ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization (original) (raw)
Abstract
We report here the identification and characterization of a protein, ERIS, an endoplasmic reticulum (ER) IFN stimulator, which is a strong type I IFN stimulator and plays a pivotal role in response to both non–self-cytosolic RNA and dsDNA. ERIS (also known as STING or MITA) resided exclusively on ER membrane. The ER retention/retrieval sequence RIR was found to be critical to retain the protein on ER membrane and to maintain its integrity. ERIS was dimerized on innate immune challenges. Coumermycin-induced ERIS dimerization led to strong and fast IFN induction, suggesting that dimerization of ERIS was critical for self-activation and subsequent downstream signaling.
Keywords: innate immunity, type I IFN, functional cDNA library screening, cytosolic RNA and dsDNA, ER retention signal
Microbial infection-induced host immune responses are initiated by the germline-encoded pattern recognition receptors, which recognize components specific to microorganisms. There are 3 major classes of such receptors: Toll-like receptors (TLRs), RIG-I–like helicases (RLHs) and NOD-like receptors (1). During infection, nucleic acids derived from microbes are recognized by TLRs and RLHs, which then trigger a series of signaling events leading to the production of type I IFNs and proinflammatory cytokines.
RLHs have recently been identified to sense the invading viruses in the cytoplasm. Unlike TLRs, which are expressed in specific cells like macrophages and dendritic cells, RLHs are found in most cell types (2). They contain caspase recruitment domain (CARD) and DExD/H helicase domain. RLHs interact with microbial nucleotides through their helicase domain. The N-terminal CARDs are responsible for activating downstream signaling pathways that mediate type I IFN production. Genetic analyses demonstrate that RIG-I and MDA5 sense distinct types of viruses (3–5). RIG-I and MDA5 use a common adaptor molecule, IPS-1 (also known as Cardif, MAVS, or VISA) (6–9). IPS-1 is found to reside on the mitochondrial membrane by its C-terminal transmembrane (TM) domain. It also contains a CARD-like domain at its N-terminus, which mediates the interaction with MDA5 or RIG-I. IPS-1 transmits the signal to TANK-binding kinase-1 (TBK1)/IκB kinase i (IKKi; also known as IKKε) and the IKK complex to activate interferon regulatory factor (IRF)-3/IRF-7 and NF-κB, respectively, collectively eliciting innate antiviral immune responses, including type I IFN production.
On the other hand, dsDNA in the cytosol, for example, genomic DNA from intracellular bacteria (e.g., Listeria, Legionella), also causes a strong host immune response independent of TLRs, leading to the induction of type I IFN. A recent report has indicated that the molecule DAI (also known as ZBP1) might serve as a cytosolic dsDNA sensor (10). However, ZBP1−/− cells showed normal type I IFN production in response to dsDNA stimulation (11). Meanwhile, reports showed that IPS-1/Cardif/MAVS/VISA was not required for dsDNA-caused innate immune activation (12). The signaling induced by cytoplasmic dsDNA leading to the activation of IRF-3/IRF-7 is largely elusive.
Although much progress has been made in understanding the antiviral immune response that leads to the induction of type I IFN, many questions still remain. For example, we do not know exactly how bifurcated anticytoplasmic RNA (dsRNA and ssRNA) and the anti-dsDNA innate immune response converge to activate TBK1/IKKi. To address these questions, other players in the pathway need to be identified. We used a functional cDNA library screening approach to search systematically for genes that could activate type I IFN production. Here, we report a protein, designated as ERIS, an endoplasmic reticulum (ER) IFN stimulator, to be an essential innate immune mediator. Overexpression of ERIS led to high type I IFN induction as well as IFN-regulated genes. Suppression of ERIS expression caused reduced response to both cytoplasmic RNA and dsDNA. ERIS resided on ER membrane, and the ER retention/retrieval sequences R76Y77R78 and R178I179R180 were found to be critical to retain this protein on the ER membrane and maintain its integrity. ERIS was dimerized/oligomerized, ubiquitinated, and phosphorylated on innate immune challenge and by TBK1/IKKi overexpression. Coumermycin-induced ERIS-Gyrase B (GyrB) dimerization led to IFN induction, suggesting that like many other molecules, oligomerization or dimerization of ERIS was critical for self-activation and subsequent downstream signaling.
Results
Overexpression of ERIS Leads to Upregulation of Type I IFNs.
To identify molecules involved in the induction of type I IFNs by virus infection, we generated a cDNA library from mouse bone marrow-derived macrophages treated with VSV for 8 h and then screened the library for its ability to activate IFN-β promoter. The cDNA library cloned into an expression plasmid was divided into pools of 100 cDNAs, which were then transfected into 293 cells, together with an IFN-β promoter-luciferase reporter plasmid. From an individual positive pool, we isolated a single clone that robustly induced the reporter plasmid over 7,500-fold compared with empty plasmid as the negative control [supporting information (SI) Fig. S1]. BLAST research identified it as identical to RIKEN full-length enriched library clone F730319K18, we named it ERIS. ERIS was a hypothetical protein, which encoded a protein of 379 aa in humans and 378 aa in mice and shared 68% identity and 81% similarity between Homo sapiens and Mus musculus, respectively. Moreover, its homologues in other species also exhibited high identity, especially at the C-terminal region, demonstrating it to be an evolutionally conserved protein (Figs. S2 and S3). By structural analysis, it was supposed to be a membrane-bound protein that contained a signal peptide at the N-terminus followed by 4 putative TM domains. Thus, ERIS was a protein with unknown function to date, leading us to investigate it further.
To make it clear that ERIS was a real IFN stimulator, more than 10 other promoters related to type I IFNs, including IFN-αs, IL-10, and IL-23, were tested. Reporter assay showed that exogenous expression of ERIS greatly up-regulated IFN-β expression, modestly up-regulated IFN-α4 and IFN-α8 expression, and up-regulated IFN-α1 and IFN-α2 expression to some extent (Fig. 1A). ERIS also activated NF-κB and interferon-sensitive response element (Fig. 1A), indicating its extensive effects on immunity.
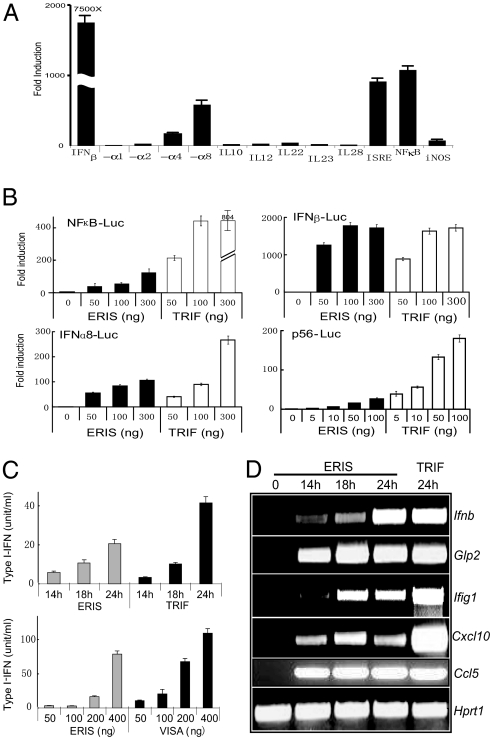
Fig. 1.
ERIS is a strong type I IFN activator. (A) 293 cells were transiently transfected with 100 ng of pMX-ERIS, along with 50 ng of indicated reporter constructs. A luciferase assay was performed after 24 h. Data represent mean ± SD. (B) 293 cells were transfected with indicated amounts of HA-ERIS or HA-TRIF, along with indicated reporters. A luciferase assay was carried out the same as in A. (C) 293 cells were transfected with 200 ng (Upper) or different dosages (Lower) of 3′HA-ERIS or TRIF/VISA as indicated. Supernatant was collected at different time points (Upper) or at 24 h (Lower) to analyze the concentration of type I IFN production by a type I IFN bioassay. (D) 293 cells were transfected with HA-ERIS or HA-TRIF. Total RNA was prepared at the indicated times after transfection and was analyzed for expression of the indicated genes by RT-PCR. Similar results were obtained in 3 independent experiments.
To confirm further its capability of activating diverse promoters, different dosages of ERIS were transiently transfected into 293 cells, together with IFN-β–Luc, NF-κB–Luc, and IFN-α8–Luc, respectively. Overexpressed ERIS activated these promoters in a dose-dependent way (Fig. 1B). ERIS-induced IFN-β activation was comparable to that induced by Toll/IL-1 receptor (TIR) domain-containing adaptor inducing IFN-beta (TRIF), a strong innate immune activator of both type I IFN and NF-κB (13, 14) (Fig. 1B). To examine whether ERIS led to type I IFN production and secretion, ERIS was overexpressed in 293 cells. The results indicated that ERIS led to the production and secretion of IFN proteins in a dose-dependent way (Fig. 1C). We also examined the effects of ERIS on type I IFNs and its regulated gene at mRNA levels. Consistent with the results of reporter assay and bioassay, IFN-β and some IFN-inducible genes were significantly up-regulated by ERIS overexpression (Fig. 1D). These data collectively demonstrate that ERIS is a potent IFN stimulator.
ERIS Is Involved in Innate Immunity.
Because ERIS was a hypothetical protein, we next tested its existence. We generated a mouse polyclonal antibody against C-terminus (amino acids 219–379) of human ERIS and found that this antibody could recognize not only overexpressed but also endogenous protein (Fig. 2A and Fig. 3), and even the mouse ERIS as well (Fig. 2B, line 7). We then assessed the expression of ERIS in several cultured human cell lines and mouse tissues. ERIS was highly expressed in immune cells such as THP1, U937, K562, and HT1080 as well (Fig. 2B). In mouse tissues, it was found that ERIS was highly expressed in thymus and spleen and modestly expressed in lung and kidney (Fig. 2C). This expression pattern further suggested that ERIS might function in the immune system.
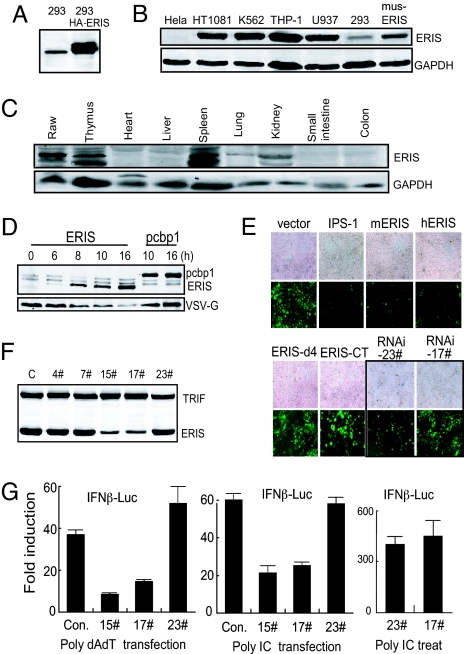
Fig. 2.
ERIS is involved in innate immunity. (A) 293 cells or 293 cells stably expressing HA-ERIS were lysed and immunoblotted with antibody against ERIS. Immunoblotting analysis was performed on several cell lines (B) or tissues derived from mouse (C) with ERIS antibody. (D) 293 cells were transfected with HA-ERIS. At indicated time points after transfection, cells were infected by VSV for 18 h and then analyzed by immunoblotting with anti-HA or anti-VSV-G. HA-pcbp1 served as a negative control. (E) 293 cells were transfected with empty vector or expression plasmids. Twenty-four or 48 h (for RNAi transfections) later, cells were infected by NDV-GFP (at a MOI of 0.1 or 0.05 for RNAi-transfected cells) for 48 h and imaged by microscopy. (F) 293 cells were transfected with HA-ERIS or HA-TRIF, along with indicated RNAi plasmids. Forty-eight hours later, cells were analyzed by immunoblotting with anti-HA. (G) 293 cells were transfected with a control or ERIS RNAi plasmid (15# or 17#) or with a nonspecific RNAi plasmid (23#), along with 50 ng of IFN-β–Luc reporter plasmid. Twenty-four hours after transfection, 1 μg/mL poly(dA:dT) (Left) or poly(I:C) (Center) was transfected for 18 h, or cells were treated with 200 μg/mL poly(I:C) for 5 h (Right). Cells were analyzed by a reporter assay. Data represent mean ± SD. Similar results were obtained in 3 independent experiments.
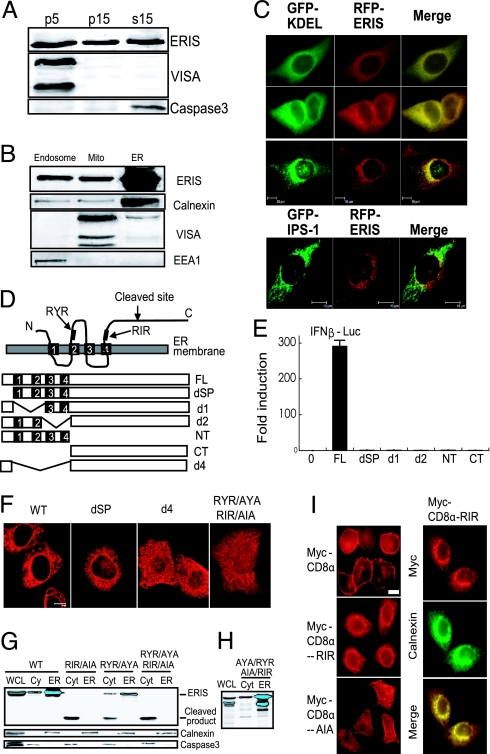
Fig. 3.
ERIS protein localized to ER membrane and ER retention signal are critical for localization. (A) HT1080 cells (1 × 107) were homogenized in an isotonic buffer and separated into pellet (P) by centrifugation at 5,000 × g (p5), and the supernatant was further centrifuged at 15,000 × g to get p15 and supernatant S15. Different fractions were analyzed with antibody against ERIS, VISA, and caspase-3. (B) HT1080 cells were homogenized and then fractionated by discontinuous sucrose gradient ultracentrifugation. Purified fractions were analyzed with antibody against calnexin (ER), IPS-1/VISA (mitochondrial membrane), and EEA1 (endosome). (C) HeLa cells were cotransfected with red fluorescent protein (RFP)-ERIS and GFP-KDEL (rows 1, 2, and 3) or GFP-VISA (row 4). Cells were fixed and then imaged by fluorescence microscopy (rows 1 and 2) or confocal microscopy (rows 3 and 4). (D) Diagram of ERIS putative ER retention/retrieval sequences and deletions and truncations. Signal peptide (SP): 1–18aa, TM1: 21–37aa, TM2: 48–67aa, TM3: 112–135aa, TM4: 158–175aa, RYR: 76–78aa, RIR: 178–180aa. (E) One hundred nanograms of indicated ERIS mutants was transiently transfected into 293 cells, along with 50 ng of IFN-β–Luc plasmid. Twenty-four hours later, a reporter assay was performed. (F) HeLa cells were transfected with indicated plasmids and then fixed and imaged by confocal microscopy. (G) 293 cells transfected with full-length ERIS or indicated ERIS mutants were subjected to fraction isolation. Fractions were analyzed with antibodies against ERIS, calnexin, or caspase-3. Cyt, cytosol; WCL, whole-cell lysate. (H) The same as G except that the reverted ERIS (AYA/AIA to RYR/RIR) was transfected into 293 cells and the localization of ERIS was analyzed by Western blotting. (I) HeLa cells were transfected with Myc-CD8α, Myc-CD8α-RIR, or Myc-CD8α-AIA. Transfected cells were labeled with antibody against Myc (Left) or were transfected with Myc-CD8α-RIR and labeled with the indicated antibodies (Right). All images are representative of at least 3 independent experiments. (Scale bar: 10 μm.)
It was believed that during viral infection, type I IFNs protect cells against viral invasion by interfering with the replication of viruses. As an inducer of type I IFNs, overexpressed ERIS depressed the synthesis of VSV G protein after VSV infection, compared with PCBP1 as a negative control (Fig. 2D). Similarly, 293 cells expressing ERIS were highly resistant to Newcastle disease virus (NDV)-GFP infection (Fig. 2E). These data, together with results in Fig. 1, further confirmed the antiviral activity of ERIS attributable to overexpression-induced type I IFN production. To clarify the involvement of endogenous ERIS in innate immunity, an RNAi approach was used to knock down ERIS expression in human cells (Fig. 2F). Suppression of ERIS expression by 15# and 17# ERIS-RNAi significantly reduced the IFN-β induction stimulated by either poly(dA:dT) or poly(I:C) transfection but not that stimulated by poly(I:C) treatment (Fig. 2G). Higher viral susceptibility to NDV infection was also observed in cells transfected with ERIS-RNAi (Fig. 2E). This indicates that ERIS may exert its immunoactivity in both cytoplasmic dsDNA- and dsRNA-triggered intracellular signaling pathways.
ERIS Localizes to ER Membrane.
Protein structural analysis implied that ERIS was a membrane-bound protein with 4 putative TM domains. We used microscopy to image HeLa cells transfected with HA-tagged ERIS. Observations on fluorescent immunostaining with anti-HA eliminated the possibility of plasma membrane distribution but showed protein accumulation in a perinuclear pattern (Fig. 3C), suggesting that ERIS might localize to intracellular membrane-coated compartments. To investigate its localization, membrane components were roughly isolated by differential centrifugation at 5,000 × g and 15,000 × g (8). Immunoblotting data showed that ERIS was found universally in P5, P15, and S15, obviously different from mitochondrial protein IPS-1 and cytosolic protein caspase-3 (Fig. 3A). We carried out refined subcellular fractionation (15, 16). HT1080 cells were homogenized in an isotonic buffer and subjected to sucrose gradient ultracentrifugation to separate and purify further endosomes, mitochondria, and ER. Our data showed that endogenous ERIS existed predominantly in purified ER fraction (Fig. 3B). This was also confirmed by fluorescence microscopy.
To determine further the region responsible for ER localization, we constructed a series of ERIS truncations and deletions (Fig. 3D). The capability of truncations and deletions of ERIS to induce type I IFN was tested. All these mutants failed to up-regulate IFN-β production (Fig. 3E). Confocal microscopy showed that ERIS mutants lacking the 4 TM domains (d4 and CT) were dispersed in the cellular cytoplasm, or even accumulated in the nucleus, whereas localization of the signal peptide deleted (dSP) appears similar to full-length ERIS (Fig. 3F). Interestingly, cloning of ERIS-dSP into a pCMV8 vector containing an ER signal peptide fully restored IFN-β induction (Fig. S4). Neither absence of the first 2 TM domains (d1) nor absence of the last 2 TM domains (d2) altered its localization (data not shown).
In addition to KDEL and KKXX (Lys-Lys-X-X), the well-known ER retention signals in most luminal ER proteins (17–19), it is widely accepted that RXR is the minimal motif of resident ER proteins for ER retention/retrieval that functions in a cytoplasmic loop of ER proteins (20). We found that ERIS contains a RYR at position 76–78aa (between TM2 and TM3) and an RIR at position 178–180aa (right after TM4) (Fig. 3D). Replacing the R residues with A residues in either (AYA or AIA) or both (AYA/AIA) of the 2 motifs reduced or eliminated the ER expression of ERIS, as shown by confocal microscopy (Fig. 3F) and also in fractionation experiments followed by immunoblotting with anti-ERIS (Fig. 3G). RIR appears to be more important for ER retention than RYR. Interestingly, in the cytosol fraction isolated from the RXR mutants, a lower band with the size of about 24 kDa was specifically detected by either anti-ERIS antibody or anti-FLAG antibody (Fig. 3G). Because the anti-ERIS antibody we made recognizes the C-terminus of ERIS (219–379aa) and the ERIS expression construct was 3′-FLAG–tagged, these data implied the cleavage of ERIS at the N-terminus. To confirm that the cleavage and relocation of ERIS-AYA/AIA was caused by the mutation, AYA/AIA was reverted to its WT version, RYR/RIR, by site-directed mutagenesis; the normal ER localization of ERIS was observed (Fig. 3H). Interestingly, similar cleaved products were also detected from d4 and CT but not from d1, d2, or dSP (data not shown), indicating that not-ER-localized-ERIS was destined for cleavage. To ascertain that RIR can function as an ER retention signal, the rat CD8α subunit, which shows distinct surface expression, was used as a model molecule to test whether this RIR sequence could induce ER localization (21). It was found that the transfected Myc-CD8α was exclusively present on the cell surface. However, when the RIR sequence (ELQA_RIR_TYN) was added to the C-terminus of Myc-CD8α, the transfected Myc-CD8α-RIR was not distributed on the cell surface but displayed a perinuclear pattern, representing a typical morphological pattern for ER localization. As expected, when the AIA sequence (ELQA_AIA_TYN) was added to the C-terminus of Myc-CD8α, the cell surface localization of Myc-CD8α was fully restored (Fig. 3I). These results clearly indicate that the RIR motif of ERIS functions as an ER retention/retrieval signal.
ERIS Is Modified on Overexpression or Innate Immune Challenges.
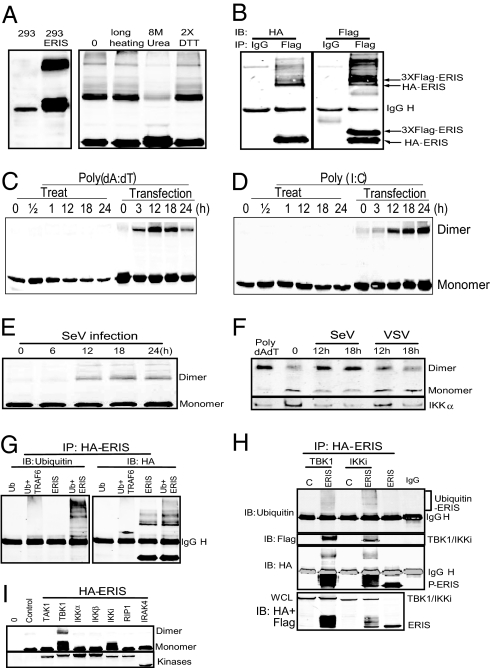
It was noteworthy that in cells transfected with ERIS, the antibody could detect 2 bands with the apparent molecular masses of 42 kDa and about 80 kDa with smear above it, which was dissolved to the 42-kDa band only when 8 M urea was added to the cell lysis buffer (Fig. 4A), indicating that the 80-kDa band was the dimer of ERIS. This was further confirmed by coimmunoprecipitation experiments (Fig. 4B). Interestingly, ERIS dimerization was induced when cells were transfected with but not treated with poly(dA:dT) or poly(I:C) by direct addition to the medium (Fig. 4 C and D). Sendai virus (SeV) infection showed similar results (Fig. 4E). We next tested the dimerization of endogenous ERIS by a native PAGE gel. The induced dimerization was detected by the anti-ERIS antibody after SV infection or poly(dA:dT) transfection but not after VSV infection, suggesting that ERIS was not activated in HT1080 cells after VSV infection (Fig. 4F). Furthermore, a coimmunoprecipitation experiment indicated that ERIS could be ubiquitinated, and the ubiquitination took place only on the dimerized ERIS (Fig. 4G).
Fig. 4.
ERIS protein is modified on overexpression or innate immune challenge. (A) 293 cells (293) or 293 cells transfected with HA-ERIS (293-ERIS; Right) were lysed and immunoblotted with anti-HA. (Right) Cell lysates were heated with SDS buffer containing 200 μM DTT for 10 min (0) or 30 min (prolonged heating), heated with SDS buffer containing 400 μM DTT (2× DTT), or heated after addition of 8 M urea into lysis buffer (8 M urea). (B) 293 cells were cotransfected with 3× Flag-ERIS and HA-ERIS. After 24 h, cell lysates underwent immunoprecipitation (IP) with anti-Flag, followed by immunoblotting (IB) with anti-HA and then anti-Flag. 293 cells stably expressing TLR3 were transfected with HA-ERIS. Thirty-six hours later, cells were treated or transfected with poly(dA:dT) (C) or poly(I:C) (D) for indicated times. Cells were lysed and immunoblotted with anti-HA. (E) 293 cells were transfected with HA-ERIS; 36 h later, cells were infected with Sendai virus (SeV) for indicated times, lysed, and immunoblotted with anti-HA. (F) HT1080 cells were infected with SeV or VSV for indicated times, and ERIS dimerization was detected by native gel electrophoresis using antibody against ERIS. (G and H) 293 cells were transfected with HA-ERIS together with empty vector (vector) or Flag-tagged expression plasmids as indicated. After 24 h, cell lysates underwent IP with anti-HA, followed by IB with indicated antibodies. (H, Lower) The whole-cell lysate (WCL) was immunoblotted with anti-HA and anti-Flag antibodies together. (I) 293 cells were transfected with HA-ERIS together with empty vector (control) or Flag-tagged expression plasmids as indicated. After 24 h, cell lysates were directly separated and immunoblotted with anti-HA and anti-Flag antibodies.
As mentioned previously, ERIS might participate in both cytoplasmic dsDNA- and dsRNA-triggered intracellular signaling pathways (Fig. 2G). Although the 2 pathways discriminated in the upstream, they converged to TBK1/IKKi to induce the activation of IRF3 (22–24). To determine whether TBK1/IKKi or IRF3 could interact with ERIS, TBK1/IKKi or IRF3 was cotransfected with ERIS into 293 cells. Coimmunoprecipitation results showed that ERIS could interact with TBK1 and IKKi but not with IRF3 (Fig. 4H and Fig. S5). Interestingly, TBK1 or IKKi overexpression led to remarkable ERIS monomer band shift and also to the dimer formation (Fig. 4H), suggesting hyperphosphorylation of the protein. In contrast, other kinases related to activation of NF-κB or type I IFNs, including IKKα, IKKβ, TAK1, RIP1, IRAK1/4, and JNK1/2, could not trigger the shift (Fig. 4I and data not shown).
ERIS Dimerization, Mediated Through Its TM Domains, Leads to Type I IFN Induction.
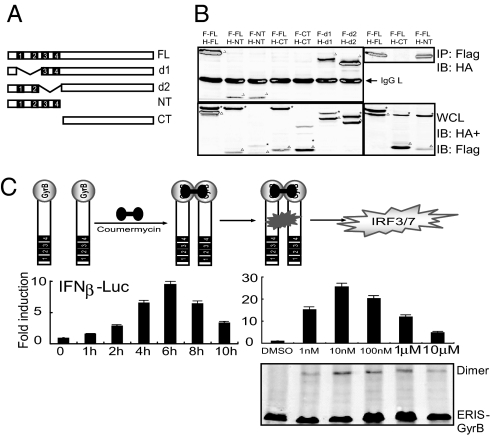
The obvious dimerization of ERIS prompted us to conduct further research. To identify domains necessary for its dimerization, truncations and deletions of ERIS were used, and the coimmunoprecipitation result showed that either Flag-tagged mutant lacking the former 2 TM domains (d1) or the latter 2 TM domains (d2) could interact with their HA-tagged counterparts (Fig. 5 A and B). The N-terminus of ERIS containing the intact TM domains (NT) could form NT homodimer and NT-FL heterodimer as well (Fig. 5B). Only absence of all 4 TM domains (d4) abolished the dimerization (Fig. 5B). These data show that the TM domains of ERIS mediate its dimerization.
Fig. 5.
ERIS dimerization is mediated by its TM domains and leads to type I IFN induction. (A) Diagram of ERIS deletions and truncations used. (B) Indicated expression plasmids (F, Flag-tagged; H, HA-tagged) were cotransfected into 293 cells. Twenty-four hours later, cell lysates underwent immunoprecipitation (IP) with anti-Flag, followed by immunoblotting with anti-HA and then anti-Flag (Upper), or whole-cell lysate (WCL) was immunoblotted with anti-HA and then anti-Flag (Lower). *, Flag-tagged protein; filled triangle, HA-tagged protein. (C) 293 cells were transiently transfected with ERIS-GyrB. After 24 h, coumermycin was added into cell culture medium for the indicated times (Left) or at increasing concentrations (Right). A luciferase assay was performed. Similar results were obtained in 3 independent experiments. Cell lysates were also separated by a native gel, and the coumermycin-induced ERIS-GyrB dimerization was analyzed by the anti-ERIS antibody (Lower).
To determine further whether dimerization was responsible for the ability of ERIS to induce type I IFN production, we took advantage of a previously developed coumermycin-induced chemical dimerization system (25, 26). Streptomyces product coumermycin binds to the B subunit of bacterial DNA GyrB with a stoichiometry of 1:2, creating natural dimerization of GyrB. We created a chimeric ERIS fusion protein in which the GyrB subunit was linked to the C-terminus of ERIS. Activation of IFN-β promoters by ERIS-GyrB fusion protein was assessed after addition of coumermycin. Reporter assay showed that passive dimerization of ERIS induced activation of IFN-β promoter, and the induction reached its peak at 6 h after coumermycin treatment at a concentration of 10 nM (Fig. 5C). Therefore, we concluded that dimerization of ERIS caused self-activation and subsequent downstream signaling, uncovering the mechanism underlying the antiviral activity of ERIS in innate immunity.
Discussion
It is believed that cells use 2 separate pathways responding to cytosolic non–self- dsRNA and -dsDNA. A protein shared by both pathways is yet to be found, except for the very downstream components like TBK1/IKKi and IRFs. Here, we report the identification and characterization of an ER protein, ERIS, that plays a pivotal role in innate response to both RNA and dsDNA in the cytoplasmic compartment of the host cells. Recently, the same molecule has been reported as “STING” by Ishikawa and Barber (27), as “MITA” by Zhong et al. (28) from Shu's group, and as “MPYS” by Jin et al. (29) from Cambier's laboratory. The function and localization of the same protein were different and controversial. In this work, we provide evidence about this protein's localization and a possible molecular mechanism for its function.
We found that (i) ERIS caused very strong type I IFN production; (ii) ERIS resided on ER membrane, and the ER retention/retrieval sequence RIR was critical to retain the protein on ER membrane and to maintain its integrity; (iii) ERIS was dimerized on innate immune challenges; and (iv) coumermycin-induced ERIS dimerization led to strong and fast IFN induction.
The modification of ERIS on innate immune challenges was closely linked to its activity to induce type I IFN production. One of the most surprising findings in our study was the induction of ERIS dimerization. It occurred when ERIS was overexpressed alone or with TBK1/IKKi or when cells were infected by virus or transfected with dsDNA or dsRNA, all of which led to strong IFN induction. The induced ERIS dimerization was not detected after VSV infection, suggesting that VSV might have ways to circumambulate the activation of cells. It was well known that VSV could infect many human cells, including 293 cells without IRF3 and NF-κB activation.
In many cases, protein oligomerization could lead to self-activation and downstream activation, such as TNF receptors (30), TRIF (31), and MyD88 (32). It was therefore hypothesized that oligomerization or dimerization of ERIS was responsible for the activation of downstream signaling. Actually, we did observe that coumermycin-induced dimerization of ERIS-GyrB fusion protein led to rapid and strong IFN production. This result clearly demonstrates that dimerization of ERIS, induced by innate immune challenges, leads to the activation of ERIS and, in turn, the downstream components, although the details remained unknown.
The ER membrane localization of ERIS was critical for its normal function. However, although d1, d2, or dSP was found to be expressed on ER membrane, none of them activated type I IFN production, suggesting that ER localization was required but not sufficient for its activity. More interestingly, a pre-protrypsin leader sequence from the expression vector fully restored the activity of ERIS to induce type I IFN production. This hydrophobic sequence was known to direct the nascent polypeptide of pre-protrypsin across the membrane of the ER (33). On the other hand, deletion of 4 TM domains (d4 or CT) or mutations on ER retention/retrieval signals altered distribution of protein in the cells. Our results provided direct evidence that ERIS is indeed an ER resident protein.
There are 2 ER retention/retrieval sequences discovered in ERIS. We showed that the RXR motif of ERIS not only functions as an ER retention/retrieval signal but is critical for protein integrity. Further investigation is required on what or which protease might be responsible for the cleavage of ERIS when it failed to be retained on the ER membrane. The size of the cleaved fragment was about 220 aa long from the C-terminus. This information would help us to analyze at which site the protein was cleaved by the protease.
Materials and Methods
Plasmids, strains, and antibodies used and detailed experimental procedures can be found in SI Text.
Large-Scale Screening and Luciferase Reporter Assay.
Escherichia coli DH5α was transformed with plasmid DNA of mouse cDNA library from bone marrow-derived macrophages treated with VSV for 8 h. Plasmid DNA was miniprepared for use as a cDNA pool. HEK293 cells were transiently transfected with 0.5 μg of pooled cDNA, along with 50 ng of the IFN-β promoter–Luc construct using standard calcium phosphate precipitation. After 24 h, cells were lysed in reporter lysis buffer, and the luciferase activity in the total cell lysate was measured with the Dual-Luciferase Reporter Assay System (Promega). As an internal control, 50 ng of TK-Renilla reporter gene was cotransfected simultaneously. Positive pools were then picked and subdivided as described previously to isolate a single clone responsible for the activity of the pool. Positive clones were sequenced and characterized by BLAST research.
RT-PCR.
Total RNA was extracted with TRIzol reagent (TianGen) and was reverse-transcribed with the Reverse Transcription System (Promega). Type I IFN induction was analyzed by RT-PCR for 28–30 cycles at 94°°C for 30 s, 58 °C for 30 s, and 72 °C for 40 s.
Type I IFN Bioassay.
Type I IFN activity was measured as previously described (34), with reference to a recombinant human IFN-β (R & D Systems) standard using a 2FTGH cell line (1 × 105 cells/mL) stably transfected with an IFN-sensitive (ISRE) luciferase construct.
RNA Interference.
Double-stranded oligonucleotides corresponding to the target sequences were cloned into the pSuper.Retro RNAi plasmid (Oligoengine, Inc.). The sequences targeting human ERIS were as follows: 15#, 5′-CATTCGCTTCCTGGATAAA-3′; 17#, 5′-GGATCGGGTTTACAGCAAC-3′; 23#, 5′-CAACTGCCGCCTCATTGCC-3′; 4#, 5′-CATCCTCCTGGGCCTCAAG-3′; and 7#, 5′-AGGGAATTTCAACGTGGCC-3′.
Fraction Isolation.
Mitochondrial and ER membranes were purified on discontinuous sucrose gradients as previously described, with some modifications (15). Briefly, cells in MTE buffer [0.27 M mannitol, 10 mM Tris-HCl, 0.1 mM EDTA (pH 7.4), 0.1 mg/mL leupeptin] were subjected to homogenization. After 40 strokes, cell homogenate was centrifuged at 700 × g for 10 min at 4 °C and the postnuclear solution (PNS) was saved. The pellet was resuspended by MTE buffer, homogenized, and centrifuged again. The 2 PNSs were collected for centrifugation at 15,000 × g for 10 min. The pellet was resuspended in MTE buffer, layered on discontinuous sucrose gradients (1.0M and 1.5 M sucrose in 10 mM Tris-HCl, pH 7.5) and centrifuged at 60,000 × g for 20 min. The mitochondrial and endosomal fraction was collected and pelleted by centrifugation at 17,000 × g for 15 min. Purified membranes were resuspended in PBS and prepared for Western blot analysis. To isolate ER fractions, postmitochondrial supernatant was layered on discontinuous sucrose gradients (1.3 M, 1.5 M, and 2.0 M sucrose in 10 mM Tris-HCl, pH 7.6) and centrifuged at 87,000 × g for 90 min. The ER fraction at the interface between the supernatant and the 1.3 M sucrose was collected and pelleted by centrifugation at 87,000 × g for 45 min. Purified ER membranes were resuspended in PBS and prepared for Western blot analysis.
Immunofluorescent Microscopy.
HeLa cells grown on glass coverslips were transfected with plasmids as indicated. For high efficiency of transfection, cells were shocked for 1 min with glycerol 4 h after transfection. After 24 h, cells were washed with PBS once and fixed in cold methyl alcohol for 15 min. Cells were then mounted onto slides directly or incubated with primary antibody and secondary antibody. Imaging of the cells was carried out using Olympus BX51 microscopy or a Leica TCS SP2 confocal system under a ×100 oil objective.
Coimmunoprecipitation, Immunoblot Analysis, and Native PAGE.
293 cells seeded on 10-cm2 dishes (1 × 106 cells/dish) were transfected with a total of 10 μg of empty plasmid or various expression plasmids. At 24–36 h after transfection, cells were lysed in lysis buffer (0.5% Triton-X-100, 150 mM NaCl, 12.5 mM β-glycerolphosphate, 1.5 mM MgCl2, 2 mM EGTA, 10 mM NaF, 1 mM Na3VO4, 2 mM DTT) containing protease inhibitors. Lysates were centrifuged and incubated with Protein A/G Sepharose (Amersham) and anti-HA or anti-Flag antibodies for at least 4 h. The beads were washed with cold PBS 4 times and eluted with DTT-containing SDS sample buffer by boiling for 10 min. Native PAGE was performed as previously described (35). Protein samples were prepared in loading buffer containing 0.125 mM Tris-HCl (pH 6.8), 30% (vol/vol) glycerol, and 2% (weight/vol) deoxycholate and were then loaded and subjected to electrophoresis at 25 mA for 1 h on ice.
Supplementary Material
Supporting Information
Acknowledgments.
We thank Dr. Lan Bao, and Dr. Xueliang Zhu (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for critical discussion and technical assistance, Lan Bao for Myc-CD8α plasmid, Chen Wang for NDV-GFP virus, and Ganes C. Sen for p56-Luc plasmid. This work was supported by grants from the National Natural Science Foundation of China (Grant 30772024), National Basic Research Program of China (Grant 2007CB914502), and Key Project of Chinese Ministry of Education (Grant 108002).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Kumagai Y, Takeuchi O, Akira S. Pathogen recognition by innate receptors. J Infect Chemother. 2008;14:86–92. doi: 10.1007/s10156-008-0596-1. [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato H, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 6.Kawai T, et al. An adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 7.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 8.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 11.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 12.Sun Q, et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Hoebe K, et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 15.Bozidis P, Williamson CD, Colberg-Poley AM. Current Protocols in Cell Biology. New York: Wiley; 2007. Isolation of endoplasmic reticulum, mitochondria, and mitochondria-associated membrane fractions from transfected cells and from human cytomegalovirus-infected primary fibroblasts. 37-3.27.1-3.27.23. [DOI] [PubMed] [Google Scholar]
- 16.Vance JE. Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem. 1990;265:7248–7256. [PubMed] [Google Scholar]
- 17.Pelham HR. Using sorting signals to retain proteins in endoplasmic reticulum. Methods Enzymol. 2000;327:279–283. doi: 10.1016/s0076-6879(00)27283-2. [DOI] [PubMed] [Google Scholar]
- 18.Teasdale RD, Jackson MR. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu Rev Cell Dev Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- 19.Townsley FM, Pelham HR. The KKXX signal mediates retrieval of membrane proteins from the Golgi to the ER in yeast. Eur J Cell Biol. 1994;64:211–216. [PubMed] [Google Scholar]
- 20.Zerangue N, Schwappach B, Jan YN, Jan LY. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 21.Zhang ZN, et al. The voltage-gated Na+ channel Nav1.8 contains an ER-retention/retrieval signal antagonized by the beta3 subunit. J Cell Sci. 2008;121:3243–3252. doi: 10.1242/jcs.026856. [DOI] [PubMed] [Google Scholar]
- 22.Ishii KJ, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 23.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 24.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 25.Farrar MA, Alberol I, Perlmutter RM. Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature. 1996;383:178–181. doi: 10.1038/383178a0. [DOI] [PubMed] [Google Scholar]
- 26.Fujii H. Receptor expression is essential for proliferation induced by dimerized Jak kinases. Biochem Biophys Res Commun. 2008;370:557–560. doi: 10.1016/j.bbrc.2008.03.095. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Jin L, et al. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wells JA. Structural and functional basis for hormone binding and receptor oligomerization. Curr Opin Cell Biol. 1994;6:163–173. doi: 10.1016/0955-0674(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 31.Funami K, Sasai M, Oshiumi H, Seya T, Matsumoto M. Homo-oligomerization is essential for Toll/interleukin-1 receptor domain-containing adaptor molecule-1-mediated NF-kappaB and interferon regulatory factor-3 activation. J Biol Chem. 2008;283:18283–18291. doi: 10.1074/jbc.M801013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hacker H, et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439:204–207. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- 33.Blobel G, Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang Z, et al. CD14 is required for MyD88-independent LPS signaling. Nat Immunol. 2005;6:565–570. doi: 10.1038/ni1207. [DOI] [PubMed] [Google Scholar]
- 35.Iwamura T, et al. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: Common and unique pathways. Genes Cells. 2001;6:375–388. doi: 10.1046/j.1365-2443.2001.00426.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information