Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila (original) (raw)
Abstract
miRNA-mediated repression affects a wide range of biological processes including development and human pathologies. The GW182 protein is a key component of miRNA repression complex, recruited by Argonaute and functioning downstream to repress translation and accelerate mRNA degradation, but little is known about how GW182 proteins act. Using both tethered function and complementation assays, we identify three independent domains of the Drosophila GW182 protein (also termed Gawky) that are sufficient to repress mRNA. Each of these domains also functions independently of poly(A) tails. These results indicate that miRNA-mediated repression is facilitated by multiple domains of GW182.
Keywords: GW182, mRNA, miRNA, translational control
INTRODUCTION
miRNAs are short noncoding RNAs that function in many eukaryotes, regulating multiple biological processes, including cell growth, division and differentiation, genome integrity, and defense against pathogens (for review, see Bushati and Cohen 2007). The prediction that more than one-third of human genes are regulated by miRNAs argues for the crucial role of miRNAs in gene expression (Lewis et al. 2005). However, the mechanisms by which miRNAs control gene expression are not well understood (for review, see Valencia-Sanchez et al. 2006; Du and Zamore 2007; Nilsen 2007; Eulalio et al. 2008a; Filipowicz et al. 2008; Wu and Belasco 2008). To exert their repressive function, miRNAs associate with Argonaute (Ago) proteins to assemble the RNA-induced silencing complex (RISC) and guide it to the mRNA targets by base complementarity. Most animal miRNAs show only partial complementarity to their target mRNAs and direct either repression of mRNA translation and/or accelerated target mRNA deadenylation.
miRNAs have been proposed to act in a variety of manners to repress mRNA translation. For example, miRNAs have been suggested to promote translation termination, interfere with the growing peptide, or block elongation (Olsen and Ambros 1999; Seggerson et al. 2002; Maroney et al. 2006; Nottrott et al. 2006 ; Petersen et al. 2006; Lytle et al. 2007). Other experiments indicate that miRNAs block translation initiation, possibly by interfering with either 7-methyl-guanosine cap function or the joining of ribosomal subunits (Humphreys et al. 2005; Pillai et al. 2005; Chendrimada et al. 2007; Kiriakidou et al. 2007; Mathonnet et al. 2007; Thermann and Hentze 2007; Wang et al. 2008).
One possible explanation of these diverse results is that miRNAs regulate translation via multiple mechanisms. Importantly, components of RISC—Ago proteins, miRNAs, and their target mRNAs—localize to so-called P-bodies, cytoplasmic foci containing untranslated mRNAs and mRNA degradation enzymes (Ding et al. 2005; Jakymiw et al. 2005; Liu et al. 2005a, b; Pillai et al. 2005; Sen and Blau 2005). Moreover, GW182 and some other P-body components play a crucial role in the miRNA pathway (Eystathioy et al. 2003; Bagga et al. 2005; Ding et al. 2005; Liu et al. 2005a; Meister et al. 2005; Rehwinkel et al. 2005; Behm-Ansmant et al. 2006; Eulalio et al. 2007; Li et al. 2008; Ding and Grosshans 2009). Drosophila GW182 (dGW182) is recruited to the miRNA repression complex via direct association with Ago1, and point mutations disrupting Ago1–dGW182 interaction abrogate repression by Ago1 (Eulalio et al. 2008b). In addition, tethering of dGW182 to an mRNA represses translation even in the absence of Ago1 (Behm-Ansmant et al. 2006). These results clearly demonstrate that dGW182 is a key player in the miRNA repression complex functioning downstream from Ago1. Thus, an important goal will be to understand the mechanism of dGW182 function.
Here we dissect the role of dGW182 protein in miRNA-mediated repression using both tethering of dGW182 deletion mutants to an mRNA reporter and complementation assay in Drosophila cultured cells. We identify three independent functional domains within dGW182 that are sufficient for mRNA repression and function independently of the mRNA poly(A) tail and endogenous dGW182 and Ago1 proteins. Existence of multiple repression mechanisms mediated by different domains of dGW182 protein might reconcile apparently contradictory models of miRNA-mediated repression reported in the current literature.
RESULTS
Three regions within dGW182 are sufficient to repress tethered mRNA
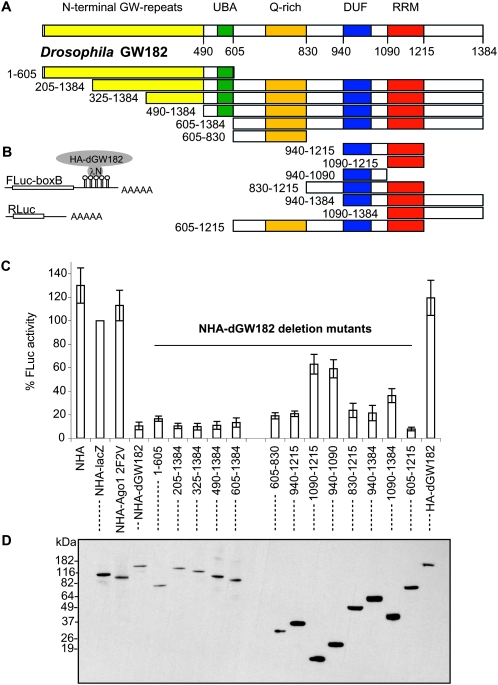
Proteins of the GW182 family are characterized by several blocks of glycine–tryptophan repeats (GW repeats), an ubiquitin-associated (UBA) domain, a glutamine-rich (Q-rich) region, and an RNA recognition motif (RRM) (Fig. 1A; Eystathioy et al. 2002; Behm-Ansmant et al. 2006; for review, see Ding and Han 2007). Previous work has shown that the N-terminal GW-repeats can bind the Argonaute protein, and the N-terminal portion of the protein including the GW-repeats, the UBA domain and the Q-rich region was reported to target dGW182 to P-bodies (Behm-Ansmant et al. 2006; Till et al. 2007). However, the domains within the GW182 proteins that function in mRNA repression are not known.
FIGURE 1.
Three separate domains of Drosophila GW182 are sufficient to repress tethered mRNA. (A) Schematic representation of Drosophila GW182 protein and generated dGW182 deletion mutants. The numbers correspond to the amino acid positions. (B) Schematic representation of reporter constructs: FLuc-boxB contains firefly luciferase coding sequence and 3′-UTR with five boxB sites specifically binding to λN peptide; RLuc contains Renilla luciferase coding sequence and no boxB sites (Rehwinkel et al. 2005). (C) Repression of FLuc-boxB mRNA by NHA-dGW182 and its deletion mutants. Drosophila S2 cells were co-transfected with plasmids encoding for FLuc-boxB, RLuc, and full-length NHA-dGW182 or one of the NHA-dGW182 deletion mutants depicted in A. As negative controls, either NHA alone, NHA fused to β-galactosidase (NHA-lacZ), or mutant NHA-Ago1 F594VF629V (designated as 2F2V) not able to recruit dGW182, or HA-dGW182 without λN peptide was transfected instead of NHA-dGW182 or NHA-dGW182 deletions. Cells were lysed 2 or 3 d post-transfection, and protein levels were measured with dual luciferase assay. Expression of firefly luciferase was normalized to that of the Renilla luciferase. Values are presented as a percentage of firefly luciferase produced in the presence of NHA-lacZ. Values represent the average of at least four experiments. The error bar shows the standard deviation. (D) Expression of different HA-fusion proteins was estimated by Western blotting with antibodies directed against HA-peptide.
To identify the domains of dGW182 that function in mRNA repression, we generated a series of Drosophila GW182 deletion mutants (Fig. 1A) and analyzed their effects on translation in transfected S2 cells using an RNA–protein tethering assay. Tethering was achieved by coexpressing firefly luciferase mRNA containing five boxB sites in its 3′-UTR (designated as FLuc-boxB) and dGW182 deletion mutants fused with HA-tag and λN peptide, which specifically recognizes the boxB hairpins (Gehring et al. 2003; Pillai et al. 2004; Rehwinkel et al. 2005). A plasmid encoding for Renilla luciferase without boxB sites was coexpressed as a transfection control. As negative controls that are not expected to repress FLuc-boxB mRNA, we used λN peptide fused to HA-tag alone (NHA) or along with β-galactosidase-coding sequence (NHA-lacZ). We also tethered to the reporter message mutant Argonaute 1 protein that fails to recruit dGW182 (Ago1 2F2V) (Eulalio et al. 2008b). Figure 1C shows the effects of tethering on FLuc-boxB expression: firefly luciferase (FLuc) activity detected in the presence of negative control NHA-lacZ was taken for 100%, and the rest was expressed accordingly. As expected, NHA-dGW182 effectively represses FLuc-boxB expression (to 10%), while none of the negative controls is able to repress when tethered to the reporter message. In addition, transfection of HA-dGW182 lacking λN peptide did not lead to repression, demonstrating that repression is a result of tethering. We then analyzed the effects of deleting different regions of dGW182 on firefly luciferase expression.
The analysis of dGW182 deletions made two important points. First, no single part of the dGW182 protein was required for repression (Fig. 1C). This demonstrates that dGW182 contains multiple domains able to repress gene expression. Second, we identified three independent domains within dGW182 protein that are sufficient for repression. The first sufficiency domain is the N-terminal portion of the protein, comprising amino acids 1–605 (1–605 fragment): it reduces firefly luciferase activity to 15%. At the same time, its function in dGW182-mediated repression must be redundant, as deletion of the N-terminal part does not significantly reduce the repressive effect of dGW182 (Fig. 1C, cf. repression by full-length protein with that of 205–1384, 325–1384, 490–1384, and 605–1384 fragments). A second domain sufficient for repression is a Q-rich region in the middle of the protein (605–830 fragment), which down-regulates FLuc-boxB expression to 19%. The third domain sufficient for repression is the C-terminal portion of the protein, comprising amino acids 940–1215 and including an RRM domain and a short conserved region upstream, which we called a domain of unknown function (DUF). The integrity of this DUF-RRM region is required to achieve efficient repression (21% FLuc activity) as neither of the domains (DUF or RRM) when tethered alone shows a strong repressive effect (Fig. 1C, cf. the effects of the 940–1215 fragment and that of the 940–1090 and 1090–1215 fragments). Further extension of the 940–1215 fragment to the C terminus or up to the Q-rich region at the N terminus (Fig. 1C, fragments including amino acids 830–1215 and 940–1384) has only a minor effect on activity of the tethered message.
Expression levels of different NHA-dGW182 deletion mutants were estimated by Western blotting (Fig. 1D). This analysis demonstrates that the observed repressive effects cannot be attributed to the differences in expression levels, as the most efficient repressors are actually least abundant (Fig. 1C, full-length dGW182, 1–605, 205–1384, 325–1384, 605–830). Moreover, we observed that although the 1–605, 605–830, and 940–1215 regions of dGW182 could all repress mRNA to a similar extent, the 940–1215 fragment was more efficiently expressed (Fig. 1D). This suggests that the 1–605 and 605–830 regions of dGW182 are stronger repression domains. We also tested how increasing amounts of dGW182 repressive domains affect the activity of tethered reporter (Supplemental Fig. 1). Again, we observed that the repression of protein expression was stronger for the 1–605 and 605–830 regions than the 940–1215. In addition, we observed that different domains reach their maximum repressive effects at different concentrations, suggesting that domains may have different affinities to their downstream targets. This experiment also demonstrates that repression can be saturated and that at the plasmid concentration used in our deletion analysis, the dGW182 effector domains exert maximal repressive properties, and a further increase of expression levels has no additional repressive effect.
The above results identify three regions of the dGW182 protein that can repress mRNA, suggesting that each of these domains can act independently and that the observed repressive effects of the full-length protein represent a combinatorial effect of these different sufficiency regions. Consistent with that view, we observed that tethered constructs that contained both the Q-rich and RRM domains (fragment including amino acids 605–1215) had an additive effect and repressed the tethered message as well as the full-length dGW182 protein.
All three dGW182 sufficiency domains increase mRNA degradation, but also repress translation
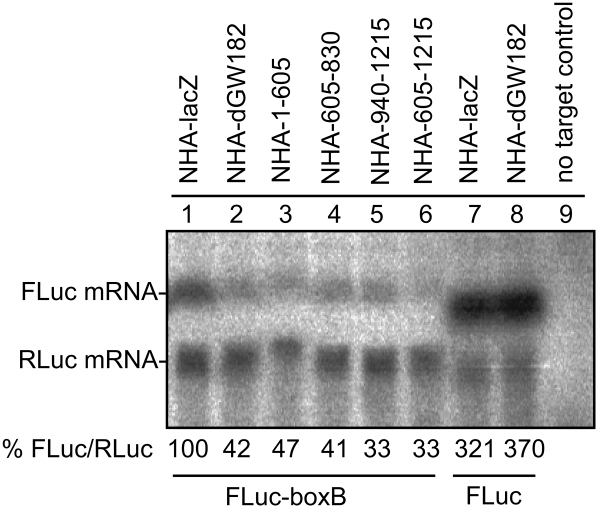
miRNAs are known to repress mRNA through both increasing mRNA degradation and repressing translation. Previous work has shown that when tethered to mRNAs, dGW182 stimulates deadenylation and mRNA degradation as well as brings about translational repression (Behm-Ansmant et al. 2006). To determine whether the different repressive regions of dGW182 would be selective for one of the pathways, i.e., affect mRNA degradation or just primarily protein production, we examined the levels of the reporter mRNA when different repressive domains were tethered to the mRNA. We observed that the full-length dGW182 protein and all three domains sufficient for repression increase degradation of the reporter mRNA approximately two- to threefold (Fig. 2, lanes 1–6). Degradation is due to tethering, as FLuc reporter without boxB sites is not affected by NHA-dGW182 (Fig. 2, lanes 7,8). These results indicate that each of the three domains is able to promote mRNA degradation.
FIGURE 2.
Drosophila GW182 and its effector domains decrease stability of tethered mRNA. Northern blotting was used to estimate the levels of firefly and Renilla luciferase mRNAs from the experiment described in Figure 1. mRNA levels were quantified by PhosphorImaging, and amounts of FLuc-boxB were normalized according to Renilla control and expressed as a percentage of FLuc-boxB level in the presence of NHA-lacZ protein (numbers below the figure represent an average of two experiments). (Lanes 1–6) FLuc-boxB and RLuc RNA levels in the presence of tethered NHA-lacZ, NHA-dGW182, and dGW182 fragments: NHA-1-605, NHA-605-830, NHA-940-1215, and NHA-605-1215, accordingly. (Lanes 7,8) To demonstrate that degradation is the effect of tethering, lanes 7 and 8 show untethered FLuc and RLuc in the presence of either (lane 7) NHA-lacZ or (lane 8) NHA-dGW182. The last lane contains total RNA from untransfected S2 cells (no target control).
These experiments provide additional evidence that dGW182 also represses protein production, independent of its effect on mRNA stability. Specifically, we observed that the reduction in mRNA levels by tethered either full-length dGW182 or its separate domains was less than the reduction in protein levels (Fig. 1C). These observations indicate that all three sufficiency domains both decreased mRNA stability and repressed protein production.
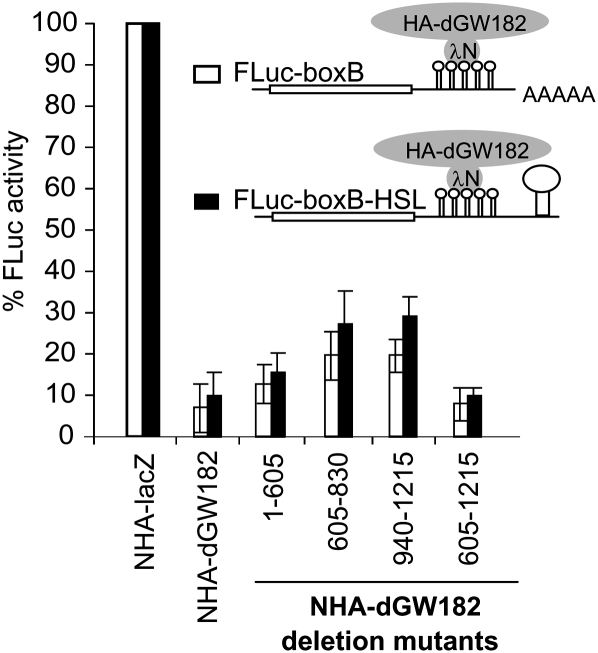
All three dGW182 sufficiency domains repress mRNA independently of poly(A) tails
miRNAs and GW182 have been shown to be able to promote deadenylation, which suggests two possible models for dGW182 function. First, it could be that one or more of the dGW182 domains are sufficient to promote deadenylation of the mRNA body. Alternatively, it could be that each domain can repress mRNA function independently of any effect on the poly(A) tail. To distinguish between these possibilities, we generated a reporter plasmid lacking a polyadenylation site and producing mRNA with the 3′-end of histone H1, which lacks a poly(A) tail and contains a specific stem–loop structure (the construct is designated as FLuc-boxB-HSL). As expected, a reporter having neither a poly(A) tail nor a histone stem–loop is not efficient in protein production; its activity is ∼104-fold lower than that of its polyadenylated counterpart (data not shown). However, addition of the histone stem–loop structure stimulates the reporter translation, so that activity of FLuc-boxB-HSL mRNA is comparable to that of polyadenylated FLuc-boxB. We then examined the effects of the different repressive domains of dGW182 on activity of FLuc-boxB-HSL mRNA.
Strikingly, we observed that tethering of the full-length dGW182 and all three different effector domains represses the FLuc-boxB-HSL mRNA almost as efficiently as the polyadenylated FLuc-boxB mRNA (Fig. 3). This demonstrates that all three dGW182 domains are able to repress mRNA independently of the poly(A) tail. This result is in line with the previous data showing that nonpolyadenylated mRNAs bearing a 3′ histone stem–loop are repressed by miRNAs and tethered dGW182 similarly to their polyadenylated counterparts (Wu et al. 2006; Eulalio et al. 2008b, 2009). We conclude that dGW182 and its individual sufficiency domains can repress mRNA translation directly, independent of a role in deadenylation.
FIGURE 3.
Repression mediated by dGW182 and its sufficiency domains is independent of poly(A) tail. Drosophila S2 cells were transfected with a boxB-containing firefly luciferase reporter, either (open bars) FLuc-boxB or (black bars) FLuc-boxB-HSL. Unlike FLuc-boxB, FLuc-boxB-HSL lacks polyadenylation signal and bears histone H1 stem–loop structure in its 3′-UTR. Together with a firefly luciferase reporter, RLuc as a normalization control and one of the NHA-fusion proteins (NHA-dGW182, or NHA-dGW182 deletion mutant, or NHA-lacZ as a negative control) were co-transfected. For the rest, the assay was performed as described in Figure 1.
All three dGW182 sufficiency domains appear to repress mRNA independently of the endogenous dGW182 and Ago1
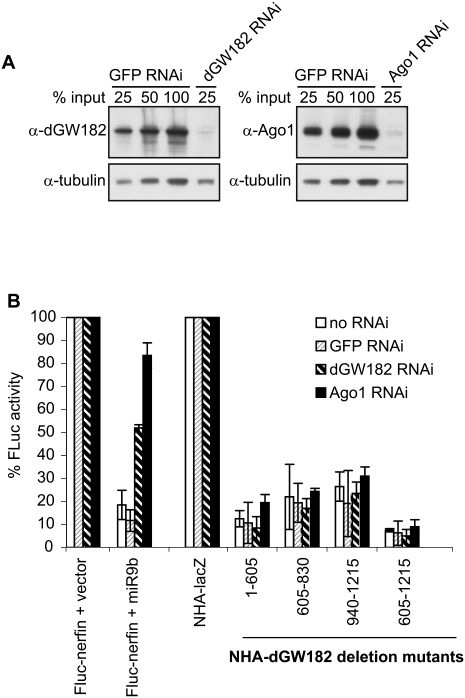
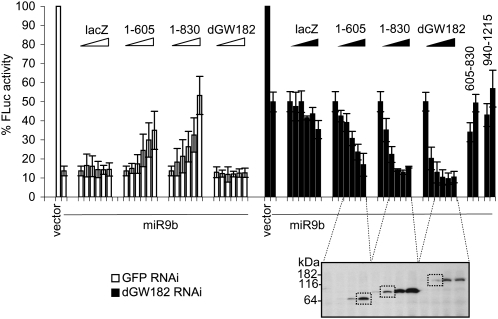
The ability of three distinct domains of dGW182 to repress mRNA function was surprising and suggested two possible models. In one view, each domain is sufficient for interacting, either directly or through additional intermediate proteins, with some target in the translation and/or decay machinery to commit the mRNA to a repressed state. Alternatively, given the known interactions between GW182 and Argonaute proteins, it could be that tethering different domains allows the recruitment of endogenous Argonaute or dGW182 proteins and then that commits the mRNA to repression. This is particularly relevant for the N-terminal domain of dGW182, which has been shown to interact with Ago1 (Behm-Ansmant et al. 2006), and the Q-rich domain, which has been hypothesized to be a self-interaction domain (Decker et al. 2007). To examine if any of the dGW182 sufficiency domains is repressing mRNA function by recruitment of the endogenous dGW182 or Ago1, we depleted endogenous proteins with gene-specific dsRNA (Worby et al. 2001) and asked if the sufficiency domains could still repress in the absence of endogenous Ago1 or dGW182 (Fig. 4).
FIGURE 4.
Repression mediated by dGW182 and its sufficiency domains appears to be independent of endogenous dGW182 and Argonaute 1. (A) S2 cells were treated with dsRNAs indicated above the lanes (dGW182, Ago1, or GFP as a negative control), and efficiencies of endogenous dGW182 and Ago1 depletions were analyzed by Western blotting with antibodies shown on the left. Expression of tubulin was estimated as a loading control. (B) dGW182 and its effector domains were transfected in S2 cells depleted of endogenous dGW182 or Ago1 and, as negative controls, in GFP-dsRNA-treated or untreated cells. Transfections were largely performed and analyzed as in Figure 1. In tethering experiments, firefly luciferase activity was expressed as a percentage of that in the presence of NHA-lacZ, for each RNAi depletion. To additionally control for efficient depletion of endogenous Ago1 and dGW182, firefly luciferase reporter containing miRNA-9b target sites (FLuc-nerfin) (Behm-Ansmant et al. 2006) was co-transfected with either miRNA-9b-encoding plasmid or the empty vector. As in all transfection experiments, RLuc was coexpressed as a transfection control. Firefly luciferase activity was normalized to that of Renilla luciferase and presented as a percentage of FLuc-nerfin expression in the presence of the empty vector.
Efficiency of endogenous dGW182 and Ago1 knockdowns was confirmed by Western blotting analysis (Fig. 4A). To additionally verify that we were able to functionally deplete endogenous dGW182 and Ago1, we transfected cells with a miRNA reporter construct, containing the firefly luciferase coding sequence fused to the nerfin 3′-UTR, which includes target sites for miRNA-9b (designated as FLuc-nerfin) (Stark et al. 2005; Behm-Ansmant et al. 2006). In untreated cells or cells treated with control GFP-specific dsRNA, coexpression of miRNA-9b leads to 10-fold down-regulation of FLuc-nerfin reporter (Fig. 4). Addition of Ago1-specific dsRNA (black bars) almost fully abrogates miRNA-mediated repression, and treatment with dGW182-specific dsRNA (striped bars) alleviates repression from 10-fold to approximately twofold. A similar partial effect of dGW182 knockdown on a FLuc-nerfin reporter was observed by Behm-Ansmant et al. (2006), despite the fact that their Western blotting analysis showed efficient depletion of dGW182. Although we cannot exclude that remaining twofold repression by miRNA-9b is due to residual dGW182, another possibility is that Ago1 may repress FLuc-nerfin via an alternative dGW182-independent mechanism.
We observed that tethering of the three dGW182 sufficiency domains in cells depleted of endogenous Ago1 or dGW182 resulted in repression comparable to that in the absence of dsRNA or in the presence of control GFP-specific dsRNA (Fig. 4). This experiment indicates that all three dGW182 sufficiency domains appear to repress tethered mRNA independently of endogenous dGW182 and Ago1.
dGW182 effector domains are able to rescue knockdown of endogenous dGW182
The results of our tethering analysis, and previous work on the dGW182–Argonaute interactions, lead to a model wherein dGW182 is recruited to mRNAs through the N-terminus-mediated interaction with Argonaute proteins and then represses translation through three different effector domains. This model predicts that the N-terminal domain of dGW182 (1–605) should be sufficient to function in actual miRNA silencing, and that the inclusion of additional repressive domains such as the 605–830 domain should increase the effectiveness of dGW182 repression. To test these predictions, we analyzed if overexpression of dGW182 deletions is able to rescue the knockdown of endogenous dGW182 in Drosophila S2 cells (Fig. 5). Note that since dGW182 is recruited to the miRNA repression complex via interaction with Ago1, in these rescue experiments we could only test N-terminal portions of dGW182 that include the N-terminal GW-repeats and are able to bind Ago1. Endogenous dGW182 was depleted by treating cells with dsRNA, and, for a negative control, a batch of cells was treated with GFP-specific dsRNA. In this experiment, we made use of FLuc-nerfin miRNA reporter construct, containing target sites for miRNA-9b. As discussed earlier, coexpression of miRNA-9b efficiently represses FLuc-nerfin mRNA in control cells (Fig. 5, first two open bars, cf. empty vector and miRNA-9b), while RNAi depletion of dGW182 partially alleviates miRNA-9b-mediated repression (Fig. 5, black bars, cf. empty vector and miRNA-9b). We then analyzed how expression of dGW182 and its deletions were able to complement the deficiency of the endogenous dGW182 protein.
FIGURE 5.
Overexpression of dGW182 and its N-terminal fragments rescues RNAi knockdown of endogenous dGW182. (Black bars) Endogenous dGW182 was depleted in Drosophila S2 cells with dsRNA as described in Figure 4; (open bars) a batch of cells was treated with GFP-specific dsRNA, as a negative control. Cells were transfected with firefly luciferase reporter containing miRNA-9b target sites (FLuc-nerfin) and with either miRNA-9b-encoding plasmid or the empty vector. As in all transfection experiments, RLuc was coexpressed as a transfection control; expression levels of firefly luciferase were normalized to Renilla luciferase activity and expressed as a percentage of the firefly luciferase activity in the presence of the empty vector. To rescue the knockdown of endogenous dGW182, increasing amounts (from 0 to 100 ng per well, in a 96-well plate) of plasmids encoding NHA-dGW182 or its deletion mutants were co-transfected: 1–605 construct, encoding N-terminal GW-repeats, and 1–830 construct, encoding GW-repeats together with the Q-rich region. Expression of NHA-dGW182 and its deletions was estimated by Western blotting with anti-HA antibody; the squared bands correspond to the minimal expression levels sufficient to rescue miRNA-9b-mediated repression to the level observed in GFP dsRNA-treated cells. Increasing amounts of NHA-lacZ were co-transfected for a negative control. To test the dependence of rescue on the N-terminal region of dGW182 required for dGW182–Ago1 interaction, we also co-transfected increasing amounts of NHA-605-830 and NHA-940-1215 plasmids encoding for Q-rich and DUF-RRM regions, accordingly (30 and 100 ng/well).
Importantly, transfection of increasing amounts of dGW182 N-terminal fragments (Fig. 5, 1–605, 1–830) was able to restore the miRNA-9b-directed repression in dGW182-depleted cells (Fig. 5, black bars), while lacZ (a negative control) has practically no effect. Note that, as predicted from the tethering results, the longer fragment of dGW182 (Fig. 5, 1–830), which contains both Q-rich and N-terminal GW-rich domains, rescues repression more efficiently than the N-terminal effector domain alone (Fig. 5, 1–605), and the full-length dGW182 rescues, in turn, more efficiently than the 1–830 dGW182 fragment. The Q-rich (Fig. 5, 605–830) and DUF-RRM (Fig. 5, 940–1215) fragments alone, without the N-terminal part, were not able to complement the deficiency of endogenous dGW182, indicating that interaction with Ago1 is required for the rescue. These results indicate that the N-terminal region of dGW182 constitutes a repressive domain, and that the Q-rich and C-terminal DUF/RRM domains can also contribute to translational repression. These observations are consistent with the tethering results and argue that the repression domains we have mapped in dGW182 by tethering experiments also function in bona fide miRNA-mediated repression of mRNAs.
Interestingly, we also observed that overexpression of the dGW182 N-terminal fragments in the presence of endogenous dGW182 (Fig. 5, open bars, GFP dsRNA-treated cells) partially prevented the miRNA-9b-induced repression. This result is consistent with the previously reported data (Eulalio et al. 2008b), suggesting a dominant-negative effect of the overexpressed N-terminal fragment. However, it is also striking that depletion of the endogenous dGW182 allowed the 1–605 fragment to effectively function in the repression mediated by miRNA-9b (Fig. 5, black bars, dGW182 dsRNA-treated cells). This latter result indicates that a full-length endogenous dGW182 acts in a dominant manner to affect the function of the 1–605 or 1–830 dGW182 fragments (see Discussion).
Similar regions within the human dGW182 homolog, TNRC6C, are sufficient to repress tethered mRNA in Drosophila cells
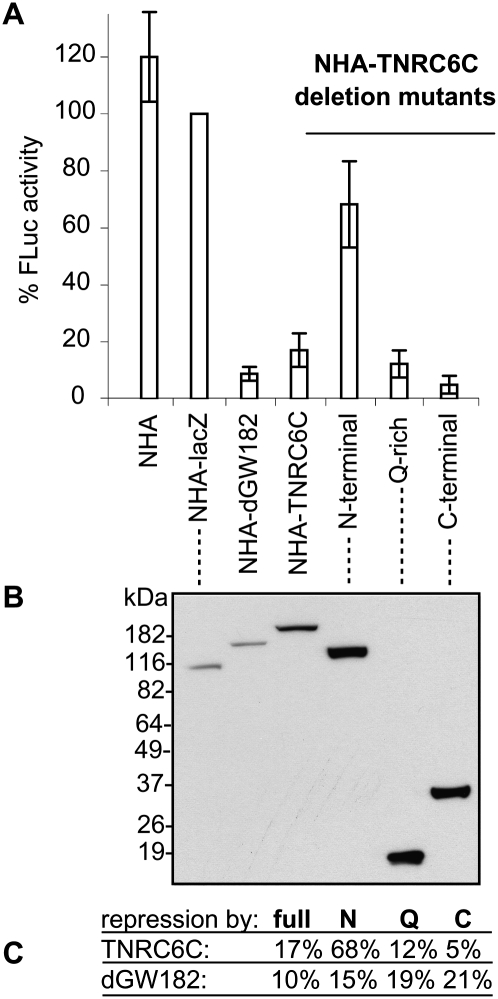
Our analysis has identified three domains within dGW182 that can function to repress mRNA. Given the conservation of domain structure between human and Drosophila GW182 orthologs, we tested whether similar domains of TNRC6C, a human dGW182 homolog, are capable of repressing mRNA activity when tethered in Drosophila S2 cells.
We observed that tethering of the full-length TNRC6C protein leads to efficient repression (17% FLuc activity), demonstrating that the mechanisms of GW182-mediated repression are at least partially conserved between human and Drosophila. Furthermore, deletion analysis of the human protein shows that similar effector domains can also repress mRNA in Drosophila (Fig. 6). The most efficient repressor domain of TNRC6C is its C-terminal region (1370–1690), comprising the conserved DUF and RRM domains, which provides down-regulation to ∼5% FLuc activity. These data are consistent with the TNRC6C mapping results obtained in human HEK293 cells (Zipprich et al. 2009) that identified the C-terminal region as the most effective in repression (more than 10-fold). The middle Q-rich domain (1080–1245) of TNRC6C is also able to repress protein synthesis in S2 cells, to ∼12% FLuc activity. Finally, the N-terminal portion of TNRC6C, containing GW-rich repeats (1–1035), is a weak repressor (68% FLuc activity). Interestingly, parallel analysis of dGW182 in mammalian cells (Zipprich et al. 2009) showed that the N-terminal domain of dGW182 is able to efficiently repress the tethered message, but the Q-rich region is a weak repressor. These results argue that efficient repression by a Q-rich domain is dependent on some aspect of the miRNA machinery in Drosophila that is different in human cells. On the contrary, the repressive effect of the N-terminal domain is observed only for the Drosophila homolog in both fly and human cells, arguing that some structural features specific for the N-terminal part of dGW182 define its role in repression.
FIGURE 6.
Human homolog of dGW182, TNRC6C, and its Q-rich and DUF-RRM regions are able to repress tethered mRNA in Drosophila S2 cells. (A) The tethering assay was performed essentially as in Figure 1C, with human protein TNRC6C and its deletion mutants tethered instead of dGW182. The N-terminal tethered fragment of TNRC6C (amino acids 1–1035) includes GW-rich repeats; the middle portion (amino acids 1080–1245) covers the Q-rich region; the C-terminal part (amino acids 1370–1690) comprises conserved DUF and RRM. Full-length NHA-dGW182 was tethered as a positive control, NHA and NHA-lacZ as negative controls. (B) Expression levels of NHA-fusion proteins were estimated by Western blotting with anti-HA antibody. (C) Summary of the repressive effects by TNRC6C, dGW182, and their deletions. The numbers show the percent translation in the presence of tethered full-length proteins, N-terminal, Q-rich, and C-terminal (DUF-RRM) fragments, accordingly.
DISCUSSION
dGW182 affects protein production upstream of effects on mRNA decay
Our results add additional support to the growing evidence that GW182 proteins decrease translation rates as well as promote mRNA degradation. Similar to earlier results (Eulalio et al. 2008b), we also demonstrate that tethered full-length dGW182 or its repressive domains reduce protein production from mRNAs without a poly(A) tail (Fig. 3), which indicates that the repressive effects of dGW182 are independent of deadenylation.
Multiple repressive domains of dGW182 and its implications
Our results demonstrate that no single region of dGW182 is required for mRNA repression, and we identify three independent domains of dGW182 that are sufficient for mRNA repression. We also observed that multiple repressive domains of TNRC6C could function in S2 cells (Fig. 6). These results suggest that GW182 proteins across multiple species will contain redundant repressive domains with a conserved mode of function.
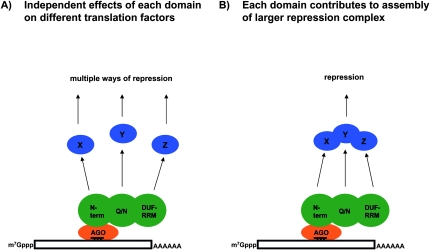
In principle, the different sufficiency domains of dGW182 could function in two manners. First, each sufficiency domain could interact with a different target protein or RNA that then impinges on mRNA function in a distinct and separable manner (Fig. 7A). The key aspect of this model is that the dGW182 protein would affect multiple and different steps in protein production and/or mRNA degradation. Alternatively, each domain of dGW182 could contribute to the assembly of a larger repression complex by recruitment of different factors into the larger assembly (Fig. 7B). The key aspect of this second model is that assembly of a single repression complex represents a limiting step, which can be influenced by multiple different interactions between members of this complex and dGW182. It should be noted that this latter model provides a possible explanation for why endogenous full-length dGW182 inhibits the function of the 1–605 or 1–830 fragments to repress mRNA in response to miRNA-9b (Fig. 5). In this view, the full-length dGW182 would bind to the downstream repression complex more efficiently, and thereby prevent the miRNA–Ago1–dGW182(1–605) assembly from recruiting the repression complex to the mRNA.
FIGURE 7.
Scheme illustrating possible modes of action of dGW182 protein in translational repression by miRNAs. See the text for details.
Although we are not yet able to rigorously distinguish between these two general possibilities, the presence of multiple interacting domains has two important implications. First, it suggests that the downstream targets of dGW182 will be multiple proteins/RNAs. This might provide an explanation for why a genome-wide RNAi screen failed to turn up additional components of the miRNA silencing pathway (Eulalio et al. 2007); there is too much redundancy downstream from dGW182 protein. A second important implication of multiple sufficiency domains in dGW182 is that this allows for GW182 orthologs and paralogs to differ in their key repressive region. For example, according to a bioinformatics search, the Caenorhabditis elegans GW182 counterparts, AIN-1 and AIN-2 (Ding et al. 2005; Zhang et al. 2007), appear to be largely missing the RRM domain and presumably then would be expected to drive repression by other domains. This also implies that in certain cell types, different domains of GW182 will be more important, and that different GW182 paralogs will show a different spectrum of repression in different cell types and in response to signaling pathways as it may change the rate-limiting interaction for assembly of the repressive complex.
Interestingly, deletion analysis of human ortholog TNRC6C in Drosophila S2 cells (this study) and, vice versa, dGW182 in mammalian cells (Zipprich et al. 2009) argues that repression by different domains may be either protein- or cell-specific. Thus, the N-terminal domain of dGW182 is a strong repressor in both fly and human cells, while the same region of TNRC6C is inefficient in repression in both cell types. This is likely to reflect different strengths of this repressive domain in the different GW182 family members. On the contrary, high repression levels by a Q-rich region, both from dGW182 and TNRC6C, are only achieved in Drosophila S2 but not human HEK293 cells, suggesting that this effect is dependent on some species-specific aspect of the miRNA repressive complex, i.e., that Drosophila cells may contain some partners of the Q-rich domain that are absent or limiting in human cells.
Function of RRM domain in repression
One of the key regions of dGW182 that can drive repression is the RRM domain and a conserved region just to the N-terminal side of this domain. The presence of the RRM domain in this region is intriguing and suggests two possible modes of function. First, it could be that the RRM domain interacts with an RNA target and thereby contributes to translation repression of the mRNA. For example, the RRM could bind the cap structure or the AUG region in the mRNA, thereby reducing translation initiation. Alternatively, the RRM could interact with an RNA involved in translation such as the initiator tRNA or the rRNA. In a second possibility, the RRM domain could normally function as part of the assembly of the miRNA repression complex on mRNAs to provide additional specificity and strength to the miRNA:mRNA interaction, which is supported by the observation that efficient repression of miRNA targets can require additional sequence information in the 3′-UTR (Vella et al. 2004; Robins et al. 2005; Zhao et al. 2005; Didiano and Hobert 2006, 2008; Grimson et al. 2007; Kertesz et al. 2007; Long et al. 2007). In this second model, the RRM might also function in repression as a platform for the recruitment of additional protein factors that then inhibit mRNA function. This model is analogous to the function of Puf proteins, which form a helical shape with one surface providing sequence-specific RNA binding and a second surface providing for direct protein–protein interactions that recruit translational repressors or mRNA degradation factors to the mRNA (Goldstrohm et al. 2006). One way of distinguishing these models is to make surface mutations on the RRM domain and ask if the predicted RNA-binding surface is required for repression when tethered, or if the opposite face contains critical interactions. This should distinguish the modes by which the RRM domain functions.
Function of Q-rich domain in repression
One possible role for the Q-rich domain is to interact with other QN-rich domains and to drive assembly of larger repression complexes. This is based on the role of QN-rich sequences in assembly of protein complexes (Decker et al. 2007). Given this, one possible way this domain might have functioned was to recruit endogenous dGW182, but this does not appear to be the case as repression by this domain is independent of endogenous dGW182. Alternative models might include this domain recruiting the Hedls/Ge-1 protein, which has a QN-rich domain and is involved in miRNA-mediated repression (Eulalio et al. 2007).
MATERIALS AND METHODS
Cell culture, transfections, and RNA interference
Transfections of Drosophila S2 cells were performed in 12-well or 96-well plates with Cellfectin Reagent (Invitrogen) according to the manufacturer's instructions. In tethering experiments, per well of a 12-well plate, we transfected 40 ng of FLuc-boxB plasmid, 100 ng of RLuc as transfection control, and 400 ng of plasmid encoding for NHA-fusion protein as indicated in figure legends. In TNRC6C tethering experiments, the amounts of NHA-fusion plasmids varied between 10 ng and 1 μg to achieve similar expression levels. In experiments with miRNA reporter, transfection mixtures contained 40 ng of FLuc-nerfin reporter plasmid, 100 ng of RLuc as transfection control, and 100 ng of either empty vector or a plasmid encoding for miRNA-9b. For the 96-well format, the amounts of plasmids were adjusted proportionally. Cells were lysed on the second or third day post-transfection. Firefly and Renilla luciferase activities were measured with the Dual-Luciferase Reporter Assay System (Promega). RNAi experiments were performed according to Worby et al. (2001). dsRNAs were 250–500 nt long and corresponded to the 3′-UTRs of targeted genes, with the exception of no-target control GFP dsRNA that was specific to the GFP coding sequence. Treatment with dsRNA was done on day 0 and repeated on day 4. Cells were transfected on day 6 and lysed on day 8 or 9.
DNA constructs
FLuc-boxB, RLuc, and miRNA-9b constructs were previously described (Rehwinkel et al. 2005). NHA-dGW182 plasmid is analogous to the λN-HA-GW182 construct described by Behm-Ansmant et al. (2006), but contains an SbfI site between the HA-tag and dGW182 coding sequence inserted for the convenience of cloning. NHA-lacZ and plasmids encoding deletion mutants of dGW182 were generated by PCR amplification of the β-galactosidase-coding region or the corresponding fragment of the dGW182-coding region and cloning it into a pAC5.1A vector containing λN and HA-tag. The NHA-Ago1 F594V F629V (2F2V) plasmid was generated from the NHA-Ago1 construct (Rehwinkel et al. 2005) by site-directed mutagenesis. To generate the NHA plasmid, NHA-dGW182 was cut with SbfI and NotI, blunt-ended, and ligated. The HA-dGW182 plasmid was generated from NHA-dGW182(940-1215) by cutting out the λN peptide region with KpnI and HindIII; in the resulting plasmid, the dGW182 940-1215 fragment was substituted with the full-length dGW182 using an SbfI–NotI digest. The FLuc-boxB-HSL construct was produced by mutating a polyadenylation site in FLuc-boxB and introducing a PCR-amplified fragment corresponding to the 3′-UTR of the histone H1 stem–loop structure into the SacI and BamHI sites. To generate the FLuc-nerfin plasmid, the nerfin 3′-UTR was PCR-amplified using F-Luc-nerfin (Stark et al. 2005; Behm-Ansmant et al. 2006) as a template and cloned into NheI–XhoI sites of the FLuc-boxB plasmid to substitute the boxB-containing 3′-UTR with the one specific to nerfin mRNA. To generate the plasmids for expression of NHA-TNRC6C and its deletion mutants in Drosophila cells, the coding region of TNRC6C and its fragments were PCR-amplified from a KIAA1582 cDNA clone and inserted into a pAC5.1A vector containing λN and HA-tag.
RNA analysis, Western blotting
The total RNA from S2 cells was isolated using Trizol LS Reagent (Invitrogen), and the levels of FLuc-boxB and RLuc mRNAs were analyzed by Northern blot. Briefly, RNA (10–20 μg) was separated on a denaturing formaldehyde agarose gel, transferred to Hybond-N+ membrane (GE Healthcare Life Sciences), and hybridized to 32P-labeled RNA probes in UltraHyb buffer (Ambion) according to the manufacturer's instructions. To estimate the expression levels of HA-fusion proteins, cell lysates were separated on a 4%–12% PAAG (Invitrogen), and the Western blot was probed using anti-HA antibodies (Roche 3F10). The amounts of loaded lysates were normalized according to expression of Renilla luciferase (transfection control). Quantification of expression levels in Northern and Western blots was done with ImageQuant software (Molecular Dynamics). The efficiency of Ago1 and dGW182 knockdowns was estimated by Western blotting with anti-Ago1 (Abcam ab5070) or anti-dGW182 (A.J. Simmonds) antibodies. For a loading control, membranes were probed with anti-tubulin antibodies (Sigma T5168).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Elisa Izaurralde for reporter plasmids FLuc-boxB, FLuc, RLuc, FLuc-nerfin, and plasmids expressing dGW182, Ago1, and miRNA-9b. We are also grateful to Elisa Izaurralde and Ana Eulalio for sharing the details of their transfection protocol. We thank Andrew J. Simmonds for a generous gift of anti-dGW182 antibodies. Drosophila S2 cells were kindly provided by Ivan Yudushkin and Oliver Bell. We are grateful to the EMBL Genomics Core Facility and Vladimir Benes for Drosophila cDNA clones. We thank Jakob Zipprich and Hansruedi Mathys for sharing their data on analysis of dGW182 and TNRC6C in mammalian cells. We are grateful to members of the Parker and Filipowicz laboratories and Nicolas Thoma for discussions. Work in the Filipowicz laboratory is partially funded by the EC FP6 “Sirocco” Program. The Friedrich Miescher Institute is supported by the Novartis Research Foundation. M.C. is a recipient of long-term EMBO and HFSP fellowships.
Footnotes
REFERENCES
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N., Cohen S.M. MicroRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Chendrimada T.P., Finn K.J., Ji X., Baillat D., Gregory R.I., Liebhaber S.A., Pasquinelli A.E., Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- Decker C.J., Teixeira D., Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae . J. Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano D., Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA–target interactions. Nat. Struct. Mol. Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- Didiano D., Hobert O. Molecular architecture of a miRNA-regulated 3′ UTR. RNA. 2008;14:1297–1317. doi: 10.1261/rna.1082708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X.C., Grosshans H. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. 2009;28:213–222. doi: 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 2007;17:411–416. doi: 10.1016/j.tcb.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Ding L., Spencer A., Morita K., Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans . Mol. Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Du T., Zamore P.D. Beginning to understand microRNA function. Cell Res. 2007;17:661–663. doi: 10.1038/cr.2007.67. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Rehwinkel J., Stricker M., Huntzinger E., Yang S.F., Doerks T., Dorner S., Bork P., Boutros M., Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes & Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008a;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008b;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Nishihara T., Rehwinkel J., Fauser M., Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T., Chan E.K., Tenenbaum S.A., Keene J.D., Griffith K., Fritzler M.J. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T., Jakymiw A., Chan E.K., Seraphin B., Cougot N., Fritzler M.J. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Gehring N.H., Neu-Yilik G., Schell T., Hentze M.W., Kulozik A.E. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- Goldstrohm A.C., Hook B.A., Seay D.J., Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys D.T., Westman B.J., Martin D.I., Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kiriakidou M., Tan G.S., Lamprinaki S., De Planell-Saguer M., Nelson P.T., Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li S., Lian S.L., Moser J.J., Fritzler M.L., Fritzler M.J., Satoh M., Chan E.K. Identification of GW182 and its novel isoform TNGW1 as translational repressors in Ago2-mediated silencing. J. Cell Sci. 2008;121:4134–4144. doi: 10.1242/jcs.036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Rivas F.V., Wohlschlegel J., Yates J.R., III, Parker R., Hannon G.J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005a;7:1161–1166. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005b;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long D., Lee R., Williams P., Chan C.Y., Ambros V., Ding Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- Lytle J.R., Yario T.A., Steitz J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney P.A., Yu Y., Fisher J., Nilsen T.W. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- Mathonnet G., Fabian M.R., Svitkin Y.V., Parsyan A., Huck L., Murata T., Biffo S., Merrick W.C., Darzynkiewicz E., Pillai R.S., et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Peters L., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T. Identification of novel Argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Nilsen T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Nottrott S., Simard M.J., Richter J.D. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- Olsen P.H., Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Petersen C.P., Bordeleau M.E., Pelletier J., Sharp P.A. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Artus C.G., Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Inhibition of translational initiation by let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Behm-Ansmant I., Gatfield D., Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins H., Li Y., Padgett R.W. Incorporating structure to predict microRNA targets. Proc. Natl. Acad. Sci. 2005;102:4006–4009. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seggerson K., Tang L., Moss E.G. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- Sen G.L., Blau H.M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Thermann R., Hentze M.W. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- Till S., Lejeune E., Thermann R., Bortfeld M., Hothorn M., Enderle D., Heinrich C., Hentze M.W., Ladurner A.G. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes & Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Vella M.C., Choi E.Y., Lin S.Y., Reinert K., Slack F.J. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes & Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Yanez A., Novina C.D. MicroRNA-repressed mRNAs contain 40S but not 60S components. Proc. Natl. Acad. Sci. 2008;105:5343–5348. doi: 10.1073/pnas.0801102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby C.A., Simonson-Leff N., Dixon J.E. RNA interference of gene expression (RNAi) in cultured Drosophila cells. Sci. STKE. 2001;2001:PL1. doi: 10.1126/stke.2001.95.pl1. [DOI] [PubMed] [Google Scholar]
- Wu L., Belasco J.G. Let me count the ways: Mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Wu L., Fan J., Belasco J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ding L., Cheung T.H., Dong M.Q., Chen J., Sewell A.K., Liu X., Yates J.R., III, Han M. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol. Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Samal E., Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Zipprich J.T., Bhattacharyya S., Mathys H., Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009 doi: 10.1261/rna.1448009. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]