategy enables multiple oncogenes and reporter elements to be introduced in a one-step procedure, thereby facilitating disease monitoring and reducing the animal husbandry associated with germline transgenic mice. To model translocation (8;21), we choose to express the AML1/ETO9a splice variant that is expressed in the majority of t(8;21) patients and has increased leukemogenic potential in mice (Yan et al. 2006; Lafiura et al. 2008). To model MLL translocations, we used the MLL/ENL variant that is among the most common and associated with a particularly poor prognosis (Schoch et al. 2003). Both fusion genes were coexpressed with enhanced green fluorescent protein (EGFP) in bicistronic constructs (Fig. 1C). Oncogenic NrasG12D was coexpressed with luciferase to enable imaging of the resulting leukemias by bioluminescence. Of note, NrasG12D was cloned downstream from an internal ribosomal entry site (IRES), which reduces expression of the ectopic Nras gene to near physiological levels (Parikh et al. 2006).
Each oncogene was introduced alone or in combination into fetal liver cells (FLCs) derived from embryonic day 13.5–15 (E13.5–E15) embryos, and the infected populations—which are highly enriched for hematopoietic stem and progenitor cells (Morrison et al. 1995)—were used to reconstitute the hematopoietic compartment of lethally irradiated syngeneic recipient mice (Fig. 1D). Recipient animals were then monitored for disease onset by analysis of peripheral blood and bioluminescence for the transduced luciferase reporter. As expected, mice receiving FLCs expressing AML1/ETO9a or MLL/ENL alone eventually developed GFP-positive leukocytosis consistent with peripheral leukemia. The survival of these animals was similar to previous reports, with AML1/ETO9a inducing leukemia with a longer latency compared with MLL/ENL (170 vs. 48 d median survival, P < 0.0001).
Although NrasG12D was not oncogenic on its own, it cooperated with both AML1/ETO9a and MLL/ENL to accelerate leukemia onset and reduce overall survival (170 d to 84 d for AML1/ETO9a, P = 0.0026; 48 d to 24 d for MLL/ENL, P < 0.0001) (Fig. 1E). Of note, leukemias arising from FLCs transduced with both retroviral vectors were invariably positive for both luciferase (Fig. 1F) and GFP (data not shown), suggesting a strong selective advantage for cells harboring a fusion protein together with Nras. Indeed, the resulting leukemias expressed the expected transgenes (Fig. 1G), and those harboring activated Nras displayed hyperactive mitogen-activated protein kinase (Mapk) signaling as assessed by flow cytometry for phospho-Erk (a downstream Ras effector in the Mapk cascade) (Fig. 1H). Thus, lesions that occur together in human AML cooperate to rapidly generate leukemia in mice.
Murine leukemias display pathological features of human AML
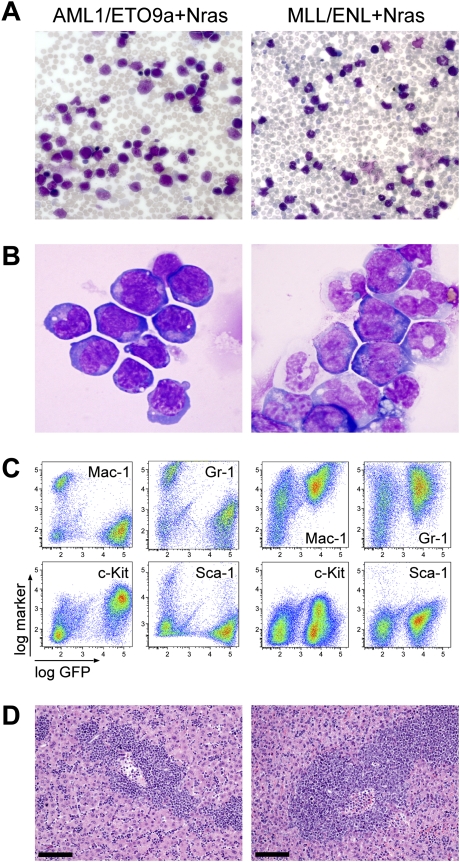
In human patients, AML is characterized by the appearance of malignant myeloid progenitors, which rapidly accumulate in bone marrow and aggressively infiltrate extramedullary tissues. Mice harboring leukemias coexpressing either AML1/ETO9a or MLL/ENL together with oncogenic Nras displayed progressive anemia and leukocytosis (Fig. 2A), with extensive hepatosplenomegaly (data not shown). Upon reaching a terminal stage, GFP-positive cells dominated their peripheral blood, bone marrow, spleen, and liver (Fig. 2C,D; data not shown). Immunophenotyping, cytologic, and histologic analyses revealed that AML1/ETO9a + Nras leukemias predominantly contain immature blasts (Mac1−, Gr1−, Ly-6C−, F4/80−/lo, CD45lo, CD3−, CD19−, B220−, TER119−, c-Kit+, Sca-1−) (Fig. 2B,C; data not shown), which parallels observations in other models of core factor-binding AML (Yan et al. 2004; Kuo et al. 2006). Although such purely immature subtypes are seen in t(8;21)-positive AML, most human cases show a significant granulocytic maturation beside immature myeloblasts, which was absent in murine AML1/ETO9a + Nras AML. In contrast, MLL/ENL + Nras leukemias predominantly consisted of more differentiated myeloid cells (Mac1+, Gr1+/lo, Ly-6C+, F4/80+/lo, CD45+, CD3−, CD19−, B220−, TER119−, c-Kit−/lo, Sca-1−) (Fig. 2B,C; data not shown) and involve the monocytic lineage—a typical feature of human 11q23-rearranged AML. As in previous MLL fusion-based mouse models (Cozzio et al. 2003), the morphology resembles myelomonocytic and monocytic subtypes, which are seen in ∼42% of human 11q23-rearranged AML (Schoch et al. 2003). Despite its potent effects on leukemia onset, Nras did not appear to influence leukemia morphology, since leukemias induced by AML1/ETO9a or MLL/ENL alone were similar to those coexpressing Nras. In summary, mosaic mouse models based on cooperation of Nras with either fusion protein recapitulate common genetic and pathologic features of human AML.
Figure 2.
Defined mosaic AML mouse models have genotype-dependent morphology consistent with human AML. May-Grünwald-Giemsa-stained peripheral blood smears (A) (original magnification 200×) and Wright-Giemsa-stained bone marrow cytocentrifugation (B) (original magnification 1000×) predominantly show immature blasts in AML1/ETO9a + Nras leukemia, while MLL/ENL + Nras leukemia is characterized by more mature myelomonocytic cells at various differentiation levels. (C) Bone marrow immunphenotyping in AML1/ETO9a + Nras leukemic mice shows infiltration of GFP+/c-Kit+/Mac-1−/Gr-1− immature blasts, while MLL/ENL + Nras bone marrow is dominated by GFP+/c-Kit−/lo/Mac-1+/Gr-1+ myelomonocytic cells. (D) Hematoxylin–eosin-stained liver sections showing massive leukemic infiltration. Bars, 100 μm.
Establishment of a murine chemotherapy regimen that mimics clinical AML induction therapy
To study factors that influence the outcome of conventional AML therapy in mice, we established a simple chemotherapy regimen involving intraperitoneal (i.p.) injection of cytarabine (100 mg/kg per day over 5 d) and doxorubicin (3 mg/kg per day over 3 d) (Supplemental Fig. S1A), which mimics the standard induction therapy used to treat AML in human patients. In initial toxicity studies, cytarabine doses of 50 and 100 mg/kg per day were myelosuppressive but well tolerated over a 7-d treatment course (Supplemental Fig. S2A). Administration of 100 mg/kg cytarabine intraperitoneally resulted in plasma levels in the range of human high-dose cytarabine regimens (Supplemental Fig. S2B; Hiddemann 1991). A consistent drug delivery in different animals and leukemia genotypes was verified using flow cytometry quantification of intracellular doxorubicin (Supplemental Fig. S2C). Of note, doxorubicin was used instead of daunorubicin, which is the more common anthracycline in human AML therapy, but resulted in severe toxicity if administered intraperitoneally in mice (data not shown).
The efficacy and safety of the combined regimen were initially evaluated in a pre-existing model based on cotransduction of MLL/ENL and FLT3-ITD (Ono et al. 2005) and were later applied to models established in this study. In order to rapidly generate homogenous treatment cohorts, we harvested bone marrow and spleen from terminally ill primary FLC recipient mice and transplanted 1 million leukemia cells into 20–30 secondary recipients. In initial studies, we observed that this combination therapy was not tolerated in mice when treatment was initiated based on a high leukemia burden in the peripheral blood; although the peripheral disease disappeared within 2 d, animals died shortly thereafter with peripheral leukopenia (data not shown).
To better monitor leukemia progression and treat animals at an earlier stage of disease, we took advantage of the luciferase reporter, which we linked to MLL/ENL in a bicistronic retrovirus (MSCV-MLL/ENL-IRES-Luciferase). Bioluminescent imaging allowed detection of MLL/ENL + FLT3-ITD leukemic cells in bone marrow, spleen, and liver ∼10 d after transplantation into secondary recipients, which corresponded to ∼30% infiltration in bone marrow and spleen in pathologic analysis, while the disease became apparent in peripheral blood smears ∼5–7 d later (Supplemental Fig. S1C; data not shown). When mice were treated when disease was detected by bioluminescent imaging, most showed some response to therapy (Supplemental Fig. S1B), while untreated controls progressed rapidly (Supplemental Fig. S1C). Although most responses were only partial (median survival benefit of 7 d, P < 0.0001) (Supplemental Fig. S1D), none of the animals appeared to succumb to treatment toxicity, but instead from advancing leukemia. These studies demonstrate the utility of bioluminescence monitoring for treatment studies and establish an effective and safe combined chemotherapy regimen that mirrors human AML induction therapy.
AMLs expressing AML1/ETO9a or MLL/ENL display differences in response to conventional chemotherapy
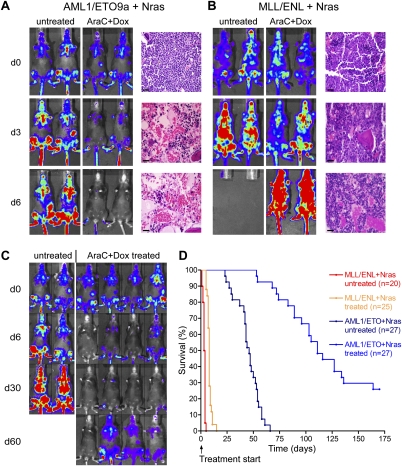
To determine whether defined AML genotypes display distinct treatment sensitivities, we transplanted multiple primary AML1/ETO9a + Nras and MLL/ENL + Nras leukemias and treated secondary recipient mice upon bioluminescent disease detection in bone marrow and spleen. Leukemias coexpressing AML1/ETO9a and Nras rapidly regressed under combined chemotherapy, leading to complete remissions. Thus, by 6 d after initiating treatment, luciferase signals disappeared completely, and histological analyses and flow cytometry revealed that the bone marrow and spleen were leukemia-free (Figs. 3A, 6C [below]; data not shown). In most instances, mice harboring these leukemias remained in remission for at least 30 d (Fig. 3C), showing an overall survival benefit of 69 d (P < 0.0001) (Fig. 3D). Although 80% of the treated mice eventually relapsed, often emanating from focal regions in bones or spleen, the rest remained leukemia-free for even 1 yr. Such behavior is reminiscent of the generally effective action of induction therapy in patients.
Figure 3.
AML1/ETO9a + Nras and MLL/ENL + Nras AMLs show dramatic differences in their response to combined chemotherapy in vivo. (A,B) Luciferase imaging and histological analysis of hematoxylin–eosin-stained bone marrow sections before (d0), during (d3), and after (d6) 5 d of chemotherapy show therapy-triggered regression and, ultimately, complete remission of AML1/ETO9a + Nras leukemia (A), while MLL/ENL + Nras leukemia (B) only shows decelerated progression, with persistence of blasts in response to treatment. Bars, 50 μm. (C) Long-term follow-up luciferase imaging of untreated and treated AML1/ETO9a + Nras leukemia. Treated mice achieve durable remissions lasting at least 30 d (d30). While most mice subsequently relapse, some mice remain in remission following chemotherapy (d60). (D) Kaplan-Meier survival curves of untreated and treated AML1/ETO9a + Nras and MLL/ENL + Nras mice following the initiation of chemotherapy.
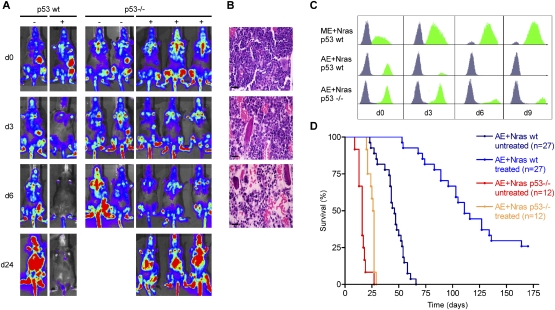
Figure 6.
Loss of p53 induces chemotherapy resistance in AML1/ETO9a + Nras AML. (A) Luciferase imaging of AML1/ETO9a + Nras leukemias generated in wild-type (left panel) or p53−/− (right panel) FLCs before (d0), during (d3), and after (d6, d24) chemotherapy. Recipient mice of p53-deficient AML1/ETO9a + Nras leukemias retain bioluminescent signal under therapy. (B) Hematoxylin–eosin-stained bone marrow sections of recipients of p53-deficient AML1/ETO9a + Nras leukemia at various time points following chemotherapy, demonstrating leukemia persistence during chemotherapy. Bars, 50 μm. (C) GFP histograms of bone marrow flow cytometry before (d0) and at various time points during (d3) and after (d6, d9) chemotherapy. While AML1/ETO9a + Nras blasts harboring wild-type p53 (AE + Nras p53 wt) rapidly clear, both MLL/ENL + Nras (ME + Nras p53 wt) and p53-deficient AML1/ETO9a + Nras leukemias (AE + Nras p53−/−) show persistence of GFP+ cells in bone marrow. (D) Kaplan-Meier survival curves of untreated and treated recipient mice of p53 wild-type and p53-deficient AML1/ETO9a + Nras leukemias. Loss of p53 impedes the long-term outcome of chemotherapy.
In stark contrast, MLL/ENL + Nras-induced AML persisted under chemotherapy, and bioluminescent imaging merely showed a decelerated progression. In addition to apoptotic cell death, histological analysis revealed refractory leukemic infiltrates in bone marrow and spleen (Fig. 3B). After a minor stagnation in disease acceleration, all treated animals progressed rapidly and died with only a minor survival benefit (5 d compared with untreated controls, P < 0.0001) (Fig. 3D). Hence, the MLL/ENL fusion protein produced leukemias that showed a very poor therapeutic response.
p53 contributes to genotype-specific differences in chemotherapy response
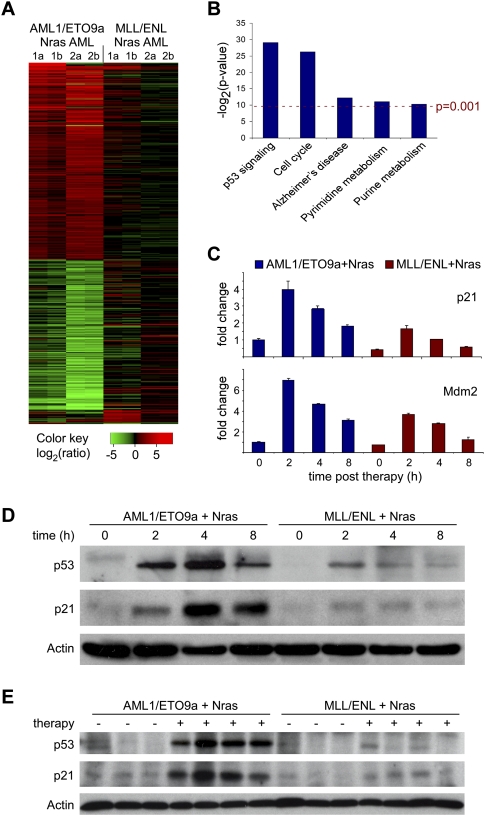
As a first step toward understanding the differential response of leukemia genotypes to therapy, we performed microarray analysis on leukemias expressing AML1/ETO9a + Nras or MLL/ENL + Nras following combined cytarabine/doxorubicin therapy in vivo. Two independent primary leukemias of each genotype were transplanted into multiple recipients, and diseased animals were either left untreated or treated with a single dose of cytarabine and doxorubicin. Two hours following treatment, spleens were isolated and subjected to transcriptional profiling using Affymetrix Mouse Genome 430 2.0 arrays. AML1/ETO9a + Nras leukemias induce complex gene expression changes immediately upon chemotherapy (greater than twofold change in 418 genes), while MLL/ENL + Nras AMLs showed much less transcriptional alterations in response to treatment (greater than twofold alteration in only 33 genes, 15 in common with AML1/ETO9a + Nras) (Fig. 4A). Using Significance Analysis of Microarrays (SAM) with the criteria of a falsediscovery rate (FDR) of ≤0.20 and an average fold change of ≥2.0 (Tusher et al. 2001), we identified an immediate drug response signature comprising 226 up-regulated and 172 down-regulated genes in AML1/ETO9a + Nras leukemias that was largely absent in treated MLL/ENL + Nras leukemia. Unsupervised clustering using this signature clearly separated both AML subtypes and identified subclusters distinguishing untreated and treated AML1/ETO9a + Nras samples; however, there was no such separation between untreated and treated MLL/ENL + Nras AMLs (Supplemental Fig. S3).
Figure 4.
In vivo expression analysis of immediate chemotherapy response programs identifies differences in p53 induction between AML1/ETO9a + Nras and MLL/ENL + Nras AMLs. (A) Chemotherapy-induced gene expression changes in AML1/ETO9a + Nras and MLL/ENL + Nras leukemias 2 h after combined chemotherapy rendered in a green–black–red pseudo color scheme for all genes with an average fold change ≥2.0 in either genotype. Samples included two independent primary leukemias (1,2) for each genotype, which were analyzed in two technical replicates (a,b). AML1/ETO9a + Nras leukemias show complex gene expression changes in response to chemotherapy, while this response profile is blunted in the MLL/ENL + Nras context. (B) KEGG pathways analysis (Ogata et al. 1999) of the AML1/ETO9a + Nras-specific chemotherapy response signature identified by SAM (FDR 0.2, fold change >2). Used _P_-values represent the EASE score (modified Fisher exact _P_-value) provided by the DAVID analysis tool (Dennis et al. 2003) available at http://david.abcc.ncifcrf.gov/summary.jsp. Two highly and three moderately significant pathway alterations are identified. (C) Quantitative real-time PCR analysis of p21 and Mdm2 at various time points after chemotherapy in vivo. Baseline expression and induction of both p53 target gene transcripts is attenuated in MLL/ENL + Nras leukemia. (D) Western blot analysis of p53 and p21 in leukemic spleens (>85% GFP+ infiltration) at various time points after i.p. administration of one dose of combined chemotherapy. AML1/ETO9a + Nras AML shows much stronger p53 induction, resulting in a stronger and more durable induction of its target p21. (E) Western blot analysis of p53 and p21 in recipient mice that were transplanted with independent primary AMLs and were either left untreated (−) or were treated with one dose of combined chemotherapy 4 h prior to sample harvest (+). Individual differences in p53 drug response programs are dependent on the AML genotype.
To further characterize this expression signature we performed KEGG pathway analysis (Ogata et al. 1999) to identify processes that are specifically altered in response to chemotherapy. Only five out of 195 analyzed pathways showed significant (P < 0.001) alteration 2 h after chemotherapy (Fig. 4B). Interestingly, the most significantly altered category involved genes associated with the p53 signaling network (P = 1.79E-09), which has been implicated as a central component in DNA damage response programs and whose integrity can contribute to the efficacy of chemotherapeutic agents (Lowe et al. 1993; Schmitt et al. 2000). Several well-characterized transcriptional p53 target genes such as Cdkn1a (p21), Mdm2, Apaf1, and Ccng1 showed strong induction in treated AML1/ETO9a + Nras leukemia, while their alteration was weaker or not detected in treated MLL/ENL + Nras AML. Through immunoblotting and/or RT–QPCR analyses on leukemia lysates from untreated and treated animals, we confirmed that the therapy-induced accumulation of p53 and two of its target genes (p21 and Mdm2) were strongly attenuated in MLL/ENL + Nras leukemias relative to those expressing AML1/ETO9a + Nras (Fig. 4C,D). In addition to these dramatic differences after chemotherapy, MLL/ENL-expressing leukemias show lower p21 levels already in the untreated state, which might indicate a lower p53 baseline activity. These differences in p53 functionality were observed in multiple primary leukemias of each genotype (Fig. 4E; data not shown). Of note, genes encoding drug transporters, drug modifiers, and efflux pumps implicated in cytarabine and doxorubicin resistance such as Abcb1 (Mdr1), Abcc1 (Mrp1), Slc29a1 (hENT1), Dck, and Cda (Galmarini et al. 2002) were not dysregulated in MLL/ENL-expressing leukemias, and we did not detect any genotype-dependent differences in the intracellular accumulation of doxorubicin in vivo (Supplemental Fig. S2C). Thus, while p53-independent mechanisms undoubtedly influence treatment sensitivity, the inability of MLL-rearranged leukemias to mount an effective p53 response may be a major factor contributing to their intrinsic chemotherapy resistance.
Impact of p53 on leukemogenesis and therapy response
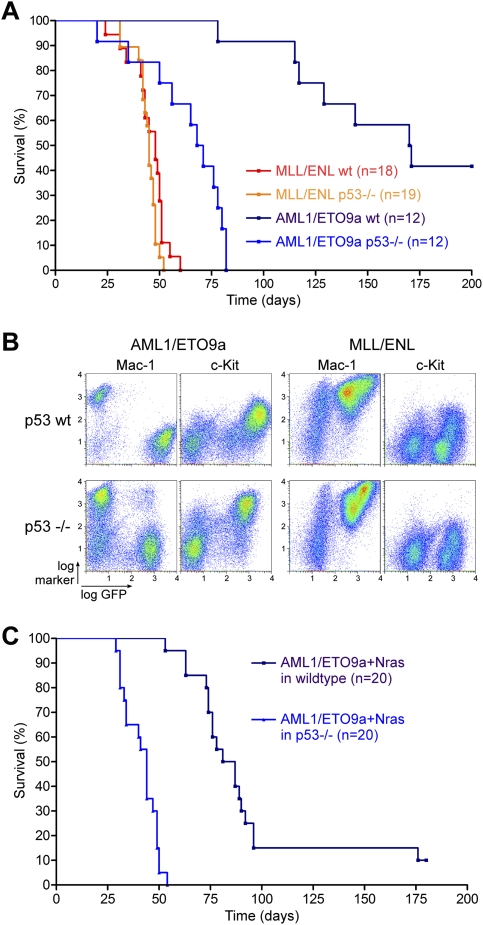
p53 is a central component in tumor suppressor networks that enable cells to evade malignant transformation by triggering fail-safe programs such as apoptosis or senescence in response to oncogene activation (for review, see Vogelstein et al. 2000; Lowe et al. 2004). To further study how p53 impacts AML leukemogenesis in the context of different oncogenic fusion proteins, we engineered murine AMLs lacking p53. Wild-type and p53−/− FLCs were transduced with retroviruses expressing AML1/ETO9a or MLL/ENL and transplanted into lethally irradiated recipients. p53 loss had a dramatic impact on leukemogenesis induced by AML1/ETO9a, leading to the appearance of AML with a significantly shortened overall survival (69 vs. 170 d, P < 0.0001) (Fig. 5A). Indeed, while the morphology and immunophenotype of AML1/ETO9a-induced leukemias were not affected by loss of p53 (Fig. 5B), their behavior paralleled the more rapid onset and aggressive nature of leukemias expressing MLL/ENL. In contrast, p53 loss had no impact on leukemogenesis induced by MLL/ENL, with leukemias showing an onset and morphology indistinguishable from wild-type MLL/ENL controls. It is noteworthy that, in our experience, MLL/ENL is the only AML oncogene that does not cooperate with p53 loss in promoting leukemia, while AML induced by lesions such as NrasG12D, KrasG12D, FLT3-ITD, full-length AML1/ETO, Pten loss, or Nf1 loss is accelerated by the absence of p53 (J. Zuber, Z. Zhao, A.R. Rappaport, and S.W. Lowe, unpubl.). Apparently, MLL/ENL enables cells to evade the p53 network during leukemogenesis.
Figure 5.
Loss of p53 dramatically accelerates AML1/ETO9a, but does not affect MLL/ENL-induced leukemogenesis. (A) Kaplan-Meier survival curves of lethally irradiated recipient mice, which were reconstituted with wild-type or p53−/− FLCs transduced with either AML1/ETO9a or MLL/ENL. Loss of p53 accelerates AML1/ETO9a-induced but not MLL/ENL-induced leukemogenesis. (B) Bone marrow immunophenotyping of wild-type and p53-deficient AML1/ETO9a- and MLL/ENL-induced leukemias. Loss of p53 does not affect the typical disease morphology induced by both fusion proteins. (C) Kaplan-Meier survival curves of lethally irradiated recipient mice, which were reconstituted with wild-type or p53−/− FLCs cotransduced with AML1/ETO9a and NrasG12D. Loss of p53 also accelerates AML1/ETO9a + Nras-induced leukemogenesis.
Beyond its role as classical tumor suppressor, studies indicate that inactivating p53 mutations confer resistance to chemotherapy in some animal models (Schmitt et al. 1999) and to cytarabine treatment of AML cell lines in vitro (Yin et al. 2006). Moreover, loss of p53 has been associated with poor treatment outcome in patients, including in AML (Wattel et al. 1994; Haferlach et al. 2008). To experimentally evaluate the impact of dysfunctional p53 on chemotherapy response, we generated AML1/ETO9a + Nras leukemias derived from p53−/− FLCs and compared their behavior with p53-expressing leukemias following induction therapy. Again, primary recipient mice developed AMLs that were morphologically similar to their p53 wild-type counterparts (data not shown), but showed accelerated disease progression (median survival 44 vs. 84 d, P < 0.0001) (Fig. 5C). Treatment of these primary leukemias in secondary recipients was performed as above, with bioluminescent imaging as a guide for initiating treatment and monitoring therapy response.
In contrast to their p53 wild-type counterparts, p53−/− leukemias expressing AML1/ETO9a + Nras responded poorly to induction chemotherapy. Using all diagnostic modalities, including luciferase imaging (Fig. 6A), histology (Fig. 6B), and bone marrow flow cytometry for GFP-positive leukemic cells (Fig. 6C), mice harboring p53-null leukemias demonstrated blast persistence and gained only a minor survival benefit from therapy (10 d survival benefit, P = 0.0001). In no cases were animals cured. These observations are in striking contrast to the strong survival benefit and 20% cure rate observed in AML1/ETO9a + Nras counterparts harboring intact p53 (Fig. 6D; see also Fig. 3). Thus, in the absence of p53, leukemias expressing AML1/ETO9a show a poor treatment response approaching that seen for MLL/ENL-expressing leukemias. These results demonstrate that modulation of p53 activity, whether through inhibition in the context of MLL/ENL or via gene deletion, has an important impact on the response of murine AMLs to conventional chemotherapy.
Discussion
In this study, we use mice harboring leukemias that accurately reflect the genetics and pathology of human AML as tractable models to study the impact of cancer heterogeneity on therapy response and to explore the molecular basis for variable leukemia behavior. We see that leukemias expressing the AML1/ETO9a oncoprotein [mimicking translocation (8;21) in humans] respond well to conventional chemotherapy, whereas those expressing MLL/ENL [recapitulating translocation (11;19) in humans] show a particularly dismal response. Remarkably, the response patterns of these two leukemia subtypes accurately mimic what is observed in human patients with the same genetic lesions. These results provide important insights into the clinical behavior of human AML and highlight the utility of genetically engineered mouse models as predictive models for therapy in patients.
Initially using genome-wide transcriptional profiling of treatment responses in vivo, we identified a gene expression signature in drug-sensitive AML1/ETO-expressing leukemias that was absent in drug-resistant leukemias expressing MLL/ENL. The most significant element of this signature involved strong alterations in genes linked to the p53 tumor suppressor network. Indeed, further evaluation of leukemia samples at various time points after chemotherapy demonstrated that the excellent response of AML1/ETO-expressing leukemias requires a robust activation of the p53 network, whereas poorly responding MLL/ENL leukemias show a dramatic attenuation of the p53 response. How this eventually leads to leukemia cell clearance remains to be determined, and, based on our gene expression and preliminary histological analyses (data not shown), may involve a combination of p53 effector functions. Although the impact of MLL/ENL on p53 may be indirect, previous cell culture studies suggest that MLL fusion proteins can associate with p53 and prevent its acetylation and activation following DNA damage (Wiederschain et al. 2005). While these observations derive from studies in cultured carcinoma and osteosarcoma cells, our data indicate that such p53-suppressive effects of MLL fusion proteins can occur in AML cells and contribute to chemotherapy resistance in an in vivo context.
In addition to its role in chemotherapy responses, p53 is a potent tumor suppressor that is lost in the majority of human epithelial cancers prior to treatment (Petitjean et al. 2007). Mutations in other genes that modulate p53 function can also occur during tumorigenesis, which allows aggressive tumors to arise in the presence of wild-type p53. Beside attenuating p53 activation following chemotherapy, MLL fusion proteins also seem to interfere with tumor-suppressive functions of p53 during myeloid leukemogenesis. In fact, in our experience, MLL fusion proteins are the only AML oncogenes that do not show increased leukemogenic potential in the absence of functional p53 (see Fig. 5; data not shown). Consistent with this, some studies report that MLL rearrangements and p53 mutations rarely co-occur in human AML (Megonigal et al. 1998; Haferlach et al. 2008). In contrast, in patients, p53 mutations exist in AML1/ETO and inv(16) AML and correlate with relapse and treatment resistance (Krejci et al. 2008). In fact, in mice we see that AML1/ETO-expressing leukemias lacking p53 show a similar aggressiveness and chemoresistance to those expressing MLL/ENL. Therefore, while p53 is mutated or deleted in only 13% of all AML patients (Haferlach et al. 2008), its functional attenuation by MLL fusion proteins represents an alternative way through which p53 networks can be impaired in AML. This inherent inhibition of p53 tumor suppressor networks may be an important element involved in the transforming potential of MLL fusion proteins.
Our studies also have implications for the clinical treatment of human AML. Although correlative studies in human patients have linked various MLL translocations (Schoch et al. 2003) and p53 mutations (Wattel et al. 1994) to poor prognosis, this information is not commonly used in guiding current AML treatment regimens. By showing that MLL/ENL expression or mutation of p53 can severely compromise therapy outcome in animal models, our studies provide direct in vivo evidence that these lesions play a causal role in promoting drug resistance. As such, the functional validation of these resistance-promoting alleles, together with the corresponding clinical data, justifies the use of these markers in patient management. For example, when extrapolated to the human condition, our data suggest that AML patients diagnosed with MLL/ENL rearrangements or p53 mutations will not show durable responses to standard chemotherapy and should be offered alternative approaches at diagnosis—perhaps more aggressive/experimental regimens or palliative care, depending on the individual case. Furthermore, our data provide functional support for the notion that, irrespective of genotype, the robustness of the p53 network activation following chemotherapy is the single most important determinant of outcome to conventional chemotherapy in AML. Indeed, studies support the technical feasibility and potential value of assessing p53 activation during the first hours of therapy in patients (Anensen et al. 2006).
Our studies also have important implications for the use of genetically engineered mouse models in preclinical studies. In principle, genetically engineered mouse models of human cancer provide the opportunity to study therapy responses of genetically defined cancers in a relevant pathophysiologic context that cannot be recapitulated in vitro or in xenotransplantation settings. Nevertheless, human and murine cells have documented differences in certain oncogenic signaling pathways, stress response programs, and drug pharmacology that may impact how tumors respond to a particular drug or drug combination (Van Dyke and Jacks 2002; Sharpless and Depinho 2006). Consequently, there has been much debate as to whether mouse models of human cancer can be predictive of therapy response in vivo.
A key impediment in “credentialing” genetically engineered mouse models as reliable preclinical systems for studying therapeutic response is the paucity of comparable mouse and clinical data for a given drug and drug combination. By studying the effects of conventional chemotherapy in genetically engineered mouse models of AML, we not only uncover factors that influence the outcome of a clinically important treatment regimen, but also address the predictability of mouse models using outcome data obtained through decades of clinical experience. Our studies find a remarkable similarity in the genotype response pattern of mouse AML models and human AML patients to standard induction chemotherapy, implying that such systems can predict the behavior of therapeutic agents in the clinic. Therefore, these and similar mouse models provide relevant systems for further studies on drug action and drug resistance and, by extrapolation, predictive test systems for evaluating new drug targets and testing new therapies in otherwise refractory malignancies.
Materials and methods
Retroviral constructs
All retroviruses were constructed in the MSCV backbone (Clontech). MSCV-IRES-Luciferase was generated by replacing GFP in MSCV-IRES-GFP with an NcoI/SalI fragment from pGL3-Basic (Promega) after excision of the 3′polyA signal. Mouse NrasG12D was amplified and mutated by PCR from IMAGE clone 6475312 (Invitrogen) and cloned downstream from the IRES; subsequently Luciferase was cloned upstream of the IRES. The AML1/ETO9a cDNA was generated by PCR from full-length human AML1/ETO-IRES-GFP and subcloned into MSCV-IRES-GFP. Human MLL/ENL was subcloned from MLL/ENL-PGK-Neo (generously provided by Irving L. Weissman) into MSCV-IRES-GFP and MSCV-IRES-Luciferase. Human FLT3-IRES-YFP and AML1/ETO-IRES-GFP were kindly provided by Christian Buske. Cloning strategies and primer sequences are available on request.
FLC isolation, retroviral transduction, and transplantation
The Cold Spring Harbor Animal Care and Use Committee approved all mouse experiments included in this work. E13.5–E15 FLCs from wild-type C57BL/6 mice were isolated, cultured, and retrovirally transduced as described previously (Schmitt et al. 2002). Retroviral cotransduction was carried out by mixing viral supernatants from independent transfections of Phoenix packaging cells in a 1:1 ratio. Retroviral transduction of GFP-expressing constructs was assessed 24 h after the last infection by flow cytometry (Guava EasyCyte, Guava Technologies), typically demonstrating transduction of ∼20% of FLCs. Transduction of luciferase-expressing constructs was confirmed qualitatively (IVIS100, Caliper LifeSciences). About 1 × 106 cells were transplanted by tail vein injection of 6- to 8-wk-old lethally irradiated C57BL/6 recipient mice (9.0 Gy as single dose administered 24 h prior to transplantation).
Monitoring and characterization of primary leukemias
Bioluminescent imaging was performed using an IVIS100 imaging system (Caliper LifeSciences). Mice were injected intraperitoneally with 150 mg/kg D-Luciferin (Caliper LifeSciences), anesthetized with isoflurane, and imaged for 1 min after a 5-min incubation following injection. Primary FLC recipient mice were sacrificed at terminal disease stage (whole-body signal in bioluminescent imaging, severe leukocytosis in peripheral blood smears, moribund appearance) by CO2 euthanasia. Statistical evaluation of overall survival was based on the log-rank (Mantel-Cox) test for comparison of the Kaplan-Meier event time format. Leukemia cells were harvested from bone marrow (by flushing tibias and femurs with DMEM) and spleen (by gently meshing enlarged spleens in DMEM between two glass slides), filtered through 100-μm cell strainers (BD Falcon) to obtain single-cell suspensions, and cryopreserved in fetal bovine serum (FBS) containing 10% DMSO. Peripheral blood smears and bone marrow cytospins were stained with Wright-Giemsa (Sigma); tissue specimens were fixed in 10% formalin and stained with hematoxylin and eosin (Sigma) using standard protocols. For immunophenotyping, single-cell suspensions of bone marrow and spleen were incubated in ACK red cell lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) for 5 min, resuspended in FACS buffer (PBS, 5%FBS, 0.1% NaN3), and preincubated with anti-CD16/CD32 (Fc-block, 1:1000; BD Pharmingen). Aliquots of 2 × 105 to 5 × 105 cells were stained for 20 min on ice with phycoerythrin-Cy5-conjugated monoclonal antibodies specific for Mac-1, Gr-1, F4/80, CD45, CD3, CD19, B220, TER119, c-Kit, Sca-1, and a phycoerythrin-conjugated Ly-6C antibody (all 1:100; Biolegend). Data were collected on an LSR-II flow cytometer (BD Biosciences) and analyzed using FACSDiva (BD Biosciences) and FlowJo (Treestar) software.
Transplantation and in vivo treatment studies
For treatment studies, each primary leukemia was transplanted into 20–30 6- to 8-wk-old sublethally irradiated recipient mice (4.5 Gy, 24 h prior to transplantation) by tail-vein injection of 1 × 106 viable GFP+ cells per recipient. Sublethal irradiation was used to achieve a more uniform disease onset in recipient animals. Recipient mice were maintained on Ciprofloxacin-supplemented drinking water (Ciprofloxacin 125 mg/L, sucrose 20 g/L; both Sigma) until 2 wk after the end of chemotherapy. This antibiotic regimen has been shown to efficiently decontaminate aerobic Gram-negative pathogens without major effects on hematopoiesis (Velders et al. 2004). Mice were monitored by bioluminescent imaging every 4 d, starting 10 d after transplantation. Chemotherapy was initiated upon detection of clear signals in pelvis, tail, and both femurs, and initial stage of hepatosplenic infiltration, which correlated with 30%–60% bone marrow infiltration as assessed by flow cytometry. Mice were treated for five consecutive days every 24 h with i.p. injections of 100 mg/kg cytarabine (Bedford Laboratories); during the first 3 d, 3 mg/kg doxorubicin (Bedford Laboratories) was administered in a separate i.p. injection. Immediate response and long-term treatment effects were monitored by weekly luciferase imaging starting the first day after treatment and by histopathological analysis of representative mice at various time points. Details of HPLC cytarabine plasma level analysis and flow cytometry measurement of intracellular doxorubicin can be found in the Supplemental Material.
Gene expression analysis (quantitative real-time PCR, microarrays, and immunoblotting)
For drug response studies, diseased mice were treated with a single dose of cytarabine (100 mg/kg) and doxorubicin (3 mg/kg) i.p. at various time points prior to harvest. Samples were obtained from enlarged spleens predominantly containing leukemia cells; infiltration of >85% GFP+ cells was confirmed by flow cytometry. Following ACK-lysis of single-cell suspensions, RNA was extracted using RNAeasy columns (Qiagen) and cDNA was synthesized using TaqMan reverse transcriptase reaction (Applied Bosystems) according to the manufacturer's protocols. Quantitative PCR analysis was performed on an iCycler mounted with an iQ5 multicolor real-time PCR detection system (Bio-Rad). Expression data were normalized against β-actin controls. For primer sequences, see the Supplemental Material. Microarray experiments were performed on Mouse Genome 430A 2.0 arrays (Affymetrix) according to the manufacturer's instructions. For Western blot analysis, ACK-treated samples were lysed in Laemmli buffer, separated by SDS-PAGE, and transferred to Immobilon PVDF membrane (Millipore). We used antibodies against p53 (IMX25, 1:1000; Leica Microsystems), p21 (C-19, 1:500; Santa Cruz Biotechnologies), Nras (F155, 1:300; Santa Cruz Biotechnologies), pan-Ras (Ras10, 1:1000; Upstate Biotechnologies), and Actin (AC-15, 1:5000; Abcam). Flow cytometry analysis of phospho-Erk in bone marrow was performed using anti-pERK (#9101, C1:100; ell Signaling) as described previously (Van Meter et al. 2007). To assess baseline phospho-Erk levels, cells were starved in serum-free medium for 30 min at 37°C. and then fixed and analyzed without cytokine stimulation.
Microarray data analysis
Raw data were processed using FARMS method (Hochreiter et al. 2006) with an up-to-date probe set definition based on Entrez gene sequences (Dai et al. 2005). Treated versus untreated samples were analyzed in biological (independent primary leukemias) and technical replicates using a one-on-one comparison scheme. We used SAM (Tusher et al. 2001) with an FDR of ≤0.20 and an average fold change of ≥2.0 as selective criteria to identify a drug response signature. Significantly perturbed KEGG pathways (Ogata et al. 1999) were identified using the functional annotation tool available at DAVID (http://david.abcc.ncifcrf.gov/summary.jsp) (Dennis et al. 2003).
Acquisition and analysis of human data
One-hundred-eleven pediatric AML cases treated at St. Jude Children's Research Hospital (SJCRH) between 1982 and 2005 were studied. Informed consent for the use of samples was obtained from patients, parents, or guardians in accordance with the Declaration of Helsinki. Study approval was obtained from the SJCRH institutional review board. Mononuclear cells were purified from diagnostic bone marrow or peripheral blood samples by density gradient centrifugation. DNA was extracted using the DNA Blood Mini Kit (Qiagen). NRAS exons 1 and 2 were amplified from genomic DNA using Accuprime GC-rich Polymerase (Invitrogen). For primer sequences see the Supplemental Material. Overall survival was defined as the time elapsed from study enrollment to death, with those still living at last follow-up considered censored. The Kaplan-Meier method was used to estimate the probability of overall survival and standard errors were determined by the method of Peto and Pike. The overall survival comparisons among subtypes were performed by Mantel-Haenszel log-rank test stratified by protocol. overall survival was studied for cases treated after 1998 and included 16 AML97, 30 AML02, and four nonprotocol patients. Significance of the association of Nras mutations with cytogenetic subtypes was analyzed by multifactor dimensionality reduction (Ritchie et al. 2001); see the Supplemental Material for details.
Acknowledgments
We thank Christian Buske and Irving Weissman for providing cDNA constructs. We gratefully acknowledge Meredith Taylor, Mei Lin Maunakea, Shannon Delaney, Eileen Earl, Lisa Bianco, and her team for excellent technical assistance; Zhenyu Xuan and Sohail Khan for microarray analysis, and Jeffrey Rubnitz, Raul Ribeiro, Stanley Pounds, and Xueyuan Cao for sharing clinical data and providing statistical analysis. We also thank Enrique Cepero, Lars Zender, Cornelius Miething, Mona Spector, Katherine McJunkin, Christof Fellmann, and other members of the Lowe lab as well as Michelle LeBeau, Kevin Shannon, and David Largaespada for constructive criticism and advice. This work was supported by a Specialized Center of Research grant from the Leukemia and Lymphoma Society, generous gifts from the Don Monti Memorial Research Foundation, and the Ambassador Felix Schnyder Memorial Grant of the Lauri Strauss Leukemia Foundation. J.Z. was supported by a research fellowship from the German Research Foundation (DFG) and is the Andrew Seligson Memorial Fellow at CSHL. A.R.R. was supported by an NIH traineeship and the Barbara McClintock fellowship. S.C.K. is the Leslie Rutherford Leukemia and Lymphoma Society Scholar; S.W.L. is a Howard Hughes Medical Institute investigator.
Footnotes
References
- Anensen N., Oyan A.M., Bourdon J.C., Kalland K.H., Bruserud O., Gjertsen B.T. A distinct p53 protein isoform signature reflects the onset of induction chemotherapy for acute myeloid leukemia. Clin. Cancer Res. 2006;12:3985–3992. doi: 10.1158/1078-0432.CCR-05-1970. [DOI] [PubMed] [Google Scholar]
- Byrd J.C., Mrozek K., Dodge R.K., Carroll A.J., Edwards C.G., Arthur D.C., Pettenati M.J., Patil S.R., Rao K.W., Watson M.S., et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: Results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- Cozzio A., Passegue E., Ayton P.M., Karsunky H., Cleary M.L., Weissman I.L. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes & Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M., Wang P., Boyd A.D., Kostov G., Athey B., Jones E.G., Bunney W.E., Myers R.M., Speed T.P., Akil H., et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G., Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. doi: 10.1186/gb-2003-4-5-p. [DOI] [PubMed] [Google Scholar]
- Dohner H. Implication of the molecular characterization of acute myeloid leukemia. Hematology (Am. Soc. Hematol. Educ. Program) 2007;2007:412–419. doi: 10.1182/asheducation-2007.1.412. [DOI] [PubMed] [Google Scholar]
- Estey E., Dohner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- Galmarini C.M., Thomas X., Calvo F., Rousselot P., El Jafaari A., Cros E., Dumontet C. Potential mechanisms of resistance to cytarabine in AML patients. Leuk. Res. 2002;26:621–629. doi: 10.1016/s0145-2126(01)00184-9. [DOI] [PubMed] [Google Scholar]
- Gilliland D.G., Jordan C.T., Felix C.A. The molecular basis of leukemia. Hematology (Am. Soc. Hematol. Educ. Program) 2004;2004:80–97. doi: 10.1182/asheducation-2004.1.80. [DOI] [PubMed] [Google Scholar]
- Grimwade D., Walker H., Oliver F., Wheatley K., Harrison C., Harrison G., Rees J., Hann I., Stevens R., Burnett A., et al. The importance of diagnostic cytogenetics on outcome in AML: Analysis of 1,612 patients entered into the MRC AML 10 trial. Blood. 1998;92:2322–2333. [PubMed] [Google Scholar]
- Haferlach C., Dicker F., Herholz H., Schnittger S., Kern W., Haferlach T. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia. 2008;22:1539–1541. doi: 10.1038/leu.2008.143. [DOI] [PubMed] [Google Scholar]
- Hiddemann W. Cytosine arabinoside in the treatment of acute myeloid leukemia: The role and place of high-dose regimens. Ann. Hematol. 1991;62:119–128. doi: 10.1007/BF01702925. [DOI] [PubMed] [Google Scholar]
- Hochreiter S., Clevert D.A., Obermayer K. A new summarization method for Affymetrix probe level data. Bioinformatics. 2006;22:943–949. doi: 10.1093/bioinformatics/btl033. [DOI] [PubMed] [Google Scholar]
- Kelly L.M., Kutok J.L., Williams I.R., Boulton C.L., Amaral S.M., Curley D.P., Ley T.J., Gilliland D.G. PML/RARα and FLT3-ITD induce an APL-like disease in a mouse model. Proc. Natl. Acad. Sci. 2002;99:8283–8288. doi: 10.1073/pnas.122233699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci O., Wunderlich M., Geiger H., Chou F.S., Schleimer D., Jansen M., Andreassen P.R., Mulloy J.C. p53 signaling in response to increased DNA damage sensitizes AML1-ETO cells to stress-induced death. Blood. 2008;111:2190–2199. doi: 10.1182/blood-2007-06-093682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo Y.H., Landrette S.F., Heilman S.A., Perrat P.N., Garrett L., Liu P.P., Le Beau M.M., Kogan S.C., Castilla L.H. Cbf β-SMMHC induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer Cell. 2006;9:57–68. doi: 10.1016/j.ccr.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Lafiura K.M., Edwards H., Taub J.W., Matherly L.H., Fontana J.A., Mohamed A.N., Ravindranath Y., Ge Y. Identification and characterization of novel AML1-ETO fusion transcripts in pediatric t(8;21) acute myeloid leukemia: A report from the Children's Oncology Group. Oncogene. 2008;27:4933–4942. doi: 10.1038/onc.2008.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley T.J., Mardis E.R., Ding L., Fulton B., McLellan M.D., Chen K., Dooling D., Dunford-Shore B.H., McGrath S., Hickenbotham M., et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe S.W., Ruley H.E., Jacks T., Housman D.E. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- Lowe S.W., Cepero E., Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- Megonigal M.D., Rappaport E.F., Nowell P.C., Lange B.J., Felix C.A. Potential role for wild-type p53 in leukemias with MLL gene translocations. Oncogene. 1998;16:1351–1356. doi: 10.1038/sj.onc.1201637. [DOI] [PubMed] [Google Scholar]
- Morrison S.J., Hemmati H.D., Wandycz A.M., Weissman I.L. The purification and characterization of fetal liver hematopoietic stem cells. Proc. Natl. Acad. Sci. 1995;92:10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan C.G., Goorha S., Radtke I., Miller C.B., Coustan-Smith E., Dalton J.D., Girtman K., Mathew S., Ma J., Pounds S.B., et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R., Nakajima H., Ozaki K., Kumagai H., Kawashima T., Taki T., Kitamura T., Hayashi Y., Nosaka T. Dimerization of MLL fusion proteins and FLT3 activation synergize to induce multiple-lineage leukemogenesis. J. Clin. Invest. 2005;115:919–929. doi: 10.1172/JCI22725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh C., Subrahmanyam R., Ren R. Oncogenic NRAS rapidly and efficiently induces CMML- and AML-like diseases in mice. Blood. 2006;108:2349–2357. doi: 10.1182/blood-2004-08-009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A., Achatz M.I., Borresen-Dale A.L., Hainaut P., Olivier M. TP53 mutations in human cancers: Functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- Ritchie M.D., Hahn L.W., Roodi N., Bailey L.R., Dupont W.D., Parl F.F., Moore J.H. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am. J. Hum. Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenk R.F., Dohner K., Krauter J., Frohling S., Corbacioglu A., Bullinger L., Habdank M., Spath D., Morgan M., Benner A., et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N. Engl. J. Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- Schmitt C.A., McCurrach M.E., de Stanchina E., Wallace-Brodeur R.R., Lowe S.W. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes & Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C.A., Rosenthal C.T., Lowe S.W. Genetic analysis of chemoresistance in primary murine lymphomas. Nat. Med. 2000;6:1029–1035. doi: 10.1038/79542. [DOI] [PubMed] [Google Scholar]
- Schmitt C.A., Fridman J.S., Yang M., Baranov E., Hoffman R.M., Lowe S.W. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- Schoch C., Schnittger S., Klaus M., Kern W., Hiddemann W., Haferlach T. AML with 11q23/MLL abnormalities as defined by the WHO classification: Incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood. 2003;102:2395–2402. doi: 10.1182/blood-2003-02-0434. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E., Depinho R.A. The mighty mouse: Genetically engineered mouse models in cancer drug development. Nat. Rev. Drug Discov. 2006;5:741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke T., Jacks T. Cancer modeling in the modern era: Progress and challenges. Cell. 2002;108:135–144. doi: 10.1016/s0092-8674(02)00621-9. [DOI] [PubMed] [Google Scholar]
- Van Meter M.E., Diaz-Flores E., Archard J.A., Passegue E., Irish J.M., Kotecha N., Nolan G.P., Shannon K., Braun B.S. K-RasG12D expression induces hyperproliferation and aberrant signaling in primary hematopoietic stem/progenitor cells. Blood. 2007;109:3945–3952. doi: 10.1182/blood-2006-09-047530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velders G.A., van Os R., Hagoort H., Verzaal P., Guiot H.F., Lindley I.J., Willemze R., Opdenakker G., Fibbe W.E. Reduced stem cell mobilization in mice receiving antibiotic modulation of the intestinal flora: Involvement of endotoxins as cofactors in mobilization. Blood. 2004;103:340–346. doi: 10.1182/blood-2002-07-2270. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Lane D., Levine A.J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Wattel E., Preudhomme C., Hecquet B., Vanrumbeke M., Quesnel B., Dervite I., Morel P., Fenaux P. p53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood. 1994;84:3148–3157. [PubMed] [Google Scholar]
- Wei J., Wunderlich M., Fox C., Alvarez S., Cigudosa J.C., Wilhelm J.S., Zheng Y., Cancelas J.A., Gu Y., Jansen M., et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell. 2008;13:483–495. doi: 10.1016/j.ccr.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederschain D., Kawai H., Shilatifard A., Yuan Z.M. Multiple mixed lineage leukemia (MLL) fusion proteins suppress p53-mediated response to DNA damage. J. Biol. Chem. 2005;280:24315–24321. doi: 10.1074/jbc.M412237200. [DOI] [PubMed] [Google Scholar]
- Yan M., Burel S.A., Peterson L.F., Kanbe E., Iwasaki H., Boyapati A., Hines R., Akashi K., Zhang D.E. Deletion of an AML1-ETO C-terminal NcoR/SMRT-interacting region strongly induces leukemia development. Proc. Natl. Acad. Sci. 2004;101:17186–17191. doi: 10.1073/pnas.0406702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Kanbe E., Peterson L.F., Boyapati A., Miao Y., Wang Y., Chen I.M., Chen Z., Rowley J.D., Willman C.L., et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat. Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- Yin B., Kogan S.C., Dickins R.A., Lowe S.W., Largaespada D.A. Trp53 loss during in vitro selection contributes to acquired Ara-C resistance in acute myeloid leukemia. Exp. Hematol. 2006;34:631–641. doi: 10.1016/j.exphem.2006.01.015. [DOI] [PubMed] [Google Scholar]