Nuclear Shuttling of She2p Couples ASH1 mRNA Localization to its Translational Repression by Recruiting Loc1p and Puf6p (original) (raw)
Abstract
The transport and localization of mRNAs results in the asymmetric synthesis of specific proteins. In yeast, the nucleocytoplasmic shuttling protein She2 binds the ASH1 mRNA and targets it for localization at the bud tip by recruiting the She3p–Myo4p complex. Although the cytoplasmic role of She2p in mRNA localization is well characterized, its nuclear function is still unclear. Here, we show that She2p contains a nonclassical nuclear localization signal (NLS) that is essential for its nuclear import via the importin α Srp1p. Exclusion of She2p from the nucleus by mutagenesis of its NLS leads to defective ASH1 mRNA localization and Ash1p sorting. Interestingly, these phenotypes mimic knockouts of LOC1 and PUF6, which encode for nuclear RNA-binding proteins that bind the ASH1 mRNA and control its translation. We find that She2p interacts with both Loc1p and Puf6p and that excluding She2p from the nucleus decreases this interaction. Absence of nuclear She2p disrupts the binding of Loc1p and Puf6p to the ASH1 mRNA, suggesting that nuclear import of She2p is necessary to recruit both factors to the ASH1 transcript. This study reveals that a direct coupling between localization and translation regulation factors in the nucleus is required for proper cytoplasmic localization of mRNAs.
INTRODUCTION
The cytoplasmic transport and localization of mRNAs is used by several eukaryotic organisms to control, in space and time, the expression of proteins involved in cell fate determination, cellular polarity, or asymmetric cell division (St. Johnston, 2005; Du et al., 2007). With more than 30 mRNAs localized at the bud tip, the budding yeast Saccharomyces cerevisiae is an excellent model system for studying the mechanisms behind mRNA transport and localization (Chartrand et al., 2001; Darzacq et al., 2003). One of these transcripts is the ASH1 mRNA, which is localized at the distal tip of daughter cells during late anaphase (Long et al., 1997; Takizawa et al., 1997). This localization promotes the asymmetric sorting of Ash1p to the daughter cell nucleus and results in the inhibition of mating-type switching in the daughter cell (Jansen et al., 1996; Sil and Herskowitz, 1996). The RNA-binding protein She2p is responsible for the recognition of bud-localized mRNAs, via its interaction with specific localization elements in these transcripts (Olivier et al., 2005). She2p also interacts with the transport machinery, constituted of the type V myosin Myo4p, via the bridging protein She3p (Bohl et al., 2000; Long et al., 2000; Takizawa and Vale, 2000). During its transport to the bud tip, the ASH1 mRNA is translationally repressed by the RNA-binding proteins Khd1p (Irie et al., 2002) and Puf6p (Gu et al., 2004), and phosphorylation of these factors at the bud tip promotes the local synthesis of the Ash1 protein (Paquin et al., 2007; Deng et al., 2008).
Previous studies have shown that nuclear factors play an important role in yeast mRNA localization. Puf6p and Loc1p are predominantly nucleolar RNA-binding proteins that bind directly the 3′ untranslated region (UTR) of ASH1 mRNA in vitro and are involved in the translational repression of this transcript (Long et al., 2001; Gu et al., 2004; Komili et al., 2007). Knockouts of these genes disrupt ASH1 mRNA localization and Ash1p sorting to the daughter cell (Long et al., 2001; Gu et al., 2004). She2p shuttles between the cytoplasm and the nucleus (Kruse et al., 2002), and recent evidence suggests that the nuclear transit of She2p is involved in the translational regulation of ASH1 mRNA (Du et al., 2008). However, the specific role of She2p in the nucleus is still unclear.
In this study, we show that She2p contains a nonclassical nuclear localization signal (NLS) that is essential for its nuclear import by the importin α Srp1p. Exclusion of She2p from the nucleus by mutagenesis of its NLS leads to defective mRNA localization and Ash1p sorting. We find that nuclear She2p is associated with Puf6p and Loc1p independently of their interaction with RNA. Exclusion of She2p from the nucleus decreases its interaction with Loc1p and Puf6p and disrupts the binding of these factors to the ASH1 mRNA. This study leads us to suggest a mechanism where She2p interacts with the translation regulation factors Puf6p and Loc1p in the nucleus and recruits these factors to the ASH1 mRNA, thereby promoting the localization of this transcript at the bud tip. This coordinated recruitment of a localization factor and translational repressors to ASH1 transcripts in the nucleus suggests that mRNA transport and translational control machineries are coupled and that this coupling in the nucleus is required for proper cytoplasmic localization and local translation of this transcript.
MATERIALS AND METHODS
Growth Media and Yeast Strains
Yeast cells were grown in either synthetic growth media lacking the nutrients indicated or rich media (Rose et al., 1990). Transformation was performed according to the protocol of Schiestl and Giestz (1989). Yeast gene disruption cassette was created by PCR amplification of the _loxP_-KAN-loxP construct in plasmid pUG6 and primers specifics for the gene of interest (Guldener et al., 1996). Specific disruption was confirmed by PCR analysis of genomic DNA. Yeasts strains and plasmids used in this study are described in Supplementary Tables S1 and S2, respectively.
Immunoprecipitation and Reverse Transcription-PCR
Fifty milliliters of yeast cells were grown to early log phase (OD600 ∼ 1) at 30°C in the appropriate medium. Formaldehyde was added to a final concentration of 1%, and cells were incubated at room temperature (RT) for 20 min. Glycine was added to a final concentration of 300 mM. Cells were washed twice in 1× PBS, harvested by centrifugation, and resuspended at an OD600 of 100 in the extraction buffer (25 mM HEPES-KOH, pH 7.5, 150 mM KCl, 2 mM MgCl2, 0.1% IGEPALCA-630, 1 mM dithiothreitol, 87.5 μg/ml phenylmethylsulfonyl fluoride, 0.5 μg/ml pepstatin, 0.5 μg/ml leupeptin, 0.5 μg/ml aprotinin, and 23 U/ml RNAguard). The cells were broken with glass beads, vortexed five times for 30 s, on ice with a 1-min pause between each vortex. The supernatant was used for immunoprecipitation and Western blot. For the immunoprecipitation of myc-tagged She2p, 10 μg of anti-myc antibody (9E10) was added to 500 μl of supernatant and incubated at 4°C with agitation for 1 h; 50 μl of protein A-Sepharose beads was then added, and the incubation at 4°C was continued for 2 h. For immunoprecipitation of tandem affinity purification (TAP)-tagged proteins, 50 μl of IgG-agarose beads was added to 500 μl of supernatant. The beads were washed four times for 3 min at 4°C with a wash buffer (25 mM HEPES-KOH, pH 7.5, 150 mM KCl, and 2 mM MgCl2). The RNA was eluted from the beads with 200 μl of 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10 mM EDTA, and 1% SDS by incubating 10 min at 65°C, followed by a phenol-chloroform extraction and ethanol precipitation. For the reverse transcription, 2 μl of RNA was incubated at 70°C for 5 min in the presence of 0.5 μg of pd(N)6 and quickly chilled on ice. The reverse transcription reaction was performed according to indications in a 1× buffer (50 mM Tris-HCl, pH 8.3, 50 mM KCl, 4 mM MgCl2, and 10 mM dithiothreitol) containing 10 mM dNTPs and 20 U of RNAguard, with 100 U reverse transcriptase for 1 h at 42°C. The cDNAs were then amplified by PCR using primers in the ASH1 sequence.
Fluorescence In Situ Hybridization and Immunofluorescence
Yeast cells were processed for fluorescence in situ hybridization (FISH) and immunofluorescence according to the protocols described in Chartrand et al. (2000). For in situ hybridization, yeast spheroplasts were hybridized with a pool of Cy3-conjugated ASH1 DNA oligonucleotide probes. For immunofluorescence, a 1:50 dilution of a mouse anti-myc 9E10 antibody (Oncogene Science, Cambridge, MA) was used as primary antibody. For the secondary antibody, a 1:1000 dilution of a anti-mouse Cy3-conjugated antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) was used.
Protein Expression and Purification
Recombinant protein GST-She2, GST-She2-M2, and GST-Srp1 were overproduced in Escherichia coli BL21 transformed with pGEX-6P1-She2, pGEX-6P1-She2-M2, and pGEX-5x3-Srp1. The cells were harvested 3 h after induction with 1 mM IPTG at 30°C, resuspended in 0.1% PBS-Triton X-100, 1 M NaCl, and 1 mg/ml lysozyme and antiproteases cocktail (PMSF + pepstatin + leupeptin + aprotinin) for 30 min on ice and sonicated. The lysate was cleared by centrifugation for 15 min at 15,000 × g at 4°C, to yield the supernatant with the overexpressed soluble protein. The glutathione _S_-transferase (GST) fusion proteins were purified by affinity chromatography with glutathione-Sepharose 4B (GE Healthcare, Waukesha, WI) and eluted with 10 mM reduced glutathione in PBS. The recombinant protein fractions were dialyzed overnight in PBS and concentrated using a 10-kDa molecular-weight cutoff filter unit (Centricon-Millipore, Bedford, MA). For the elution of Srp1p, the GST tag was cleaved with Factor Xa overnight at room temperature.
GST Pulldown Assays
For the recombinant protein interactions of Srp1p with She2p and She2-M2, purified Srp1p was incubated with 5 μg of GST-She2p (wild type or mutant) bound to glutathione-Sepharose 4B (GE Healthcare). The binding was performed at room temperature for 3 h in 500 μl of binding buffer (50 mM HEPES-KOH, pH 7.3, 20 mM potassium acetate, 2 mM EDTA, 0.1% Triton X-100, and 5% glycerol). The matrix was recovered by centrifugation and washed four times with 500 μl of binding buffer. The bound proteins were eluted by boiling in Laemmli buffer and separated on a 10% SDS-PAGE. For the interactions between recombinant GST-Srp1p and endogenous She2p-myc, She2-M2-myc, She2-M2-M5A-myc, and She2-M5A-myc, 5 μg of recombinant GST-Srp1p was bound to glutathione-Sepharose 4B and incubated with yeast extract for 2.5 h at 18°C. The matrix was recovered by centrifugation and washed four times with 500 μl of binding buffer. The bound proteins were eluted with preheated SDS sample buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, 12.5 mM EDTA, and 0.02% bromophenol blue). Eluted proteins were analyzed by Western blot.
RESULTS
Monomeric She2p Interacts Directly with the Importin α Srp1p in Order to Enter the Nucleus
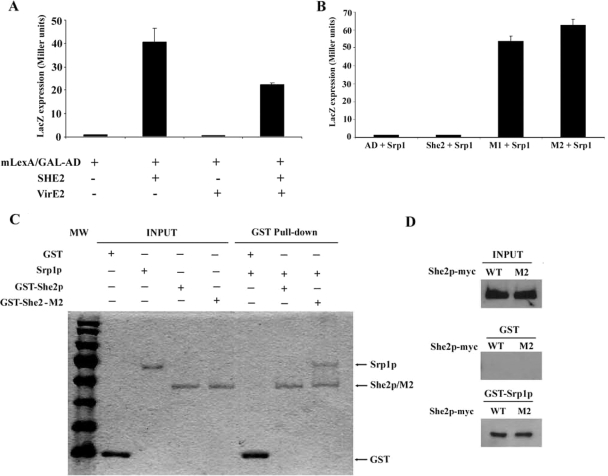
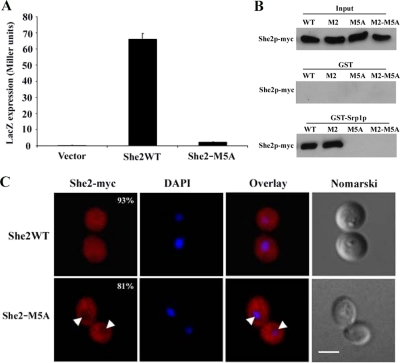
Previous work has shown that She2p transits through the nucleus (Kruse et al., 2002; Du et al., 2008). Because of its size (26 kDa), which is below the nuclear pore diffusion limit (40 kDa in yeast), a She2p monomer may enter passively through the nuclear pores. However, because the functional structure of She2p is that of a homodimer of more than 50 kDa (Niessing et al., 2004), it is possible that active nuclear import is required. To determine if She2p shuttles between the nucleus and the cytoplasm actively or passively, a yeast genetic assay was used (Rhee et al., 2000). In this assay, the protein of interest is fused to a chimera made of a modified LexA protein (mLexA), containing a disrupted NLS, and of the GAL4 activation domain (mLexA-GAL4AD). If the protein of interest contains a NLS, it promotes the nuclear import of the mLexA-GAL4AD chimera, which activates the expression of reporter genes (LacZ and HIS3). In this assay, a fusion of She2p with the mLexA-GAL4AD resulted in the activation of LacZ, whereas the mLexA-GAL4AD itself induced little β-galactosidase activity, suggesting that She2p promotes the nuclear import of the fusion protein (Figure 1A). It was possible that the She2p-mLexA-GAL4AD fusion protein could diffuse passively through the nuclear pores. To eliminate this possibility, the 70-kDa protein VirE2, from Agrobacterium tumefaciens, was added to the She2p-mLexA-GAL4AD in order to increase the size of the fusion protein (Rhee et al., 2000). Even this large fusion protein was actively imported in the nucleus in a She2p-dependent manner (Figure 1A), suggesting that She2p contains an active NLS.
Figure 1.
Monomeric She2p interacts directly with the importin α Srp1p and is actively imported into the nucleus. (A) Yeast nuclear import assay. Plasmids expressing the chimeric protein mLexA/GAL4AD alone or in fusion with She2p, VirE2, or She2p+VirE2 were transformed in L40 yeast strain, and their nuclear import efficiency was determined by measuring β-galactosidase activity. (B) Yeast two-hybrid assay. Srp1p was used as bait, and coexpressed with She2 wild-type or the mutants She2-M1 and She2-M2 in pJ69-4A strain. Protein interactions were determined by measuring β-galactosidase activity. (C) GST pulldown assay to detect interaction between Srp1p and She2p in vitro. Recombinant proteins GST, Srp1p, GST-She2p, and GST-She2-M2 expressed and purified from bacteria were loaded on gel for input levels. Equal amount of Srp1p were loaded on glutathione beads bound with GST, GST-She2p, or GST-She2-M2. After washes, proteins on the beads were eluted by boiling, loaded on SDS-PAGE gel, and detected by Coomassie Blue staining. (D) GST pulldown assay to detect interaction between Srp1p and She2p-myc or She2p-M2-myc from yeast extracts. Input: total She2p-myc or She2p-M2-myc in yeast extracts; GST: She2p from yeast extracts interacting with GST alone; GST-Srp1p: She2p from yeast extracts interacting with GST-Srp1p.
The main nuclear import pathway in yeast depends on the importin α Srp1p (Lange et al., 2007). A previous large scale two-hybrid screen in yeast found that Srp1p interacts with She2p (Ito et al., 2001), suggesting that nuclear import of She2p may depend on Srp1p. To explore this possibility, the interaction between She2p and Srp1p was tested in a yeast two-hybrid assay. However, no interaction between these two proteins could be detected in this assay (Figure 1B). Although She2p forms a homodimer and dimerization is important for its RNA-binding capacity (Niessing et al., 2004), it is possible that Srp1p interacts only with the She2p monomer. Two mutants of She2p that have been shown to disrupt She2p dimerization, She2p-M1 and She2p-M2, containing the mutations Cys68→Tyr and Ser120→Tyr, respectively, were generated (Niessing et al., 2004; and data not shown). When these monomeric mutants were tested in the yeast two-hybrid assay, both interacted strongly with Srp1p (Figure 1B), suggesting that it is indeed the She2p monomer which interacts with Srp1p. To confirm these results, recombinant Srp1p, GST-She2p, and GST-She2p-M2 were purified from bacteria and the direct interaction between Srp1p and the She2p variants was tested by GST pulldown. As shown in Figure 1C, although the GST-She2 protein was unable to pull down Srp1p, GST-She2p-M2 and Srp1p did bind in this assay, suggesting a direct interaction between Srp1p and a She2p monomer.
The observation that only monomeric She2p interacted with Srp1p raised the possibility that a fraction of endogenous yeast She2p may be able to interact with Srp1p. To investigate this question, wild-type She2p and mutant M2 tagged with 9xMyc were expressed at endogenous levels in a she2 yeast strain. Recombinant GST-Srp1p was used to pull down the She2p-myc variants from yeast protein extracts. Interestingly, using this GST pulldown assay, both wild-type She2p-myc and She2p-M2-myc from yeast extracts interacted with GST-Srp1p at similar levels (Figure 1D). Although one would have expected that more She2p-M2-myc than wild-type She2p-myc would be pulled down by GST-Srp1p, equal amount of both proteins were repeatedly pulled down. This is possibly due to the saturation of GST-Srp1p by the numerous NLS-containing proteins in the extract, so that only a small but equal amount of She2p wild-type and M2 mutant may be pulled down by GST-Srp1p. Nevertheless, these results suggest that a significant fraction of She2p-myc can interact with Srp1p in vivo.
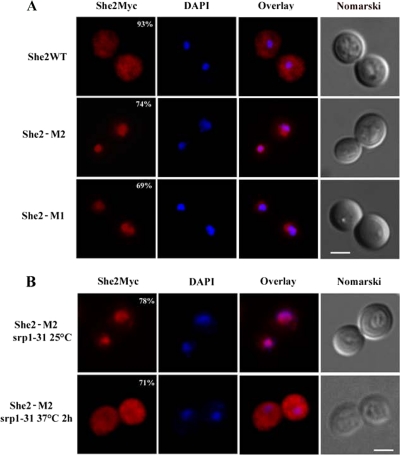
Because the monomeric She2p interacts with Srp1p better than the She2p dimer, this raises the possibility that the monomeric protein may accumulate in the nucleus. The myc-tagged She2 wild-type, M1 and M2 proteins were expressed at endogenous levels in a she2 yeast strain and their intracellular distribution was determined by immunofluorescence. As shown in Figure 2A, although the wild-type She2p-myc was present in both cytoplasm and nucleus of yeast cells, She2p-M1-myc and She2p-M2-myc accumulated only in the nucleus. To determine if this nuclear accumulation of the monomeric She2p depends on Srp1p, She2p-M2-myc was expressed in a temperature-sensitive mutant strain of SRP1, the srp1-31 strain (Loeb et al., 1995), and its distribution at permissive (25°C) and restrictive (37°C) temperatures was measured. Although She2p-M2-myc was mostly nuclear in the srp1-31 strain at 25°C, its nuclear import was impaired when the strain was shifted to nonpermissive temperature for 2 h (Figure 2B). Altogether, these data suggest that the nuclear import of She2p depends on Srp1p.
Figure 2.
Nuclear import of monomeric She2p depends on Srp1p. (A) Immunofluorescence detection of wild-type She2p-myc (WT) and mutants She2p-M1-myc (SHE2-M1) and She2p-M2-myc (SHE2-M2) in yeast cells. Percentage of cells with the displayed phenotype is indicated. Scale, 2 μm. (B) Immunofluorescence on She2p-M2-myc in srp1-31 strain at permissive (top panels) or restrictive (bottom panels) temperature. Percentage of cells with the displayed phenotype is indicated. Scale, 2 μm.
A Nonclassical NLS Promotes the Nuclear Import of She2p
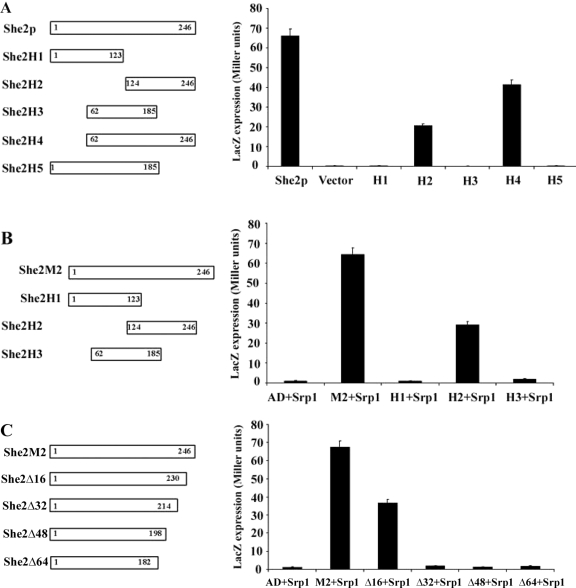
As mentioned previously, no NLS had yet been identified in She2p. To map this NLS, we used the genetic nuclear import and yeast two-hybrid assays described above. Five different deletions were generated in She2p (She2p-H1 to -H5, Figure 3A), fused to the mLexA-GAL4AD protein and tested in the nuclear import assay. As shown in Figure 3A, only the deletions containing the last 61 amino acids of She2p (She2p-H2 and -H4) were able to activate the expression of LacZ. In the yeast two-hybrid assay using Srp1p as bait, only the deletion that contains the C-terminal end of She2p (She2p-H2) interacted with Srp1p (Figure 3B), confirming the results from the nuclear import assay. To define the She2p NLS more precisely, successive deletions of 16 amino acids were performed from the C-terminal end of the She2p-M2 protein (She2p-Δ16 to -Δ64) and tested for their interaction with Srp1p in the yeast two-hybrid assay. As shown in Figure 3C, whereas a deletion of the last 16 amino acids of She2p still interacted with Srp1p (She2-Δ16), deletion of the last 32 amino acids completely disrupted this interaction (mutant She2-Δ32), suggesting that part of the NLS lies between amino acids 214 and 230 of She2p.
Figure 3.
Identification of an NLS at the C-terminal end of She2p. (A) Yeast nuclear import assay. Diagram of the She2p deletions used in this assay (H1–H5). She2p deletions were fused to the LexA/GAL4AD chimera, and their nuclear import efficiency was determined by measuring β-galactosidase activity. Vector: plasmid expressing LexA/GAL4AD chimera alone. (B) Yeast two-hybrid assay between Srp1p and She2p-M2 deletions. Diagram of the She2p-M2 deletions used in this assay (H1–H3). Interaction between Srp1p and a She2p-M2 deletion was determined by measuring β-galactosidase activity. AD: plasmid expressing Gal4 activation domain only. (C) Yeast two-hybrid assay between Srp1p and She2p-M2 deletions at its C-terminus. Diagram of the She2p-M2 deletions used in this assay (Δ16–Δ64). Interaction between Srp1p and a She2p-M2 deletion was determined by measuring β-galactosidase activity. AD, plasmid expressing Gal4 activation domain only.
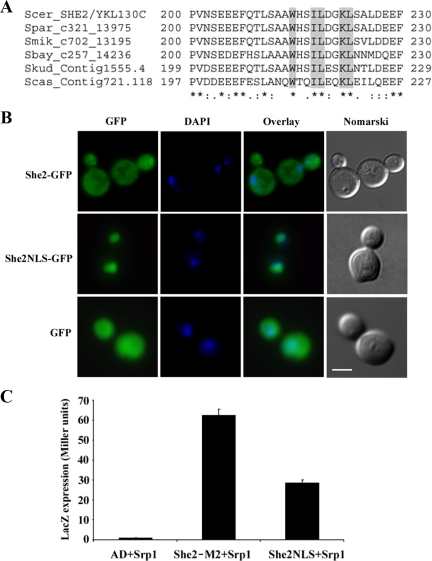
Surprisingly, the amino acid sequence of this region of She2p is very poor in basic amino acids (arginine and lysine), which are commonly found in classical NLS sequences (Lange et al., 2007). An alignment of amino acids 200–230 of She2p from Saccharomyces sensu stricto species showed the presence of only one highly conserved lysine (position 222) and no arginine (Figure 4A). To determine if this region of She2p can independently act as a NLS, a 30-amino acid peptide encompassing positions 200–230 of She2p was fused at the amino-terminal end of green fluorescent protein (GFP; She2NLS-GFP), and the distribution of this fusion protein was determined by epifluorescence microscopy. As shown in Figure 4B, GFP alone was distributed in both the cytoplasm and the nucleus of yeast cells, as a She2p-GFP fusion protein (which shuttles between the nucleus and cytoplasm). However, She2NLS-GFP accumulated in the nucleus of yeasts. This She2NLS peptide was also found to interact with Srp1p in the yeast two-hybrid assay (Figure 4C), suggesting that this peptide has the properties of a true NLS.
Figure 4.
A nonclassical NLS mediates nuclear import of She2p. (A) Sequence alignment of a 30-amino acid peptide containing the NLS of She2p. Sequences from She2p homologues from S. cerevisiae (Scer), S. paradoxus (Spar), S. mikatae (Smik), S. bayanus (Sbay), S. kuderii (Skud), and S. casei (Scas) are shown. Amino acids mutated are underlined in gray. (B) Epifluorescence microscopy on yeast cells expressing She2-GFP (top panels), GFP alone (bottom panels), or GFP fused to a 30-amino acid NLS of She2p (middle panels). Scale, 2 μm. (C) Srp1p interacts with the 30-amino acid NLS peptide from She2p in a yeast two-hybrid assay. Interaction between Srp1p and She2p-M2 protein or the She2NLS peptide was determined by measuring β-galactosidase activity. AD, plasmid expressing Gal4 activation domain only.
To better define the NLS in this peptide, the K222 residue and four other highly conserved amino acids surrounding this lysine (W215, I219, L220, and L223) were mutated to alanine in She2p to generate the mutant She2-M5A (Figure 4A). When tested in the nuclear import assay, the She2-M5A protein failed to promote the nuclear accumulation of the mLexA-GAL4AD fusion protein and to activate the expression of LacZ (Figure 5A). A myc-tagged She2-M5A protein was expressed at endogenous levels in a she2 strain and its interaction with Srp1p was tested using a GST pulldown assay. Mutation of these five residues in the NLS disrupted the interaction between She2p and GST-Srp1p (Figure 5B). To eliminate the possibility that the M5A mutation favors the dimeric conformation of She2p over the monomeric form and therefore inhibits its interaction with Srp1p, the Ser120→Tyr mutation, which produces a She2p monomer, was introduced in She2p-M5A. The resulting protein, She2p-M2-M5A-myc, was still unable to bind GST-Srp1p (Figure 5B), suggesting that the M5A mutation disrupts the binding interface between She2p and Srp1p. Finally, the distribution of this mutant She2p was determined by immunofluorescence. As shown in Figure 5C, although the wild-type She2p-myc was found in both cytoplasm and nucleus, the She2-M5A-myc protein was excluded from the yeast nucleus. Altogether, these results show that the NLS of She2p is present between amino acids 214–222 at the C-terminal end of this protein and that mutation of five specific residues in this NLS disrupts the nuclear targeting of She2p.
Figure 5.
Mutations in NLS of She2p impair interaction with Srp1p and nuclear import of this factor. (A) Yeast nuclear import assay. She2p wild-type (WT) or NLS mutant (She2p-M5A) were fused to the LexA/GAL4AD chimera and their nuclear import efficiency was determined by measuring β-galactosidase activity. Vector: plasmid expressing LexA/GAL4AD chimera alone. (B) GST pulldown assay to detect interaction between Srp1p and She2p-myc, She2p-M2-myc, She2p-M5A-myc, or She2p-M2-M5A-myc from yeast extracts. Input: total She2p-myc, She2p-M2-myc, She2p-M5A-myc or She2p-M2-M5A-myc in yeast extracts; GST: She2p from yeast extracts interacting with GST alone; GST-Srp1p: She2p from yeast extracts interacting with GST-Srp1p. (C) Immunofluorescence on yeast cells expressing wild-type She2p-myc (top panels) or She2p-M5A-myc (bottom panels). White arrows point to low She2p-myc level in the nuclei, and numbers reflect extent of the phenotype observed. Scale, 2 μm.
Nuclear Import of She2p Is Required for Proper Localization of the ASH1 mRNA at the Bud Tip and for the Sorting of Ash1p
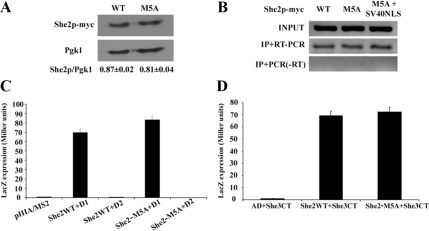
The generation of a mutant She2 protein that cannot be targeted to the nucleus opened the possibility of exploring the nuclear function of She2p in terms of ASH1 mRNA localization and Ash1p asymmetric distribution. First, the She2p-M5A mutant was tested to determine if the point mutations in the NLS had any effect on its RNA-binding capacity or its interaction with She3p. Expression levels of She2p-myc and She2p-M5A-myc in yeast were similar, as shown by Western blot (Figure 6A). Immunoprecipitation of the myc-tagged She2p variants, followed by RT-PCR amplification of the ASH1 mRNA, showed no significant difference in ASH1 mRNA binding between the wild-type and the M5A mutant in vivo (Figure 6B). Because only a small amount of ASH1 mRNA is present in the nucleus, this result shows that even when She2p is excluded from the nucleus, it can still efficiently interact with this transcript in the cytoplasm. This was confirmed in a yeast three-hybrid assay that measures the interaction between She2p and the ASH1 mRNA zip codes (Long et al., 2000; Olivier et al., 2005). In this assay, one of the ASH1 zip code (the domain 1 of element E2B or D1), was used as bait for the interaction with She2p. No difference between wild-type She2 and She2-M5A proteins was observed in binding of this ASH1 localization element (Figure 6C). Finally, to determine if this mutation cause any disruption in the binding of She2p with She3p, which bridges She2p to the myosin Myo4p, a yeast two-hybrid assay was used (Long et al., 2000). In this assay, She2p-M5A showed no defect in its interaction with the C-terminal domain of She3p compared with wild-type She2p (Figure 6D).
Figure 6.
The NLS-mutated She2 protein is as functional as the wild-type She2p. (A) Expression levels of wild-type She2p-myc and She2p-M5A-myc proteins determined by Western blot. Expression levels were normalized with Pgk1 protein. (B) Immunoprecipitation of She2p-myc wild-type (WT), M5A mutant (M5A), or M5A mutant+SV40NLS, followed by reverse transcription and PCR amplification of endogenous ASH1 mRNA. Input: ASH1 mRNA from total yeast extract; IP+RT-PCR: ASH1 mRNA from immunoprecipitate, reverse-transcribed, and amplified by PCR; IP + PCR (−RT): ASH1 mRNA from immunoprecipitate, amplified by PCR without reverse transcriptase. (C) Three-hybrid assay using yeast strain YBZ1 she2 transformed with plasmid expressing either wild-type (She2WT) or NLS mutant (She2M5A) of She2p. This strain also expressed the GAL4 activation domain fused with the C-terminal domain of She3p, along with a plasmid expressing a chimeric RNA containing the localization E2B-D1 fused to the MS2 stem-loop (D1). Controls: empty vector (pIIIA/MS2-2) and an inactive fragment of the localization element E2B (D2; Olivier et al., 2005). (D) Interaction between the C-terminal domain of She3p and wild-type (She2WT) or NLS-mutated (She2M5A) She2 proteins in a yeast two-hybrid assay. AD, plasmid expressing Gal4 activation domain only.
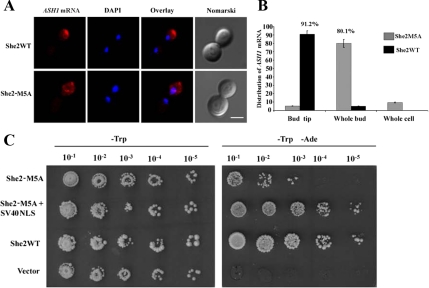
To determine the effect of the nuclear exclusion of She2p on ASH1 mRNA localization, myc-tagged She2p or She2p-M5A were expressed in a she2 yeast strain and the localization of ASH1 mRNA in these strains was visualized by FISH. As shown in Figure 7A, although the wild-type She2p promoted the localization of the ASH1 mRNA at the bud tip of cells in anaphase, the She2p-M5A had a reduced efficiency in ASH1 mRNA localization, as it accumulated in the whole bud of the cells. Indeed, although more than 90% of late-anaphase cells expressing wild-type She2p had bud tip localization of the ASH1 mRNA, this percentage dropped to below 10% in yeasts expressing She2p-M5A (Figure 7B). The effect of the mutation of the She2p NLS on the asymmetric distribution of the Ash1 protein and HO promoter activity was determined using the yeast strain K5547, in which the ADE2 gene is under the control of the HO promoter and which contains a deletion of the SHE2 gene (Jansen et al., 1996). In this strain, symmetric distribution of Ash1p leads to repression of the ADE2 gene and absence of growth on plate lacking adenine (−Ade). If She2p function is restored, Ash1p accumulates in the daughter cell, so expression of the ADE2 gene in the mother cell allows growth on −Ade plates. When transformed with a plasmid expressing the wild-type She2p, the K5547 strain grew on −Ade plates, whereas the same strain transformed with the empty vector did not grow (Figure 7C). When transformed with a plasmid expressing She2p-M5A, a slower growth on −Ade plates was observed compared with the strain expressing wild-type She2p, suggesting that the M5A mutation partially disrupt the asymmetric distribution of Ash1p. However, expression of a She2-M5A protein containing the SV40 NLS (She2p-M5A+SV40NLS), which restores the nuclear localization of She2p-M5A and maintains its interaction with ASH1 mRNA in vivo (Figure 6B), resulted in the same growth on −Ade plates as the strain expressing wild-type She2p (Figure 7C). This suggests that it was the defective nuclear import of She2p-M5A that disrupted the asymmetric localization of Ash1p. Altogether, these results show that the nuclear import of She2p is required for the localization of the ASH1 mRNA at the bud tip and the sorting of the Ash1 protein to the daughter cell nucleus.
Figure 7.
Nuclear import of She2p is required for proper ASH1 mRNA localization and Ash1p sorting. (A) Fluorescent in situ hybridization on ASH1 mRNA in yeast cells expressing either wild-type She2p-myc (top panels) or She2p-M5A-myc (bottom panels). Scale, 2 μm. (B) Scores on mRNA localization phenotypes from A. (C) Yeast genetic assay for Ash1p asymmetric distribution. Tenfold dilutions of exponentially growing K5547 (HO-ADE2, she2) transformed either with the empty YCPlac22 plasmid (vector), YCP22-She2-myc (SHE2), YCP22-She2-M5A+SV40NLS, or YCP22-She2-M5A-myc were spotted on plates lacking tryptophan (−Trp) or lacking tryptophan and adenine (−Trp −Ade) and incubated at 30°C.
Nuclear Import of She2p Is Essential for the Recruitment of Loc1p and Puf6p to the ASH1 mRNA
The work presented so far shows that the nuclear import of She2p is important for its function in cytoplasmic mRNA localization, but the reason is not clear. Two other nuclear RNA-binding proteins are known to play a role in ASH1 mRNA localization and Ash1p sorting: Puf6p and Loc1p. Puf6p is predominantly nucleolar, binds the localization element E3 in the 3′UTR of ASH1 and is involved in the translational repression of this transcript (Gu et al., 2004; Deng et al., 2008). Its deletion results in defects in both ASH1 mRNA localization and Ash1p asymmetric distribution. Loc1p is also a nucleolar protein, and it binds the 3′UTR of the ASH1 mRNA (Long et al., 2001). Its deletion disrupts ASH1 mRNA localization, and recent data suggest that it is also involved in the translational regulation of this transcript (Long et al., 2001; Komili et al., 2007). Because the She2p-M5A mutant displayed phenotypes similar to the PUF6 and LOC1 deletions (whole bud accumulation of ASH1 mRNA, partial asymmetric distribution of Ash1p), we raised the hypothesis that nuclear She2p might be involved in the binding of Puf6p and Loc1p to the ASH1 mRNA. Hence, the absence of She2p in the nucleus might disrupt the recruitment of Puf6p and Loc1p to the ASH1 mRNA, leading to phenotypes similar to PUF6 and LOC1 knockouts.
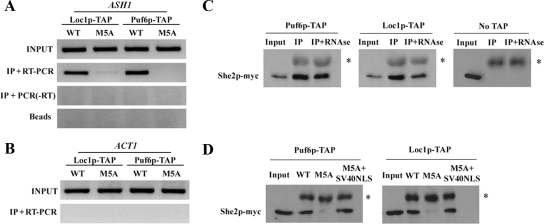
To explore this possibility, She2p and She2p-M5A were expressed at endogenous levels in strains deleted of the endogenous SHE2 gene and containing a TAP-tag integration at the C-terminus of either PUF6 or LOC1 open reading frames. Expression of She2p-M5A had no effect on Loc1p-TAP and Puf6p-TAP expression levels (data not shown). The interaction between the ASH1 mRNA and the Puf6-TAP and Loc1-TAP proteins in vivo was determined by RNA immunoprecipitation. In this assay, the RNP complexes were cross-linked in vivo with formaldehyde, followed by immunoprecipitation of the Puf6-TAP and Loc1-TAP proteins. After de-crosslinking, the associated mRNAs were purified and reverse-transcribed, and the ASH1 cDNA was detected by PCR amplification. In a yeast strain expressing the wild-type She2p, both Puf6p-TAP and Loc1p-TAP interacted with the ASH1 mRNA in vivo (Figure 8A), but not with ACT1 mRNA (Figure 8B). However, when She2p-M5A was expressed, no ASH1 mRNA was found associated with Puf6p-TAP and very little with Loc1p-TAP (Figure 8A), suggesting that the presence of She2p in the nucleus is essential for Puf6p and Loc1p to bind the ASH1 mRNA in vivo.
Figure 8.
Nuclear She2p recruits Puf6p and Loc1p on the ASH1 mRNA. (A) Immunoprecipitation of Loc1p-TAP and Puf6p-TAP from strains expressing either She2p-myc wild-type (WT) or She2-M5A-myc (M5A), followed by RNA purification and RT-PCR amplification of ASH1 mRNA. Input: ASH1 mRNA from total yeast extract; IP+RT-PCR: ASH1 mRNA from immunoprecipitate, reverse-transcribed, and amplified by PCR; IP+PCR (−RT): ASH1 mRNA from immunoprecipitate, amplified by PCR without reverse transcriptase; Beads: mRNA from yeast extract incubated with protein A-Sepharose beads, without antibody. (B) Detection of ACT1 mRNA from immunoprecipitated Puf6p-TAP and Loc1p-TAP. Input: ACT1 mRNA from total yeast extract; IP+RT-PCR: ACT1 mRNA from immunoprecipitate, reverse-transcribed, and amplified by PCR. These results are representative of three independent experiments. (C) She2p-myc coimmunoprecipitates with Loc1p-TAP and Puf6p-TAP. No TAP: yeast strain without TAP-tagged Loc1p or Puf6p; Input: She2p-myc from total yeast extract; IP: immunoprecipitated She2p-myc; IP+RNAse: immunoprecipitated She2p-myc after treatment with RNAse A. The asterisk corresponds to the IgG heavy chain. (D) Nuclear import of She2p is required for its interaction with Loc1p and Puf6p. Immunoprecipitation of Loc1p-TAP and Puf6p-TAP from strains expressing either wild-type She2p-myc (WT), She2-M5A-myc (M5A), or She2-M5A+SV40NLS (M5A+SV40NLS). Input: wild-type She2p-myc from total yeast extract. IP, immunoprecipitation. The asterisk corresponds to the IgG heavy chain.
These results raise the possibility that She2p interacts with Puf6p and Loc1p, and recruit them to the ASH1 mRNA. Therefore, interaction between She2p, Puf6p, and Loc1p in vivo was explored using coimmunoprecipitation. Pulldown of both TAP-tagged Puf6p and Loc1p resulted in the coimmunoprecipitation of She2p-myc (Figure 8C), suggesting an interaction between these factors in vivo. This interaction was independent of RNA because treatment of yeast extracts with RNAse A before immunoprecipitation still resulted in an efficient pulldown of She2p-myc by both Puf6p-TAP and Loc1p-TAP (Figure 8C). Finally, to determine if the presence of She2p in the nucleus is important for its interaction with Puf6p and Loc1p, the coimmunoprecipitation was repeated with She2p-M5A-myc mutant. In this experiment, cross-linking of protein complexes with formaldehyde before immunoprecipitation was performed in order to avoid reconstitution of complexes in the yeast extract after breaking the cells. As shown in Figure 8D, a clear reduction in the amount of She2p-M5A-myc that coimmunoprecipitated with either Puf6p-TAP or Loc1p-TAP was observed compared with wild-type She2p-myc. Wild-type levels of interaction were recovered when the SV40 NLS was fused to the She2p-M5A protein (Figure 8D), suggesting that nuclear import of She2p is required for its interaction with Loc1p and Puf6p. Altogether, these results suggest that nuclear import of She2p is required for its interaction with Puf6p and Loc1p and for their recruitment to the ASH1 mRNA.
DISCUSSION
A Nonclassical NLS Promotes the Nuclear Import of She2p by Binding Importin α
In this work, we show that She2p is actively imported into the nucleus via its interaction with the importin α Srp1p. Our data suggest that She2p is not imported as a native dimer, which is the conformation that binds RNA (Niessing et al., 2004), because the She2p dimer was unable to interact with Srp1p in vitro. She2p from yeast extracts was able to interact with Srp1p, suggesting that a fraction of She2p in vivo may adopt a conformation that is different from the native dimer. However, it is still unclear if this population of She2p corresponds to monomers. A possibility is that this population of She2p may contain posttranslational modifications. Phosphorylation is known to regulate the nuclear import of proteins, such as Gln3p in yeast (Carvalho et al., 2001), and She2p has been reported to be a phosphoprotein in vivo (Gonsalvez et al., 2003). Once in the nucleus, She2p adopts a dimeric conformation because it can bind RNA. Intriguingly, wild-type She2p-myc was not excluded from the nucleus of the srp1-31 strain at nonpermissive temperature (data not shown). This is possibly due to the rapid shutdown of transcription when this strain is shifted at 37°C (Liu et al., 1999). Because She2p nuclear export depends on the nuclear export of newly synthesized mRNAs (Kruse et al., 2002), the shutdown of transcription would explain the absence of nuclear depletion of wild-type She2p in the srp1-31 strain.
Using the interaction between Srp1p and She2p, a 30-amino acid sequence with NLS properties was identified. This NLS promotes the nuclear import of GFP and interacts with Srp1p. Interestingly, its sequence is very divergent from classical monopartite and bipartite NLS because it contains only one lysine and is rich in hydrophobic residues. To our knowledge, all the currently reported NLS that bind directly importin α contain at least two essential basic amino acids (Chen et al., 2005; Lange et al., 2007), suggesting that the repertoire of nuclear localization signals may be larger than suggested. Mutation of five conserved residues in this NLS disrupted the nuclear targeting of She2p and its interaction with Srp1p, confirming its role as a nuclear localization sequence. The defective ASH1 mRNA localization and poor asymmetric sorting of Ash1p seen in the She2p-M5A mutant strain seem to result from the nuclear exclusion of this protein and not from secondary effects of this mutation. Indeed, the RNA-binding capacity of the NLS-mutated She2p was similar to wild-type She2p, as was its interaction with She3p. More important, adding an heterologous classical NLS (like the SV40 NLS) to the She2p-M5A completely restored the function of this protein in vivo.
Nuclear She2p Couples mRNA Localization and Translational Repression
Disrupting the nuclear import of She2p affects the localization of the ASH1 transcript and the asymmetric distribution of Ash1p. We provide evidence that this localization defect is linked to the disrupted interaction between Puf6p and Loc1p with the ASH1 mRNA because 1) She2p interacts in vivo with both Puf6p and Loc1p; 2) the presence of She2p in the nucleus is important for this interaction; 3) exclusion of She2p from the nucleus disrupts the binding of Loc1p and Puf6p to the ASH1 mRNA, and 4) nuclear exclusion of She2p phenocopies the knockouts of LOC1 and PUF6 in term of ASH1 mRNA localization and Ash1p distribution.
Altogether, these data suggest a direct coupling between the mRNA transport and translational control machineries. A role of nuclear She2p in translational control is indeed supported by recent data from the Jansen lab, which showed that nuclear exclusion of She2p accelerates Ash1p synthesis (Du et al., 2008). Intriguingly, Du et al. did not report any defect in ASH1 mRNA localization when She2p was excluded from the nucleus, unlike what we observed (see Figure 7). They used a She2 protein fused to the Myo4p-binding domain of She3p, which resulted in a fusion protein that remained anchored on the actin cytoskeleton via its binding to Myo4p and is excluded from the nucleus. However, it is possible that such tight association of She2p, and of the ASH1 mRNA, to the localization machinery and to the actin cytoskeleton suppresses the localization defects caused by the nuclear exclusion of She2p.
Because She2p binds several localization elements within the coding sequence of the ASH1 mRNA (Chartrand et al., 2002), this coupling may reduce the possibility that elongating ribosomes could displace She2p from this transcript. Such coupling is supported by the finding that She2p coimmunoprecipitates with Puf6p and Loc1p, suggesting an interaction between these proteins in vivo. The mechanism by which She2p promotes the recruitment of Loc1p and Puf6p on the ASH1 mRNA in vivo is not known, because all three proteins can bind the 3′UTR of this transcript independently in vitro (Bohl et al., 2000; Long et al., 2001; Gu et al., 2004). One possibility is that, being in the nucleolus, Puf6p and Loc1p are spatially restricted from polyA+ mRNAs. Because She2p has been recently shown to transit through the nucleolus (Du et al., 2008), it may either bring the ASH1 mRNA in the nucleolus, where Puf6p and Loc1p can bind this transcript, or She2p may recruit these two factors in the nucleolus and bring them to the ASH1 mRNA in the nucleoplasm.
Roles of Nuclear Proteins in Cytoplasmic mRNA Localization
The importance of nuclear events in cytoplasmic mRNA localization is a well-described phenomenon. Several RNA-binding proteins implicated in cytoplasmic mRNA localization are known to be exclusive residents of the nucleus or to shuttle between the cytoplasm and the nucleus (Farina and Singer, 2002). Reports from several model systems have shown that mRNA processing in the nucleus affects its cytoplasmic fate. For instance, proper splicing of the oskar mRNA is required for its localization at the posterior pole of the Drosophila embryo (Hachet and Ephrussi, 2004). In this case, members of the exon junction complex, such as Y14-Mago and eIFIIIA, are assembled on the oskar mRNA in the nucleus and are involved in the cytoplasmic localization of this transcript. In Xenopus oocyte, the nucleocytoplasmic shuttling proteins hnRNP I and Vg1RBP/Vera initiate a localization complex with the Vg1 mRNA in the nucleus (Cote et al., 1999; Kress et al., 2004). Remodeling of this ribonucleoprotein complex has been shown to occur after its nuclear export (Kress et al., 2004). In fibroblasts, both ZBP1 and ZBP2/KSRP proteins can bind the β-actin mRNA in the nucleus (Gu et al., 2002; Oleynikov and Singer, 2003). A handover mechanism has been proposed where the predominantly nuclear ZBP2 binds the nascent β-actin transcript and facilitates the subsequent recruitment of ZBP1, the factor involved in the cytoplasmic localization of the β-actin mRNA (Pan et al., 2007).
Our study reveals another function for the nucleo-cytoplasmic shuttling of RNA-binding proteins involved in mRNA localization. By promoting the recruitment of Puf6p and Loc1p on the nuclear ASH1 mRNA, She2p initiates the translational repression of the localized mRNA before its export in the cytoplasm and prevents premature translation of this transcript. Coupling mRNA localization and translational control constitutes an efficient way to ensure that the translation of transcripts targeted for localization will be properly regulated. This raises the possibility that other transcripts that are localized at the bud tip of yeasts may also be translationally repressed by Puf6p and/or Loc1p via their recruitment with She2p.
Supplementary Material
[Supplemental Materials]
ACKNOWLEDGMENTS
We thank Drs. Gerry Fink (Whitehead Institute) and Michael Culbertson (University of Wisconsin–Madison) for reagents and strains. We also thank Emmanuelle Querido for critical reading of the manuscript. This work was supported by a grant from the Canadian Institutes for Health Research. P.C. is a Chercheur-Junior 2 fellow from the Fond de Recherche en Santé du Québec.
Abbreviations used:
NLS
nuclear localization signal.
Footnotes
REFERENCES
- Bohl F., Kruse C., Frank A., Ferring D., Jansen R. -P. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000;19:5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho J., Bertram P. G., Wente S. R., Zheng X.F.S. Phosphorylation regulates the interaction between Gln3p and the nuclear import factor Srp1p. J. Biol. Chem. 2001;276:25359–25365. doi: 10.1074/jbc.M103050200. [DOI] [PubMed] [Google Scholar]
- Chartrand P., Meng X., Huttelmaier S., Donato D., Singer R. H. Asymmetric sorting of Ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol. Cell. 2002;10:1319–1330. doi: 10.1016/s1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- Chartrand P., Singer R. H., Long R. M. RNP localization and transport in yeast. Annu. Rev. Cell Dev. Biol. 2001;17:297–310. doi: 10.1146/annurev.cellbio.17.1.297. [DOI] [PubMed] [Google Scholar]
- Chartrand P., Singer R. H., Long R. M. Sensitive and high-resolution detection of RNA in situ. Methods Enzymol. 2000;318:493–506. doi: 10.1016/s0076-6879(00)18072-3. [DOI] [PubMed] [Google Scholar]
- Chen M. -H., Ben-Efraim I., Mitrousis G., Walker-Kopp N., Sims P. J., Cingolani G. Phospholipid scramblase 1 contains a nonclassical nuclear localization signal with unique binding site in importin α. J. Biol. Chem. 2005;280:10599–10606. doi: 10.1074/jbc.M413194200. [DOI] [PubMed] [Google Scholar]
- Cote C. A., Gautreau D., Denegre J. M., Kress T. L., Terry N. A., Mowry K. L. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol. Cell. 1999;4:431–437. doi: 10.1016/s1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- Darzacq X., Powrie E., Gu W., Singer R. H., Zenklusen D. RNA asymmetric distribution and daughter/mother differentiation in yeast. Curr. Opin. Microbiol. 2003;6:614–620. doi: 10.1016/j.mib.2003.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Singer R. H., Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 2008;22:1037–1050. doi: 10.1101/gad.1611308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T. -G., Schmid M., Jansen R. -P. Why cells move messages: the biological functions of mRNA localization. Semin. Cell Dev. Biol. 2007;18:171–177. doi: 10.1016/j.semcdb.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Du T. G., Jellbauer S., Müller M., Schmid M., Niessing D., Jansen R. P. Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO Rep. 2008;9:781–787. doi: 10.1038/embor.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina K. L., Singer R. H. The nuclear connection in RNA transport and localization. Trends Cell Biol. 2002;12:466–472. doi: 10.1016/s0962-8924(02)02357-7. [DOI] [PubMed] [Google Scholar]
- Gonsalvez G. B., Lehmann K. A., Ho D. K., Stanitsa E. S., Williamson J. R., Long R. M. RNA-protein interactions promote asymmetric sorting of the ASH1 mRNA ribonucleoprotein complex. RNA. 2003;9:1383–1399. doi: 10.1261/rna.5120803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Deng Y., Zenklusen D., Singer R. H. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004;18:1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Pan F., Zhang H., Bassell G. J., Singer R. H. A predominantly nuclear protein affecting cytoplasmic localization of β-actin mRNA in fibroblasts and neurons. J. Cell Biol. 2002;156:41–52. doi: 10.1083/jcb.200105133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids. Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O., Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- Irie K., Tadauchi T., Takizawa P. A., Vale R. D., Matsumoto K., Herskowitz I. The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J. 2002;21:1158–1167. doi: 10.1093/emboj/21.5.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. P., Dowzer C., Michaelis C., Galova M., Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin Myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- Komili S., Farny N. G., Roth F. P., Silver P. A. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress T. L., Yoon Y. J., Mowry K. L. Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J. Cell Biol. 2004;165:203–211. doi: 10.1083/jcb.200309145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse C., Jaedicke A., Beaudouin J., Bohl F., Ferring D., Guttler T., Ellenberg J., Jansen R. P. Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J. Cell Biol. 2002;159:971–982. doi: 10.1083/jcb.200207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. Classical nuclear localization signals: definition, function, and interaction with importin α. J. Biol. Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Guo W., Tartakoff P. Y., Tartakoff A. M. A Crm1p-independent nuclear export path for the mRNA-associated protein, Npl3p/Mtr13p. Proc. Natl. Acad. Sci. USA. 1999;96:6739–6744. doi: 10.1073/pnas.96.12.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb J. D., Schlenstedt G., Pellman D., Kornitzer D., Silver P. A., Fink G. R. The yeast nuclear import receptor is required for mitosis. Proc. Natl. Acad. Sci. USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R. M., Gu W., Lorimer E., Singer R. H., Chartrand P. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J. 2000;19:6592–6601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R. M., Gu W., Meng X., Gonsalvez G., Singer R. H., Chartrand P. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J. Cell Biol. 2001;153:307–318. doi: 10.1083/jcb.153.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R. M., Singer R. H., Meng X., Gonzalez I., Nasmyth K., Jansen R. -P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- Niessing D., Huttelmaier S., Zenklusen D., Singer R. H., Burley S. K. She2p is a novel RNA binding protein with a basic helical hairpin motif. Cell. 2004;119:491–502. doi: 10.1016/j.cell.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Oleynikov Y., Singer R. H. Real-time visualization of ZBP1 association with β-actin mRNA during transcription and localization. Curr. Biol. 2003;13:199–207. doi: 10.1016/s0960-9822(03)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier C., Poirier G., Gendron P., Boisgontier A., Major F., Chartrand P. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol. Cell. Biol. 2005;25:4752–4766. doi: 10.1128/MCB.25.11.4752-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Huttelmaier S., Singer R. H., Gu W. ZBP2 facilitates binding of ZBP1 to β-actin mRNA during transcription. Mol. Cell. Biol. 2007;27:8340–8351. doi: 10.1128/MCB.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin N., Ménade M., Poirier G., Donato D., Drouet E., Chartrand P. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol. Cell. 2007;26:795–809. doi: 10.1016/j.molcel.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Rhee Y., Gurel F., Gafni Y., Dingwall C., Citovsky V. A genetic system for detection of protein nuclear import and export. Nat. Biotech. 2000;18:433–437. doi: 10.1038/74500. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P. Methods in Yeast Genetics. A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Schiestl R., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Sil A., Herskowitz I. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell. 1996;84:711–722. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- St. Johnston D. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Takizawa P. A., Sil A., Swedlow J. R., Herskowitz I., Vale R. D. Actin-dependent localization of an mRNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- Takizawa P. A., Vale R. D. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc. Natl. Acad. Sci. USA. 2000;97:5273–5278. doi: 10.1073/pnas.080585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Materials]