The PRC1 Polycomb group complex interacts with PLZF/RARA to mediate leukemic transformation (original) (raw)
Abstract
Ectopic repression of retinoic acid (RA) receptor target genes by PML/RARA and PLZF/RARA fusion proteins through aberrant recruitment of nuclear corepressor complexes drives cellular transformation and acute promyelocytic leukemia (APL) development. In the case of PML/RARA, this repression can be reversed through treatment with all-trans RA (ATRA), leading to leukemic remission. However, PLZF/RARA ectopic repression is insensitive to ATRA, resulting in persistence of the leukemic diseased state after treatment, a phenomenon that is still poorly understood. Here we show that, like PML/RARA, PLZF/RARA expression leads to recruitment of the Polycomb-repressive complex 2 (PRC2) Polycomb group (PcG) complex to RA response elements. However, unlike PML/RARA, PLZF/RARA directly interacts with the PcG protein Bmi-1 and forms a stable component of the PRC1 PcG complex, resulting in PLZF/RARA-dependent ectopic recruitment of PRC1 to RA response elements. Upon treatment with ATRA, ectopic recruitment of PRC2 by either PML/RARA or PLZF/RARA is lost, whereas PRC1 recruited by PLZF/RARA remains, resulting in persistent RA-insensitive gene repression. We further show that Bmi-1 is essential for the PLZF/RARA cellular transformation property and implicates a central role for PRC1 in PLZF/RARA-mediated myeloid leukemic development.
Keywords: Leukemia, PLZF/RARA, Polycomb group, gene repression, retinoic acid
Regulation of gene expression is achieved through the control of transcriptional activators or repressors and through the epigenetic regulation of chromatin structure. Dysfunction of either of these two regulatory components disturbs the cellular homeostasis causing numerous diseases. The Polycomb group (PcG) family of proteins are chromatin factors whose role is to maintain the repressed transcriptional state of their target genes. The stable and heritable epigenetic gene repression afforded by the PcG regulates not only body patterning, but epigenetic cellular memory, stem cell renewal, and cancer development (for review, see Sparmann and van Lohuizen 2006). At the molecular lever, PcG proteins function as Polycomb-repressive complexes (PRCs), of which the best studied are termed PRC1 and PRC2. PRC2 is involved in the initiation of gene repression, through trimethylation of Lys 27 of histone H3 (H3K27me3). This epigenetic mark is recognized by the chromodomain of Polycomb (Pc) in the PRC1 complex, an event that is believed to aid in the recruitment of PRC1 for the maintenance of gene repression. How the PcG is initially recruited to silence specific target genes is unclear, although certain sequence-specific DNA-binding proteins are capable of recruiting PcG complexes to specific target loci leading to PcG-dependent repression. Thus, the concerted actions of sequence-specific DNA-binding factors and epigenetic chromatin modification are likely to play a fundamental role for stable and heritable gene repression by the PcG (Grimaud et al. 2006), with noncoding RNAs contributing an additional layer of control (Lempradl and Ringrose 2008).
Deregulation of this essential PcG control system contributes to aberrations in development and can lead to different types of cancer. Gene amplification or overexpression of PRC1 or PRC2 components results in a broad spectrum of cancers believed to arise from aberrant silencing of PcG target genes (for review, see Sparmann and van Lohuizen 2006; Rajasekhar and Begemann 2007). However, cancer development can also result from the ectopic recruitment of the PcG to genes not normally under its control. Recently, the ectopic recruitment of PRC2 by the oncogenic transcription factor PML/RARA to retinoic acid (RA)-responsive genes was found to play a fundamental role in the development of acute promyelocytic leukemia (APL) (Villa et al. 2007). Ectopic repression of genes by a PcG complex leading to cancer development is an interesting oncogenic mechanism, particularly in the case of APL. APL originates from illegitimate recombination of the RA receptor α (RARA) gene with one of five different partner genes, creating an oncogenic X/RARA fusion protein (Licht 2006). APL patients presenting the chimeric fusion protein PML/RARA are remarkably sensitive to pharmacological doses of all_-trans_ RA (ATRA) and undergo complete remission, while the PLZF/RARA-associated APL is more severe, presenting a poor prognosis due to a nonresponse to ATRA treatment (Licht et al. 1995). In the case of PML/RARA, therapeutic doses of ATRA leads to the release of corepressor complexes and to the loss of ectopic PRC2 recruitment to RA target genes, reinducing their expression and redifferentiating the leukemic blast (Villa et al. 2007). In the case of PLZF/RARA, however, it is still not understood how the fusion protein induces ATRA-insensitive stable and heritable gene repression.
To better understand the molecular mechanisms underlying PcG involvement in ATRA-insensitive APL, we studied the role of PcG complexes in ATRA-insensitive PLZF/RARA transformation. It has been reported previously that the DNA-binding protein PLZF is involved in the stable repression of Hox genes during mouse development and recruits the PcG protein Bmi-1 and the associated PRC1 complex to the HoxD locus (Barna et al. 2002). These observations prompted us to investigate whether there was a role for PRC1 in PLZF/RARA-mediated repression. We show that, unlike PML/RARA, PLZF/RARA is capable of interacting with Bmi-1 and can form an integral component of PRC1. These interactions lead to the PLZF/RARA-dependent in vitro and in vivo recruitment of PRC1 to RA response elements (RAREs) in an ATRA-insensitive manner, leading to PcG-dependent transformation of the cell. The interaction between PLZF/RARA and PRC1 provides new insight into how the fusion protein may induce leukemogenesis, and how the ectopic recruitment of PRC1 can play a role in determining cellular transformation. Identifying the factors capable of targeting PcG complexes and the molecular differences between ATRA-sensitive and ATRA-insensitive gene repression by RARA fusion proteins is essential to understand disease progression.
Results
PLZF/RARA interacts with Bmi-1 through its BTB-POZ domain
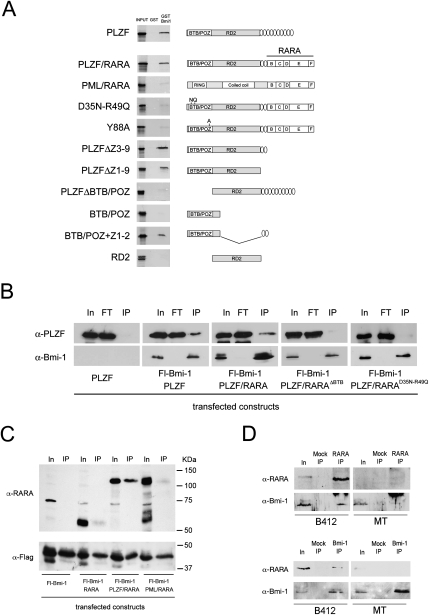
The domains mediating all of the known interactions between PLZF and its partners are located in the N-terminal half of the protein, which is retained in the PLZF/RARA chimera. As Bmi-1 interacts with PLZF (Barna et al. 2002), we tested whether PLZF/RARA retained the ability to associate with Bmi-1. GST “pull-down” assays showed that PLZF/RARA interacts with Bmi-1, whereas PML/RARA, the other major APL oncogenic fusion protein, does not (Fig. 1A). Through domain deletion experiments, we found that the interaction is mediated by the PLZF BTB domain, in conjunction with the two first PLZF zinc finger domains (Fig. 1A). While the exact function of these two zinc fingers is unknown, they are dispensable for DNA binding but essential for repression (Dong et al. 1996).
Figure 1.
PLZF/RARA interacts with Bmi-1 through its BTB-POZ domain. (A) GST and GST-Bmi-1 pull-down of 35S-labeled wild-type and mutant isoforms of PLZF, PLZF/RARA, and PML/RARA. Structures of PLZF, PLZF/RARA, and PML/RARA isoforms used in GST pull-down and co-IP experiments are shown. (B) Nuclei of 293T cells cotransfected with Flag-Bmi-1 and the PLZF or PLZF/RARA constructs were immunoprecipitated with anti-Flag beads and were immunoblotted with either an anti-PLZF or anti-Bmi-1 antibody. (IP) 5% of the input; (FT) the flow-through. (C) Nuclei of 293T cells cotransfected with Fl-Bmi-1 and RARA, PLZF/RARA, or PML/RARA were immunoprecipitated with anti-Flag beads and immunoblotted with either an anti-RARA or anti-Flag antibody. (IP) 2.5% of the input. (D) Nuclei of U937-MT (control) of U937-B412 (PLZF/RARA) cells pretreated with zinc were immunoprecipitated with anti-RARA (top panels) or anti-Bmi-1 antibodies (bottom panels) and immunoblotted for PLZF/RARA (α-RARA) or Bmi-1 (α-Bmi-1). Mock IP represents immunoprecipitation without antibody; RARA IP and Bmi-1 IP represent 2.5% of input.
BTB-POZ domains are linked to the assembly of macromolecular transcriptional repression complexes that are often involved in hematological cancers (Kelly and Daniel 2006). Point mutations (D35N-R49Q and Y88A) within the PLZF BTB domain diminish its dimerization properties and abolish the transformation potential of PLZF/RARA (Puccetti et al. 2005; Kwok et al. 2006). However, these mutations do not perturb its interaction between HDAC and SMRT–NCoR corepressors, suggesting that transformation by PLZF/RARA is not due solely to its interactions with these corepressor complexes. We found that disrupting PLZF/RARA dimerization through the Y88A or the D35N-R49Q mutations abrogates Bmi-1 interaction (Fig. 1A,B). The interaction between PLZF/RARA and Bmi-1 was further investigated in vivo through coimmunoprecipations (co-IPs) from transiently transfected mammalian cell extracts. Western blot analysis on proteins coprecipitating with Flag-tagged Bmi-1 demonstrates that PLZF/RARA associates with Bmi-1 in vivo and that, like in vitro, this interaction requires the dimerized form of PLZF/RARA (Fig. 1B). This interaction is PLZF/RARA-specific, since no interaction was observed with RARA or PML/RARA (Fig. 1C), and is also observed with endogenous Bmi-1 when PLZF/RARA is expressed conditionally using the U937-B412 cell line (Fig. 1D; Ruthardt et al. 1997). These results demonstrate that PLZF/RARA interacts directly with Bmi-1 and correlates loss of PLZF/RARA transforming potential, through the D35N-R49Q mutation, to loss of Bmi-1 interaction.
PLZF/RARA incorporates in PRC1 and increases its inhibitory activity on chromatin containing RAREs
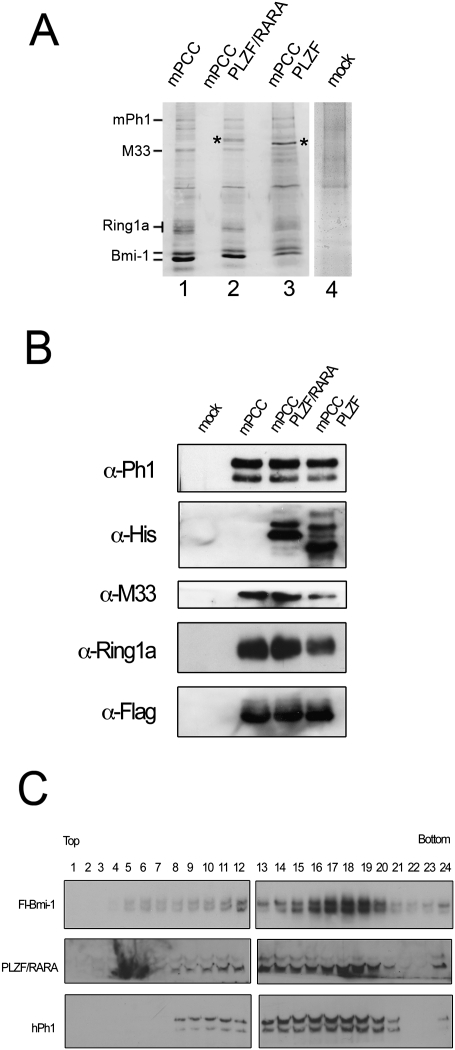
Bmi-1 is a core component of the mammalian PRC1 complex (Lavigne et al. 2004). Thus, to address whether PLZF/RARA is capable of recruiting the PRC1 complex to chromatin, we reconstituted the mammalian PRC1 core complex (mPCC) (Fig. 2A) from baculovirus-infected Sf9 cells (Lavigne et al. 2004). Coinfecting the cells with a recombinant virus encoding either PLZF or PLZF/RARA, we found that both PLZF and PLZF/RARA associate efficiently with the PcG proteins in the mPCC complex, creating recombinant mPCC–PLZF and mPCC–PLZF/RARA PcG complexes (Fig. 2). These data demonstrate that both PLZF and PLZF/RARA are integral components of a mammalian PRC1 complex.
Figure 2.
PLZF and PLZF/RARA associate with the reconstituted PRC1 complex. (A) Colloidal Coomassie blue-stained gel of purified mPCC (lane 1), mPCC–PLZF/RARA (lane 2), mPCC–PLZF (lane 3), and mock (lane 4) complexes. Asterisks highlight the bands corresponding to PLZF/RARA (lane 2) and PLZF (lane 3) verified by Western analyses. The mock complex corresponds to anti-Flag purification of extracts from Sf9 cells infected with mPh1, PLZF/RARA, M33, and Ring1a viruses. Densitometry analyses of the Coomassie-stained complexes show that the various mPCC complexes are ∼55%–65% pure, when compared with the mock purified complex. (B) Western blots of the complexes shown in A. PLZF/RARA and PLZF were detected by an anti-His tag antibody, and Bmi-1 was revealed by an anti-Flag tag antibody. (C) Western analyses of the sedimentation profiles of Bmi-1, PLZF/RARA, and mPh1 following fraction of the mPCC–PLZF/RARA complex by glycerol gradient sedimentation, showing that PLZF/RARA remains associated with the mPCC complex.
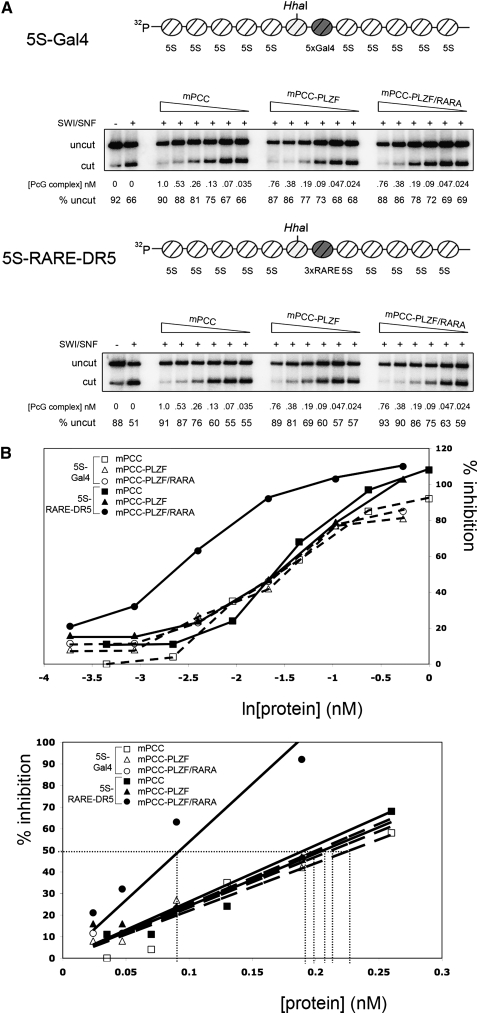
The recruitment of PRC1 and PCC to chromatin renders the chromatin refractory to chromatin remodeling, creating a transcriptionally nonpermissive environment (Francis et al. 2001). Direct association of sequence-specific DNA-binding proteins with PCC enhances this effect when the recognition sequence of the DNA-binding protein is present in the chromatin DNA sequence, demonstrating sequence-specific targeting of PCC complexes in vitro (Mulholland et al. 2003). To determine whether the RARA moiety of PLZF/RARA was capable of increasing the chromatin-repressing activity of mPCC through recruitment of the complex to RARE-containing chromatin, we performed restriction enzyme accessibility (REA) assays on in vitro reconstituted 5S chromatin arrays (Fig. 3A). Using reconstituted 5S-Gal4 control chromatin, containing repetitive Gal4-binding sites but no RARE (Fig. 3A), we found that mPCC, mPCC–PLZF, and mPCC–PLZF/RARA all maximally inhibited chromatin remodeling at ∼1 nM active protein concentration, with 50% maximal inhibition observed at 0.2 nM (Fig. 3B), which is in accordance with published results for mPCC (Lavigne et al. 2004). On a 5S-RARE-DR5 array, these values remained largely unchanged for mPCC and mPCC–PLZF (∼0.75 nM) (Fig. 3B). In contrast, we observed a marked increase in inhibitory activity when using mPCC–PLZF/RARA complex. In this case, maximal inhibition of chromatin remodeling by mPCC–PLZF/RARA was observed at ∼0.2 nM active complex, with 50% inhibition achieved using twofold less complex than for mPCC and mPCC–PLZF (see Fig. 3B). No protein or complex had any effect on HhaI activity on naked DNA (Supplemental Fig. S1A), and the individual DNA-binding proteins had no effect on SWI/SNF remodeling on either template (Supplemental Fig. S1B,C). It is noteworthy, however, that while the inclusion of PLZF/RARA into the complex increases the inhibitory activity of PCC by at least twofold, simply adding purified PLZF/RARA to mPCC appears to have little additional effect (see Supplemental Fig. S1B,C), suggesting that PLZF/RARA is required to be incorporated into the PRC1 complex to achieve enhanced inhibitory activity on SWI/SNF chromatin remodeling in the presence of the RARE-DR5 sequence, which is similar in effect to the Drosophila PCC–Zeste complex (Mulholland et al. 2003)
Figure 3.
PLZF/RARA recruits PRC1 to chromatin containing RARE elements. (A) REA assays on in vitro reconstituted 5S chromatin arrays. PhosphorImager scans of HhaI digestion products following chromatin incubation in the presence or absence of SWI/SNF and PCC complexes with the 5S-Gal4 template (top panel) or the 5S-RARE-DR5 template (bottom panel). Percentages of template undigested by HhaI are given. The concentrations of PCC complexes (in nanomolar) used in the reactions are the measured concentration of active DNA-binding molecules. Schematic representation of 5S array templates used in the REA assays are shown above the data. “5S-Gal4” contains five Gal4-binding sites downstream from the unique HhaI restriction site, whereas “5S-RARE-DR5” contains three RARE DR5 elements. (B) Data from REA assays shown in A are graphically represented as percentage of inhibition (Francis et al. 2001) as a function of ln (active protein concentration) (top graph) and active protein concentration (bottom graph), showing increased efficiency of the PCC–PLZF/RARA complex to inhibit chromatin remodeling when RARE DR5 elements are present in the chromatin. (Bottom graph) The linear section of the data is shown, where the quantities of active complex required to achieve 50% inhibition of chromatin remodeling are highlighted (dotted line), demonstrating that PCC–PLZF/RARA is approximately twofold more efficient at inhibition of chromatin remodeling than PCC and PCC–PLZF when RARE DR5 elements are present.
Thus, PLZF/RARA can directly associate with PRC1, and this association is sufficient to recruit the complex to RAREs, which renders the chromatin refractory to chromatin remodeling events.
PLZF/RARA recruits PRC1 to RAREs in vivo
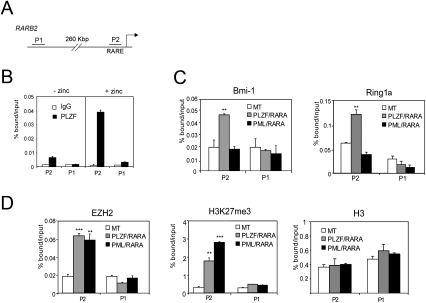
The RARA moiety of PLZF/RARA and PML/RARA retains its ability to bind RAREs, leading to recruitment of PML/RARA and PLZF/RARA to RARA target genes in vivo. Chromatin immunoprecipitation (ChIP) of PLZF/RARA conditionally expressed in U937-B412 cells showed that PLZF/RARA is present at the upstream RARE of the RARβ2 promoter (P2), a model target of RARA, but not in the P1 region, which is not RA-responsive (Fig. 4B). Recently, it was shown that PML/RARA interacts with and recruits the PRC2 complex to the P2 promoter (Villa et al. 2007). To compare the recruitment of PcG complexes to P2 by PLZF/RARA and PML/RARA fusion proteins, we examined the presence of PRC1 and PRC2 complexes at the P2 promoter in cells conditionally expressing PLZF/RARA (U937-B412) or PML/RARA (U937-PR9) from a zinc-inducible promoter (Ruthardt et al. 1997). Expression of either PML/RARA or PLZF/RARA leads to PRC2 enrichment, identified by EZH2 and its trimethylated Lys 27 of histone H3 (H3K27me3) modification (Fig. 4D). However, we found that Bmi-1 and Ring1, major components of PRC1, were specifically enriched on the P2 promoter only upon expression of PLZF/RARA and not PML/RARA (Fig. 4C). These data demonstrate that expression of either PML/RARA or PLZF/RARA leads to PRC2 enrichment at an endogenous target gene, whereas PLZF/RARA expression distinguishes itself by the additional recruitment of PRC1.
Figure 4.
PLZF/RARA targets PRC1 and PRC2 to RARβ2 promoter. (A) Representation of the RARβ2 promoter. P1 and P2 indicate the regions amplified by PCR. (B) PLZF/RARA on RARβ2 promoter was analyzed by quantitative ChIP (qChIP) with an anti-PLZF antibody on chromatin fragments prepared from U937-B412 cells pretreated with zinc or untreated. (**) P < 0.01 (C,D) qChIP analyses of the PRC1 subunits Bmi-1 and Ring1a (C), or EZH2, H3K27me3, and histone H3 (D) at RARβ2 promoter in the absence (MT) or in the presence of either PLZF/RARA or PML/RARA. Error bars represent standard deviations obtained from at least two independent experiments. (**) P < 0.01; (***) P < 0.001.
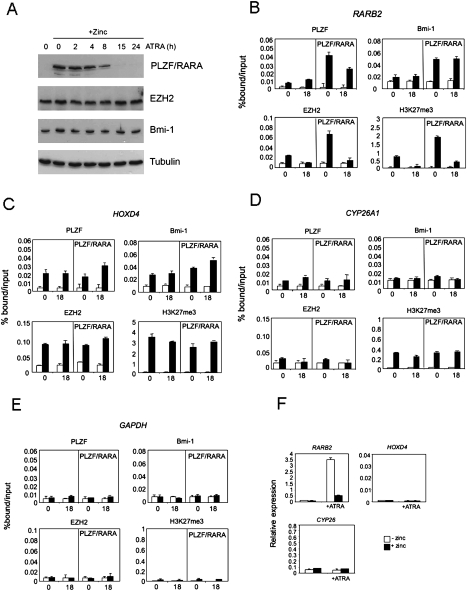
PRC1 recruitment is ATRA-insensitive
The recruitment of PRC2 to P2 by PML/RARA can be reversed upon treating the cells with ATRA (Villa et al. 2007). To assess the effect of ATRA treatment on PLZF/RARA-mediated recruitment of PcG complexes, we performed ChIP analyses on the RARβ2 promoter in U937-B412 cells treated with ATRA or untreated in the presence or absence of PLZF/RARA (Fig. 5B). Treatment of PLZF/RARA-expressing cells leads to degradation of the fusion protein (Fig. 5A; Koken et al. 1999; Rego et al. 2000), although PLZF/RARA can still be detected at the P2 promoter (Fig. 5B), indicating that some chromatin-bound PLZF/RARA is protected from degradation. We observed that ATRA treatment strongly reverses the levels of EZH2 and H3K27me3 at the P2 promoter without affecting the PLZF/RARA-mediated Bmi-1 recruitment (Fig. 5B). PRC1 and PRC2 recruitment was not affected by PLZF/RARA expression at either the PcG-targeted HOXD4 promoter (Fig. 5C), the CYP26A1 promoter that has been described as a RARA and a PcG target in F9 embryonal carcinoma cells (Fig. 5D; Gillespie and Gudas 2007), the RARβ2 P1 promoter (data not shown), or the unrelated GAPDH promoter (Fig. 5E). No changes in expression patterns of either HOXD4 or CYP26A1 were observed under any conditions tested (Fig. 5F). These data correlate the ATRA-insensitive recruitment of PRC1 to the RARβ2 promoter with inhibition of the ATRA-stimulated expression of RARβ2 (Fig. 5F). Consistent with repression of RARβ2, no increase in histone H3 acetylation at the RARβ2 promoter was observed following ATRA treatment in the presence of PLZF/RARA (Supplemental Fig. S2). These data implicate PRC1 in the PLZF/RARA-mediated repression of ATRA-stimulated gene expression.
Figure 5.
Effect of ATRA treatment on PLZF/RARA-mediated PRC1 and PRC2 recruitment. (A) Western analyses of nuclear extracts prepared from PLZF/RARA-expressing cells (+Zinc) at various times of ATRA treatment. Proteins were blotted and probed with antibodies to PLZF, EZH2, Bmi-1, and β-tubulin. (B–E) Analyses of the effects no stimulation (0) or 18 h of ATRA stimulation (18) on the enrichment of PLZF/RARA, Bmi-1, EZH2, and H3K27me3 in untreated B412 cells or B412 cells expressing PLZF/RARA (PLZF/RARA panels) at the RARβ2 promoter (B), the HOXD4 promoter (C), the CYP26A1 promoter (D), or the GAPDH promoter (E). Data are presented as percentage of bound/input and error bars indicate the standard deviation obtained from two or three independent experiments. White bars represent mock immunoprecipitation; black bars represent specific immunoprecipitation with the indicated antibodies. (F) The expression levels of RARβ2, HOXD4, and CYP26A1 were analyzed by quantitative real time PCR (qRT–PCR). RNA from U937-B412 cells (white bars) or zinc-pretreated U937-B412 cells (black bars; +zinc), either unstimulated or stimulated with 1 μM ATRA for 18 h (+ATRA) was extracted and 1–2 μg of RNA was reverse-transcribed. Gene expression of each gene is shown relative to HPRT expression.
Bmi-1 is required for PLZF/RARA-mediated transformation
In APL, the expression pattern of PLZF/RARA mimics that of PLZF, since PLZF/RARA expression is under the control of the PLZF promoter. Thus, to determine whether PRC1 can play a role in the PLZF/RARA-mediated diseased state, we first determined in which hematopoietic compartments PRC1 components and PLZF are expressed. Expression profiling of PRC1 PcG components and PLZF in hematopoietic stem cells showed that all of these components are expressed in the same bone marrow (BM) compartment (Supplemental Fig. S3), in accordance with previously published data for PcG protein expression patterns (Iwama et al. 2004).
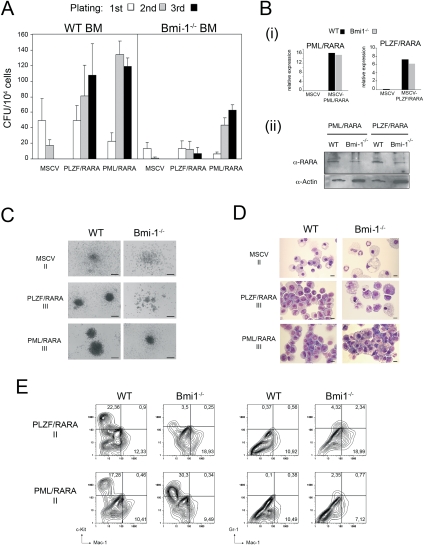
Next, we assessed whether Bmi-1 is required for transformation of hematopoietic progenitors by PLZF/RARA. In methylcellulose culture, normal progenitors exhaust their proliferative capacity after the second passage, while transformed progenitors form colonies for more than three passages (Lavau et al. 1997). We used this assay of transformation to compare the replating capacity of wild-type and Bmi-1−/− BM progenitor cells (Bel et al. 1998) when transduced by PLZF/RARA or PML/RARA. Even though Bmi-1−/− hematopoietic progenitor compartments are reduced (Supplemental Fig. S4A) due to defects in proliferation and self-renewal capacity (Lessard and Sauvageau 2003; Park et al. 2003), they can still be transformed (Supplemental Fig. S4). Empty vector control-infected wild-type cells lose colony-forming capacity after the second plating, while PLZF/RARA- or PML/RARA-infected wild-type cells continue to form colonies (Fig. 6A). In Bmi-1−/− progenitor cells, the two fusion proteins display differences in colony formation: While PML/RARA transduction yields a significant number of third-round colonies, PLZF/RARA transduction cannot impose extended colony formation capacity on Bmi-1−/− cells (Fig. 6A).
Figure 6.
Bmi-1 is required for PLZF/RARA transformation. (A) Bar charts indicate the number of colonies formed in methylcellulose cultures of lin BM transduced with the empty vector (MSCV), MSCV-PLZF/RARA (PLZF/RARA), or MSCV-PML/RARA (PML/RARA). Data represent the number of colonies from wild-type and Bmi-1−/− BM during three successive replatings. Error bars are standard deviation of the mean from five (PLZF/RARA) or two (PML/RARA) independent replating experiments. (B) Transgene expression was assessed by qRT-PCR (panel i) and immunoblotting (panel ii) from wild-type (WT) or Bmi-1−/− BM cells infected with either PLZF/RARA or PML/RARA viruses harvested after the third passage. Fusion proteins were detected in panel ii using antibodies against PLZF (PLZF/RARA) or RARA (PML/RARA). (C,D) Typical morphology (C) and MGG staining (D) of the second-round (II) or third-round (III) colonies generated from wild-type or Bmi-1−/− cells transduced with empty vector, PLZF/RARA, or PML/RARA. Bars: C, 200 μm; D, 10 μm. (E) Surface marker expression analyzed by FACS of the second-round colonies generated from wild-type or Bmi-1−/− cells transduced with PLZF/RARA or PML/RARA.
The tumor suppressor locus Ink4a/Arf can be repressed by Bmi-1, and its derepression in the absence of Bmi-1 results in defects in proliferation (Jacobs et al. 1999). To test whether the loss of PLZF/RARA transformation potential in Bmi-1−/− progenitors was the result of expression of Ink4a/Arf, we infected BM progenitors isolated from Bmi-1−/−/Ink4a−/−/Arf−/− mice with PLZF/RARA. We found that PLZF/RARA-infected Bmi-1−/−/Ink4a−/−/Arf−/− progenitors gave only a small number of colonies that were diffuse and small in size, and that could not sustain successive replatings (Supplemental Fig. S5). These data suggest that the requirement of Bmi-1 for PLZF/RARA transformation of BM cells is independent of Ink4a/Arf expression. Additionally, these effects are not due to loss of PLZF/RARA expression in Bmi-1−/− cells, as quantitative RT–PCR (qRT–PCR) and Western blot analysis show that PLZF/RARA expression is maintained after the third passage in Bmi-1−/− cells (Fig. 6B).
Morphological analyses of BM cells from third-round colonies from both PLZF/RARA- and PML/RARA-transduced Bmi-1−/− progenitors showed that whereas colonies from PML/RARA-transduced Bmi-1−/− cells were as large and compact as wild-type-transduced cells, the rarely seen PLZF/RARA-infected Bmi-1−/− colonies remained diffuse and smaller in size (Fig. 6C). Giemsa and FACS analyses showed that cells from PLZF/RARA-transduced _Bmi-1−/−_-derived colonies were fully mature, reflected by their morphology (Fig. 6D) and their pronounced reduction in expression of the c-kit progenitor marker, together with increased expression of the myeloid differentiation marker Gr-1 (Fig. 6E). In contrast, cells from PML/RARA-transduced _Bmi-1−/−_-derived colonies, while morphologically more mature than the wild-type-transduced counterparts, display more blast-like features compared with PLZF/RARA (Fig. 6D) and retain expression of c-kit progenitor marker (Fig. 6E).
Taken together, these data show that Bmi-1 is necessary for transformation of myeloid progenitors by PLZF/RARA, but not by the related PML/RARA oncogenic fusion, supporting our model that PRC1 plays a key role in PLZF/RARA-mediated transformation.
Discussion
PLZF/RARA associates with PRC1
X/RARA APL fusion proteins represent a model of altered transcription factors that function as dominant-acting oncoproteins. Their ability to form oligomers and to recruit corepressors is critical for inducing oncogenic transcriptional repression (So and Cleary 2004), which is the result of ectopic epigenetic modification of RA target genes (Di Croce et al. 2002). Forced oligomerization of RARA, which in APL is due to the fusion partner protein, is necessary and sufficient to induce cellular transformation (Kwok et al. 2006; Sternsdorf et al. 2006). Besides inducing oligomerization, the fusion partner has a decisive impact on the diseased state and sensitivity to the therapeutic effects of ATRA, due to ectopic recruitment of its interacting proteins to RA-responsive genes. We show that, through the PLZF moiety, PLZF/RARA can interact with the PcG protein Bmi-1. Bmi-1 is a member of the PRC1 complex that can inhibit chromatin remodeling (Francis et al. 2001; Mulholland et al. 2003) and transcription (King et al. 2002) through the compaction of chromatin structure (Francis et al. 2004). The role of sequence-specific binding factors in targeting PRC1 to chromatin passes by one of two mechanisms: binding of the transcription factor to DNA, thereby recruiting the complex, as is the case for the GAGA factor; or incorporation of the transcription factor in the protein complex, thereby recruiting the complex to DNA, as is the case for Zeste (Mulholland et al. 2003) and Pho (Mohd-Sarip et al. 2005). We show that PLZF/RARA is incorporated into the mPCC (mPCC–PLZF/RARA) (Fig. 2). The DNA-binding domains of RARA are preserved in PLZF/RARA, which serves to recruit mPCC–PLZF/RARA to RARE-containing chromatin in vitro (Fig. 3) and in vivo (Fig. 4), providing essential mechanistic insight into how recruitment is achieved. The REA assays (Fig. 3) demonstrate that the effect of incorporation of PLZF/RARA into PRC1 leads to the RARA-mediated recruitment of the complex to RAREs, and that the recruitment is not occurring solely via some histone mark.
Divergence in PRC recruitment between PLZF/RARA and PML/RARA
Recently, PRC2 activity was shown to play a key role in PML/RARA-mediated oncogenesis, in which ectopic recruitment and methylation activities of PRC2 and DNA methyltransferases by PML/RARA lead to oncogenic repression of RARA-controlled genes (Villa et al. 2007). The histone H3K27me3 modification laid down by PRC2 is believed to aid in the recruitment of PRC1 (Fischle et al. 2003; Min et al. 2003). This idea fits well with the model of hierarchical recruitment of PcG complexes, in which the establishment of robust epigenetic silencing of Hox genes requires a transient interaction between PRC2 and PRC1 components that is then maintained by PRC1 complexes (Poux et al. 2001). However, while H3K27me3 serves a role in aiding in PRC1 binding, it is not the sole deciding factor for PRC1 recruitment, since a large number of genes displaying H3K27me3 do not have bound PRC1 (for review, see Ringrose and Paro 2007), and in Drosophila cells, PRC1 binds to discrete sites, whereas H3K27me3 covers large genomic domains (Beisel et al. 2007). Indeed, while expression of PML/RARA leads to ectopic PRC2 recruitment at RA response genes (Villa et al. 2007), the H3K27me3 mark alone is insufficient to recruit PRC1 (Fig. 4). Conversely, we found that expression of PLZF/RARA leads to ectopic recruitment of both PRC1 and PRC2. While reconstitution experiments and immunoprecipitations demonstrate that recruitment of PRC1 by PLZF/RARA is through interactions between PLZF/RARA and Bmi-1, recruitment of PRC2 upon expression of PLZF/RARA is likely to be due to the known interactions between RARA and PRC2 (see Epping et al. 2005; Villa et al. 2007). Thus, we observe a fundamental difference in recruitment of PcG complexes between PML/RARA and PLZF/RARA, due to the additional interactions between PLZF/RARA and PRC1.
PRC1 is insensitive to RA treatment and is essential for PLZF/RARA transformation
Treatment of PML/RARA-expressing cells with ATRA leads to degradation of the fusion protein (Rego et al. 2000), loss of PRC2 recruitment, and derepression of RA response genes (Villa et al. 2007). Treatment of PLZF/RARA cells with ATRA also leads to degradation of the fusion protein, but with very little change in the RA response gene repressed state (He et al. 1998; Lin et al. 1998). Our data show that while the soluble nuclear pool of PLZF/RARA is degraded upon ATRA treatment (Fig. 5A), some of the chromatin-bound fraction remains protected from degradation (Fig. 5B)—protection that we attribute to either the incorporation of PLZF/RARA into the PRC1 complex (Fig. 2), or the highly compact nature of the chromatin once targeted by PRC1 (Francis et al. 2004). Like PML/RARA, we found that treatment of PLZF/RARA cells with ATRA leads to a stark reversal in recruited PRC2 and its H3K27me3 mark, whereas PRC1 recruited by PLZF/RARA was unaffected. This suggests that PRC1 plays a role in the continued maintenance of ectopic silencing of PLZF/RARA, helping us to understand why PLZF/RARA-targeted genes respond poorly to ATRA.
If the recruitment of PRC1 by PLZF/RARA to its target genes plays a direct role in ectopic gene repression, then circumventing its recruitment would have profound effects on PLZF/RARA oncogenic potential. Bmi-1 is a major component of the PRC1 complex (Levine et al. 2002), and likely mediates the liaison between PRC1 and PLZF/RARA (see Fig. 1). When this interaction can no longer occur in myeloid progenitors lacking Bmi-1, PLZF/RARA loses its ability to transform. PML/RARA, however, is still able to transform Bmi-1−/− progenitors, which is in agreement with our model that PRC1 is required for PLZF/RARA-mediated but not PML/RARA-mediated transformation.
PRCs: a deciding factor in the efficiency of a therapeutic RA response?
While PcG complexes play a central role in epigenetic gene repression, the repressive state imposed can either be stable, as in Hox repression, or more dynamic, as with certain lineage-determining transcription factors in stem cells. We found that both PRC2 and PRC1 are ectopically recruited to RA response genes through PLZF/RARA expression, whereas only PRC2 is recruited when PML/RARA is expressed. This difference is due to a unique interaction between PLZF/RARA and Bmi-1 that recruits the PRC1 complex to RA response genes and is essential for the transformation ability of PLZF/RARA. Thus, while PRCs play a major role in both PLZF/RARA-mediated and PML/RARA-mediated transformation, we propose a model explaining how the different PcG complexes involved have a decisive impact on RA response: PML/RARA interacts with and ectopically recruits PRC2 to RA response genes, whereas PLZF/RARA interacts with and ectopically recruits both PRC1 and PRC2. Through the effect of the PcG on chromatin, these recruitments participate toward repression of the RA response gene. Upon treatment with pharmacological doses of RA, PRC2 recruitment by either PML/RARA or PLZF/RARA is lost. In the case of PML/RARA, this leads to reactivation of the response gene. However, PRC1 recruited by PLZF/RARA is unaffected by RA treatment and continues to provide neomorphic Bmi-1-dependent repression of PLZF/RARA target genes. Due to the requirement of Bmi-1 in mediating the interaction between PLZF/RARA and the PRC1 complex, experiments designed to disrupt the PLZF/RARA–PRC1 interaction in vivo will prove critical in understanding the molecular mechanisms driving this persistent leukemic state and toward developing a therapeutic response.
Materials and methods
Antibodies and constructs
Antibodies used were against Bmi (H-99), M2 (Sigma), His tag (BD Biosciences), M33 (Abcam), PLZF (H-300), RARA (C-20) (Santa Cruz Biotechnologie), β-Tubulin1 (Sigma), and Ring1 and Ph1 (Satijn et al. 1997) for Western blot; and Bmi-1 (1.T.21, Abcam), PLZF (H-300, Santa Cruz Biotechnologies), EZH2 (AC22), H3ac (06599) (Upstate Biotechnologies), H3K27me3 (ab8898), H3 (ab1791) (Abcam), Flag (M2, Sigma), and Ring1 (Satijn et al. 1997) for immunoprecipitation or ChIP. cDNAs used in this study are described in the Supplemental Material.
Protein interaction assays
GST pull-downs were performed as described previously (Kwok et al. 2006). For co-IP experiments, 293T cells were cotransfected with Flag-Bmi-1 and the following constructs: PLZF, PLZF/RARA, RARA, or PML/RARA. Nuclear protein was extracted and complexes were immunoprecipitated with M2 anti-Flag beads (Sigma Aldrich), washed three times with PBS, and eluted with Flag peptide (0.5 mg/mL). For endogenous co-IP in U937 cells, PLZF/RARA was induced with zinc for 24 h, nuclear proteins were extracted, and endogenous Bmi-1 was immunoprecipitated with anti-Bmi-1 and PLZF/RARA was immunoprecipitated with anti-RARA overnight at 4°C. Immunoprecipitates were washed with PBS and eluted with SDS sample buffer.
Purification of mPCC complexes
Sf9 cells were coinfected simultaneously with recombinant baculovirus encoding Flag-Bmi1, M33, mPh1, Ring1A (mPCC; Lavigne et al. 2004) and either His-PLZF (mPCC–PLZF) or His-PLZF/RARA (mPCC–PLZF/RARA). Volumes of each high-titer virus were determined empirically as the minimum required to infect all cells determined by cell cycle arrest. Forty-eight hours post-infection, cells were harvested, nuclear extracts were performed, and complexes were purified according to Lavigne et al. (2004) using 15 column volumes of 1.2 M KCl buffer for the high-stringency wash, and including 10 μM ZnCl2 in all buffers (PCCB). Complexes were quantitated by Bradford analysis using BSA as the standard, and active DNA-binding protein concentrations were determined by filter-binding analyses using linearized DNA (Francis et al. 2001). A mock purification was performed by M2 anti-Flag purification of cells infected with M33, mPh1, Ring1A, and His-PLZF/RARA viruses.
mPCC–PLZF/RARA was further fractionated using glycerol gradient sedimentation. One-hundred microliters of mPCC–PLZF/RARA complex were applied to a 10%–40% glycerol gradient in PCCB200 and centrifuged for 21 h at 30,000 rpm in a Beckman SW50 rotor. Two-hundred-microliter fractions were collected and resolved by 8% SDS-PAGE, blotted to nitrocellulose membranes, and probed with either anti-mPh1, anti-His (BD Biosciences; PLZF/RARA), or M2 anti-Flag (Sigma; Bmi−) antibodies.
5S array assemblies and assays
5S-RARE-DR5 DNA was constructed by replacing the PstI–XbaI fragment containing the Gal4 DNA-binding sites from the 5S-G5E4 vector with a 91-base-pair (bp) fragment containing three RA receptor-binding sites (DR5T-GGTTCA_CCGAA_AGTTCA). Nucleosome arrays were assembled and analyzed by gradient salt dialysis and REA assays were performed as described (Francis et al. 2001). Radioactive 5S nucleosome arrays (1 nM) were incubated with varying concentrations of mPCC complexes in 12 mM HEPES, 0.1 mM EDTA, 10 mM Tris, 100 ng/μL BSA, 0.2 mM DTT, 4 mM MgCl2, and 10 mM ZnCl2 for 20 min at 30°C prior to addition of purified SWI/SNF (Sif et al. 1998) at concentrations determined to be saturating for chromatin remodeling (typically 250 ng), and 4 U HhaI (New England Biolabs). Reactions were allowed to proceed for 60 min at 30°C and were stopped by the addition of 2 μL of DSB (50 mM Tris, 100 mM EDTA, 1% SDS, 25% glycerol). Proteins were digested with 10 μg of proteinase K for 15 min at 50°C, and DNA was resolved on 0.8% TAE agarose gels. Gels were vacuum dried and exposed to a PhosphorImager (Fuji, SLA-5000) for quantitation (Image Gauge, Fuji).
Cell lines
Human myelomonoblastic cell lines U937-MT, U937-PR9, or U937-B412 were maintained at exponential growth in RPMI supplemented with 10% fetal calf serum. U937-PR9 and U937-B412 contain, respectively, PML/RARA cDNA and PLZF/RARA cDNA under the control of the zinc-inducible human metallothionein promoter (Ruthardt et al. 1997). For protein induction, cells were prestimulated with 0.1 mM ZnSO4 for at least 24 h. For ATRA stimulation, cells were treated with 1 μM ATRA for the indicated times.
ChIP assays
ChIP was performed as described before in Batsche et al. (2006) with some modifications. Cells were treated with 1% formaldehyde (10 min, room temperature), and the reaction was stopped by the addition of glycine at the final concentration of 0.125 M. After two washes in PBS, cells were resuspended in 10 mM Tris-HCl (pH 8), 10 mM EDTA, 0.5 mM EGTA, 10 mM, and 0.25% Triton X-100; the soluble fraction was eliminated by centrifugation; and chromatin was extracted with 250 mM NaCl, 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 0.5 mM EGTA for 20 min on ice. Chromatin was resuspended in 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, and 0.5% SDS, and sonicated for 15 min using Diagenode Bioruptor (full power, 20 sec on 30 sec off). DNA fragment size (<1 kb) was verified by agarose gel electrophoresis. For ChIP using Bmi-1, Ring1a, PLZF, or Ezh2 antibodies, chromatin extracted from 1 × 107 cells per condition was used. For ChIP using H3 or H3K27me3, chromatin from 2 × 106 cells per condition was used. Chromatin was diluted five times in 150 mM NaCl, 10 mM Tris-HCl (pH 8.0), 1% Triton X-100, 0.1% sodium deoxycholate (NaDOC), 1 mM EDTA, and 0.5 mM EGTA. Chromatin, precleared 1 h with protein G-coupled magnetic beads, was incubated overnight at 4°C using the different antibodies followed by an incubation for 2 h with blocked protein G-coupled magnetics beads (Dynabeads, Invitrogen). Immunocomplexes were washed with 1× buffer 1 (10 mM Tris-HCl at pH 8.0, 1% Triton X-100, 0.1% NaDOC, 150 mM NaCl), 1× buffer 2 (10 mM Tris-HCl at pH 8, 1% NP-40, 0.1% NaDOC, 150 mM KCl), 1× buffer 3 (10 mM Tris-HCl at pH 8.0, 0.5% Triton X-100, 0.1% NaDOC, 300 mM NaCl), 1× buffer 4 (20 mM Tris-HCl at pH 8.0, 0.5% NP-40, 0.5% NaDOC, 250 mM LiCl, 1 mM EDTA), and 1× buffer 5 (20 mM Tris-HCl at pH 8.0, 0.1% NP-40, 150 mM NaCl, 20 mM Tris-HCl at pH 8.0, 1 mM EDTA). Elution, reverse cross-linking, and purification were performed using Chelex (Bio-Rad). Beads were eluted in 100 μL of water containing of 10% Chelex by boiling for 10 min, then were incubated with proteinase K for 30 min at 55°C and reboiled for 10 min.
Amplifications (40 cycles) were performed using quantitative real-time PCR using Brilliant SYBR Green Master Mix (Stratagene) according to the manufacturer's instructions. IgG control “cycle over the threshold” Ct values were subtracted to Input or immunoprecipitation Ct values and converted into bound value by 2(−[IP Ct or input Ct − IgG IP Ct]). Data are expressed as percentage of bound/input.
Primers used are given in the Supplemental Material.
Statistical analyses
Statistical analyses were performed with the Student's t test using GraphPad and two-tailed P values are given as follows: (*) P < 0.1; (**) P < 0.01; and (***)P < 0.001.
Retroviral infection and replating assay
BM cells were harvested from 3-wk-old littermate wild-type or Bmi-1−/− mice. Hematopoietic progenitors and stem cells were negatively selected for lineage expression by magnetic activated cell sorting (Miltenyi) and prestimulated for 48 h before infection. Viral supernatants were used to infect hematopoietic progenitors and stem cells by spinoculation at 2000 rpm for 2 h at 32°C. Infections were repeated twice and transduced cells were plated in methylcellulose supplemented with mSCF (50 ng/mL; R&D), mIL6 (20 ng/mL), mIL3, and GM-CSF (10 ng/mL; Abcys) in the presence of selective drugs. Colonies were counted after 7–9 d and replated three times at 10,000 cells when possible. One independent experiment represents the mean obtained from parallel platings of infected cells taken from a pool of BM cells obtained from up to two animals. After the second replating, cellular morphology of the colonies were analyzed by Wright-Giemsa staining of cytospin. Immunophenotypic analyses were performed by FACS using fluorochrome-conjugated antibodies to c-Kit (clone eB149), Mac-1 (clone M1; eBioscience), and Gr-1 (clone RB6-8C5; Becton Dickinson).
Acknowledgments
We thank A. Pietersen and C. Pritchard for isolating the Ink4a−/−/Arf−/−/Bmi1−/−-deficient BM, A. Iwama for the Bmi-1 expression vector, M. Ruthard for the gift of U937 and derivative cells and PLZF/RARA mutants, M. Djabali for providing the Bmi−/+ mice and helpful discussions, R. Kingston and M. Lavigne for providing the mPCC baculovirus, and H. de Thé for MSCV-PML/RARA DNA. This work was supported by CNRS, INSERM, and grants from Association pour la Recherche sur le Cancer (ARC) to E.D., A.S., J.P., and M.S., from AICR to M.S., and from CEFIPRA to A.S. and J.P. H.B. was supported by a fellowship from the CNRS and the société Française d'hématologie (SFH).
Footnotes
References
- Barna M, Merghoub T, Costoya JA, Ruggero D, Branford M, Bergia A, Samori B, Pandolfi PP. Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev Cell. 2002;3:499–510. doi: 10.1016/s1534-5807(02)00289-7. [DOI] [PubMed] [Google Scholar]
- Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- Beisel C, Buness A, Roustan-Espinosa IM, Koch B, Schmitt S, Haas SA, Hild M, Katsuyama T, Paro R. Comparing active and repressed expression states of genes controlled by the Polycomb/Trithorax group proteins. Proc Natl Acad Sci. 2007;104:16615–16620. doi: 10.1073/pnas.0701538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bel S, Coré N, Djabali M, Kieboom K, Van der Lugt N, Alkema MJ, Van Lohuizen M. Genetic interactions and dosage effects of Polycomb group genes in mice. Development. 1998;125:3543–3551. doi: 10.1242/dev.125.18.3543. [DOI] [PubMed] [Google Scholar]
- Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, Fuks F, Lo Coco F, Kouzarides T, Nervi C, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- Dong S, Zhu J, Reid A, Strutt P, Guidez F, Zhong HJ, Wang ZY, Licht J, Waxman S, Chomienne C, et al. Amino-terminal protein–protein interaction motif (POZ-domain) is responsible for activities of the promyelocytic leukemia zinc finger–retinoic acid receptor-α fusion protein. Proc Natl Acad Sci. 1996;93:3624–3629. doi: 10.1073/pnas.93.8.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005;122:835–847. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes & Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol Cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Gillespie RF, Gudas LJ. Retinoic acid receptor isotype specificity in F9 teratocarcinoma stem cells results from the differential recruitment of coregulators to retinoic response elements. J Biol Chem. 2007;282:33421–33434. doi: 10.1074/jbc.M704845200. [DOI] [PubMed] [Google Scholar]
- Grimaud C, Negre N, Cavalli G. From genetics to epigenetics: The tale of Polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- He LZ, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi PP. Distinct interactions of PML–RARα and PLZF–RARα with co-repressors determine differential responses to RA in APL. Nat Genet. 1998;18:126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- Iwama A, Oguro H, Negishi M, Kato Y, Morita Y, Tsukui H, Ema H, Kamijo T, Katoh-Fukui Y, Koseki H, et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 2004;21:843–851. doi: 10.1016/j.immuni.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Kelly KF, Daniel JM. POZ for effect—POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16:578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- King IF, Francis NJ, Kingston RE. Native and recombinant polycomb group complexes establish a selective block to template accessibility to repress transcription in vitro. Mol Cell Biol. 2002;22:7919–7928. doi: 10.1128/MCB.22.22.7919-7928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koken MH, Daniel MT, Gianni M, Zelent A, Licht J, Buzyn A, Minard P, Degos L, Varet B, de The H. Retinoic acid, but not arsenic trioxide, degrades the PLZF/RARα fusion protein, without inducing terminal differentiation or apoptosis, in a RA-therapy resistant t(11;17)(q23;q21) APL patient. Oncogene. 1999;18:1113–1118. doi: 10.1038/sj.onc.1202414. [DOI] [PubMed] [Google Scholar]
- Kwok C, Zeisig BB, Dong S, So CW. Forced homo-oligomerization of RARα leads to transformation of primary hematopoietic cells. Cancer Cell. 2006;9:95–108. doi: 10.1016/j.ccr.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigne M, Francis NJ, King IF, Kingston RE. Propagation of silencing; Recruitment and repression of naive chromatin in trans by polycomb repressed chromatin. Mol Cell. 2004;13:415–425. doi: 10.1016/s1097-2765(04)00006-1. [DOI] [PubMed] [Google Scholar]
- Lempradl A, Ringrose L. How does noncoding transcription regulate Hox genes? Bioessays. 2008;30:110–121. doi: 10.1002/bies.20704. [DOI] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht JD. Reconstructing a disease: What essential features of the retinoic acid receptor fusion oncoproteins generate acute promyelocytic leukemia? Cancer Cell. 2006;9:73–74. doi: 10.1016/j.ccr.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Licht JD, Chomienne C, Goy A, Chen A, Scott AA, Head DR, Michaux JL, Wu Y, DeBlasio A, Miller WH, Jr, et al. Clinical and molecular characterization of a rare syndrome of acute promyelocytic leukemia associated with translocation (11;17) Blood. 1995;85:1083–1094. [PubMed] [Google Scholar]
- Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes & Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A, Cleard F, Mishra RK, Karch F, Verrijzer CP. Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex. Genes & Dev. 2005;19:1755–1760. doi: 10.1101/gad.347005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland NM, King IF, Kingston RE. Regulation of Polycomb group complexes by the sequence-specific DNA binding proteins Zeste and GAGA. Genes & Dev. 2003;17:2741–2746. doi: 10.1101/gad.1143303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Poux S, Melfi R, Pirrotta V. Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes & Dev. 2001;15:2509–2514. doi: 10.1101/gad.208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccetti E, Zheng X, Brambilla D, Seshire A, Beissert T, Boehrer S, Nurnberger H, Hoelzer D, Ottmann OG, Nervi C, et al. The integrity of the charged pocket in the BTB/POZ domain is essential for the phenotype induced by the leukemia-associated t(11;17) fusion protein PLZF/RARα. Cancer Res. 2005;65:6080–6088. doi: 10.1158/0008-5472.CAN-04-3631. [DOI] [PubMed] [Google Scholar]
- Rajasekhar VK, Begemann M. Concise review: Roles of polycomb group proteins in development and disease: A stem cell perspective. Stem Cells. 2007;25:2498–2510. doi: 10.1634/stemcells.2006-0608. [DOI] [PubMed] [Google Scholar]
- Rego EM, He LZ, Warrell RP, Jr, Wang ZG, Pandolfi PP. Retinoic acid (RA) and As2O3 treatment in transgenic models of acute promyelocytic leukemia (APL) unravel the distinct nature of the leukemogenic process induced by the PML–RARα and PLZF–RARα oncoproteins. Proc Natl Acad Sci. 2000;97:10173–10178. doi: 10.1073/pnas.180290497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Ruthardt M, Testa U, Nervi C, Ferrucci PF, Grignani F, Puccetti E, Grignani F, Peschle C, Pelicci PG. Opposite effects of the acute promyelocytic leukemia PML–retinoic acid receptor α (RAR α) and PLZF–RAR α fusion proteins on retinoic acid signalling. Mol Cell Biol. 1997;17:4859–4869. doi: 10.1128/mcb.17.8.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satijn DP, Gunster MJ, van der Vlag J, Hamer KM, Schul W, Alkema MJ, Saurin AJ, Freemont PS, van Driel R, Otte AP. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol Cell Biol. 1997;17:4105–4113. doi: 10.1128/mcb.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes & Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So CW, Cleary ML. Dimerization: A versatile switch for oncogenesis. Blood. 2004;104:919–922. doi: 10.1182/blood-2004-03-0992. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Sternsdorf T, Phan VT, Maunakea ML, Ocampo CB, Sohal J, Silletto A, Galimi F, Le Beau MM, Evans RM, Kogan SC. Forced retinoic acid receptor α homodimers prime mice for APL-like leukemia. Cancer Cell. 2006;9:81–94. doi: 10.1016/j.ccr.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Villa R, Pasini D, Gutierrez A, Morey L, Occhionorelli M, Vire E, Nomdedeu JF, Jenuwein T, Pelicci PG, Minucci S, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11:513–525. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]