Proteolipid Protein Is Required for Transport of Sirtuin 2 into CNS Myelin (original) (raw)
Abstract
Mice lacking the expression of proteolipid protein (PLP)/DM20 in oligodendrocytes provide a genuine model for spastic paraplegia (SPG-2). Their axons are well myelinated but exhibit impaired axonal transport and progressive degeneration, which is difficult to attribute to the absence of a single myelin protein. We hypothesized that secondary molecular changes in PLP_null_ myelin contribute to the loss of PLP/DM20-dependent neuroprotection and provide more insight into glia-axonal interactions in this disease model. By gel-based proteome analysis, we identified >160 proteins in purified myelin membranes, which allowed us to systematically monitor the CNS myelin proteome of adult PLP_null_ mice, before the onset of disease. We identified three proteins of the septin family to be reduced in abundance, but the nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase sirtuin 2 (SIRT2) was virtually absent. SIRT2 is expressed throughout the oligodendrocyte lineage, and immunoelectron microscopy revealed its association with myelin. Loss of SIRT2 in PLP_null_ was posttranscriptional, suggesting that PLP/DM20 is required for its transport into the myelin compartment. Because normal SIRT2 activity is controlled by the NAD+/NADH ratio, its function may be coupled to the axo-glial metabolism and the long-term support of axons by oligodendrocytes.
Keywords: myelin, oligodendrocyte, cytoskeleton, neurodegeneration, Pelizaeus–Merzbacher disease, hereditary spastic paraplegia, NAD+/NADH, acetylation, metabolism
Introduction
Axonal degeneration is a common feature of many neurological diseases. Various hereditary leukodystrophies and demyelinating neuropathies exhibit axonal involvement, and axonal loss within demyelinated lesions is a major cause of clinical disability in patients with multiple sclerosis (Bjartmar et al., 1999; Popko, 2003). Thus, a primary perturbation of myelinating glial cells can have profound secondary effects on axonal function and survival.
Myelination and axonal preservation are two independent functions of oligodendrocytes. This was revealed by the analysis of mouse mutants that lack expression of either myelin proteolipid protein (PLP) (Griffiths et al., 1998b) or 2′,3′-cyclic nucleotide phosphodiesterase (CNP) (Lappe-Siefke et al., 2003). Both mutants produce normal amounts of myelin and lack neurological impairments characteristic for dysmyelination. Only adult PLP_null_ and CNP_null_ develop ataxia and hindlimb paralysis, resulting from a length-dependent axonal loss preceded by decelerated fast axonal transport (Edgar et al., 2004) and axonal swellings (spheroids). Also, human patients with a null mutation of the PLP1 gene develop a length-dependent CNS axonopathy in the absence of major myelin abnormalities [i.e., spastic paraplegia type 2 (SPG-2)] (Garbern et al., 1997; Inoue, 2005). Hypomyelination by itself does not invariably lead to axonal loss, as revealed by the histopathology of conditional cholesterol-deficient mutants (Saher et al., 2005).
PLP1 encodes two splice isoforms (PLP and DM20) of the major membrane-spanning CNS myelin proteins (Nave et al., 1987), four-helix bundle proteins of unknown function in vivo (Griffiths et al., 1998a). PLP_null_ myelin appears fully functional with respect to nerve conduction, despite its reduced physical stability and minor differences at the ultrastructural level (Klugmann et al., 1997; Rosenbluth et al., 2006). In contrast to null mutations, many point mutations in the human and mouse gene cause cytotoxic PLP/DM20 misfolding, the unfolded protein response, and oligodendrocyte death, resulting in severe dysmyelination and premature death in Pelizaeus–Merzbacher disease (PMD) or corresponding mouse models (Werner et al., 1998; Inoue, 2005). Whereas the role of PLP/DM20 in PMD is gradually understood, the detrimental axonal changes in SPG-2 patients and in aged PLP_null_ mice have remained unexplained. Although the primary function of oligodendrocytes in maintaining axonal integrity is plausible, it is speculative how a single structural myelin protein such as PLP is tied into providing “trophic support” to myelinated axons (Yin et al., 2006).
We considered the possibility that the genetic loss of one myelin protein may have secondary molecular consequences in the myelin compartment. Recently, proteome analysis has been used to catalog myelin-associated proteins and identify novel myelin components (Taylor et al., 2004; Huang et al., 2005; Vanrobaeys et al., 2005; Roth et al., 2006). Here, we identify, by two-dimensional (2D) differential fluorescence intensity gel electrophoresis (DIGE) (Unlu et al., 1997) combined with protein identification by mass spectrometry (MS), alterations of the PLP_null_ myelin proteome that precede widespread axonal degeneration. Most strikingly, we demonstrate the virtual absence of sirtuin 2 (SIRT2), a nicotinamide adenine dinucleotide (NAD+)-dependent protein deacetylase of unknown function in vivo. PLP-dependent transport of SIRT2 into CNS myelin may contribute to the role of oligodendrocytes in maintaining axonal integrity.
Materials and Methods
Animals.
PLP_null_ mice (Klugmann et al., 1997) were bred for 15 generations into the C57BL/6 background using mice from the breeding colony of the Max Planck Institute of Experimental Medicine. Wild-type animals were obtained from the same colony. Genotyping was performed as described previously (Klugmann et al., 1997). Experiments were in compliance with the animal policies of the Max Planck Institute of Experimental Medicine, approved by the German Federal State of Niedersachsen. Only male mice at the indicated ages were used.
Myelin purification.
A light-weight membrane fraction enriched for myelin was purified from mouse brains homogenized in 0.32 m sucrose according to the study by Norton and Poduslo (1973). The protein concentration was determined using the 2-D Quant kit according to the manufacturer (GE Healthcare, Piscataway, NJ). Mice for myelin proteome analysis were 75 d of age.
2D gel electrophoresis.
For large-scale 2D-isoelectric focusing (IEF)/SDS-PAGE, 400 μg of myelin 2 was first solubilized in 2× IEF sample buffer (7 m urea, 2 m thiourea, 2% ASB-14, 2% ampholytes, 2% DTT), diluted in rehydration buffer (7 m urea, 2 m thiourea, 2% ASB-14, 1% ampholytes, 0.2% DTT), centrifuged (2 min, 16,000 × g), and the supernatant was subjected to IEF in immobilized pH gradient (IPG, 18 cm Immobiline nonlinear pH 3–10; GE Healthcare) for ∼45 kVh. The use of the zwitterionic detergent amidosulfobetaine-14 (ASB-14) substantially improved myelin protein solubilization (Taylor et al., 2004) but led to a slightly decreased resolution in IEF as indicated by horizontal smearing. After completion of IEF, the proteins were reduced in SDS equilibration buffer (50 mm Tris-HCl, pH 8.8, 6 m urea, 30% glycerol, 2% SDS, and 0.002% bromophenol blue) containing 10 mg/ml DTT and subsequently alkylated in SDS equilibration buffer containing 25 mg/ml iodoacetamide. In the second dimension, the proteins were separated on 20 × 20 cm gradient gels (8–16% acrylamide) under denaturing conditions. For small-scale 2D-IEF/SDS-PAGE (protein load, 100 μg), myelin 2 was first delipidated and precipitated with methanol/chloroform according to (Wessel and Flugge, 1984). IEF and SDS-PAGE were performed as before using 7 cm IPG strips (Immobiline nonlinear, pH 3–11, ∼10 kVh) and precast NuPAGE 8–16% Tris-Glycine mini-gels.
For large-scale 2D-16-benzyldimethyl-_n_-hexadecylammonium chloride (16-BAC)/SDS-PAGE, 400 μg of myelin 2 was resuspended in 2 mm EGTA, pH 7.0, and centrifuged (10 min, 16,000 × g, 4°C). To deplete soluble and membrane-associated proteins, this pellet was additionally subjected to six consecutive washing/centrifugation cycles (2 × 50 mm Tris, 150 mm NaCl, 1 × 1 m KCl, 1 × 0.1 m Na2CO3, 2 × 2 mm EGTA). The final pellet was resuspended in 100 μl of 16-BAC sample buffer (22.5% urea, 5% 16-BAC, 5% glycerol, 37.5 mm DTT, 0.025% Pyronin Y) and separated on 7.5% acrylamide 16-BAC gels (20 × 20 cm) as described previously (Hartinger et al., 1996). After the first dimension, gels were stained with Coomassie brilliant blue (CBB) R250. Whole lanes were excised, equilibrated in 100 mm Tris/HCl pH 8.8, and transferred onto 20 × 20 cm SDS gels (8–16% acrylamide unless stated otherwise). 2D-16-BAC/SDS-PAGE in the mini-gel scale (protein load, 50 μg) was performed as before with the exception that the washing/centrifugation cycles were omitted and equilibration after the first dimension was in 100 mm Tris/HCl pH 7.0. Precast NuPAGE 4–12% Bis-Tris gels (Invitrogen, San Diego, CA) were used for the second dimension. All final gels were blotted or stained overnight with colloidal CBB G250. Gel plugs were excised manually.
2D-DIGE.
For 2D-DIGE, myelin 2 was first delipidated and precipitated with methanol/chloroform according to Wessel and Flugge (1984). The pellet was solubilized in labeling buffer (20 mm Tris/HCl pH 8.5, 7 m urea, 2 m thiourea, 2% ASB-14), centrifuged (5 min, 16,000 × g), and the protein concentration of the supernatant was determined using the 2-D Quant kit (GE Healthcare). Fifty micrograms each of wild-type and PLP_null_ myelin were minimally labeled with 400 pmol of Cy3 and Cy5, respectively, following the manufacturer's protocol (GE Healthcare). To avoid dye-specific protein labeling, every pair of protein samples from two independent myelin preparations was processed in duplicate while swapping the dyes. Thereby, four replicate gels were obtained, allowing to monitor regulation factors down to twofold changes (Karp et al., 2005). Fifty micrograms of an internal standard consisting of a mixture of all four myelin samples under investigation were labeled with 400 pmol Cy2 and included on all gels to facilitate gel matching, thereby eliminating artifacts from experimental variation. The three differentially labeled fractions were mixed, diluted in rehydration buffer, and subjected to large-scale 2D-IEF/SDS-PAGE as described above. Labeled Proteins were visualized with a Typhoon 9400 imager using the default settings recommended by the manufacturer (GE Healthcare). Spot matching across gels and normalization based on the internal standard was performed with the multifluorescence analysis tool of the Proteomweaver 3.1 software (Definiens/Bio-Rad). To analyze the significance of regulation, a Student's t test was performed, and spots were accepted as significantly different for p < 0.01. Differentially regulated proteins were excised directly from the analytical gels poststained with colloidal Coomassie and subjected protein identification by MS.
Protein identification.
Excised gel plugs were subjected to an automated platform for the identification of gel-separated proteins as described previously (Jahn et al., 2006). Briefly, the robotic liquid handling system Genesis ProTeam 150 Advanced Digest (Tecan, Männedorf, Switzerland) was used to perform the tryptic in-gel digest with previous reduction/carboxamidomethylation of the proteins and to subsequently prepare the extracted tryptic peptides for matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) on prestructured sample supports (AnchorChip; Bruker Daltonics, Billerica, MA) according to the thin layer affinity method. Using an Ultraflex I MALDI-TOF/TOF mass spectrometer (Bruker Daltonics), peptide mass fingerprint spectra were automatically acquired, postprocessed, and subjected to database searches (Jahn et al., 2006). For confirmation and to cover proteins difficult to identify by peptide mass fingerprinting such as proteins in mixtures, posttranslationally modified proteins, and small proteins, the mass spectrometer was operated in the MS/MS mode within the same automated analysis loop to record fragment ion spectra of up to four selected precursor ions in a result-dependent manner. Data base searches in the Swiss-Prot or NCBInr primary sequence databases restricted to the taxonomy Mus musculus were performed using the Mascot Software 2.0 (Matrix Science, Boston, MA) licensed in-house. Carboxamidomethylation of cysteines was specified as fixed and oxidation of methionines as variable modification. The monoisotopic mass tolerance was set to 100 ppm, and one missed cleavage was allowed. Database searches of MS/MS data sets were performed as above with the fragment mass tolerance set to 0.7 Da. Only proteins represented by at least one peptide sequence above significance threshold in combination with the presence of at least four peptide masses assigned in the peptide mass fingerprint were considered as identified.
Western blotting.
Proteins were blotted onto polyvinylidene difluoride membranes (Hybond P; GE Healthcare), blocked with 5% milk powder in TBS and Triton X-100 (TBST) (150 mm NaCl, 10 mm Tris/HCl, pH 7.4; 0.1% Tween 20), and incubated with primary antibodies in the same solution overnight at 4°C. Blots were washed with TBS/0.05% Tween 20, incubated with appropriate secondary horseradish peroxidase-conjugated antibodies (Dianova, Hamburg, Germany), washed with TBS/0.05% Tween 20 and once with TBS, and developed by enhanced chemiluminescence (Pierce, Rockford, IL). Antibodies for Western blot analysis were specific for SIRT2 (Santa Cruz H-95, 1:2000, Santa Cruz Biotechnology, Santa Cruz, CA; or kindly provided by Olga Shakhova, Institute of Clinical Pathology, University Hospital, Zurich, Switzerland, 1:2000, yielding identical results), PLP/DM20 (A431; 1:5000) (Jung et al., 1996), CNP [Sigma (St. Louis, MO) 11-5B, 1:1000], myelin basicprotein (MBP) (A0623, 1:1000; Dako, High Wycombe, UK), myelin-oligodendrocyte glycoprotein (MOG) (8–18C5, 1:2500) (Linnington et al., 1984), or α-tubulin (Sigma T5168, 1:2000). For the detection of acetylated proteins, blots were blocked and washed as above and incubated with acetyl-lysine antibodies (Cell Signaling #9441, 1:500; Cell Signaling Technology, Beverly, MA) in TBST containing 5% BSA.
Quantitative real time-PCR.
Total RNA was extracted using Qiazol Reagent according to the manufacturer (Qiagen, Hilden, Germany). The integrity of purified RNA was confirmed using the Agilent 2100 Bioanalyser (Agilent Technologies, Santa Clara, CA). For real-time (RT)-PCR analysis, cDNA was synthesized from total RNA using random nonamer primers and Superscript III RNase H reverse transcriptase (Invitrogen). Real-time PCR was performed using the ABI Prism 7700 Sequence Detection System and SYBR Green Master Mix according to the manufacturer (Applied Biosystems, Foster City, CA). All reactions were performed in duplicate resulting in an almost complete overlap of the amplification plots (data not shown). PCR primers were specific for β-actin (forward 5′-TGACAGGATGCAGAAGGAGA and reverse 5′-CGCTCAGGAGGAGCAATG), PLP (forward 5′- TCAGTCTATTGCCTTCCCTAGC and reverse 5‘-AGCATTCCATGGGAGAACAC), or SIRT2 (forward 5′-CACTACTTCATCCGCCTGCT and reverse 5′-CCAGCGTGTCTATGTTCTGC).
Immunostaining.
Primary oligodendrocyte cultures were gained by isolation of oligodendrocyte progenitor cells (OPCs) from rat postnatal day 1 (P1) cortices and cultured according to the study by McCarthy and de Vellis (1980). Briefly, after overnight incubation in an orbitary shaker at 180 rpm, progenitor cells were immunolabeled with A2B5 antibodies and selected using magnetic beads (Miltenyi Biotec, Auburn, CA). Progenitors were maintained proliferating by adding basic fibroblast growth factor (20 ng/ml) and PDGF (10 ng/ml) to Sato medium (DMEM, 100 μg/ml albumin, 100 μg/ml apo-transferrin, 16 μg/ml putrescine, 0.06 ng/ml progesterone, 40 ng/ml selenium, 5 μg/ml insulin, 1 mm sodium pyruvate, 2 mm l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin). Differentiation was obtained by culturing cells in the same medium, devoid of mitogens.
For immunocytochemistry, primary OPCs were incubated live with hybridoma cell supernatant A2B5 (1:5) or galactosyl cerebroside (1:10) (30 min, 37°C), followed by very gentle washes with PBS and fixation with 4% paraformaldehyde (PFA) in 0.1 m PBS. Incubation with rhodamine-conjugated secondary antibodies (1:100; Jackson ImmunoResearch, West Grove, PA) was conducted for 1 h at room temperature. After immunolabeling of surface antigens, the cells were permeabilized with blocking buffer [0.1 m phosphate buffer, 5% normal goat serum (Vector Laboratories, Burlingame, CA), 0.5% Triton X-100] and then incubated with anti-SIRT2 antibodies (1:200; overnight at room temperature). Immunoreactive cells were visualized using biotinylated secondary antibodies (1:200; GE Healthcare) followed by avidin-conjugated FITC (1:500; GE Healthcare). Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (1:1000; Invitrogen). Images were captured using a Hamamatsu (Bridgewater, NJ) CCD camera connected to an inverted fluorescence microscope (DM RA; Leica, Nussloch, Germany) and interfaced to a G4 Mac computer (Apple Computers, Cupertino, CA).
For immunohistochemistry (IHC), wild-type and PLP_null_ mice (n = 3 each) were anesthetized with Avertin (Sigma-Aldrich) and transcardially perfused with HBSS (Cambrex, Walkersville, MD) followed by fixative (4% PFA in 0.1 m PBS). Paraffin-embedded brains and dorsal roots were sectioned on a slide microtome (4 μm; Microm HM400). For IHC, we used the LSAB2 system according to the manufacturer (Dako, Hamburg, Germany). Sections were counterstained with HE (Merck, Darmstadt, Germany). Equivalent sections were analyzed by light microscopy with a Leica DM RXA microscope. Antibodies for immunostaining were specific for CNP (SMI-91; Covance Research Products, Berkeley, CA); MBP (SMI-99; Covance Research Products); GalC (Cedarlane Laboratories, Burlington, Ontario, Canada); A2B5 supernatant from hybridoma cells provided by Dr. R. Bansal (University of Connecticut, Farmington, CT); or SIRT2 [immunocytochemistry provided by Dr. M. Tainsky (Wayne State University School of Medicine, Detroit, MI); IHC, Santa Cruz H-95].
Electron microscopy of myelin fractions.
Myelin fractions were immersion-fixed with 4% formaldehyde (Serva, Heidelberg, Germany) and 2.5% glutaraldehyde (Science Services, Munich, Germany) in PBS and embedded in 2% low melting point-agarose in H2O. Agarose blocks were postfixed in 2% OsO4 (ChemPur), dehydrated with ethanol and propylenoxide, and embedded in Epon (Serva). Ultrathin sections (50 nm) were cut using a Leica Ultracut S ultramicrotome (Leica, Vienna, Austria) and stained with an aqueous solution of 4% uranyl acetate (Merck) followed by lead citrate (Reynolds, 1963). The sections were viewed in a LEO EM 912AB electron microscope (Zeiss, Oberkochen, Germany), and pictures were taken with an on-axis 2048 × 2048 CCD camera (Proscan, Scheuring, Germany).
Immunoelectron microscopy.
Wild-type mice (C57BL/6) were anesthetized with Avertin and transcardially perfused with HBSS followed by fixative (4% formaldehyde and 0.2% glutaraldehyde in 0.1 m phosphate buffer containing 0.5% NaCl). The CNS was dissected, and pieces of the dorsal spinal cord white matter at the level of the thorax were infiltrated with 2.3 m sucrose in 0.1 m phosphate buffer overnight, mounted onto aluminum pins for cryo-ultramicrotomy, and frozen in liquid nitrogen. Cultured primary oligodendrocytes were infiltration-fixed with 2% formaldehyde and 0.2% glutaraldehyde. After washing, the cells were scraped from the dish in 0.1 m phosphate buffer containing 1% gelatin, spun down and suspended in 10% gelatin in 0.1 m phosphate buffer at 37°C. The pellets in gelatin were cooled on ice and cut in small blocks. The blocks were infiltrated with 2.3 m sucrose in 0.1 m phosphate buffer, mounted onto aluminum pins and frozen in liquid nitrogen. Ultrathin cryosections were cut with a diamond knife (Diatome, Biel, Switzerland) using a Leica UC6 cryo-ultramicrotome (Leica, Vienna, Austria). The sections were picked up in a 1:1 mixture of 2% methylcellulose (Sigma) and 2.3 m sucrose (Liou et al., 1996). After five washes in PBS with 0.1% glycine (Serva), sections were blocked (3 min with 1% BSA in PBS). For immunolabeling, spinal cord sections were incubated with antiserum specific for SIRT2 in blocking buffer, followed by five washes with PBS and 20 min incubation with protein A-gold (10 nm) in blocking buffer. For double-immunolabeling, sections of cultured primary oligodendrocytes were first incubated with antiserum specific for SIRT2 followed by protein-A-gold (10 nm). To denature Fc-tails of antibodies, sections were fixed with 1% glutaraldehyde (5 min), followed by five washes and blocking. Then sections were incubated with anti-PLP antiserum followed by protein-A-gold (15 nm) in blocking buffer. After fixation with 1% glutaraldehyde (3 min), sections were washed with H2O and contrasted (5 min) with neutral uranyl acetate (2% in 1.5 m oxalic acid, pH adjusted to 7.0 with ammonium hydroxide). Next, grids were transferred to ice-cold droplets of 1.8% methylcellulose containing 0.4% uranyl acetate, incubated 5 min, and picked up with a wire loop. Excess fluid was drained from the loop by gentle tapping to Whatman filter paper, and the sections were embedded in the remaining thin film by air-drying (Liou et al., 1996). Colloidal gold conjugated to protein A was obtained from the Cell Microscopy Center (Department of Cell Biology, University Medical Center at Utrecht, The Netherlands) and prepared as done previously (Roth et al., 1978; Slot and Geuze, 1985). Antibodies for immunoelectron microscopy were specific for SIRT2 (1:100; provided by Dr. O. Shakhova), or PLP (A431; 1:250) (Jung et al., 1996). Sections were analyzed with a LEO EM912 Omega (Zeiss, Oberkochen, Germany), and digital micrographs were obtained with an on-axis 2048 × 2048-CCD camera (Proscan, Scheuring, Germany).
Results
Myelin purification
For “myelin purification” of adult (P75) wild-type (C57BL/6) mouse brains, we followed the commonly used method of sequential enrichment by two discontinuous sucrose density gradient centrifugations (Norton and Poduslo, 1973), with the myelin-enriched fraction obtained at the interphase between 0.32 and 0.85 m sucrose (supplemental Fig. 1_A_, available at www.jneurosci.org as supplemental material). In the following, we refer to “myelin 1” as the interphase fraction from the first centrifugation and “myelin 2” as the interphase from the second centrifugation. By electron microscopy, myelin 1 contained a range of cellular membranes, including myelinated axonal fragments (supplemental Fig. 1_B_,B′, available at www.jneurosci.org as supplemental material). Myelin 2 contained mostly multilamellar myelin sheaths (supplemental Fig. 1_C_,C′, available at www.jneurosci.org as supplemental material), but we also noticed mitochondria (supplemental Fig. 1_C″_, available at www.jneurosci.org as supplemental material), suggesting that the sucrose-density floatation of mitochondria and myelin is similar. For comparison, myelin 2 isolated from PLP_null_ mice also contained mostly multilamellar myelin sheaths, but the number of membrane layers was lower (supplemental Fig. 1_D_,D′, available at www.jneurosci.org as supplemental material), reflecting reduced adhesive properties (Klugmann et al., 1997; Bizzozero et al., 2001). For simplicity, we will use the term “myelin-associated” for all proteins detected in myelin 2, including myelin-associated axonal proteins.
Myelin proteome analysis
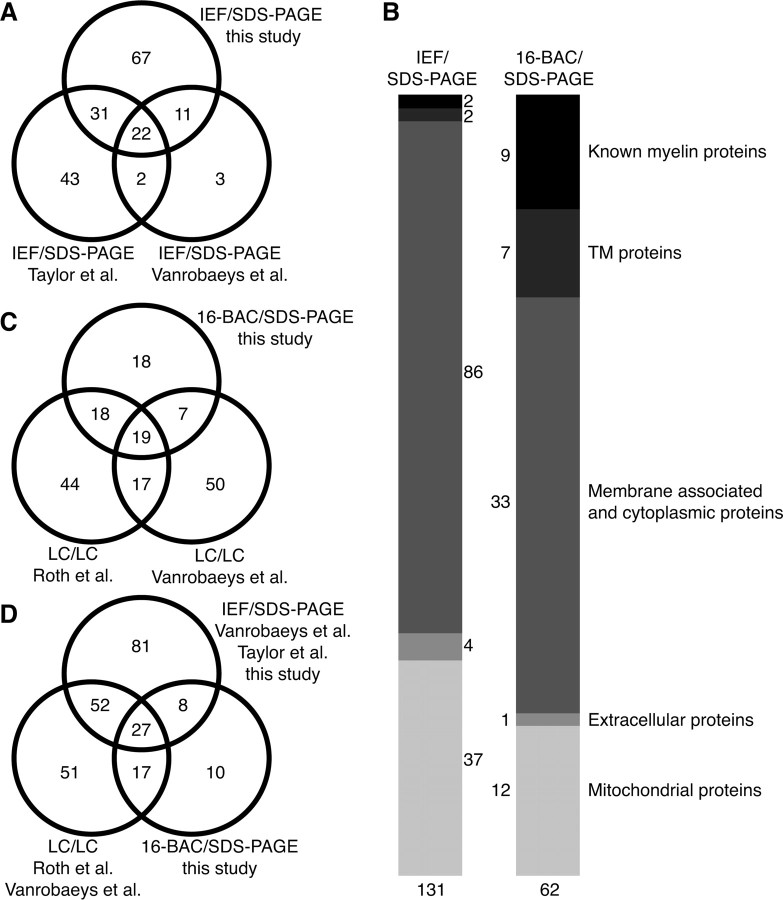
To establish a gel-based reference map of the myelin proteome for subsequent mouse mutant analysis, we subjected purified myelin to conventional 2D gel electrophoresis comprising IEF in immobilized pH gradients as the first dimension and SDS-PAGE as the second dimension (2D-IEF/SDS-PAGE). The protein profile of myelin was highly reproducible, as evidenced by the overlay of several 2D-IEF/SDS-PAGE replicates, each visualized by colloidal Coomassie stain (Fig. 1A) (data not shown). Gel plugs (protein spots) were excised and subjected to tryptic in-gel digest. For high-confidence protein identification on the basis of peptide mass and sequence information, we used automated protein identification based on MALDI-TOF-MS (Jahn et al., 2006). This allowed the identification of 217 proteins from 250 picked spots. Because of alternative splicing and posttranslational modifications, several proteins were found in more than one spot, leading to a library of 131 nonredundant myelin-associated proteins (Table 1). Among the 131 proteins, 67 were newly reported as myelin-associated, and 64 confirmatory of previous studies (Taylor et al., 2004; Vanrobaeys et al., 2005). Interestingly, 66 myelin-associated proteins have now been identified by 2D-IEF/SDS-PAGE in different laboratories and under different experimental conditions (e.g., mouse or rat) (Fig. 2A).
Figure 1.
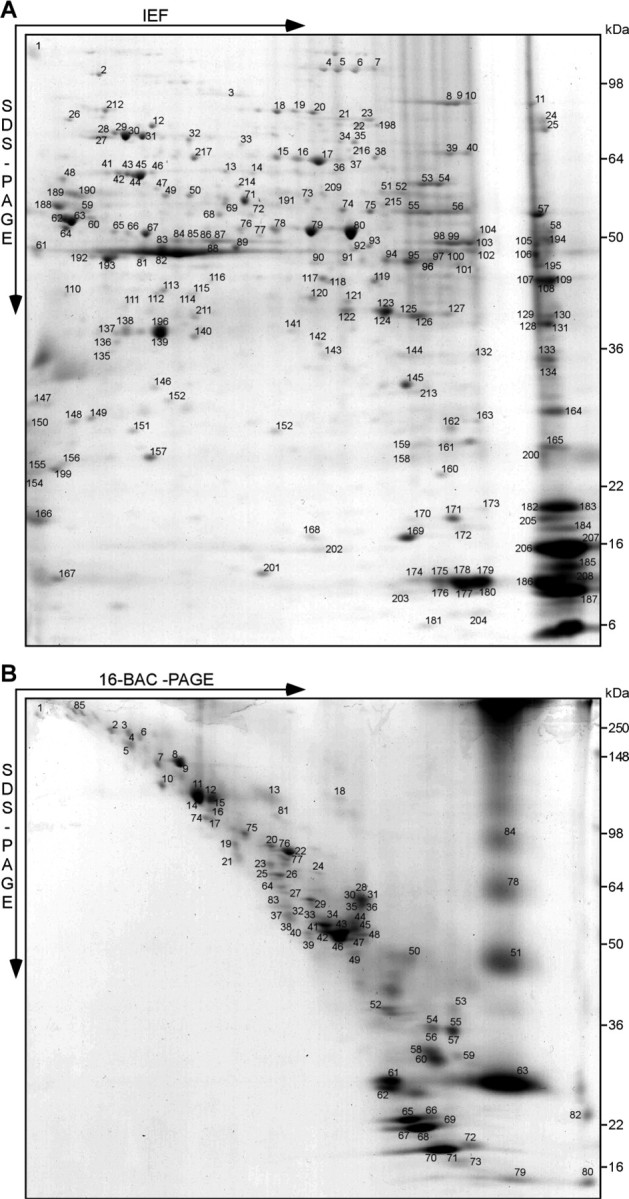
Gel-based myelin proteome maps. A, B, Myelin 2 from wild-type animals was two-dimensionally separated in different gel systems, and proteins visualized by colloidal Coomassie staining were identified by MALDI-TOF-MS. The spot numbers correspond to proteins listed in Table 1. A, 2D-IEF/SDS-PAGE with IEF in a nonlinear pH gradient (pH 3–10) as the first dimension and SDS-PAGE as the second dimension. B, 2D-16-BAC/SDS-PAGE with separation in a 16-BAC gel as the first and SDS-PAGE as the second dimension. Representative gels are shown.
Table 1.
The CNS myelin proteome
| Protein name | Accession | Acronym | IEF spot number | 16BAC spot number | IEF-T | IEF-V | LC-V | LC-R | Allen |
|---|---|---|---|---|---|---|---|---|---|
| Known myelin proteins | |||||||||
| Claudin-11, OSP | Q60771 | CLD11 | nd | 82 | nd | nd | Yes | nd | + |
| CNP | P16330 | CN37 | 61, 82, 84–88, 97–100, 102–106 | 18,34,41–43, 45,46 | nd | nd | Yes | Yes | + |
| Contactin 1 | P12960 | CNTN1 | nd | 7 | Yes | nd | Yes | Yes | + |
| Myelin basic protein | P04370 | MBP | 182–185, 187, 200, 202, 205–207 | 61,62,65–73 | nd | Yes | Yes | Yes | + |
| Myelin proteolipid protein | P60202 | MYPR | nd | 51,63,78,84 | nd | nd | Yes | Yes | + |
| Myelin associated glycoprotein | P20917 | MAG | nd | 13 | nd | nd | Yes | Yes | + |
| Myelin oligodendrocytic basic protein | NP032640 | (NCBI) | nd | 79 | nd | nd | Yes | nd | + |
| Myelin oligodendrocyte glycoprotein | Q61885 | MOG | nd | 6.58 | nd | nd | Yes | Yes | + |
| Neurofascin | Q810U3 | NFASC | nd | 5 | nd | nd | nd | Yes | + |
| Transmembrane proteins | |||||||||
| Calnexin | P35564 | CALX | nd | 81 | nd | nd | nd | Yes | + |
| Contactin-associated protein 1, Caspr | O54991 | CNTP1 | nd | 3, 4 | nd | nd | nd | nd | +/− |
| Immunoglobulin superfamily member 8 | Q8R366 | IGSF8 | nd | 19 | nd | nd | Yes | Yes | + |
| Na+K+-ATPase α 1 | Q8VDN2 | AT1A1 | nd | 12.15 | nd | nd | Yes | Yes | +/− |
| Na+K+-ATPase α 2 | Q6PIE5 | AT1A2 | nd | 11 | nd | nd | nd | Yes | + |
| Na+K+-ATPase α 3 | Q6PIC6 | AT1A3 | nd | 11.14 | nd | nd | nd | Yes | + |
| Na+K+-ATPase β 1 | P14094 | AT1B1 | nd | 44 | nd | nd | Yes | Yes | +/− |
| N-CAM 120 | P13594 | NCA12 | 1 | nd | Yes | nd | nd | Yes | + |
| Thy 1 membrane glycoprotein | P01831 | THY1 | 150 | nd | nd | nd | Yes | Yes | +/− |
| Cytoplasmic proteins | |||||||||
| 14–3-3 protein gamma, YWHAG | P61982 | 1433G | 147 | nd | Yes | Yes | nd | Yes | +/− |
| Actin β | P60710 | ACTB | 192.193 | nd | nd | Yes | Yes | Yes | + |
| Actin γ | P63260 | ACTG | nd | 48 | nd | nd | nd | nd | + |
| ADP ribosylation factor 6, ARF6 | P62331 | ARF6 | 166 | nd | nd | nd | nd | nd | + |
| Aldolase A, Fructose-bisphosphate | P05064 | ALDOA | 108 | nd | Yes | Yes | Yes | Yes | nd |
| Argininosuccinate synthase | P16460 | ASSY | nd | 40 | nd | nd | nd | nd | nd |
| Cannabinoid receptor interacting | Q5M8N0 | CB032 | 173 | nd | nd | nd | nd | nd | nd |
| Carbonic anhydrase 2 | P00920 | CAH2 | 145 | nd | Yes | nd | Yes | nd | + |
| Cell division control protein 42, cdc42 | P60766 | CDC42 | 155 | nd | nd | nd | nd | nd | + |
| Actin related protein α, Centractin | P61164 | ACTZ | 92 | nd | nd | nd | nd | nd | + |
| Clathrin heavy chain | Q68FD5 | CLH | nd | 6 | nd | nd | nd | Yes | nd |
| Creatine kinase brain | Q04447 | KCRB | 83 | nd | Yes | Yes | Yes | Yes | + |
| Crystallin α2 | P23927 | CRYAB | 160 | nd | Yes | nd | Yes | nd | + |
| Dihydropyrimidinase related 2, CRMP-2 | O08553 | DPYSL2 | 15–17, 33, 36, 47, 110 | 23 | Yes | Yes | Yes | Yes | + |
| Dynamin 1 | P39053 | DYN1 | 4, 5, 6, 7 | 12 | Yes | nd | nd | Yes | + |
| EH-domain containing protein 1 | Q9WVK4 | EHD1 | nd | 24 | Yes | nd | Yes | nd | + |
| EH-domain-containing protein 3 | Q9QXY6 | EHD3 | nd | 24 | nd | nd | nd | nd | + |
| Elongation factor 1-α 1 | P10126 | EF1A1 | 194 | 38 | nd | nd | Yes | Yes | + |
| Elongation factor 1-α 2 | P62631 | EF1A2 | 58 | 32 | nd | nd | nd | nd | nd |
| Enolase 1 | P17182 | ENOA | 76, 78–80, 112, 114, 118, 119 | 33 | Yes | Yes | Yes | nd | + |
| Enolase 2, neuron-specific, NSE | P17183 | ENOG | 64 | nd | Yes | Yes | Yes | nd | + |
| Ezrin | P26040 | EZRI | 18 | nd | Yes | nd | nd | nd | + |
| Fascin | Q61553 | FSCN1 | 74.75 | nd | nd | nd | nd | nd | + |
| Glial fibrillary acidic protein | P03995 | GFAP | 65 | 33 | nd | nd | nd | nd | + |
| Glucose-6-phosphate isomerase | P06745 | G6PI | nd | 64 | nd | nd | nd | Yes | nd |
| Glutamine synthetase | P15105 | GLNA | 94–96 | nd | Yes | Yes | Yes | nd | + |
| Glutathione S-transferase Mu 1 | P10649 | GSTM1 | 217 | nd | nd | nd | nd | nd | nd |
| GAPDH | P16858 | G3P | 128.131 | nd | Yes | Yes | Yes | nd | + |
| Guanine nucleotide-binding α2 | P18873 | GNAO2 | nd | 50 | Yes | nd | nd | nd | nd |
| Guanine nucleotide-binding β1 | P62874 | GBB1 | 196 | nd | Yes | Yes | Yes | nd | + |
| Guanine nucleotide-binding β2 | P62880 | GBB2 | 138.139 | nd | nd | Yes | nd | Yes | + |
| Guanine nucleotide-binding β4 | P29387 | GBB4 | 140 | nd | nd | nd | nd | nd | + |
| Guanine nucleotide-binding β5 | P62881 | GBB5 | 116 | nd | nd | nd | nd | nd | + |
| Hsp 90-α, HSP89 | P07901 | HS90A | nd | 16 | nd | nd | nd | nd | + |
| Hspa2, Hsp70.2, Hsp72 | P17156 | HSP72 | 28, 29 | 20 | nd | nd | nd | nd | + |
| Hspa5, Hsp70, Grp78, BiP | P20029 | GRP78 | 26 | nd | Yes | nd | nd | nd | + |
| Hsp70/Hsp90 organizing, STIP1 | Q60864 | STIP1 | 38 | nd | Yes | nd | nd | nd | + |
| Hspa8, Hsp73 | P63017 | HSP7C | 29 | 20 | nd | Yes | Yes | Yes | + |
| Neurofilament 66 kDa, internexin α | P46660 | AINX | 41–43, 45, 46 | 25–27 | Yes | Yes | nd | Yes | nd |
| Lactate dehydrogenase B | P16125 | LDHB | 211 | nd | Yes | nd | nd | nd | nd |
| Malate dehydrogenase, cytoplasmic | P14152 | MDHC | 141 | nd | Yes | Yes | Yes | nd | +/− |
| Moesin | P26041 | MOES | 22.198 | nd | nd | nd | nd | nd | + |
| Munc 18, syntaxin-binding protein 1 | O08599 | STXB1 | nd | 77 | Yes | nd | nd | Yes | + |
| Myosin 1d | Q5SYD0 | MYO1D | nd | 10 | nd | nd | nd | Yes | |
| Myosin 14 | Q6URW6 | MYH14 | nd | 1 | nd | nd | nd | nd | nd |
| NDRG1, N-myc downstream regulated | Q62433 | NDRG1 | 89 | nd | Yes | Yes | Yes | nd | + |
| Neurofilament triplet H | P19246 | NFH | 197 | 2 | nd | nd | nd | nd | +/− |
| Neurofilament triplet L | P08551 | NFL | 27.6 | 22 | Yes | Yes | nd | Yes | +/− |
| Neurofilament triplet M | P08553 | NFM | nd | 8.9 | nd | nd | nd | Yes | nd |
| NSF, _N_-ethylmaleimide sensitive fusion | P46460 | NSF | 21.23 | 75 | Yes | nd | nd | Yes | + |
| Nucleoside diphosphate kinase A | P15532 | NDKA | 168.169 | nd | Yes | nd | Yes | nd | nd |
| Nucleoside diphosphate kinase B | Q01768 | NDKB | 170.171 | nd | Yes | nd | Yes | nd | nd |
| Pacsin 1 | Q61644 | PACN1 | 59 | nd | nd | nd | nd | nd | +/− |
| Peptidyl-prolyl _cis_-trans isomerase A | P17742 | PPIA | 172 | nd | nd | Yes | Yes | nd | + |
| Peroxiredoxin 1 | P35700 | PRDX1 | 159.165 | nd | Yes | Yes | nd | Yes | + |
| Peroxiredoxin 2 | Q61171 | PRDX2 | 199 | nd | nd | Yes | nd | nd | +/− |
| Phosphatidylethanolamine binding | P70296 | PEBP | 156 | nd | nd | Yes | nd | nd | + |
| Phosphatidylinositol transfer α | P53810 | PIPNA | 142 | nd | nd | nd | nd | nd | +/− |
| Phosphoglycerate dehydrogenase | Q61753 | SERA | 209 | nd | nd | nd | nd | nd | nd |
| Phosphoglycerate mutase 1 | Q9DBJ1 | PGAM1 | 213 | nd | Yes | nd | Yes | nd | +/− |
| PLCb1, phospholipase Cb1 | Q9Z1B3 | PLCB1 | 66 | nd | Yes | nd | nd | nd | +/− |
| Programmed cell death 6-interacting | Q9WU78 | PDC6I | 4 | nd | nd | nd | nd | nd | + |
| Protein disulfide isomerase Grp58 | P27773 | PDIA3 | 214 | nd | Yes | nd | nd | nd | + |
| Pyruvate kinase isozyme M2 | P52480 | KPYM | 53.54 | nd | Yes | Yes | Yes | nd | + |
| Rab GDP dissociation inhibitor α | P50396 | GDIA | 48.148 | nd | Yes | nd | Yes | Yes | + |
| Rab GDP dissociation inhibitor β | Q61598 | GDIB | 77 | nd | Yes | nd | nd | nd | + |
| Radixin | P26043 | RADI | 19.2 | nd | nd | nd | nd | nd | + |
| Ras-like protein Ral-A | P63321 | RALA | nd | 59 | nd | nd | nd | nd | + |
| Ras-related protein Rap-1A | P62835 | RAP1A | 154 | nd | Yes | nd | Yes | Yes | + |
| Septin 2 | P42208 | SEPT2 | 90, 91, DIGE | 39 | Yes | nd | Yes | nd | + |
| Septin 4 | P28661 | SEPT4 | DIGE | nd | nd | nd | nd | nd | + |
| Septin 7 | O55131 | SEPT7 | 194 | 37 | Yes | nd | Yes | Yes | + |
| Septin 8 | Q8CHH9 | SEPT8 | 69, 70, 71, DIGE | 83 | nd | Yes | Yes | Yes | + |
| Sirtuin 2, NAD-dependent | Q8VDQ8 | SIRT2 | 120–127, DIGE | nd | Yes | Yes | Yes | Yes | + |
| Soluble NSF attachment protein α | Q9DB05 | SNAA | 135 | nd | nd | nd | nd | nd | + |
| Soluble NSF attachment protein β | P28663 | SNAB | 136 | nd | nd | nd | nd | nd | +/− |
| Soluble NSF attachment protein γ | Q9CWZ7 | SNAG | 111 | nd | nd | nd | nd | nd | +/− |
| Spectrin α 2 | P16546 | SPTA2 | nd | 85 | Yes | nd | nd | nd | +/− |
| Superoxide dismutase 1, Cu/Zn | P08228 | SODC | 168 | nd | nd | nd | Yes | nd | +/− |
| Synapsin 1 | O88935 | SYN1 | 24.25 | nd | nd | nd | nd | nd | +/− |
| Synapsin 2 | Q64332 | SYN2 | 215 | nd | nd | nd | nd | nd | + |
| Synaptosomal-associated protein 25 | P60879 | SNP25 | 150 | nd | nd | Yes | Yes | Yes | nd |
| T-complex 1 α B | P11983 | TCPA2 | 14 | nd | nd | nd | nd | nd | nd |
| T-complex 1 β | P80314 | TCPB | 73 | nd | nd | nd | nd | nd | nd |
| T-complex 1 ϵ | P80316 | TCPE | 13 | nd | nd | nd | nd | nd | nd |
| T-complex 1 γ | P80318 | TCPG | 37 | nd | nd | nd | nd | nd | nd |
| Transgelin 3 | Q9R1Q8 | TAGL3 | 158 | nd | nd | nd | nd | nd | + |
| Transitional ER ATPase | Q01853 | TERA | 2 | nd | Yes | nd | nd | nd | nd |
| Transketolase | P40142 | TKT | 39.4 | 21 | Yes | nd | Yes | nd | + |
| Tubulin α 1 | P68369 | TBA1 | 190 | 3.28 | nd | nd | nd | Yes | + |
| Tubulin α 2 | P05213 | TBA2 | 189 | nd | Yes | Yes | Yes | nd | nd |
| Tubulin β 2C | P68372 | TBB2C | 191 | 35.36 | Yes | nd | Yes | Yes | nd |
| Tubulin β 4 | Q9D6F9 | TBB4 | 137.188 | 31 | nd | Yes | Yes | Yes | + |
| Tubulin polymerization-promoting | Q7TQD2 | P25A | 164 | nd | nd | Yes | nd | nd | + |
| Ubiquitin | P62991 | UBIQ | 181 | nd | nd | nd | nd | nd | nd |
| Ubiquitin hydrolase L1 | Q9R0P9 | UCHL1 | 149 | nd | Yes | nd | nd | nd | +/− |
| Vacuolar ATP synthase A, ubiquitous | P50516 | VATA1 | 31 | nd | nd | nd | nd | nd | nd |
| Vacuolar ATP synthase B, brain | P62814 | VATB2 | 49.5 | nd | nd | nd | nd | nd | nd |
| WD-repeat protein 1 | O88342 | WDR1 | 216 | nd | nd | nd | nd | nd | + |
| Extracellular proteins | |||||||||
| Hemoglobin α | P01942 | HBA | 163, 176, 177, 180, 186, 204 | nd | nd | nd | Yes | Yes | nd |
| Hemoglobin β | P02088 | HBB1 | 174, 175, 178, 179 | 80 | nd | nd | Yes | Yes | nd |
| Macrophage migration inhibitory factor | P34884 | MIF | 203 | nd | nd | nd | Yes | nd | + |
| Serum albumin | P07724 | ALBU | 32 | nd | nd | nd | nd | Yes | +/− |
| Mitochondrial proteins | |||||||||
| Aconitate hydratase | Q99KI0 | ACON | 8, 9–11 | 17 | Yes | nd | nd | Yes | |
| ADP/ATP translocase 1, SLC25A4 | P48962 | ADT1 | nd | 55.57 | nd | nd | nd | Yes | |
| ADP/ATP translocase 2, SLC25A5 | P51881 | ADT2 | nd | 54.56 | nd | nd | nd | nd | |
| Aspartate aminotransferase | P05202 | AATM | 107–109 | 49 | nd | nd | Yes | Yes | |
| Aspartate glutamate carrier 1, SLC25A12 | Q8BH59 | CMC1 | nd | 76 | nd | nd | nd | nd | |
| ATP synthase F0 complex B | Q9CQQ7 | AT5F1 | nd | 60 | nd | nd | nd | nd | |
| ATP synthase F0 complex D | Q9DCX2 | ATP5H | 157 | nd | Yes | nd | nd | nd | |
| ATP synthase F1 complex α | Q03265 | ATPA | 55–57 | 29 | Yes | Yes | Yes | Yes | |
| ATP synthase F1 complex β | P56480 | ATPB | 62, 63 | nd | Yes | Yes | Yes | Yes | |
| ATP synthase F1 complex γ | Q91VR2 | ATPG | 134 | nd | nd | nd | Yes | Yes | |
| ATP synthase O subunit | Q9DB20 | ATPO | 165 | nd | nd | nd | nd | nd | |
| Creatine kinase ubiquitous | P30275 | KCRU | 101 | nd | Yes | Yes | nd | Yes | |
| Cytochrome c oxidase Va | P12787 | COX5A | 167 | nd | Yes | nd | nd | nd | |
| Cytochrome c oxidase Vb | P19536 | COX5B | 201 | nd | nd | nd | nd | nd | |
| Cytochrome c oxidase Via | P43024 | CX6A1 | 176 | nd | nd | nd | nd | nd | |
| Cytochrome c somatic | P62897 | CYC | 208 | nd | nd | nd | Yes | Yes | |
| Elongation factor Tu | Q8BFR5 | EFTU | 93 | nd | nd | nd | nd | nd | |
| Glucose regulated protein 75 kDa | P38647 | GRP75 | 12 | nd | Yes | nd | nd | nd | |
| Hspd1, Hsp60 | P63038 | CH60 | 44 | nd | Yes | Yes | nd | Yes | |
| Isocitrate dehydrogenase 3 α | Q9D6R2 | IDH3A | 113, 115 | nd | nd | nd | Yes | nd | |
| Malate dehydrogenase, mitochondrial | P08249 | MDHM | 129.13 | nd | Yes | nd | Yes | Yes | |
| Mitochondrial inner membrane protein | Q8CAQ8 | IMMT | 3 | 74 | nd | nd | nd | nd | |
| NADH-ubiquinone oxidoreductase 24 kDa | Q9D6J6 | NUHM | 151 | nd | nd | nd | nd | nd | |
| NADH-ubiquinone oxidoreductase 30 kDa | Q9DCT2 | NUGM | 152 | nd | nd | nd | nd | nd | |
| NADH-ubiquinone oxidoreductase 42 kDa | Q99LC3 | NUDM | 117 | nd | nd | nd | nd | nd | |
| NADH-ubiquinone oxidoreductase 75 kDa | Q91VD9 | NUAM | 212 | nd | nd | nd | nd | Yes | |
| Oxoglutarate dehydrogenase | Q9D2G2 | ODO2 | 68.72 | nd | nd | nd | nd | nd | |
| Phosphate carrier protein, Slc25A25 | Q8VEM8 | MPCP | nd | 53 | nd | nd | nd | nd | |
| Prohibitin | P67778 | PHB | 146 | 56 | nd | nd | nd | nd | |
| Pyruvate dehydrogenase E1 α | P35486 | ODPA | 94 | nd | nd | nd | nd | nd | |
| Pyruvate dehydrogenase E2 | NP663589 | (NCBI) | 217 | nd | nd | nd | nd | nd | |
| Pyruvate dehydrogenase E3 | O08749 | DLDH | 51.52 | nd | nd | nd | nd | nd | |
| Succinate dehydrogenase subunit A | Q8K2B3 | DHSA | 34.35 | nd | nd | nd | nd | nd | |
| Succinyl-CoA ligase β | Q9Z2I9 | SUCB1 | 81 | nd | nd | nd | nd | nd | |
| Superoxide dismutase 2, Mn | P09671 | SODM | 161 | nd | Yes | nd | Yes | nd | |
| Peroxide reductase | P20108 | PRDX3 | 153 | nd | nd | nd | nd | nd | |
| Ubiquinol-cytochrome c reductase Fe-S | Q9CR68 | UCRI | 162 | nd | nd | nd | nd | nd | |
| Ubiquinol-cytochrome c reductase core 2 | Q9DB77 | UQCR2 | 195 | 47 | nd | nd | Yes | Yes | |
| Ubiquinol-cytochrome c reductase core 1 | Q9CZ13 | UQCR1 | 66.67 | nd | Yes | nd | nd | Yes | |
| Voltage-dependent anion channel 1 | Q60932 | VDAC1 | 132–144 | 52 | Yes | nd | nd | Yes | |
| Voltage-dependent anion channel 2 | Q60930 | VDAC2 | 143 | nd | nd | nd | nd | Yes | |
| 131 proteins | 62 | 98 | 38 | 93 | 98 |
Figure 2.
Comparison of myelin proteome libraries. A, Venn diagram comparing the number of myelin-associated proteins identified by MS after separation by 2D-IEF/SDS-PAGE in the present study and those reported previously based on the same technique (Taylor et al., 2004; Vanrobaeys et al., 2005). B , Comparison of myelin-associated proteins identified in this study by 2D-IEF/SDS-PAGE or 2D-16-BAC/SDS-PAGE. The classification corresponds to Table 1. Classification as mitochondrial or extracellular proteins was based on Swiss-Prot data entries. Note that known myelin proteins and proteins with a transmembrane (TM) domain were more efficiently identified in the 2D-16-BAC/SDS-PAGE approach. C, Venn diagram comparing the number of myelin-associated proteins identified in the present study by gel-based 2D-16-BAC/SDS-PAGE, and those previously reported based on gel-free LC/LC-MS (Vanrobaeys et al., 2005; Roth et al., 2006). D, Venn diagram comparing myelin proteome libraries by technical approaches. Numbers of proteins detected by 2D-IEF/SDS-PAGE (Taylor et al., 2004; Vanrobaeys et al., 2005; this study) were compared with gel-free LC/LC (Vanrobaeys et al., 2005; Roth et al., 2006) or 2D-16-BAC/SDS-PAGE (this study). Note the high overlap of proteins identified independent of the technique used.
We grouped the identified proteins of myelin-2 as follows: (1) “known” myelin proteins, (2) membrane-spanning, (3) cytoplasmic, (4) extracellular, and (5) mitochondrial proteins. Consistent with the limited ability of 2D-IEF/SDS-PAGE to resolve hydrophobic and basic proteins, several known myelin proteins were not identified. Interestingly, this group included the most abundant CNS myelin protein, PLP/DM20, a highly hydrophobic membrane-spanning protein, of which the identification required an alternative gel system (see below). The category “membrane-spanning proteins” was based on calculations using TMHMM (version 2.0) (http://smart.embl-heidelberg.de). It includes neural cell adhesion molecules (NCAM-120, Thy-1), previously detected in oligodendrocytes (Almqvist et al., 1986; Kramer et al., 1999; Roth et al., 2006). The majority of myelin-associated proteins were simply designated as “cytoplasmic” to avoid additional subclassification by ambiguous or speculated functions. We used gene ontology terms (http://david.abcc.ncifcrf.gov) to denote previously annotated biological functions: a large fraction was implicated in cellular physiology (73%), catalytic activities (47%), purine nucleotide (ATP or GTP) binding (35%), or transport (31%) but also in acetylation (19%) and the cytoskeleton (17%). The category “cytoplasmic” also included proteins previously detected in oligodendrocytes (or myelin) such as cdc42 (Liang et al., 2004), CRMP-2/dihydropyrimidinase-like (Smith et al., 2001), elongation factor 1a (Barbarese et al., 1995), and synapsin (Madison et al., 1999). Other proteins were described previously in Schwann cells, such as ERM proteins (ezrin, radixin, and moesin) in growing processes (Scherer et al., 2001). In confirmation of previous myelin proteome analyses, we found the NAD+-dependent deacetylase SIRT2 and small cytoskeleton-associated GTPases of the septin family (SEPT2, 7, 8) (Taylor et al., 2004; Vanrobaeys et al., 2005; Roth et al., 2006) and newly identified also SEPT4 as a myelin-associated protein.
To identify myelin proteins that escape identification by conventional 2D-IEF/SDS-PAGE, we used 2D 16-BAC SDS-PAGE as an alternative gel system, which also resolves membrane-spanning and highly basic proteins (Hartinger et al., 1996). This led to the identification of 85 proteins from 100 picked spots, yielding a library of 62 nonredundant proteins (Fig. 1B, Table 1), including 18 novel myelin-associated proteins and 44 confirmations. As expected, many known myelin proteins and membrane-spanning proteins were better represented by 16-BAC/SDS gels (Fig. 2B), including MOG (Johns and Bernard, 1999), myelin-associated glycoprotein (Schachner and Bartsch, 2000), claudin-11 (Bronstein et al., 2000), contactin (Falk et al., 2002), neurofascin (Tait et al., 2000), and PLP/DM20 (Klugmann et al., 1997) (Table 1). Also, two highly basic proteins, MBP (Boggs, 2006) and MOBP (Montague et al., 2006), were easily identified. Among the membrane-spanning proteins, the expression of Na+/K+-ATPase subunits in oligodendrocytes reportedly depends on neuronal activity (Knapp et al., 2000). The role of other proteins in myelin remains to be determined, including immunoglobulin superfamily member 8 (IgSF8) (Vanrobaeys et al., 2005; Roth et al., 2006), Munc-18 (Madison et al., 1999), ras-like protein A (Ral-A), the Eps15 homology domain-containing proteins (EHD1 and EHD3), argininosuccinate synthase, dynamin 1, and various septins.
Together, our gel-based approaches have yielded two-dimensional maps of CNS myelin proteins and a myelin proteome library of the mouse brain. In contrast to two-dimensional liquid-chromatography (LC/LC) “shotgun” analyses (Vanrobaeys et al., 2005; Roth et al., 2006) (Fig. 2C,D), our approach retained myelin protein integrity and thereby information on protein abundance and isoforms. All published studies have operationally defined the term “myelin protein” without an independent verification. Although the identification of new myelin proteins by more than one study increases confidence, some of these proteins may only have copurified with myelin, such as proteins associated with axonal membranes or mitochondria. Nevertheless, the high degree of reproducibility of the 2D maps allowed us to systematically monitor altered protein expression profiles in the context of myelin disease.
Myelin proteome of PLP_null_ mice
PLP_null_ mice provide a genuine model for SPG-2 in humans, with progressive axonal degeneration in the presence of normal amounts of CNS myelin. We hypothesized that PLP and DM20 themselves are unlikely to mediate protection of axons. To identify candidate proteins that could be implicated in the failure of oligodendrocytes to support myelinated axons, we systematically screened PLP_null_ mice for (secondary) alterations in the myelin proteome that exist before the onset of disease. We used 2D-DIGE, comprising the labeling of purified myelin proteins from control and PLP_null_ mice with two different amine-reactive cyanine dyes. To avoid labeling of amino head groups (in phosphatidylethanolamine and phosphatidylserine) and to further improve resolution in 2D-IEF/SDS-PAGE (Rabilloud, 1996), myelin was delipidated and precipitated by a methanol/chloroform treatment (Wessel and Flugge, 1984) before dye addition. We note that this 2D-DIGE-specific pretreatment of samples limits comparability with the reference map (Fig. 1A).
As expected from the morphology of young adult PLP_null_ mice (Klugmann et al., 1997), the protein profile of myelin was very similar in wild-type and mutant mice (Fig. 3A). PLP/DM20 itself could not be resolved by 2D-IEF/SDS-PAGE (see above). In the representative 2D gel (Fig. 3A), blue spots indicate proteins reduced in PLP_null_ myelin, all of which were identified by MS. The only protein virtually absent from mutant myelin was SIRT2, a NAD+-dependent deacetylase (Fig. 3B). A total of four spots represented SIRT2 (Fig. 3B, I–IV), most likely two splice isoforms (see below) and isomers differing in net charge. The spot intensities indicate that SIRT2 constitutes an abundant protein in wild-type myelin (Fig. 3B), and that the abundant SIRT2 isoforms (III and IV) were most dramatically reduced in PLP_null_ myelin (Fig. 3C). After 2D-16-BAC/SDS-PAGE, SIRT2 was represented by one elliptic spot (Fig. 3D), presumably because the first dimension of protein separation is by molecular mass rather than charge. The absence of SIRT2 from PLP_null_ myelin was confirmed (Fig. 3D). On 2D-16-BAC/SDS-PAGE gels, monomeric and dimeric PLP was readily visualized (and lacking from PLP_null_ myelin) (Fig. 3D). By 2D-DIGE, we also identified SEPT2, SEPT4, and SEPT8 to be reduced by ∼40% in PLP_null_ myelin (Fig. 3B,E). Septins form filamentous heterotrimers with GTPase activity (Kinoshita, 2006) and may contribute to the myelin cytoskeleton. The analysis of myelin-associated septins will be described elsewhere.
Figure 3.
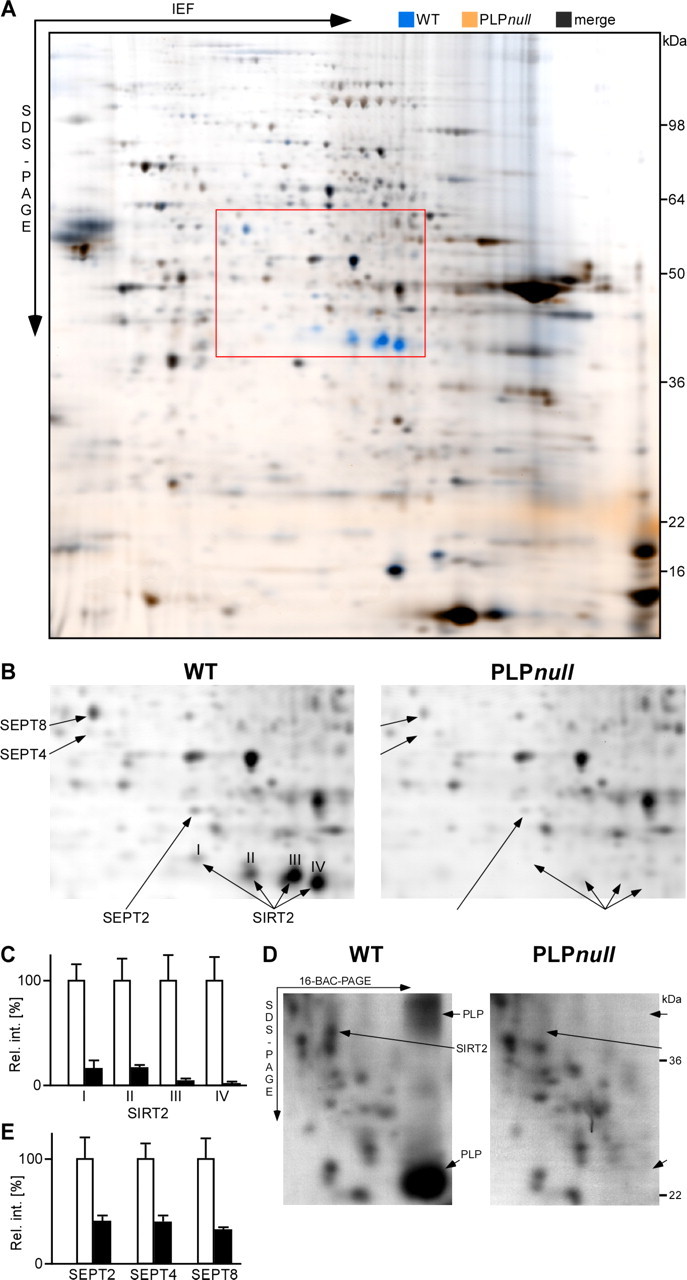
Differential myelin proteome analysis of the PLP_null_ mouse model of hereditary spastic paraplegia (SPG-2). A, 2D-DIGE of wild-type (WT; false-colored in blue) and PLP_null_ (false-colored in orange) myelin 2. Proteins less abundant in PLP_null_ myelin 2 constituted blue spots and were identified by MALDI-TOF-MS. B, Separation of detection channels and enlargement of the gel region marked by the red frame in A. Proteins associated with wild-type myelin (left; blue channel from A) included spots constituted by the septins SEPT2, SEPT4, and SEPT8 and four spots constituted by SIRT2 (I–IV, arrows). The principal spot pattern is similar in PLP_null_ myelin (right; orange channel from A), but spots constituted by SEPT2, SEPT4, and SEPT8 are diminished, and SIRT2 is virtually lacking (arrows). C, Relative intensities of spots constituted by SIRT2 (I–IV). Wild-type spots were expressed as 100% (open bars), and the SD is given. Note that spots constituted by SIRT2 are virtually absent from PLP_null_ myelin (closed bars). D, Comparison of wild-type and PLP_null_ myelin by 2D-16-BAC/SDS-PAGE. Note that spots constituted by PLP and SIRT2 (left, arrows) are lacking from PLP_null_ myelin (right). E, Relative intensities of spots constituted by SEPT2, SEPT4, and SEPT8 in wild-type myelin (open bars) or PLP_null_ myelin (closed bars).
SIRT2 is a myelin protein only in the presence of PLP/DM20
To independently confirm SIRT2 as a novel myelin protein, we analyzed normal mouse brain lysates and the membrane fractions myelin-1 and myelin-2 by Western blotting. Two bands were detected, corresponding to alternatively spliced isoforms of 43.2 kDa (variant 1, 389 residues) and 39.5 kDa (variant 2, 352 residues) (Voelter-Mahlknecht et al., 2005), depicted schematically in Figure 4A. The amount of SIRT2 in the sequentially myelin-enriched brain fractions was considerably higher than in brain lysates (Fig. 4B) (i.e., comparable with PLP/DM20 or CNP), demonstrating true copurification with myelin membranes. Interestingly, only variant 2 of SIRT2 was enriched in wild-type myelin. The absence of SIRT2 from PLP_null_ myelin was confirmed by Western blotting (Fig. 4C), demonstrating the lack of both PLP/DM20 and SIRT2 but not of MBP. Importantly, SIRT2 was already absent from myelin in 15-d-old mutant mice (data not shown), demonstrating that this molecular defect precedes axonal degeneration by several weeks. We also examined the abundance of SIRT2 in brain homogenates by Western blotting (Fig. 4D). SIRT2 variant 2 (Fig. 4A) (enriched in wild-type myelin) was virtually undetectable in PLP_null_ brain homogenates, whereas variant 1 was equally abundant in brains from wild-type and PLP_null_ mutants.
Figure 4.
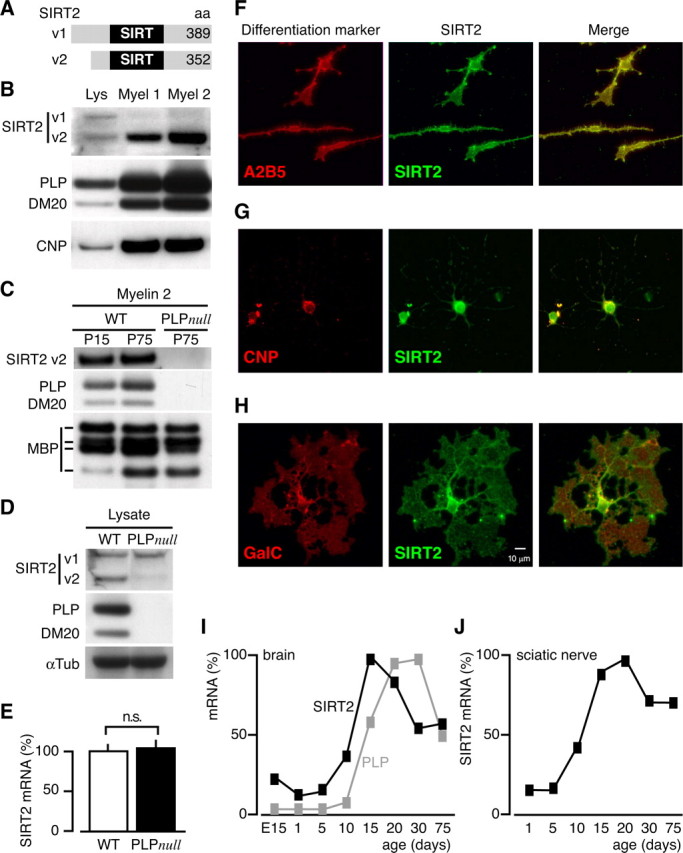
SIRT2 expression profile in myelinating glia. A, Two SIRT2 isoforms emerge by alternative splicing. Variant (v) 1 and v2 differ by a 37 amino acids domain at the N terminus, and both contain the enzymatically active SIRT-domain. B, 1D-PAGE/Western blot analysis of brain lysate (Lys) and myelin-enriched fractions (Myel 1 and Myel 2). The enrichment of SIRT2 with myelin purification was equivalent to that of the classical myelin proteins PLP/DM20 and CNP. Note that the two bands detected in brain lysate most likely represent the two splice isoforms, and that only v2 was enriched in myelin. C, 1D-PAGE/Western blot analysis of wild type (WT) and PLP_null_ myelin 2. SIRT2 and PLP were undetectable in PLP_null_ myelin 2, whereas MBP was unaltered. SIRT2 and PLP were equally abundant in wild-type myelin of young animals (P15) and adults (P75), whereas the abundance of certain MBP splice isoforms was developmentally regulated. D, 1D-PAGE/Western blot analysis of SIRT2 in wild-type and PLP_null_ brain lysates. Variant 1 was equally abundant in wild-type and PLP_null_, whereas variant 2 was virtually undetectable in PLP_null_ brains. E, Quantitative RT-PCR analysis of SIRT2 mRNA expression in adult (P75) wild-type (open bar) and PLP_null_ brains (closed bar). SIRT2 mRNA expression was unaltered. F–H, Immunodetection of SIRT2 (green) and differentiation markers (red) in cultured primary oligodendrocytes. A2B2 was used for oligodendrocyte precursors (F), CNP for differentiated oligodendrocytes (G), and GalC for more mature oligodendrocytes (H). I–J, Quantitative RT-PCR analysis of SIRT2 mRNA expression during brain development (I) or during sciatic nerve development (J). Maximum mRNA levels were arbitrarily set at 100%.
On the basis of in situ_-hybridization in the brain (www.ncbi.nlm.nih.gov/projects/gensat/), SIRT2 is most abundant in white matter. In PLP_null mice, the absence of SIRT2 from myelin could be a result of reduced RNA transcription or posttranscriptional regulation. SIRT2 transcript levels were quantified in PLP_null_ and control brains by qRT-PCR, and steady state mRNA levels were the same (Fig. 4E). Thus, SIRT2 mRNA is available for translation in PLP_null_ brains, but SIRT2 protein is not transported into myelin.
To resolve at which stages of oligodendrocyte development SIRT2 is expressed, we cultured rat oligodendrocyte progenitors in the presence of mitogens and then allowed them to differentiate by mitogen withdrawal. The presence of SIRT2 during oligodendrocyte lineage progression was defined by double immunocytochemistry with stage-specific markers (Fig. 4F–H). Additionally, we performed RT-PCR at corresponding stages of development (data not shown). With both techniques, SIRT2 was detected throughout the oligodendrocyte lineage, with higher abundance in mature oligodendrocytes. We also used RT-PCR to monitor SIRT2 mRNA during brain development. Overall, CNS expression revealed a peak of SIRT2 transcript level abundance that coincided with the peak of myelination (Fig. 4I), and a similar profile was seen during development of the sciatic nerve (Fig. 4J).
To address the localization of SIRT2 in vivo, we immunolabeled the protein on paraffin sections from adult control and PLP_null_ brains. SIRT2 was abundant throughout the wild-type white matter (Fig. 5A) but strongly reduced in PLP_null_ mice (Fig. 5C). In the wild-type CNS, SIRT2 was associated with myelin along the entire internodal region. Paranodal enrichment was only seen in the PNS (Fig. 5B,D), where it was independent of PLP. Cytoplasmic SIRT2 labeling was also prominent in white matter astrocytes in both wild-type and PLP_null_ mice (data not shown). Together, the enzyme is expressed in glia of the white matter and transported into myelin, whereas in the absence of PLP/DM20, SIRT2 is not incorporated into myelin and essentially degraded (Benjamins and Morell, 1978).
Figure 5.
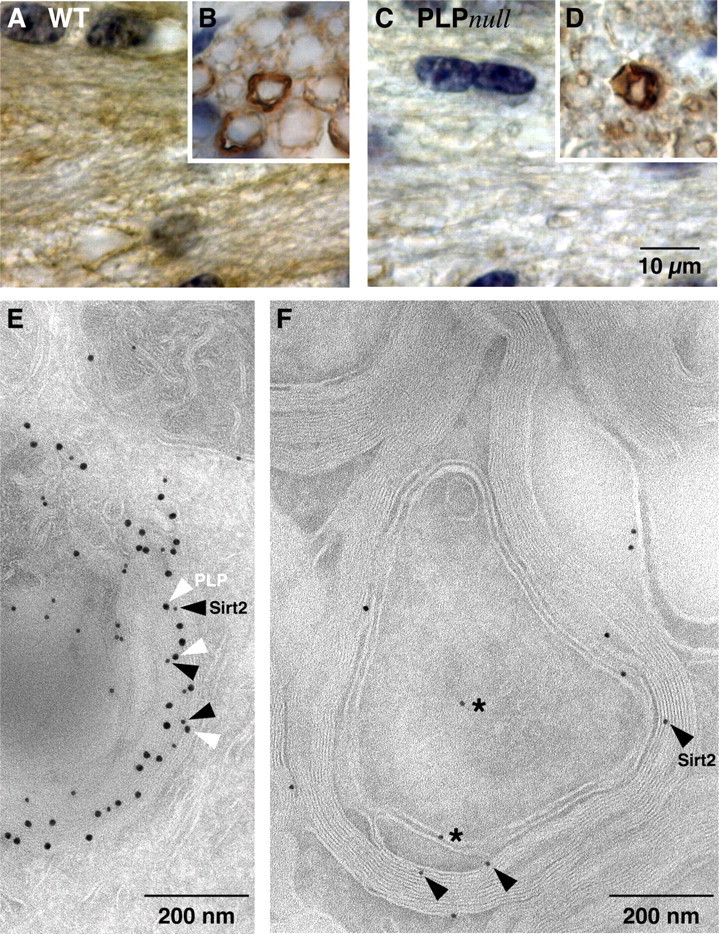
SIRT2 is a component of wild-type CNS myelin. A–D, Immunodetection of SIRT2 (brown; nuclei were counterstained in blue). Shown are representative images of the CNS white matter (coronal sections of the corpus callosum in A and C) or PNS (cross sections of the dorsal roots in B and D) from wild-type (A, B) and PLP_null_ (C, D) mice. Note that SIRT2 labeling marks all myelinated fibers in wild-type (A) but is essentially absent from mutant CNS myelin (C). Also note that SIRT2 expression in myelinating Schwann cells and enrichment in paranodes of PNS myelin is not dependent on PLP (B, D). E, Immunodetection of SIRT2 (10 nm gold particles; black arrowheads) together with PLP (15 nm gold particles; white arrowheads) on sections of cultured primary oligodendrocytes. Note that most SIRT2 labeling was in close proximity to PLP labeling and attached to the same membrane of a myelin-like multimembrane stack, possibly of endosomal origin. F, Immunodetection of SIRT2 visualized with gold particles in spinal cord cross sections. SIRT2 labeling was mostly confined to compact and adaxonal myelin (arrowheads). Axons and the extracellular space between myelin and axon were occasionally labeled (stars).
To define the exact localization of SIRT2 in oligodendrocytes and in CNS myelin, SIRT2 was visualized by cryo-immunoelectron microscopy of cultured mouse oligodendrocyte progenitors and spinal cord sections, using specific antibodies and protein-A coupled to 10 nm gold particles. We also performed double immunolabeling with PLP, which was visualized with 15 nm gold particles. In cultured oligodendrocytes, PLP was readily detectable in myelin-like multimembrane stacks, possibly of endosomal origin (Trajkovic et al., 2006), and a subfraction of PLP was colabeled with SIRT2 (Fig. 5E). This suggests that SIRT2 may be transported into CNS myelin by attachment to a membrane microdomain defined by PLP/DM20. We detected SIRT2 in compact CNS myelin and also prominent at the inner tongue (Fig. 5F). The occasional labeling of cells other than oligodendrocytes (mainly white matter astrocytes and axons) was expected, because SIRT2 is expressed by a wide range of tissues and cell types (Yang et al., 2000). Importantly, SIRT2 was not detected in PLP_null_ myelin (data not shown).
Acetylated myelin proteins
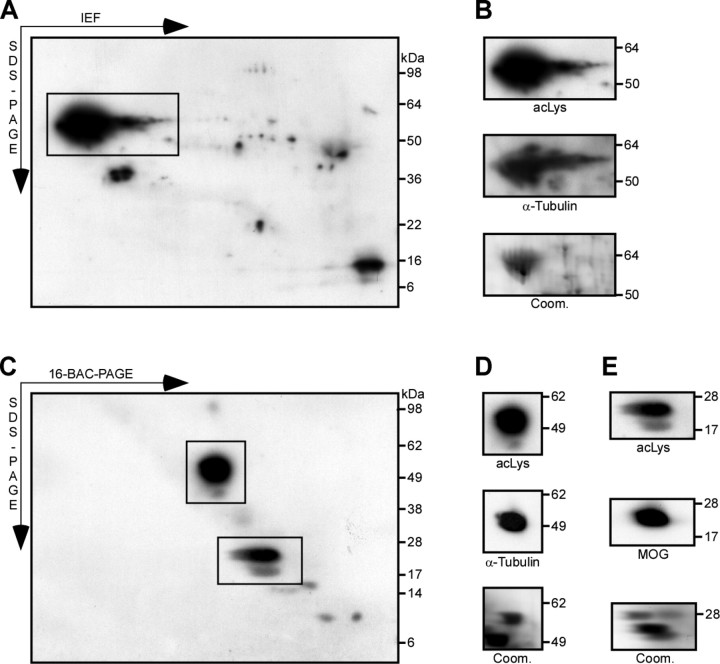
Does CNS myelin contain acetylated proteins that could be substrates for SIRT2 enzyme activity? We analyzed the membrane fraction myelin-2 from PLP_null_ and control mice by Western blot, using antibodies directed against lysine residues that are acetylated at their ε-amino groups. To achieve high resolution, myelin proteins were separated by 2D-IEF/SDS-PAGE (Fig. 6A,B) or 2D-16-BAC/SDS-PAGE (Fig. 6C–E). The spot pattern indicated the presence of several acetylated proteins in CNS myelin. A comparison with the reference maps of the myelin proteome suggested that the major spot corresponded to α-tubulin (Fig. 6A,C), which is a known in vitro substrate of SIRT2 (North et al., 2003; Li et al., 2007). Its identity was confirmed by reprobing the membranes with antibodies specific for α-tubulin (Fig. 6B,D), and by mass spectrometric identification from parallel Coomassie-stained gels. Comparison with the 2D-16-BAC/SDS-PAGE reference map, protein identification by MS, and Western blotting experiments suggested that also MOG is an acetylated protein (Fig. 6E). Consistent with the myelin proteome analysis (see above), MOG was only detected without previous isoelectric focusing. There was no obvious hyperacetylation of α-tubulin or MOG in PLP_null_ myelin when compared with controls (data not shown). However, Western blotting with anti-acetyl-lysine antibodies revealed several spots on 2D-IEF/SDS-PAGE gels (supplemental Fig. 2, available at www.jneurosci.org as supplemental material), representing myelin-associated proteins of minor abundance that appeared hyperacetylated in PLP_null_ myelin when compared with equivalent spots from control myelin. Although their identity could not be determined yet, they are potential SIRT2 substrates and candidate molecules that could play a role in a neuroprotective function dysregulated in PLP_null_ mice.
Figure 6.
Acetylated proteins in the myelin-enriched fraction. A, Western blot analysis of PLP_null_ myelin 2 with antibodies specific for acetylated lysine after 2D-IEF/SDS-PAGE. The major acetylated protein is boxed. B, The identity of the major acetylated protein (from A) as α-tubulin was suggested by spot pattern comparison with the myelin proteome reference map. The membrane was stripped and reprobed with antibodies specific for α-tubulin. Myelin was also separated in parallel for visualization by colloidal Coomassie stain (bottom). The spots at the corresponding positions were identified by MALDI-TOF-MS as α-tubulin. Note that several acetylated proteins were of too low abundance to constitute spots on Coomassie gels and could not be identified. C, Western blot analysis of PLP_null_ myelin 2 with antibodies specific for acetylated lysine after 2D-16-BAC/SDS-PAGE. Major acetylated proteins are boxed. D, E, The identity of the major acetylated proteins (from C) as α-tubulin and MOG was suggested by spot pattern comparison with the myelin proteome reference map. The membrane was stripped and reprobed with antibodies specific for α-tubulin (D) or MOG (E). Myelin was also separated in parallel for colloidal Coomassie stain, and the spots at the corresponding positions were identified by MALDI-TOF-MS as α-tubulin (D) and MOG (E). Note that membrane-spanning proteins (such as MOG) were only detected without previous isoelectric focusing (IEF). acLys, Acetylated lysine; Coom., colloidal Coomassie staining.
Discussion
We performed a gel-based proteome analysis to determine the molecular composition of CNS myelin, which has been experimentally more challenging for this than for other cellular systems because of unusual biophysical properties of myelin membranes and their hydrophobic proteins. More importantly, we have shown that the CNS myelin proteome is subject to unexpected changes in the mouse model of a human myelin disease genetically defined by the absence of a single myelin protein, PLP/DM20. Specifically, SIRT2, a newly identified myelin component, is posttranscriptionally excluded from CNS myelin in PLP_null_ mice. This reveals that PLP/DM20 is required for the transport of distinct proteins into the myelin compartment. It is possible that SIRT2 contributes to the protective function of oligodendrocytes preventing the secondary axonal degeneration observed in human patients with PMD and SPG-2.
Myelin proteomics
For the application in mouse mutants, a refinement of myelin proteome analysis was necessary. The significant overlap between myelin protein libraries established by 2D-IEF/SDS-PAGE (Taylor et al., 2004; Vanrobaeys et al., 2005) suggests that myelin purification by sequential sucrose gradient centrifugation (Norton and Poduslo, 1973) is very reproducible across different laboratories. However, previously known myelin proteins were underrepresented in myelin proteome libraries established after isoelectric focusing. Therefore, we compared conventional 2D-IEF/SDS-PAGE with an alternative gel system. The efficient identification of membrane-spanning and known myelin proteins suggests that 2D-16-BAC/SDS-PAGE is better suited (than 2D-IEF/SDS-PAGE) to systematically analyze the myelin proteome. For this task, it was as efficient as the technically more demanding gel-free proteome analyses based on two-dimensional liquid chromatography (LC/LC) coupled to MS (Vanrobaeys et al., 2005; Roth et al., 2006). Moreover, LC/LC has the principal disadvantage that separation occurs at the level of proteolytic peptides, often eliminating information on protein abundance, isoforms, and post-translational modifications. Thus, gel-based protein maps allow differential myelin proteome analysis in mouse models and with disease-associated autopsy material.
The overlap of the myelin proteome libraries mentioned in this paper is substantial, considering different starting material (mouse, rat), separation technique (IEF, 16-BAC, LC/LC), and instrumentation for protein identification by MS. This is an important issue because in other (“nonmyelin”) applications, very little overlap was found for the identical starting material when analyzed by different laboratories (Chamrad and Meyer, 2005). Thus, complete overlap of myelin proteome libraries is unlikely to be achieved, and laboratories in the field may have to establish their individual reference library.
Changes of the myelin proteome in PLP_null_ mice
Differential myelin proteome analysis promises to be crucial in identifying molecular changes associated with myelin diseases. We anticipate that it will primarily rely on gel-based quantification, in particular on the upcoming combination of differential fluorescence labeling (as in 2D-DIGE) with 2D-PAGE using a cationic detergent in the first dimension (as in 2D-16-BAC/SDS-PAGE). Such technical refinements have been reported recently (Helling et al., 2006). Using the 2D-DIGE techniques available at present, we detected alterations of the myelin proteome in the PLP_null_ mouse, a bona fide model for human SPG-2 with secondary axonal degeneration (Klugmann et al., 1997; Griffiths et al., 1998b; Edgar et al., 2004). The lack of PLP/DM20 resulted in diminished abundance of several proteins in CNS myelin, well before the onset of neurological symptoms. The abundance of three small GTPases of the septin family was reduced, and the NAD+-dependent deacetylase SIRT2 was virtually absent. SIRT2 was independently identified as myelin constituent by several proteome analyses (Taylor et al., 2004; Vanrobaeys et al., 2005; Roth et al., 2006). SIRT2 mRNA was detected in the white matter by _in situ_-hybridization (http://www.ncbi.nlm.nih.gov/projects/gensat/), and we found that SIRT2 transcription follows the approximate time course of myelination. We detected the protein at all stages of oligodendrocyte development. Thus, in agreement with very recent reports (Li et al., 2007; Southwood et al., 2007), SIRT2 is an oligodendroglial protein. Our biochemical and histological data suggest that SIRT2 is targeted to CNS myelin, where it may contribute to the glial support of axonal integrity.
Importantly, not SIRT2 gene expression but the abundance of this protein in CNS myelin is PLP/DM20 dependent. How might PLP/DM20 facilitate SIRT2 delivery to myelin? Our attempts to coimmunoprecipitate the two proteins from brain lysates have failed (data not shown). Although this would not exclude a direct molecular interaction, we suggest that SIRT2 is indirectly associated with PLP/DM20 in membrane lipid rafts that are important for oligodendroglial membrane sorting (Kramer et al., 2001; Lee, 2001).
SIRT2 in myelinating oligodendrocytes
The function of SIRT2 in myelin is unknown. Mammalian SIRT2 is a member of the sirtuin family of proteins related to the silent information regulator 2 (Sir2) of yeast, a nuclear histone deacetylase (Tanny et al., 1999; Imai et al., 2000). Sirtuins are modulators of cellular functions, including cell type specification and differentiation of oligodendrocyte progenitors (Marin-Husstege et al., 2002; Shen et al., 2005; Cunliffe and Casaccia-Bonnefil, 2006). SIRT2 exhibits 27% sequence identity to mammalian SIRT1, a nuclear NAD+-dependent deacetylase implicated in the delay of Wallerian degeneration when activated by excess NAD+ synthesis (Araki et al., 2004). Sirtuins are NAD+-dependent enzymes that catalyze the acetyl transfer from the ε-amino group of a lysine (in a protein substrate) to ADP-ribose in NAD+. This reaction results in a deacetylated protein, nicotinamide, and the metabolite 2′-_O_-acetyl-ADP-ribose (OAADPr) (Blander and Guarente, 2004; Denu, 2005). Depending on the molecular context, the main biological function of sirtuins is deacetylation or ADP-ribosylation, and additional regulatory roles were suggested for nicotinamide and OAADPr. In addition to gene regulation, sirtuins have been implicated in microtubule dynamics, cellular metabolism, apoptosis, and aging. Mammalian sirtuins are differentially localized in the nucleus (SIRT1, SIRT6), mitochondria (SIRT3, SIRT4), and the cytoplasm (SIRT2) (Anekonda and Reddy, 2006; Haigis et al., 2006). In the mammalian brain, oligodendrocytes are the major cell type expressing SIRT2, and its transport into myelin depends on the coexpression with PLP/DM20.
Several mammalian proteins undergo reversible acetylation (Polevoda and Sherman, 2002; Shimazu et al., 2007). SIRT2 deacetylates α-tubulin, at least in vitro, and deacetylation amplifies microtubule dynamics (North et al., 2003). In cultured oligodendrocytes, process outgrowth in response to intracellular signals involves cytoskeletal reorganization (Klein et al., 2002). Indeed, a very recent report suggests that SIRT2 regulates microtubule dynamics also in normal oligodendrocyte development (Li et al., 2007). Mice lacking CNP from myelin develop length-dependent axonal loss in the presence of normal amounts of myelin, leading to ataxia and hindlimb paralysis (Lappe-Siefke et al., 2003; Rasband et al., 2005), similar in phenotype to PLP_null_ mice. Whereas CNP_null_ myelin does not lack SIRT2 (data not shown), it is intriguing that CNP itself is a microtubule assembly protein (Bifulco et al., 2002; Lee et al., 2005). It thus appears that spatiotemporally controlled microtubule stability in oligodendrocytes (by SIRT2, CNP, and likely other factors) may be critical for normal axon-glia interaction and for the prevention of axonal degeneration.
On the other hand, α-tubulin was not obviously hyperacetylated in PLP_null_ myelin. It is possible that the acetylation status of a protein is not preserved during biochemical myelin purification (because of the presence of acetylases/deacetylases). Alternatively, α-tubulin is not a relevant substrate of SIRT2 in CNS myelin. In fact, we consider all acetylated myelin-associated proteins as candidate substrates for SIRT2. With respect to known myelin proteins, MOG was also acetylated in CNS myelin (but not hyperacetylated in PLP_null_ myelin). Interestingly, MOG was suggested to regulate oligodendroglial microtubule stability (Johns and Bernard, 1999), and spatiotemporally controlled (de)acetylation may influence this function. In preliminary MS experiments, it has been difficult to map the lysine residues in MOG that undergo acetylation, likely because the sequence coverage after tryptic in-gel digests is never complete (MacCoss et al., 2002), in particular for membrane-spanning proteins. Moreover, little is known on the dynamic range of acetylation of nonhistone proteins, raising the possibility that acetylated peptides are only present as low-abundant components after proteolytic cleavage and therefore escape identification by MS. Future experiments will aim at the identification of acetylation sites by using additional (less specific) proteases and enriching acetylated polypeptides. Additional acetylated proteins are present in CNS myelin but remain to be identified.
Together, our results demonstrate a novel function of the major CNS myelin protein PLP/DM20, which is required for the transport of SIRT2 into CNS myelin. This metabolically controlled enzyme may in turn contribute to the PLP-dependent neuroprotection provided to axons by oligodendrocytes (Griffiths et al., 1998b; Yin et al., 2006). The NAD+/NADH redox levels of neurons and glial cells were suggested to be transcellularly coupled (Cerdan et al., 2006). In the normal brain, SIRT2 activation in oligodendrocytes and myelin (resulting from increased axonal NAD+/NADH ratios) may reduce the acetylation status of myelin-associated proteins with consequences for their net charge and function. We speculate that the competence of oligodendrocytes to sense and react to NAD+ levels in white matter tracts is required for their normal role in maintaining long-term axonal integrity.
Footnotes
This work was supported by National Institutes of Health Grant NS042925 (P.C.B.), National Multiple Sclerosis Society Grant 3957 (P.C.B.), the Max Planck Society, the Deutsche Forschungsgemeinschaft Center for Molecular Physiology of the Brain, Bundesministerium für Bildung und Forschung (Leukonet), the Hertie Foundation, the del Marmol Foundation, and the Myelin Project. Antibodies were kindly provided by M. Tainsky, O. Shakhova, R. Bansal, and C. Linington. Primary cultured oligodendrocytes for electron microscopy were a kind gift from C. Winterstein and E.-M. Krämer-Albers. We thank M. C. Motta, H. Kratzin, and G. Saher for helpful discussions and A. Fahrenholz, D. Hesse, M. Zander, T. Ruhwedel, and K. Claus for technical assistance.
References
- Almqvist P, Carlsson SR, Hardy JA, Winblad B. Regional and subcellular distribution of Thy-1 in human brain assayed by a solid-phase radioimmunoassay. J Neurochem. 1986;46:681–685. doi: 10.1111/j.1471-4159.1986.tb13025.x. [DOI] [PubMed] [Google Scholar]
- Anekonda TS, Reddy PH. Neuronal protection by sirtuins in Alzheimer's disease. J Neurochem. 2006;96:305–313. doi: 10.1111/j.1471-4159.2005.03492.x. [DOI] [PubMed] [Google Scholar]
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Barbarese E, Koppel DE, Deutscher MP, Smith CL, Ainger K, Morgan F, Carson JH. Protein translation components are colocalized in granules in oligodendrocytes. J Cell Sci. 1995;108:2781–2790. doi: 10.1242/jcs.108.8.2781. [DOI] [PubMed] [Google Scholar]
- Benjamins JA, Morell P. Proteins of myelin and their metabolism. Neurochem Res. 1978;3:137–174. doi: 10.1007/BF00964057. [DOI] [PubMed] [Google Scholar]
- Bifulco M, Laezza C, Stingo S, Wolff J. 2′,3′-Cyclic nucleotide 3′-phosphodiesterase: a membrane-bound, microtubule-associated protein and membrane anchor for tubulin. Proc Natl Acad Sci USA. 2002;99:1807–1812. doi: 10.1073/pnas.042678799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzozero OA, Bixler HA, Davis JD, Espinosa A, Messier AM. Chemical deacylation reduces the adhesive properties of proteolipid protein and leads to decompaction of the myelin sheath. J Neurochem. 2001;76:1129–1141. doi: 10.1046/j.1471-4159.2001.00116.x. [DOI] [PubMed] [Google Scholar]
- Bjartmar C, Yin X, Trapp BD. Axonal pathology in myelin disorders. J Neurocytol. 1999;28:383–395. doi: 10.1023/a:1007010205037. [DOI] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Boggs JM. Myelin basic protein: a multifunctional protein. Cell Mol Life Sci. 2006;63:1945–1961. doi: 10.1007/s00018-006-6094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronstein JM, Tiwari-Woodruff S, Buznikov AG, Stevens DB. Involvement of OSP/claudin-11 in oligodendrocyte membrane interactions: role in biology and disease. J Neurosci Res. 2000;59:706–711. doi: 10.1002/(SICI)1097-4547(20000315)59:6<706::AID-JNR2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Cerdan S, Rodrigues TB, Sierra A, Benito M, Fonseca LL, Fonseca CP, Garcia-Martin ML. The redox switch/redox coupling hypothesis. Neurochem Int. 2006;48:523–530. doi: 10.1016/j.neuint.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Chamrad D, Meyer HE. Valid data from large-scale proteomics studies. Nat Methods. 2005;2:647–648. doi: 10.1038/nmeth0905-647. [DOI] [PubMed] [Google Scholar]
- Cunliffe VT, Casaccia-Bonnefil P. Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mech Dev. 2006;123:24–30. doi: 10.1016/j.mod.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Denu JM. The Sir 2 family of protein deacetylases. Curr Opin Chem Biol. 2005;9:431–440. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Edgar JM, McLaughlin M, Yool D, Zhang SC, Fowler JH, Montague P, Barrie JA, McCulloch MC, Duncan ID, Garbern J, Nave KA, Griffiths IR. Oligodendroglial modulation of fast axonal transport in a mouse model of hereditary spastic paraplegia. J Cell Biol. 2004;166:121–131. doi: 10.1083/jcb.200312012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk J, Bonnon C, Girault JA, Faivre-Sarrailh C. F3/contactin, a neuronal cell adhesion molecule implicated in axogenesis and myelination. Biol Cell. 2002;94:327–334. doi: 10.1016/s0248-4900(02)00006-0. [DOI] [PubMed] [Google Scholar]
- Garbern JY, Cambi F, Tang XM, Sima AA, Vallat JM, Bosch EP, Lewis R, Shy M, Sohi J, Kraft G, Chen KL, Joshi I, Leonard DG, Johnson W, Raskind W, Dlouhy SR, Pratt V, Hodes ME, Bird T, Kamholz J. Proteolipid protein is necessary in peripheral as well as central myelin. Neuron. 1997;19:205–218. doi: 10.1016/s0896-6273(00)80360-8. [DOI] [PubMed] [Google Scholar]
- Griffiths I, Klugmann M, Anderson T, Thomson C, Vouyiouklis D, Nave KA. Current concepts of PLP and its role in the nervous system. Microsc Res Tech. 1998a;41:344–358. doi: 10.1002/(SICI)1097-0029(19980601)41:5<344::AID-JEMT2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Griffiths I, Klugmann M, Anderson T, Yool D, Thomson C, Schwab MH, Schneider A, Zimmermann F, McCulloch M, Nadon N, Nave KA. Axonal swellings and degeneration in mice lacking the major proteolipid of myelin. Science. 1998b;280:1610–1613. doi: 10.1126/science.280.5369.1610. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Hartinger J, Stenius K, Hogemann D, Jahn R. 16-BAC/SDS-PAGE: a two-dimensional gel electrophoresis system suitable for the separation of integral membrane proteins. Anal Biochem. 1996;240:126–133. doi: 10.1006/abio.1996.0339. [DOI] [PubMed] [Google Scholar]
- Helling S, Schmitt E, Joppich C, Schulenborg T, Mullner S, Felske-Muller S, Wiebringhaus T, Becker G, Linsenmann G, Sitek B, Lutter P, Meyer HE, Marcus K. 2-D differential membrane proteome analysis of scarce protein samples. Proteomics. 2006;6:4506–4513. doi: 10.1002/pmic.200600169. [DOI] [PubMed] [Google Scholar]
- Huang JK, Phillips GR, Roth AD, Pedraza L, Shan W, Belkaid W, Mi S, Fex-Svenningsen A, Florens L, Yates JR, III, Colman DR. Glial membranes at the node of Ranvier prevent neurite outgrowth. Science. 2005;310:1813–1817. doi: 10.1126/science.1118313. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Inoue K. PLP1-related inherited dysmyelinating disorders: Pelizaeus-Merzbacher disease and spastic paraplegia type 2. Neurogenetics. 2005;6:1–16. doi: 10.1007/s10048-004-0207-y. [DOI] [PubMed] [Google Scholar]
- Jahn O, Hesse D, Reinelt M, Kratzin HD. Technical innovations for the automated identification of gel-separated proteins by MALDI-TOF mass spectrometry. Anal Bioanal Chem. 2006;386:92–103. doi: 10.1007/s00216-006-0592-1. [DOI] [PubMed] [Google Scholar]
- Johns TG, Bernard CC. The structure and function of myelin oligodendrocyte glycoprotein. J Neurochem. 1999;72:1–9. doi: 10.1046/j.1471-4159.1999.0720001.x. [DOI] [PubMed] [Google Scholar]
- Jung M, Sommer I, Schachner M, Nave KA. Monoclonal antibody O10 defines a conformationally sensitive cell-surface epitope of proteolipid protein (PLP): evidence that PLP misfolding underlies dysmyelination in mutant mice. J Neurosci. 1996;16:7920–7929. doi: 10.1523/JNEUROSCI.16-24-07920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp NA, Griffin JL, Lilley KS. Application of partial least squares discriminant analysis to two-dimensional difference gel studies in expression proteomics. Proteomics. 2005;5:81–90. doi: 10.1002/pmic.200400881. [DOI] [PubMed] [Google Scholar]
- Kinoshita M. Diversity of septin scaffolds. Curr Opin Cell Biol. 2006;18:54–60. doi: 10.1016/j.ceb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Klein C, Kramer EM, Cardine AM, Schraven B, Brandt R, Trotter J. Process outgrowth of oligodendrocytes is promoted by interaction of fyn kinase with the cytoskeletal protein tau. J Neurosci. 2002;22:698–707. doi: 10.1523/JNEUROSCI.22-03-00698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klugmann M, Schwab MH, Puhlhofer A, Schneider A, Zimmermann F, Griffiths IR, Nave KA. Assembly of CNS myelin in the absence of proteolipid protein. Neuron. 1997;18:59–70. doi: 10.1016/s0896-6273(01)80046-5. [DOI] [PubMed] [Google Scholar]
- Knapp PE, Itkis OS, Mata M. Neuronal interaction determines the expression of the alpha-2 isoform of Na, K-ATPase in oligodendrocytes. Brain Res Dev Brain Res. 2000;125:89–97. doi: 10.1016/s0165-3806(00)00125-5. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Klein C, Koch T, Boytinck M, Trotter J. Compartmentation of Fyn kinase with glycosylphosphatidylinositol-anchored molecules in oligodendrocytes facilitates kinase activation during myelination. J Biol Chem. 1999;274:29042–29049. doi: 10.1074/jbc.274.41.29042. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Schardt A, Nave KA. Membrane traffic in myelinating oligodendrocytes. Microsc Res Tech. 2001;52:656–671. doi: 10.1002/jemt.1050. [DOI] [PubMed] [Google Scholar]
- Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, Griffiths IR, Nave KA. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- Lee AG. Myelin: delivery by raft. Curr Biol. 2001;11:R60–R62. doi: 10.1016/s0960-9822(01)00008-2. [DOI] [PubMed] [Google Scholar]
- Lee J, Gravel M, Zhang R, Thibault P, Braun PE. Process outgrowth in oligodendrocytes is mediated by CNP, a novel microtubule assembly myelin protein. J Cell Biol. 2005;170:661–673. doi: 10.1083/jcb.200411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang B, Tang J, Cao Q, Wu Y, Wu C, Guo J, Ling EA, Liang F. Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating α-tubulin. J Neurosci. 2007;27:2606–2616. doi: 10.1523/JNEUROSCI.4181-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Draghi NA, Resh MD. Signaling from integrins to Fyn to Rho family GTPases regulates morphologic differentiation of oligodendrocytes. J Neurosci. 2004;24:7140–7149. doi: 10.1523/JNEUROSCI.5319-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnington C, Webb M, Woodhams PL. A novel myelin-associated glycoprotein defined by a mouse monoclonal antibody. J Neuroimmunol. 1984;6:387–396. doi: 10.1016/0165-5728(84)90064-x. [DOI] [PubMed] [Google Scholar]
- Liou W, Geuze HJ, Slot JW. Improving structural integrity of cryosections for immunogold labeling. Histochem Cell Biol. 1996;106:41–58. doi: 10.1007/BF02473201. [DOI] [PubMed] [Google Scholar]
- MacCoss MJ, McDonald WH, Saraf A, Sadygov R, Clark JM, Tasto JJ, Gould KL, Wolters D, Washburn M, Weiss A, Clark JI, Yates JR., III Shotgun identification of protein modifications from protein complexes and lens tissue. Proc Natl Acad Sci USA. 2002;99:7900–7905. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison DL, Krueger WH, Cheng D, Trapp BD, Pfeiffer SE. SNARE complex proteins, including the cognate pair VAMP-2 and syntaxin-4, are expressed in cultured oligodendrocytes. J Neurochem. 1999;72:988–998. doi: 10.1046/j.1471-4159.1999.0720988.x. [DOI] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague P, McCallion AS, Davies RW, Griffiths IR. Myelin-associated oligodendrocytic basic protein: a family of abundant CNS myelin proteins in search of a function. Dev Neurosci. 2006;28:479–487. doi: 10.1159/000095110. [DOI] [PubMed] [Google Scholar]
- Nave KA, Lai C, Bloom FE, Milner RJ. Splice site selection in the proteolipid protein (PLP) gene transcript and primary structure of the DM-20 protein of central nervous system myelin. Proc Natl Acad Sci USA. 1987;84:5665–5669. doi: 10.1073/pnas.84.16.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Norton WT, Poduslo SE. Myelination in rat brain: method of myelin isolation. J Neurochem. 1973;21:749–757. doi: 10.1111/j.1471-4159.1973.tb07519.x. [DOI] [PubMed] [Google Scholar]
- Polevoda B, Sherman F. The diversity of acetylated proteins. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-5-reviews0006. 0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popko B. Myelin: not just a conduit for conduction. Nat Genet. 2003;33:327–328. doi: 10.1038/ng0303-327. [DOI] [PubMed] [Google Scholar]
- Rabilloud T. Solubilization of proteins for electrophoretic analyses. Electrophoresis. 1996;17:813–829. doi: 10.1002/elps.1150170503. [DOI] [PubMed] [Google Scholar]
- Rasband MN, Tayler J, Kaga Y, Yang Y, Lappe-Siefke C, Nave KA, Bansal R. CNP is required for maintenance of axon-glia interactions at nodes of Ranvier in the CNS. Glia. 2005;50:86–90. doi: 10.1002/glia.20165. [DOI] [PubMed] [Google Scholar]
- Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth J, Nave KA, Mierzwa A, Schiff R. Subtle myelin defects in PLP-null mice. Glia. 2006;54:172–182. doi: 10.1002/glia.20370. [DOI] [PubMed] [Google Scholar]
- Roth AD, Ivanova A, Colman DR. New observations on the compact myelin proteome. Neuron Glia Biol. 2006;2:15–21. doi: 10.1017/S1740925X06000068. [DOI] [PubMed] [Google Scholar]
- Roth J, Bendayan M, Orci L. Ultrastructural localization of intracellular antigens by the use of protein A-gold complex. J Histochem Cytochem. 1978;26:1074–1081. doi: 10.1177/26.12.366014. [DOI] [PubMed] [Google Scholar]
- Saher G, Brugger B, Lappe-Siefke C, Mobius W, Tozawa R, Wehr MC, Wieland F, Ishibashi S, Nave KA. High cholesterol level is essential for myelin membrane growth. Nat Neurosci. 2005;8:468–475. doi: 10.1038/nn1426. [DOI] [PubMed] [Google Scholar]
- Schachner M, Bartsch U. Multiple functions of the myelin-associated glycoprotein MAG (siglec-4a) in formation and maintenance of myelin. Glia. 2000;29:154–165. doi: 10.1002/(sici)1098-1136(20000115)29:2<154::aid-glia9>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Scherer SS, Xu T, Crino P, Arroyo EJ, Gutmann DH. Ezrin, radixin, and moesin are components of Schwann cell microvilli. J Neurosci Res. 2001;65:150–164. doi: 10.1002/jnr.1138. [DOI] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Horinouchi S, Yoshida M. Multiple histone deacetylases and the CREB-binding protein regulate pre-mRNA 3′-end processing. J Biol Chem. 2007;282:4470–4478. doi: 10.1074/jbc.M609745200. [DOI] [PubMed] [Google Scholar]
- Slot JW, Geuze HJ. A new method of preparing gold probes for multiple-labeling cytochemistry. Eur J Cell Biol. 1985;38:87–93. [PubMed] [Google Scholar]
- Smith R, Kavanagh E, Morrison HG, Gould RM. Messenger RNAs located in spiny dogfish oligodendrocyte processes. Biol Bull. 2001;201:255–256. doi: 10.2307/1543352. [DOI] [PubMed] [Google Scholar]
- Southwood CM, Peppi M, Dryden S, Tainsky MA, Gow A. Microtubule deacetylases, SirT2 and HDAC6, in the nervous system. Neurochem Res. 2007;32:187–195. doi: 10.1007/s11064-006-9127-6. [DOI] [PubMed] [Google Scholar]
- Tait S, Gunn-Moore F, Collinson JM, Huang J, Lubetzki C, Pedraza L, Sherman DL, Colman DR, Brophy PJ. An oligodendrocyte cell adhesion molecule at the site of assembly of the paranodal axo-glial junction. J Cell Biol. 2000;150:657–666. doi: 10.1083/jcb.150.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Taylor CM, Marta CB, Claycomb RJ, Han DK, Rasband MN, Coetzee T, Pfeiffer SE. Proteomic mapping provides powerful insights into functional myelin biology. Proc Natl Acad Sci USA. 2004;101:4643–4648. doi: 10.1073/pnas.0400922101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K, Dhaunchak AS, Goncalves JT, Wenzel D, Schneider A, Bunt G, Nave KA, Simons M. Neuron to glia signaling triggers myelin membrane exocytosis from endosomal storage sites. J Cell Biol. 2006;172:937–948. doi: 10.1083/jcb.200509022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unlu M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- Vanrobaeys F, Van Coster R, Dhondt G, Devreese B, Van Beeumen J. Profiling of myelin proteins by 2D-gel electrophoresis and multidimensional liquid chromatography coupled to MALDI TOF-TOF mass spectrometry. J Proteome Res. 2005;4:2283–2293. doi: 10.1021/pr050205c. [DOI] [PubMed] [Google Scholar]
- Voelter-Mahlknecht S, Ho AD, Mahlknecht U. FISH-mapping and genomic organization of the NAD-dependent histone deacetylase gene, Sirtuin 2 (Sirt2) Int J Oncol. 2005;27:1187–1196. [PubMed] [Google Scholar]
- Werner H, Jung M, Klugmann M, Sereda M, Griffiths IR, Nave KA. Mouse models of myelin diseases. Brain Pathol. 1998;8:771–793. doi: 10.1111/j.1750-3639.1998.tb00200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Yang YH, Chen YH, Zhang CY, Nimmakayalu MA, Ward DC, Weissman S. Cloning and characterization of two mouse genes with homology to the yeast Sir2 gene. Genomics. 2000;69:355–369. doi: 10.1006/geno.2000.6360. [DOI] [PubMed] [Google Scholar]
- Yin X, Baek RC, Kirschner DA, Peterson A, Fujii Y, Nave KA, Macklin WB, Trapp BD. Evolution of a neuroprotective function of central nervous system myelin. J Cell Biol. 2006;172:469–478. doi: 10.1083/jcb.200509174. [DOI] [PMC free article] [PubMed] [Google Scholar]