The Biosynthetic Pathway for Myxol-2′ Fucoside (Myxoxanthophyll) in the Cyanobacterium Synechococcus sp. Strain PCC 7002 (original) (raw)
Abstract
Synechococcus sp. strain PCC 7002 produces a variety of carotenoids, which comprise predominantly dicylic β-carotene and two dicyclic xanthophylls, zeaxanthin and synechoxanthin. However, this cyanobacterium also produces a monocyclic myxoxanthophyll, which was identified as myxol-2′ fucoside. Compared to the carotenoid glycosides produced by diverse microorganisms, cyanobacterial myxoxanthophyll and closely related compounds are unusual because they are glycosylated on the 2′-OH rather than on the 1′-OH position of the ψ end of the molecule. In this study, the genes encoding two enzymes that modify the ψ end of myxoxanthophyll in Synechococcus sp. strain PCC 7002 were identified. Mutational and biochemical studies showed that open reading frame SynPCC7002_A2032, renamed cruF, encodes a 1′-hydroxylase and that open reading frame SynPCC7002_A2031, renamed cruG, encodes a 2′-O-glycosyltransferase. The enzymatic activity of CruF was verified by chemical characterization of the carotenoid products synthesized when cruF was expressed in a lycopene-producing strain of Escherichia coli. Database searches showed that homologs of cruF and cruG occur in the genomes of all sequenced cyanobacterial strains that are known to produce myxol or the acylic xanthophyll oscillaxanthin. The genomes of many other bacteria that produce hydroxylated carotenoids but do not contain crtC homologs also contain cruF orthologs. Based upon observable intermediates, a complete biosynthetic pathway for myxoxanthophyll is proposed. This study expands the suite of enzymes available for metabolic engineering of carotenoid biosynthetic pathways for biotechnological applications.
A wide variety of organisms produce carotenoid glycosides, which act as natural surfactants, stabilize membranes, and possibly contribute to regulating the permeability of membranes to oxygen (4, 41, 51). The first carotenoid glycosides were isolated from saffron in 1818 (6). However, the structures of the glycosylated carotenoids phleixanthophyll and 4-keto-phleixanthophyll were the first to be completely determined, in 1967, after their isolation from Mycobacterium phlei (15, 34). Tertiary glycosides are relatively rare in nature but seem to be common in carotenoid biosynthesis (34). These include the glycosylated carotenoids of the myxobacteria, which have characteristic C-3′,4′ desaturation and C-1′ glycosylation (18, 19). Acylated carotenoid C-1′-glycosides are broadly distributed among bacteria, including Salinibacter ruber and members of the Chloroflexi and Chlorobi (24, 46, 47).
The glycosylated carotenoids of cyanobacteria differ from the examples mentioned above in that glycosylation characteristically occurs on the C-2′-hydroxyl group rather than that at the C-1′ position of the ψ end of myxol (3′,4′-didehydro-1′,2′-dihydro-β,ψ-carotene-3,1′,2′-triol) or oscillol (3,4,3′,4′-tetradehydro-1,2,1′,2′-ψ,ψ-carotene-1,2,1′,2′-tetrol) to form myxoxanthophyll or oscillaxanthin, respectively (16, 17). Myxoxanthophyll is thus far found uniquely in members of the phylum Cyanobacteria, and this compound is named after the synonym for this group of organisms, i.e., Myxophyceae (14, 42). As carotenoids from increasingly diverse bacteria are characterized, the apparent uniqueness of myxoxanthophyll to cyanobacteria will probably not persist. For example, the aglycone myxol occurs in marine flavobacteria, along with saporaxanthin (38, 49). Oscillaxanthin, which was once thought to be unique to the Nostophyceae, was recently identified as the major pigment of three strains of Methylobacterium spp. (20). Moreover, oscillol appears to be a precursor of the glycosylated and acylated carotenoids of Thermomicrobium roseum (52).
Several variations on the myxoxanthophyll pathway, which lead to a variety of possible compounds, are known to occur in cyanobacteria. A number of different sugars commonly occur in myxoxanthophyll derivatives found in different strains. Strains of Oscillatoria and Spirulina spp. produce compounds that are chinovosides, fucosides, or methylfucosides (1, 7). Derivatives containing fucose have been found in Nostoc punctiforme strain PCC 73102 and Nostoc sp. strain PCC 7120 (45), whereas myxoxanthophyll dimethylfucoside has been found in Synechocystis sp. strain PCC 6803 (42). Variations in the ring oxidation level of the basal compound, with the addition of a keto group at the C-4 position or of an additional hydroxyl group at the C-2 or C-4 position, may also lead to several related compounds.
Myxol is presumably synthesized from lycopene, the acyclic precursor of all carotenoids in cyanobacteria (see Fig. 1) (27). Because of the occurrence of a β-ionone ring in the final product, monocyclic γ-carotene is also presumed to be an intermediate in this pathway (21, 41). The question of what enzyme is responsible for the formation of the β-ionone ring from the linear ψ end of γ-carotene has been contentious but has recently been resolved. Although CrtL-type lycopene cyclases occur in some cyanobacteria, genes for lycopene cyclases of this family do not occur in the genomes of sequenced cyanobacteria that produce myxoxanthophyll (11, 26). Mohamed and Vermaas reported that the open reading frame (ORF) sll0254 encodes an enzyme thought to be involved simultaneously in the cyclization and ψ-end hydroxylation of lycopene in Synechocystis sp. strain PCC 6803 (31). However, subsequent studies of both Synechococcus sp. strain PCC 7002 and Synechocystis sp. strain PCC 6803 showed that this is not the case (11, 26). Like nearly all other cyanobacteria lacking CrtL homologs, both of these cyanobacteria contain two genes, cruA and cruP, which encode lycopene cyclases (26). A third class of organisms, which so far includes only Synechococcus sp. strains PCC 7942 and PCC 6301, have CrtL and CruP homologs.
FIG. 1.
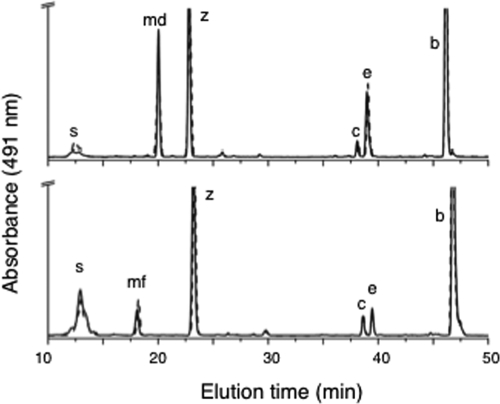
HPLC elution profiles for pigments from two cyanobacteria. (A) HPLC elution profiles (obtained by the jegpsu method) for pigments extracted from the wild type (solid line) and the slr1293 mutant (dotted line) of Synechocystis sp. strain PCC 6803. (B) HPLC elution profiles (obtained by the jegpsu method) for pigments extracted from the wild type (solid line) and SynPCC7002_A1623 mutant (dotted line) of Synechococcus sp. strain PCC 7002. Peak identities: s, synechoxanthin; md, myxol-2′ dimethylfucoside; mf, myxol-2′ fucoside; z, zeaxanthin; c, cryptoxanthin; e, echinenone; and b, β-carotene.
The well-conserved β-hydroxylase CrtR functions in the C-3 hydroxylation of myxoxanthophyll, and CrtR also functions in the synthesis of zeaxanthin, cryptoxanthin, and 3′-hydroxy-echinenone (11, 21, 27, 28). CrtR seems to be responsible for all C-3 hydroxylation reactions in Synechocystis sp. strain PCC 6803, and it is notable that CrtR seems to be extremely highly conserved among cyanobacteria (11, 27). Considerable confusion has existed concerning the remaining reactions of this biosynthetic pathway. Mohamed and Vermaas reported that ORF slr1293 encodes the 3′,4′ desaturase in Synechocystis sp. strain PCC 6803 (30). Furthermore, Mohamed and Vermaas additionally reported that the product of ORF sll0254 plays a dual role as a lycopene cyclase and a mediator of ψ-end hydroxylation during myxoxanthophyll biosynthesis in Synechocystis sp. strain PCC 6803 (31). However, subsequent studies have shown that the sll0254 product and its orthologs, renamed CruE, are carotene desaturases/methyltransferases that participate in the synthesis of the aromatic carotenoid synechoxanthin (12, 13).
In this study, we identified two genes, cruF and cruG, which encode the C-1′-hydroxylase and 2′-O-glycosyltransferase, respectively, that are uniquely required for mxyoxanthophyll biosynthesis in Synechococcus sp. strain PCC 7002. Orthologs of these genes are found in all sequenced genomes of cyanobacteria that synthesize myxoxanthophyll. Additionally, in contrast to the data in a previous report (30), we show that ORF slr1293 of Synechocystis sp. strain PCC 6803 and its ortholog SynPCC7002_A2031 in Synechococcus sp. strain PCC 7002 do not play an essential role in myxoxanthophyll biosynthesis.
MATERIALS AND METHODS
Strains and growth conditions.
The wild type and mutants of Synechococcus sp. strain PCC 7002 were routinely grown under standard growth conditions (at 38°C with continuous illumination at 250 μmol of photons m−2 s−1; sparged with 1% [vol/vol] CO2 in air) in 25-ml test tube cultures containing medium A supplemented with 1 mg of NaNO3 ml−1 (medium A+) (40). Media were additionally supplemented with 50 μg of gentamicin ml−1, 100 μg of kanamycin ml−1, or 100 μg of spectinomycin ml−1 as required. Freshwater strains, including Synechocystis sp. strain PCC 6803, Anabaena variabilis strain ATTC 29413, Nostoc sp. strain PCC 7120, and Synechococcus sp. strain PCC 7942, were grown in B-HEPES medium, prepared by supplementing BG-11 medium (39) with 4.6 mM HEPES-KOH and 18 mg of ferric ammonium citrate liter−1, or on solid B-HEPES medium supplemented with 15 g of Bacto agar (Difco; Voigt Global Distribution, Lawrence, KS) liter−1. Plates were incubated at 30°C with light at an intensity of 150 μmol of photons m−2 s−1. Colleagues from other laboratories provided other cyanobacterial cells.
Identification of genes and phylogenetic comparisons.
Analyses of cyanobacterial genomes were conducted using databases, the blastp algorithm (3), and the phylogenetic profiling tools at the Integrated Microbial Genomes website (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi [Joint Genome Institute, Walnut Creek, CA]). For the construction of phylogenetic trees, amino acid sequences were aligned with the ClustalW tool in MacVector 9.0 (MacVector, Inc., Cary, NC) and neighbor-joining trees were generated from the resulting alignments with PAUP 4.0 (Sinauer Associates, Inc., Sunderland, MA).
Insertional inactivation of genes by interposon mutagenesis.
The upstream and downstream flanking sequences of genes selected for insertional inactivation were amplified by PCR by using genomic DNA templates isolated from wild-type Synechococcus sp. strain PCC 7002 or wild-type Synechocystis sp. strain PCC 6803 and the primers described in Table S1 in the supplemental material. The reverse primers for the left flank and the forward primers for the right flank contained BamHI, EcoRI, HindIII, or XbaI restriction sites as indicated in Table S1 in the supplemental material. PCR products were digested with the appropriate restriction enzymes and purified from agarose gels by using a Perfectprep gel cleanup kit (Eppendorf; catalog no. 0032 007.759). For insertional inactivation of slr1293 and SynPCC7002_A2031, the aadA gene, conferring resistance to streptomycin and spectinomycin, was excised from plasmid pSRA2 (10) with BamHI or EcoRI as required and purified by agarose gel electrophoresis. For insertional activation of SynPCC7002_A1623, the aphAII gene, conferring resistance to kanamycin, was excised from plasmid pRL170 (9) with HindIII and purified by electrophoresis on an agarose gel. For insertional inactivation of SynPCC7002_A2032, the aacC1 gene, conferring resistance to streptomycin and spectinomycin, was excised from plasmid pMS255 (10) with XbaI and purified in the same manner. The fragments were mixed at a 3:1:3 ratio of the left flank to the antibiotic resistance cassette to the right flank and ligated with T4 DNA ligase. The ligation products were separated on agarose gels, and products of the desired size were excised, purified as described above, and used to transform Synechococcus sp. strain PCC 7002 or Synechocystis sp. strain PCC 6803 as described previously by Frigaard et al. (9).
Expression of cruF in a lycopene-producing strain of Escherichia coli.
The cruF gene (SynPCC7002_A2032) from Synechococcus sp. strain PCC 7002 was amplified by PCR using primers cruFxf and cruFxR (see Table S1 in the supplemental material), digested by the enzymes NdeI and BamHI, and cloned into the plasmid pET3atr (48) via the NdeI and BamHI restriction sites. E. coli cells producing lycopene were transformed with the expression construct via electroporation as described previously (26). For the purification of pigments, ∼10 Luria-Bertani (LB) plates were inoculated with ∼500 CFU per plate. The plates were incubated for 3 days at 25°C, and the cells were then washed off with liquid LB medium. The cell suspension was poured into centrifuge tubes, the cells were pelleted, and carotenoids were extracted and purified by preparative high-performance liquid chromatography (HPLC).
HPLC analysis of pigment extracts.
Cyanobacterial cells were harvested in late exponential phase (optical density at 730 nm, ∼1.0). Pigments were extracted from whole cells by sonication in 7:2 acetone-methanol or methanol alone when indicated, and the cellular debris was removed by centrifugation. The supernatant was filtered through a 0.2-μm polytetrafluoroethylene syringe filter (catalog no. 6783-0402; Whatman, Clifton, NJ). Filtered extracts were analyzed immediately without further manipulation. The HPLC system consisted of an analytical 5-μm Discovery C18 column (25 cm by 4.6 mm; Supelco, Bellefonte, PA) fitted to a binary pump (model no. G1312A) and solvent degasser (1100 series, model no. 1379A; Agilent Technologies, Palo Alto, CA). Eluates were monitored with a 1,024-element diode array detector (1100 series, model no. G1315B; Agilent Technologies, Palo Alto, CA), and the system was controlled with Agilent ChemStation software for HPLC. For C18 chromatography, method jegpsu was used. For this method, solvent A was methanol-acetonitrile-water (21:16.5:62.5, vol/vol/vol) and contained 10 mM ammonium acetate; solvent B was methanol-acetonitrile-ethyl acetate (50:20:30, vol/vol/vol). The solvent gradient program (time in minutes, percent solvent B, and flow rate in milliliters per minute) was as follows: 0, 20, and 0.750; 10, 70, and 1.0; 40, 100, and 1.0; and 50, 100, and 1.0. The same solvent system but with the following gradient (time in minutes and percent solvent B) was utilized for separating carotenoids produced in E. coli (Ecolicar method): 0 and 20, 3 and 30, 30 and 100, and 35 and 100. An alternate method, jC303, used a 25-cm by 4.6-mm by 5.0-μm ProntoSil C30 column. For this method, solvent A was methanol-acetonitrile-water (70:20:10), solution B was methyl-_tert_-butyl ether-methanol-acetonitrile (50:30:20), and the gradient was as follows (time in minutes and percent solution B), with a flow rate of 1 ml min−1: 0 and 20, 10 and 40, 20 and 50, 30 and 50, 35 and 100, 65 and 100, and 66 and 20.
Pigment purification and analysis.
For larger-scale purification of pigments, a semipreparative 5-μm Discovery C18 column (25 cm by 10 mm; Supelco, Bellefonte, PA) was used with the same HPLC equipment, gradient, and conditions as described above for C18 chromatography except that the flow rate was increased to 3.5 ml min−1. Pigment-containing fractions were collected and dried under a stream of nitrogen gas. Chemical analyses of compounds were performed as described previously (5, 11). To test for the presence of hydroxyl groups, a small (∼200-ng) sample of the carotenoid was dissolved in anhydrous pyridine and treated with Sigma-Sil-A (trimethylchlorosilane-1,1,1,3,3,3-hexamethyldisilazane-pyridine [1:3:9, vol/vol/vol; 10 to 100 μl]; Sigma, St. Louis, MO). Alternatively, samples were dried under nitrogen and resuspended in acetic anhydride (100 μl) containing a trace amount of pyridine. Reactions were quenched with excess methanol, the solution was dried under nitrogen, and the products were resuspended in methanol prior to HPLC analysis. Eric Snyder and Li Zhang at the Proteomics and Mass Spectrometry Core Facility of the Huck Institutes of the Life Sciences, The Pennsylvania State University, performed the mass spectrometric analyses.
RESULTS
Identification of myxol-2′ fucoside from Synechococcus sp. strain PCC 7002.
Nostoc sp. strain PCC 7120 has been shown previously to produce myxol-2′ fucoside as a major carotenoid (45). A carotenoid with a retention time and an absorption spectrum similar to those of myxol-2′ fucoside was purified from Synechococcus sp. strain PCC 7002 by preparative HPLC. In the C18 (jegpsu) and C30 HPLC (jC303) analyses described above, this compound had elution times that were identical to those of the authentic myxol-2′ fucoside that had been purified from Nostoc sp. strain PCC 7120 (see Fig. S1 in the supplemental material). Furthermore, as expected for myxol-2′ fucoside, liquid chromatography-mass spectrometry (MS) analysis showed that this compound had a molecular mass of 730 Da. Together, these data established that Synechococcus sp. strain PCC 7002 synthesizes myxol-2′ fucoside.
Bioinformatic and phylogenetic analyses of putative genes encoding enzymes of the myxoxanthophyll biosynthesis pathway.
A diagnostic feature of myxoxanthophyll and related carotenoids is that, compared to lycopene or β-carotene, these compounds have an extended polyene chromophore, which results from an additional double bond at the 3′,4′ position. Such extended chromophores are commonly observed in carotenoids of purple bacteria, in which they occur in spirilloxanthin and spheroidenone (43). In purple bacteria, the desaturation of the 3′,4′ bond is performed by CrtD, a carotenoid 3′,4′ desaturase (2); however, most cyanobacteria apparently do not have an orthologous enzyme.
Mohamed and Vermaas (30) proposed that the product of ORF slr1293 in Synechocystis sp. strain PCC 6803 is the carotenoid 3′,4′ desaturase of myxoxanthophyll biosynthesis. To test this suggestion, the distribution of slr1293 orthologs in sequenced strains of cyanobacteria was examined. As reported by Mohamed and Vermaas (30), the slr1293 enzyme was found to be distantly similar to the CrtI phytoene desaturase family, which includes CrtI, CrtD, and CrtH. However, the slr1293 product is actually most similar (29% identical and 45% similar) to the carotenoid isomerase CrtH (the sll0033 product in Synechocystis sp. strain PCC 6803). Furthermore, the distribution of genes orthologous to slr1293 also suggested that it is highly conserved and found in nearly all sequenced cyanobacterial genomes (see Table S2 in the supplemental material). Although both Synechocystis sp. strain PCC 6803 and Synechococcus sp. strain PCC 7002 in fact synthesize myxoxanthophyll (11) (see Table S3 in the supplemental material), only two cyanobacteria, Synechococcus sp. strain JA-2-3B′a(2-13) and Synechococcus sp. strain JA-3-3Ab, lack orthologs of slr1293. Because many cyanobacteria do not synthesize myxoxanthophyll, the nearly universal occurrence of genes orthologous to slr1293 is inconsistent with the role that was postulated for the product of this gene by Mohamed and Vermaas (30). Furthermore, although the green sulfur bacterium Chlorobaculum tepidum does not synthesize carotenoids that are desaturated at the 3′,4′ position, its genome also contains an apparent ortholog of slr1293, CT0180. The product of ORF CT0180 is 34% identical and 44% similar to the product of slr1293 (8). To clarify the role of slr1293 in carotenoid biosynthesis, null mutants of ORF slr1293 in Synechocystis sp. strain PCC 6803 and its ortholog SynPCC7002_A1623 in Synechococcus sp. strain PCC 7002 were constructed.
Construction and characterization of disruption mutants of slr1293 and SynPCC7002_A1623.
A fully segregated null mutation of slr1293 in Synechocystis sp. strain PCC 6803 was obtained and verified by PCR (see Fig. S2B in the supplemental material). A fully segregated knockout of SynPCC7002_A1623, which is the Synechococcus sp. strain PCC 7002 ortholog of slr1293, was similarly obtained and verified by PCR (see Fig. S2D in the supplemental material). Methanolic extracts of cells of both mutant strains were analyzed by HPLC, and the results were compared with those for extracts from cells of the respective wild-type strains. Both mutants continued to synthesize myxoxanthophyll at levels similar to those synthesized by the respective wild-type strains (Fig. 1). Mohamed and Vermaas had reported that their slr1293 mutant failed to produce myxoxanthophyll but that it continued to synthesize other glycosylated carotenoids, which had similar HPLC retention times but were saturated at the 3′,4′ position (30). Because the absorbance maxima of such compounds would be blue shifted by about 10 nm, the resulting spectra should have been similar to that of γ-carotene (see Fig. S3 in the supplemental material). No compounds with these properties were observed among the elution products from the HPLC analysis. Because the myxoxanthophylls found in extracts from both mutants were identical to those from the corresponding wild-type strains in both HPLC retention time and absorption properties, the products of slr1293 in Synechocystis sp. strain PCC 6803 and of its ortholog SynPCC7002_A1623 in Synechococcus sp. strain PCC 7002 are clearly not essential for myxoxanthophyll biosynthesis. This finding is in agreement with the data in a recent report by Mochimaru et al. (29).
Phylogenetic identification of the cruF and cruG genes.
Mohamed and Vermaas (31) also reported that the product of Synechocystis sp. strain PCC 6803 ORF sll0254 serves as a bifunctional lycopene cyclase/dioxygenase responsible for ψ-end oxygenations in myxoxanthophyll biosynthesis. This conclusion has previously been shown to be incorrect (12, 26). sll0254 instead encodes an enzyme, designated CruE, which is responsible for the conversion of β-rings to χ-rings during the synthesis of the aromatic xanthophyll synechoxanthin (12, 13). Because all previous studies failed to identify genes uniquely required for myxoxanthophyll biosynthesis, we sought to identify the genes that encode the enzymes responsible for the two ψ-end hydroxylations and the glycosylation that must occur during myxoxanthophyll biosynthesis.
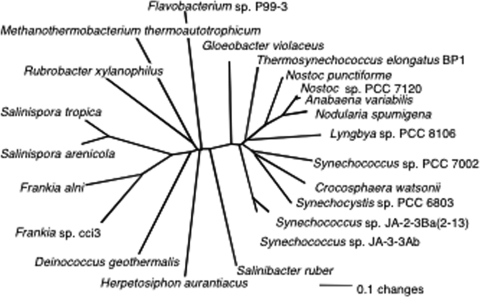
The marine Flavobacterium sp. strain P99-3 has been shown previously to synthesize myxol (49, 53). Analysis of the carotenoid biosynthetic cluster from this organism revealed a gene, designated orf1, which encodes an enzyme with 26% identity and 44% similarity to the product of ORF sll0814 of Synechocystis sp. strain PCC 6803. Orthologs of this gene, here renamed cruF, occur in many cyanobacteria but are notably absent from all Prochlorococcus spp. and various marine Synechococcus spp., as well as Trichodesmium erythraeum, none of which synthesize myxoxanthophyll (see Table S3 in the supplemental material) (11). Homologs are also found in a wide variety of carotenoid-synthesizing bacteria, but interestingly, this gene never occurs in strains that have crtC, which encodes an alternative enzyme, hydroxyneurosporene synthase, that can also hydroxylate the 1′ position of carotenoids (37). The cyanobacterial proteins homologous to the orf1 and sll0814 products are roughly 60% identical and 70% similar, and they form a distinctive clade in a phylogenetic analysis (Fig. 2). Interestingly, these gene products have a distribution that is nearly identical to that of CruA among the cyanobacteria whose genomes were analyzed for this study. T. erythraeum, which has cruA but not cruF, was the only exception identified (see Table S2 in the supplemental material).
FIG. 2.
Neighbor-joining distance tree showing relationships among homologs of CruF found in cyanobacteria and other bacteria.
Phylogenetic profiling also identified a gene that encodes a type 2 glycosyltransferase; this gene has a distribution among cyanobacteria that precisely matches that of cruF and expresses an enzyme with the ability to synthesize myxoxanthophyll. This gene, herein designated cruG, is apparently orthologous to genes found in the carotenoid biosynthetic gene clusters of other bacteria, including S. ruber (24). CruG (the sll1004 product) from Synechocystis sp. strain PCC 6803 was 63% identical and 74% similar to CruG (the SynPCC7002_A2031 product) from Synechococcus sp. strain PCC 7002 and 50% identical and 65% similar to its ortholog (the glr1357 product) in Gloeobacter violaceus strain PCC 7421. CruG was 30% identical and 44% similar to an apparent homolog in S. ruber, and it was 28% identical and 45% similar to its homolog in C. tepidum. Interestingly, cruG is clustered with cruF to form an apparent dicistronic operon in 11 of 16 cyanobacteria that have both genes. This organization is not found in Synechocystis sp. strain PCC 6803, Lyngbya sp. strain PCC 8106, Thermosynechococcus elongatus strain BP-1, and Synechococcus sp. strains JA-3-3aB and JA-2-3B′a(2-13).
Characterization of a cruF mutant and recombinant CruF.
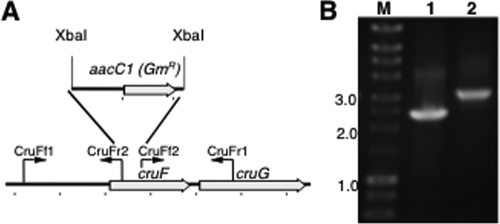
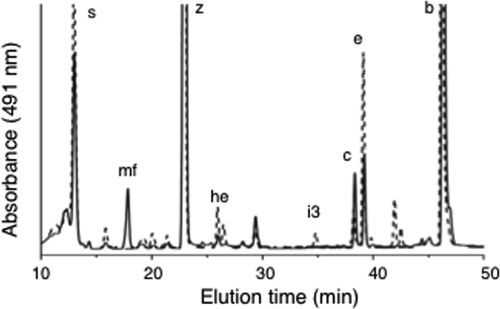
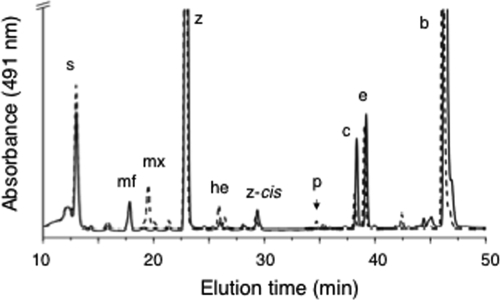
A fully segregated disruption mutation of cruF in Synechococcus sp. strain PCC 7002 was obtained. The mutation was generated as described in Materials and Methods by using the primer pairs CruFf1/CruFr2 and CruFf2/CruFr1, and segregation was verified using the primer pair CruFf1/CruFr1 (see Table S1 in the supplemental material). In the construction used for mutagenesis, 225 bp in the 5′ region of cruF was replaced with the aacC1 cartridge from pMS255 via XbaI restriction sites (Fig. 3). Methanolic extracts from the resulting null mutant were analyzed by HPLC, and no myxol-2′ fucoside or 3-dehydroxymyxol-2′ fucoside was detected (Fig. 4). Furthermore, no compound whose spectrum was similar to that of myxoxanthophyll could be detected in extracts from the mutant cells. Torulene, a proposed 3′,4′ desaturated intermediate in myxoxanthophyll synthesis (42), was not detected.
FIG. 3.
Restriction map and PCR verification of the cruF mutant of Synechococcus sp. strain PCC 7002. (A) Restriction map showing the construction of the cruF mutant of Synechococcus sp. strain PCC 7002. (B) Agarose gel electrophoresis analysis of amplicons from the cruF loci of the wild type (lane 1) and cruF::aacC1 mutant (lane 2). The data show that the cruF and cruF::aacC1 alleles in the mutant strain are fully segregated. Selected sizes of markers (lane M) in kilobases are indicated to the left.
FIG. 4.
HPLC elution profiles (obtained by the jegpsu method) of pigments extracted from cells of the wild type (solid line) and the cruF mutant (dotted line) of Synechococcus sp. strain PCC 7002. Peak identities: s, synechoxanthin; mf, myxol-2′ fucoside; z, zeaxanthin; he, hydroxy-echinenone; i3, 18-hydroxyrenierapurpurin; c, cryptoxanthin; e, echinenone; and b, β-carotene.
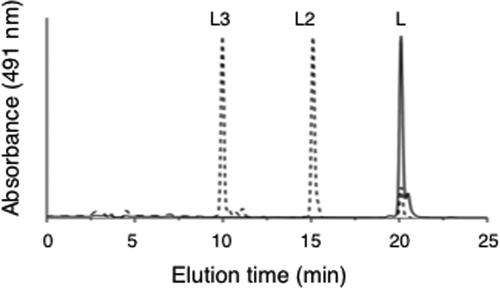
To verify the activity of CruF, the cruF gene of Synechococcus sp. strain PCC 7002 was cloned into the pET3atr expression vector to produce plasmid pSLF, and an E. coli strain that synthesizes lycopene, BL21(DE3) harboring pAClyc (26), was transformed with the plasmid. Transformants were grown with antibiotic selection on LB plates at 25 to 28°C. Colonies harboring plasmid pSLF were small and slow growing compared to control colonies harboring empty vector pET3atr. When cruF was expressed in the lycopene-producing strain of E. coli, cell extracts contained two carotenoid products with lycopene chromophores, and both of these products were more hydrophilic than lycopene (Fig. 5).
FIG. 5.
HPLC elution profiles (obtained by the Ecolicar method) of pigments extracted from E. coli BL21(DE3) strains harboring pAClyc, along with empty vector pET3atr (solid line) or plasmid pSLF (dotted line). Peak identities: L, lycopene; L2, 1-hydroxylycopene; and L3, 1,1′-dihydroxylycopene.
The two compounds produced in E. coli were subjected to additional chemical analyses. The molecular masses of these two compounds were determined to be 544 and 562 Da. These masses are consistent with the addition of one or two water molecules across the 1,2 and 1′,2′ double bonds of lycopene. A sample of 1-hydroxylycopene was not modified by acetic anhydride but was modified by Sigma-Sil-A (see Fig. S4 in the supplemental material). A sample of 1,1′-dihydroxylycopene was mixed with zeaxanthin, and the mixture was then treated with acetic anhydride. Zeaxanthin was rapidly and completely converted to the diacetyl compound, but none of the 1,1′-dihydroxylycopene was acetylated (data not shown). The acetylation of tertiary hydroxyl groups by acetic anhydride, in which solvent acetylation occurs quite slowly compared with the rapid modification that occurs for both primary and secondary alcohols (33), occurs only in the presence of pyridine. Because the hydroxyl groups of the lycopene derivatives from E. coli behaved like tertiary hydroxyl groups, these hydroxyl groups must be located at the C-1 and C-1′ positions. Thus, these two compounds were inferred to be 1-hydroxy-1,2-dihydro-ψ,ψ-carotene and 1,1′-dihydroxy-1,2,1′,2′-tetrahydro-ψ,ψ-carotene based on elution times, spectral properties, MS results, and the chemical modification of hydroxyl groups. Because 1-hydroxylycopene accumulated in a cruA mutant (26), it was concluded that the hydroxylation of one of the ψ ends of lycopene or possibly of γ-carotene is the first committed step in the branch of the carotenoid pathway that leads to myxoxanthophyll (or oscillaxanthin) and that CruF is responsible for this activity.
Characterization of a cruG mutant.
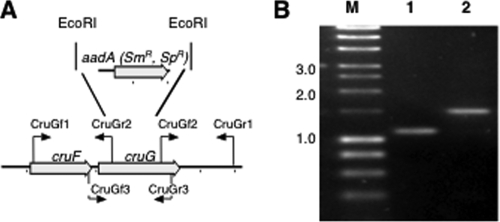
A fully segregated disruption mutation of cruG in Synechococcus sp. strain PCC 7002 was also obtained. This mutation was generated as described above using the primer pairs CruGf1/CruGr2 and CruGf2/CruGr1. In the construction employed for transformation, the aadA cartridge from pSRA2 replaced 724 bp of cruG via introduced EcoRI sites (Fig. 6A). Complete segregation of alleles was verified via the primer pair CruGf3/CruGr3 (Fig. 6B). Methanolic extracts of the cruG null mutant were analyzed by HPLC, and these analyses showed that the peak corresponding to myxol-2′ fucoside was absent in the mutant profile. However, a new compound (designated mx) with a later elution time appeared in profiles of extracts from the mutant; the absorption spectrum of this compound was nearly identical to that of myxol-2′ fucoside (Fig. 7). A small peak corresponding to 3-deoxy-myxol (plectaniaxanthin), the same compound that accumulated in a crtR mutant (11), was also detected (Fig. 7).
FIG. 6.
Restriction map and PCR verification of the cruG mutant of Synechococcus sp. strain PCC 7002. (A) Restriction map showing the construction of the cruG mutant of Synechococcus sp. strain PCC 7002. (B) Agarose gel electrophoresis analysis of amplicons from the cruG loci of the wild type (lane 1) and the cruG::aadA mutant (lane 2). The data show that the cruG and cruG::aadA alleles in the mutant strain are fully segregated. The sizes of selected marker fragments (lane M) in kilobases are indicated to the left.
FIG. 7.
HPLC elution profiles (obtained by the jegpsu method) of pigments extracted from cells of the wild type (solid line) and the cruG mutant (dotted line) of Synechococcus sp. strain PCC 7002. Peak identities: s, synechoxanthin; mf, myxol-2′ fucoside; mx, myxol; z, zeaxanthin; he, hydroxy-echinenone; z-cis, cis isomer of zeaxanthin; p, plectaniaxanthin; c, cryptoxanthin; e, echinenone; and b, β-carotene.
Compound mx was purified from the cruG mutant by preparative HPLC, and hydroxyl-counting experiments were conducted by treating samples with Sigma-Sil-A reagent as described previously (5, 12). Compound mx could be modified up to three times by trimethylsilylation, which verified that three hydroxyl groups were present (see Fig. S5 in the supplemental material). Thus, it was tentatively concluded that compound mx was free myxol. Verification of this assignment was obtained by comparing this compound with the carotenoids of A. variabilis strain ATCC 29413, which was previously shown to synthesize myxol (44). Myxol and plectanixanthin were purified from A. variabilis strain ATCC 29413, and these authentic standards were compared with the compounds isolated from the cruG mutant by two different HPLC methods (see Materials and Methods). The elution times of authentic myxol and plectanixanthin were identical to those of the compounds produced in the cruG mutant; moreover, in each case the compounds coeluted as single, sharp peaks when the appropriate compounds were mixed and analyzed by the two HPLC methods (see Fig. S6 in the supplemental material). CruG is the first carotenoid glycosyltransferase that specifically modifies the C-2′-OH position of carotenoid ψ ends. Based on compounds produced by these organisms and the findings of recent mutational studies with C. tepidum, the homologs of CruG in S. ruber and C. tepidum are C-1′-O-glycosyltransferases (24, 25).
Attempts to identify the remaining step(s) of myxoxanthophyll biosynthesis.
The synthesis of the 2′-hydroxyl moiety of myxol-2′ fucoside requires at least one additional enzyme that can recognize the hydroxylated ψ end and perform the 3′,4′ desaturation and 2′ hydroxylation reactions. The overall reaction is similar to the reaction catalyzed by CrtA, spheroidene monooxygenase, in purple nonsulfur bacteria in that it requires the addition of an oxygen atom and the removal of two hydrogen atoms (22). Because no apparent CrtA ortholog exists in Synechococcus sp. strain PCC 7002 or any other cyanobacterium, an alternative enzyme (or enzymes) must be responsible for the 2′ hydroxylation reaction. Phylogenetic profiling identified genes homologous to SynPCC7002_A1949 of Synechococcus sp. strain PCC 7002 as candidates to encode this activity. Homologs of SynPCC7002_A1949 occur in all sequenced cyanobacteria that have cruF, but homologs are found additionally in Synechococcus sp. strains PCC 7942 and PCC 6301, which do not have cruF and do not synthesize myxoxanthophyll. Although the distribution of this gene did not precisely match that of cruF, the homologs of SynPCC7002_A1949 were still considered to be attractive candidates to express the missing activity, because they have both dehydrogenase and oxidase domains and the reaction requires a 3′,4′ dehydrogenase activity and 2′-hydroxylase activity. An attempt was made to inactivate the SynPCC7002_A1949 gene, but only stable merodiploids were obtained, and the carotenoid compositions of the merodiploids were similar to that of the wild type. Thus, the product of this gene is essential to Synechococcus sp. strain PCC 7002 under standard growth conditions. Because myxol-2′ fucoside is clearly not required for viability, as demonstrated by the construction of crtR, cruF, and cruG null mutants (11, 21; this study), it is unlikely that the product of SynPCC7002_A1949 is involved in myxoxanthophyll production. Another candidate gene, SynPCC7002_A2446 of Synechococcus sp. strain PCC 7002, was also identified by phylogenetic profiling. A fully segregated mutation was obtained, but no change in myxol-2′ fucoside biosynthesis was observed. Further attempts to identify the gene or genes involved in 3′,4′ desaturation and 2′ hydroxylation in the myxol-2′ fucoside biosynthetic pathway in Synechococcus sp. strain PCC 7002 have not yet been successful.
DISCUSSION
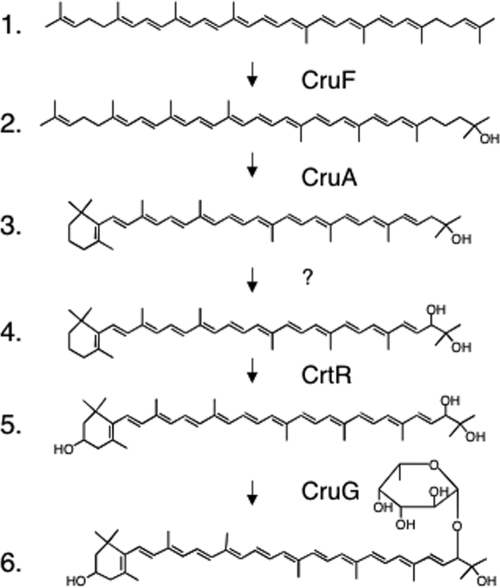
The results reported here and elsewhere allow one to propose the complete pathway for myxoxanthophyll biosynthesis from lycopene. Three different intermediates (1-hydroxylycopene, plectaniaxanthin, and myxol) specific to the myxol-2′ fucoside branch of the carotenoid biosynthetic pathway of Synechococcus sp. strain PCC 7002 accumulated in a mutant described previously (26) or in some of the mutant strains described herein. We have shown previously that 1-hydroxylycopene accumulates in a lycopene cylase mutant (26), and in this study, we have shown that CruF can produce this compound from lycopene. Additionally, plectaniaxanthin and myxol accumulate in a cruG null mutant. Furthermore, all of these intermediates were absent in a cruF mutant strain. Thus, we propose that the first committed step in myxol-2′ fucoside synthesis is the hydroxylation of the tertiary carbon of a ψ end of lycopene or γ-carotene by CruF. This hydroxylation reaction would sequester the intermediate product from the pool of substrate molecules available to the CruA and CruP lycopene cyclases. The formation of the β-ionone ring by lycopene cyclase either before or after hydroxylation would lead to 1′-hydroxy-γ-carotene. This compound would next be converted into 3-dehydroxymyxol (plectaniaxanthin) by an as-yet-unidentified enzyme(s). Because no intermediate corresponding to 1′,2′-dihydroxy-1′,2′-dihydro-β,ψ-carotene or 1′-hydroxy-3′,4′-didehydro-1′,2′-dihydro-β,ψ-carotene was observed in the wild type or any mutant strains, it is highly likely that the desaturation at the 3′,4′ position and the hydroxylation at the 2′ position are carried out by a single enzyme or are very tightly coupled reactions. Plectaniaxanthin (or its glycosylated analog) has been observed in crtR mutants of Synechocystis sp. strain PCC 6803 and Synechococcus sp. strain PCC 7002, as well as in desiccated field samples of Nostoc commune (11, 21, 35). In Synechococcus sp. strain PCC 7002, CrtR apparently acts on 3-dehydroxymyxol (plectaniaxanthin) to form myxol. Finally, CruG glycosylates the hydroxyl group at the 2′ position to form myxoxanthophyll (Fig. 8).
FIG. 8.
Proposed pathway for the biosynthesis of myxol-2′ fucoside. The roles of CruF, CruA, CrtR, and CruG are indicated. The order of cyclization and 1′ hydroxylation may be reversed (see Fig. S7 in the supplemental material). Compound identities: 1, lycopene; 2, 1-hydroxylycopene; 3, 1′-hydroxy-γ-carotene; 4, plectaniaxanthin; 5, myxol; and 6, myxol-2′ fucoside. The enzyme(s) for the introduction of the 3′,4′ double bond and the 2′-OH group is currently unknown.
Because Synechococcus sp. strain PCC 7002 has an ORF (SynPCC7002_A2832) encoding a product with strong sequence similarity to GDP-fucose synthetase (32), it seems likely that CruG directly transfers a fucosyl moiety to myxol to produce myxol-2′ fucoside. In Synechocystis sp. strain PCC 6803, one or two methyltransferases are additionally required to produce the myxol-2′ dimethylfucoside found in this species (42). The production of oscillaxanthin would proceed similarly, with CruF first acting on both ends of lycopene; this step would be followed by 3,4 and 3′,4′ desaturation, 2,2′ hydroxylation, and finally, the glycosylation of both the 2- and 2′-hydroxyl groups. Interestingly, although only myxol-2′ fucoside is produced in Synechococcus sp. strain PCC 7002, the studies of expression in E. coli reported herein show that CruF has the ability to modify both ψ ends of lycopene. Some cyanobacteria, such as Arthrospira maxima, produce both myxoxanthophyll and oscillaxanthin simultaneously, while others, such as G. violaceus strain PCC 7421, produce only oscillaxanthin (see Table S3 in the supplemental material) (36, 50). The observation that CruF from a strain that makes myxol-2′ fucoside but not oscillaxanthin can nevertheless modify both ψ ends of lycopene in E. coli suggests that this distribution could be achieved simply by modulating the competition between CruF and the CruA and CruP carotenoid cyclases for substrate ψ ends. This interpretation is supported by the observation that, with the exception of previously described lycopene cyclase mutants (26), cyanobacteria generally do not accumulate carotenoids with unmodified ψ ends. The observation that, among sequenced strains, all cyanobacteria that have CruF also have a CruA-type lycopene cyclase raises the possibility that the corresponding genes may have been coinherited by lateral gene transfer when the CruA-type lycopene cyclase was acquired by some ancestral cyanobacterium. Alternatively, it is possible that these two genes have been inherited vertically but that the CruF gene has subsequently been lost by some strains.
Based upon the results reported herein and those recently reported elsewhere (29), the slr1293 enzyme is not required for myxoxanthophyll biosynthesis as reported previously (30). The function of the slr1293 protein in carotenoid biosynthesis, if indeed it acts in this pathway at all, is likely as an isomerase that acts early in the pathway. This enzyme is both broadly conserved among bacteria and nonessential for the production of any major carotenoid in Synechococcus sp. strain PCC 7002 or Synechocystis sp. strain PCC 6803 under normal laboratory conditions. Among the other known cyanobacterial carotenoid biosynthetic enzymes, only CrtH, a carotenoid isomerase to which the slr1293 product has high sequence similarity, shares these properties (8). It has recently been shown that plants have an enzyme responsible for the isomerization of 15-_cis_-ζ-carotene, and it is therefore possible that cyanobacteria do as well (23). The product of Synechocystis sp. strain PCC 6803 ORF sll0254, now renamed cruE, is not involved in lycopene cyclization or myxoxanthophyll biosynthesis as reported previously (31), nor is it essential in Synechocystis sp. strain PCC 6803 or Synechococcus sp. strain PCC 7002 (12, 26). Instead, CruE is a carotene desaturase/methyltransferase that converts β-rings into χ-rings during the synthesis of synechoxanthin (12).
The genes and enzyme(s) responsible for the 3′,4′ desaturation and 2′ hydroxylation remain to be identified. If one draws parallels to other carotenoid biosynthetic pathways, it is tempting to suggest that a single oxygenase, analogous to but distinct from CrtA (22), may be responsible for the remaining activity. In fact, although no obvious homolog of CrtA exists in cyanobacteria, the 2′-hydroxylase activity has long been predicted to be carried out by a CrtA-like enzyme (42). Because it is likely that only cyanobacteria with CruF and CruG produce myxoxanthophyll, the discovery of these enzymes will hopefully aid in the identification of the remaining enzyme(s). It is probable that as the genomes of additional organisms are sequenced, a gene cluster that includes the cruF gene and that for the missing enzyme(s) as well may eventually be identified.
Because it has been reported previously that A. variabilis strain ATCC 29413 produces free myxol (44), it is intriguing that the genome of this organism encodes a CruG homolog, which is 59% identical and 74% similar to Synechocystis sp. strain PCC 6803 CruG. It had even been suggested by Takaichi et al. (44) that this organism would be a useful negative out-group for the identification of the glycosyltransferase by phylogenetic profiling. It is possible that CruG activity has been lost during laboratory cultivation. Alternatively, this finding may indicate that some cyanobacteria alter myxol glycosylation in response to environmental conditions as an adaptive mechanism. The identification of the cruG gene will promote more detailed comparative biochemical studies of the glycosylation of myxol and similar compounds in cyanobacteria and other bacteria. Homologs of CruG in other bacteria are 1′-O-glycosyltransferases. S. ruber makes a compound, salinixanthin, which is very similar to myxoxanthophyll. Salinixanthin has a 4-keto group instead of the 3-OH moiety and carries an acyl-sugar moiety attached to the C-1′ position instead of C-2′ (24). S. ruber has homologs of both CruF (30% identical and 45% similar) and CruG (30% identical and 44% similar). A detailed comparison of cyanobacterial CruG (2′-O-glycosyltransferase) with the S. ruber homolog would be informative, because the two enzymes obviously glycosylate different hydroxyl groups on substrates that are hydroxylated at both the 1′ and 2′ positions (24).
The identification of CruF and the ability to eliminate myxol-2′ fucoside production completely at the first step of the pathway will now allow studies of the role(s) of myxoxanthophylls within cyanobacteria. Results from preliminary experiments with the cruF mutant suggest that myxoxanthophyll helps to protect against reactive oxygen species during growth under high-intensity light (G. Shen and D. A. Bryant, unpublished results). Since myxoxanthophyll is considered to be highly important to the functionality of the cytoplasmic and outer membranes, its presence in a subset of cyanobacteria raises questions about the structural role(s) of myxoxanthophyll (and related glycosides) in the cell wall and outer membrane. The observation that myxoxanthophylls (and the related enzymes) are missing from the Prochlorococcus spp. and strains of open-water, marine Synechococcus spp. implies a distinct structural role that is completely missing in the open-water marine strains. The structural differences in and carotenoid contents of the cytoplasmic membranes, cell walls, and outer membranes of a broad range of cyanobacteria should be examined in detail to address this issue. With the identification of the genes encoding the enzymes responsible for the synthesis of zeaxanthin (11, 21, 28), synechoxanthin (12), and myxoxanthophyll (this study), it is now possible for the first time to construct mutants with completely defined xanthophyll compositions. CruF and CruG also provide new tools for tracing the evolution of carotenoid biosynthesis in cyanobacteria and other bacteria and provide new opportunities for metabolic engineering of carotenoid biosynthesis pathways in other organisms.
Supplementary Material
[Supplemental material]
Acknowledgments
This research was supported by National Science Foundation grants MCB-0077586 and MCB-0519743 to D.A.B.
We gratefully acknowledge Eric Snyder and Li Zhang (The Pennsylvania State University) for assistance with the MS. We thank Richard W. Castenholz (University of Oregon), John B. Waterbury (Woods Hole Oceanographic Institution), Theresa Thiel (University of Missouri—St. Louis), and Carlos Gomez-Lojero (Centro de Investigación y Estudios Avanzados del IPN, Mexico), who provided cyanobacterial cells or strains used in the analyses described in this study.
Footnotes
▿
Published ahead of print on 20 March 2009.
REFERENCES
- 1.Ackermann, T., O. Skulberg, and S. Liaan-Jensen. 1992. A comparison of the carotenoids of strains of Oscillatoria and Spirulina (cyanobacteria). Biochem. Syst. Ecol. 20761-769. [Google Scholar]
- 2.Albrecht, M., H. Linden, and G. Sandmann. 1996. Biochemical characterization of purified ζ-carotene desaturase from Anabaena PCC 7120 after expression in Escherichia coli. Eur. J. Biochem. 236115-120. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 4.Dembitsky, V. M. 2005. Astonishing diversity of natural surfactants. 3. Carotenoid glycosides and isoprenoid glycolipids. Lipids 40535-557. [DOI] [PubMed] [Google Scholar]
- 5.Eugester, C. 1995. Chemical derivatization: microscale tests for the presence of common functional groups in carotenoids, p. 71-80. In G. Britton, S. Liaan-Jensen, and H. Pfander (ed.), Carotenoids, vol. 1A. Isolation and analysis. Birkhäuser, Basel, Switzerland. [Google Scholar]
- 6.Eugester, C. 1995. History: 175 years of carotenoid chemistry, p. 1-11. In G. Britton, S. Liaan-Jensen, and H. Pfander (ed.), Carotenoids, vol. 1A. Isolation and analysis. Birkhäuser, Basel, Switzerland. [Google Scholar]
- 7.Foss, P., O. M. Skulberg, L. Kilaas, and S. Liaan-Jensen. 1986. The carbohydrate moieties bound to the carotenoids myxol and oscillol and their chemosystematic applications. Phytochemistry 251127-1132. [Google Scholar]
- 8.Frigaard, N.-U., J. A. Maresca, C. E. Yunker, A. D. Jones, and D. A. Bryant. 2004. Genetic manipulation of carotenoid biosynthesis in the green sulfur bacterium Chlorobium tepidum. J. Bacteriol. 1865210-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frigaard, N.-U., Y. Sakuragi, and D. A. Bryant. 2004. Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum using _in vitro_-made DNA constructs and natural transformation. Methods Mol. Biol. 274324-330. [DOI] [PubMed] [Google Scholar]
- 10.Frigaard, N.-U., G. D. Voigt, and D. A. Byant. 2002. Chlorobium tepidum mutant lacking bacteriochlorophyll c made by inactivation of the bchK gene, encoding bacteriochlorophyll c synthase. J. Bacteriol. 1843368-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham, J. E. 2008. Carotenoid biosynthesis in Synechococcus sp. PCC 7002: identification of the enzymes and the carotenoids. Ph.D. dissertation. The Pennsylvania State University, University Park.
- 12.Graham, J. E., and D. A. Bryant. 2008. The biosynthetic pathway for synechoxanthin, an aromatic carotenoid synthesized by the euryhaline, unicellular cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 1907966-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham, J. E., J. T. Lecomte, and D. A. Bryant. 2008. Synechoxanthin: an aromatic C40 xanthophyll is a major carotenoid in the cyanobacterium Synechococcus sp. PCC 7002. J. Nat. Prod. 711647-1650. [DOI] [PubMed] [Google Scholar]
- 14.Heilbron, I. M. 1942. Some aspects of algal chemistry. Nature 149398-400. [Google Scholar]
- 15.Hertzberg, S., and S. Liaan-Jensen. 1967. Bacterial carotenoids. 20. Carotenoids of Mycobacterium phlei strain Vera. 2. Structures of phleixanthophylls—two novel tertiary glucosides. Acta Chem. Scand. 2115-41. [PubMed] [Google Scholar]
- 16.Hertzberg, S., and S. Liaan-Jensen. 1969a. The structure of myxoxanthophyll. Phytochemistry 81259-1280. [Google Scholar]
- 17.Hertzberg, S., and S. Liaan-Jensen. 1969b. The structure of oscillaxanthin. Phytochemistry 81281-1292. [Google Scholar]
- 18.Kleinig, H., H. Rechenback, H. Achenbach, and J. Stadler. 1971. Carotenoid pigments of Sorangium compositum (Myxobacterales) including two new carotenoid glucoside esters and two new carotenoid rhamnosides. Arch. Microbiol. 78224-230. [DOI] [PubMed] [Google Scholar]
- 19.Kleinig, H., and H. Rechenback. 1973. New glucoside ester from Chondromyces apiculatus. Phytochemistry 122483-2485. [Google Scholar]
- 20.Konovalova, H., S. Shvlin, and P. Rokvtko. 2007. Characteristics of carotenoids of methylotrophic bacteria of Methylobacterium genus. Mikrobiol. Z. 6935-41. (In Ukrainian.) [PubMed] [Google Scholar]
- 21.Lagarde, D., and W. Vermaas. 1999. The zeaxanthin biosynthesis enzyme β-carotene hydroxylase is involved in myxoxanthophyll synthesis in Synechocystis sp. PCC 6803. FEBS Lett. 454247-251. [DOI] [PubMed] [Google Scholar]
- 22.Lang, H. P., R. J. Cogdell, S. Takaichi, and C. N. Hunter. 1995. Complete DNA sequence, specific Tn_5_ insertion map, and gene assignment of the carotenoid biosynthesis pathway of Rhodobacter sphaeroides. J. Bacteriol. 1772064-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, F., C. Murillo, and E. T. Wurtzel. 2007. Maize Y9 encodes a product essential for the 15-cis-ζ-carotene isomerization. Plant Physiol. 1441181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lutnaes, B. F., A. Oren, and S. Liaaen-Jensen. 2002. New C40-carotenoid acyl glycoside as principal carotenoid in Salinibacter ruber, an extremely halophilic eubacterium. J. Nat. Prod. 651340-1343. [DOI] [PubMed] [Google Scholar]
- 25.Maresca, J. A., and D. A. Bryant. 2006. Two genes encoding new carotenoid-modifying enzymes in the green sulfur bacterium Chlorobium tepidum. J. Bacteriol. 1886217-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maresca, J. A., J. E. Graham, M. Wu, J. A. Eisen, and D. A. Bryant. 2007. Identification of a fourth family of lycopene cyclases in photosynthetic bacteria. Proc. Natl. Acad. Sci. USA 10411784-11789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maresca, J. A., J. E. Graham, and D. A. Bryant. 2008. Carotenoid biosynthesis in chlorophototrophs: the biochemical and genetic basis for structural diversity. Photosynth. Res. 97121-140. [DOI] [PubMed] [Google Scholar]
- 28.Masamoto, K., N. Misawa, T. Kaneko, R. Kikuno, and H. Toh. 1998. β-Carotene hydroxylase gene from the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 39560-564. [DOI] [PubMed] [Google Scholar]
- 29.Mochimaru, M., H. Masukawa, T. Maoka, H. E. Mohamed, W. F. Vermaas, and S. Takaichi. 2008. Substrate specificities and availability of fucosyltransferase and β-carotene hydroxylase for myxol 2′-fucoside synthesis in Anabaena sp. strain PCC 7120 compared with Synechocystis sp. strain PCC 6803. J. Bacteriol. 1906726-6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohamed, H. E., and W. F. Vermaas. 2004. Slr1293 in Synechocystis sp. strain PCC 6803 is the C-3′,4′ desaturase (CrtD) involved in myxoxanthophyll bioynthesis. J. Bacteriol. 1865621-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohamed, H. E., and W. F. Vermaas. 2006. Sll0254 (CrtLdiox) is a bifunctional lycopene cyclase/dioxygenase in cyanobacteria producing myxoxanthophyll. J. Bacteriol. 1883337-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohamed, H. E., A. M. van de Meene, R. W. Roberson, and W. F. Vermaas. 2005. Myxoxanthophyll is required for normal cell wall structure and thylakoid organization in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 1876883-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naves, Y., and P. Ardizio. 1954. Acetylation of tertiary alcohols in the presence of pyridine. Ann. Pharm. Fr. 12471-476. (In French.) [PubMed] [Google Scholar]
- 34.Pfander, H. 1976. Carotenoid glycosides. Pure Appl. Chem. 47121-128. [Google Scholar]
- 35.Potts, M., J. J. Olie, J. S. Nickels, J. Parsons, and D. C. White. 1987. Variation in phospholipid ester-linked fatty acids and carotenoids of desiccated Nostoc commune (cyanobacteria) from different geographic locations. Appl. Environ. Microbiol. 534-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt, K., and R. Schburr. 1970. Die carotinoide der grunen schwefelbakterien: carotinoidzusammensetzung in 18 staemmen. Arch. Microbiol. 74350-355. [Google Scholar]
- 37.Scolnik, P. A., M. A. Walker, and B. L. Marrs. 1980. Biosynthesis of carotenoids derived from neurosporene in Rhodopseudomonas capsulata. J. Biol. Chem. 2552427-2432. [PubMed] [Google Scholar]
- 38.Shindo, K., K. Kikuta, A. Suzuki, A. Katsuto, H. Kasai, M. Ysumoto-Hirose, Y. Matsuo, N. Misawa, and S. Tachaichi. 2007. Rare carotenoids, (3R)-saproxanthin and (3R,2′S)-myxol, isolated from novel marine bacteria (Flavobacteriacae) and their antioxidant activities. Appl. Microbiol. Biotechnol. 741350-1357. [DOI] [PubMed] [Google Scholar]
- 39.Stanier, R., R. Kunisawa, M. Mandel, and G. Cohen-Bazire. 1971. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 35171-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevens, S. E., Jr., and R. D. Porter. 1980. Transformation of Agmenellum quadruplicatum. Proc. Natl. Acad. Sci. USA 776052-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sunbzynksi, W. K., E. Markowska, and J. Sielewiesiuk. 1991. Effect of polar carotenoids on the oxygen diffusion-concentration product in lipid bilayers. An EPR label study. Biochim. Biophys. Acta 106868-72. [DOI] [PubMed] [Google Scholar]
- 42.Takaichi, S., T. Maoka, and K. Masamoto. 2001. Myxoxanthophyll in Synechocystis sp. PCC 6803 is myxol 2′-dimethyl-fucoside, (3R,2S)-myxol 2′-(2,4-di-O-methyl-α-L-fucoside), not rhamnoside. Plant Cell Physiol. 42756-762. [DOI] [PubMed] [Google Scholar]
- 43.Takaichi, S., and M. Mochimaru. 2007. Carotenoids and carotenogenesis in cyanobacteria: unique ketocarotenoids and carotenoid glycosides. Cell. Mol. Life Sci. 642607-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takaichi, S., M. Mochimaru, and T. Maoka. 2006. Presence of free myxol and 4-hydroxymyxol and the absence of myxol glycosides in Anabaena variabilis ATCC 29413, and proposal of a biosynthetic pathway of carotenoids. Plant Cell Physiol. 47211-216. [DOI] [PubMed] [Google Scholar]
- 45.Takaichi, S., M. Mochimaru, T. Maoka, and H. Katoh. 2005. Myxol and 4-ketomyxol 2′-fucosides, not rhamnosides, from Anabaena sp. PCC 7120 and Nostoc punctiforme PCC 73102, and proposal for the biosynthetic pathway of carotenoids. Plant Cell Physiol. 46497-504. [DOI] [PubMed] [Google Scholar]
- 46.Takaichi, S., K. Tsuji, K. Matsuura, and K. Shimada. 1995. A monocyclic carotenoid glucoside ester is a major carotenoid in the filamentous bacterium Chloroflexus aurantiacus. Plant Cell Physiol. 36773-778. [Google Scholar]
- 47.Takaichi, S., Z.-W. Wang, M. Umetsu, T. Nozawa, K. Shimada, and M. T. Madigan. 1997. New carotenoids from the thermophilic green sulfur bacterium Chlorobium tepidum: 1′,2′-dihydro-γ-carotene, 1′,2′-dihydrochlorobactene, and OH-chlorobactene glucoside ester and the carotenoid composition of different strains. Arch. Microbiol. 168270-276. [DOI] [PubMed] [Google Scholar]
- 48.Tan, S. 2001. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 21224-234. [DOI] [PubMed] [Google Scholar]
- 49.Teramoto, M., S. Takaichi, Y. Inomata, H. Ikenaga, and N. Misawa. 2003. Structural and functional analysis of a lycopene β-monocyclase gene isolated from a unique marine bacterium that produces myxol. FEBS Lett. 545120-126. [DOI] [PubMed] [Google Scholar]
- 50.Tsuchiya, T., S. Takaichi, N. Misawa, T. Maoka, H. Miyashita, and M. Mimuro. 2005. The cyanobacterium Gloeobacter violaceus PCC 7421 uses bacterial-type phytoene desaturase in carotenoid biosynthesis. FEBS Lett. 5792125-2129. [DOI] [PubMed] [Google Scholar]
- 51.Woodall, A. A., G. Britton, and M. J. Jackson. 1997. Carotenoids and protection of phospholipids in solution or in liposomes against oxidation by peroxyl radicals: relationship between carotenoid structure and protective ability. Biochim. Biophys. Acta 1336575-586. [DOI] [PubMed] [Google Scholar]
- 52.Wu, D., J. Raymond, M. Wu, S. Chatterj, Q. Ren, J. E. Graham, D. A. Bryant, F. Robb, A. Colman, L. J. Tallon, J. H. Badger, R. Madupa, N. L. Ward, and J. A. Eisen. 2009. Complete genome sequence of the aerobic CO-oxidizing thermophile Thermomicrobium roseum. PLOS ONE 4e4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yokoyama, A., and W. Miki. 1995. Isolation of myxol from a marine bacterium Flavobacterium sp. associated with marine sponge. Fish. Sci. 61684-686. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]