Neurodegenerative diseases target large-scale human brain networks (original) (raw)
. Author manuscript; available in PMC: 2010 Apr 16.
Abstract
During development, the healthy human brain constructs a host of large-scale, distributed, function-critical neural networks. Neurodegenerative diseases have been thought to target these systems, but this hypothesis has not been systematically tested in living humans. We used network-sensitive neuroimaging methods to show that five different neurodegenerative syndromes cause circumscribed atrophy within five distinct healthy human intrinsic functional connectivity networks. We further discovered a direct link between intrinsic connectivity and gray matter structure. Across healthy individuals, nodes within each functional network exhibited tightly correlated gray matter volumes. The findings suggest that human neural networks can be defined by synchronous baseline activity, a unified corticotrophic fate, and selective vulnerability to neurodegenerative illness. Future studies may clarify how these complex systems are assembled during development and undermined by disease.
Introduction
Recent functional magnetic resonance imaging (fMRI) advances have helped researchers delineate the human brain's intrinsic functional network architecture (Fox and Raichle, 2007; Fox et al., 2005; Fransson, 2005; Greicius et al., 2003; Seeley et al., 2007). These studies have shown that, during task-free conditions, correlated spontaneous activity occurs within spatially distinct, functionally related groups of cortical and subcortical regions (Beckmann et al., 2005; Seeley et al., 2007; Vincent et al., 2007). As predicted by principles governing network-based synaptic physiology (Bi and Poo, 1999; Katz and Shatz, 1996), regions with synchronous baseline activity feature direct or indirect anatomical connections (Greicius et al., 2008; Seeley et al., 2007; Vincent et al., 2007). Blood oxygen level dependent (BOLD) signal fluctuations within these intrinsic connectivity networks (ICNs) occur at low frequencies (0.01-0.08 Hz), exist in non-human primates, and continue during general anesthesia and sleep, suggesting that ICNs cannot be explained by ongoing conscious mentation alone (Fox and Raichle, 2007). On the other hand, ICNs remain detectable during mental effort (Fransson, 2006), and ICN strength and variation influence task performance inside (Fox et al., 2007; Hesselmann et al., 2008) and outside (Seeley et al., 2007) the scanner. How ICN patterns and fluctuations relate to gray matter structure in health and disease, however, remains unknown.
Neurodegenerative diseases cause progressive, incapacitating cognitive, behavioral, and motor dysfunction. Early on, misfolded disease proteins aggregate within small, selectively vulnerable neuron populations that reside in specific brain regions (Graveland et al., 1985; Hyman et al., 1984; Seeley et al., 2006). Synapses falter, and damage spreads to new regions, accompanied by worsening clinical deficits (Selkoe, 2002). Often, later-affected regions bear known anatomical connections with the sites of earlier injury (Seeley et al., 2008a). Based on neuropathology (Braak and Braak, 1991), neuroimaging (Buckner et al., 2005; Greicius et al., 2004), and evidence from transgenic animal models (Palop et al., 2007), some authors have suggested that neurodegeneration may relate to neural network dysfunction (Buckner et al., 2005; Palop et al., 2006). In human spongiform encephalopathies, which cause rapidly progressive dementia through conformational changes in misfolded prion protein, direct evidence supports disease propagation along transsynaptic connections (Scott et al., 1992). For all other neurodegenerative diseases, limited human experimental data support the “network degeneration hypothesis”. If demonstrated as a class-wide phenomenon, however, this framework would have major mechanistic significance, predicting that the spatial patterning of disease relates to some structural, metabolic, or physiological aspect of neural network biology. Confirming the network degeneration hypothesis would also have clinical impact, stimulating development of new network-based diagnostic and disease-monitoring assays.
To test the network degeneration hypothesis in living humans, we studied patients with five distinct neurodegenerative syndromes and two healthy control groups (Figure 1). Only early age-of-onset dementia syndromes were included, enabling us to better match patient groups for age and other demographic variables (Table S1). Patients were diagnosed with Alzheimer's disease (AD, n = 24), behavioral variant frontotemporal dementia (bvFTD, n = 24), semantic dementia (SD, n = 24), progressive nonfluent aphasia (PNFA, n = 13), or corticobasal syndrome (CBS, n = 17), based on standard research criteria. Diagnoses were made on clinical grounds, therefore neuroimaging and pathological data did not influence group membership. To weight our analyses toward the distinctive, early-stage neuroanatomic features of each syndrome, we excluded patients with moderate or severe dementia (defined by Clinical Dementia Rating scale scores > 1). After defining the patient groups, we conducted a series of quantitative structural and functional imaging analyses in patients and controls to test the hypothesis that syndrome-associated regional degeneration patterns reflect distinct human neural network architectures.
Figure 1. Study design schematic.
Patient groups were compared to HC1 subjects to determine syndromic atrophy patterns. From these maps, distinct seed ROIs were extracted (see Table S2) and used in functional (HC2) and structural (HC1) correlation analyses. These experiments determined the functional intrinsic connectivity networks (ICNs 1-5) and structural covariance networks (SCNs 1-5) associated with each of the five syndrome-related seeds. ICN and SCN maps were then compared to all five syndromic atrophy maps to derive GOF scores, yielding 50 total scores ((5 ICN maps×5 atrophy maps) + (5 SCN maps×5 atrophy maps)), which are summarized in Figure 5.
Results
Each neurodegenerative syndrome features a distinct regional vulnerability pattern
First, we established each syndrome's functional and anatomical deficit profiles compared to 65 healthy, age-matched controls (Table S1 and Figure 2A). A standard neuropsychological battery was administered, and MR voxel-based morphometry (VBM) facilitated whole-brain statistical parametric grey matter comparisons between each patient group and controls. The findings replicated previous work, performed by our group and others, on the five syndromes (Boccardi et al., 2005; Gorno-Tempini et al., 2004; Josephs et al., 2006; Josephs et al., 2008; Seeley et al., 2005; Seeley et al., 2008a). In summary, AD was associated with episodic memory dysfunction and prominent medial temporal, posterior cingulate/precuneus, and lateral temporoparietal atrophy. BvFTD, SD, and PNFA, which together make up the clinical frontotemporal dementia (FTD) spectrum, each showed a unique deficit signature. BvFTD featured prominent behavioral deficits with anterior cingulate, frontoinsular, striatal, and frontopolar degeneration. SD resulted in loss of word and object meaning accompanied by left-predominant temporal pole and subgenual cingulate involvement. PNFA presented with nonfluent, effortful, and agrammatic speech and was associated with left frontal operculum, dorsal anterior insula, and precentral gyrus atrophy. Patients with CBS had prominent, asymmetric sensorimotor impairment, with akinesia, rigidity, apraxia, and cortical sensory loss or other cortical cognitive deficits; accordingly, CBS gray matter loss was confined to dorsal frontoparietal sensorimotor association areas, primary motor and sensory cortices, and dorsal insula. The early-stage, syndrome-specific anatomical patterns provided seed regions for our subsequent network analyses in healthy controls.
Figure 2. Convergent syndromic atrophy, healthy ICN, and healthy structural covariance patterns.
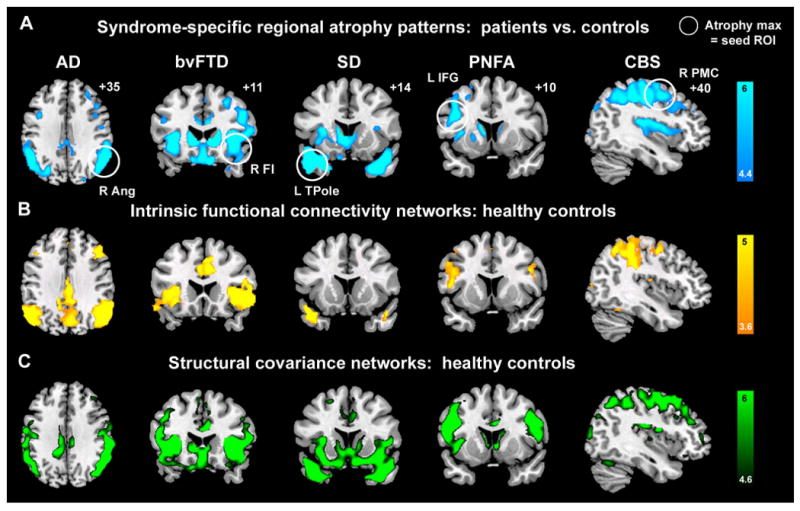
(A) Five distinct clinical syndromes showed dissociable atrophy patterns, whose cortical maxima (circled) provided seed ROIs for ICN and SC analyses. (B) ICN mapping experiments identified five distinct networks anchored by the five syndromic atrophy seeds. (C) Healthy subjects further showed gray matter volume covariance patterns that recapitulated results shown in (A) and (B). For visualization purposes, results are shown at P < 0.00001 uncorrected (A & C) and P < 0.001 corrected height and extent thresholds (B). In A-C, results are displayed on representative sections of the MNI template brain. In coronal and axial images, the left side of the image corresponds to the left side of the brain. ANG = angular gyrus; FI = frontoinsula; IFGoper = inferior frontal gyrus, pars opercularis; PMC = premotor cortex; TPole = temporal pole.
Syndromic atrophy foci anchor large-scale functional networks in the healthy brain
The network degeneration hypothesis predicts that syndromic atrophy patterns should recapitulate healthy functional network architectures. To evaluate this possibility, we identified the most atrophied cortical region in each patient group (Figure 2A, Table S2) and used these regions of interest (ROIs) to seed ICN mapping experiments in a separate group of 19 healthy controls (HC2). These controls, also age-matched to the patient groups, underwent six minutes of task-free fMRI scanning. From these data, we extracted the mean BOLD signal timeseries from the five syndrome-associated ROIs and entered these timeseries into five separate whole brain intrinsic functional connectivity analyses. The resulting ROI-based network maps then served as spatial templates for independent component analysis (ICA), following previous approaches (Greicius et al., 2004; Seeley et al., 2007). Next, we identified a best-fit ICA-generated component for each network template for each subject and combined these components to produce group-level network maps for each seed ROI. As anticipated, the five disease-vulnerable ROIs anchored five distinct ICNs in healthy controls (Figure 2B). Remarkably, as predicted by the network degeneration hypothesis, these distributed network maps, though generated from isolated cortical seed ROIs, closely mirrored the atrophy patterns seen in the five neurodegenerative syndromes (Figures 2 and 3).
Figure 3. Detailed network mapping of the right frontal insula, a focus of neurodegeneration in bvFTD.
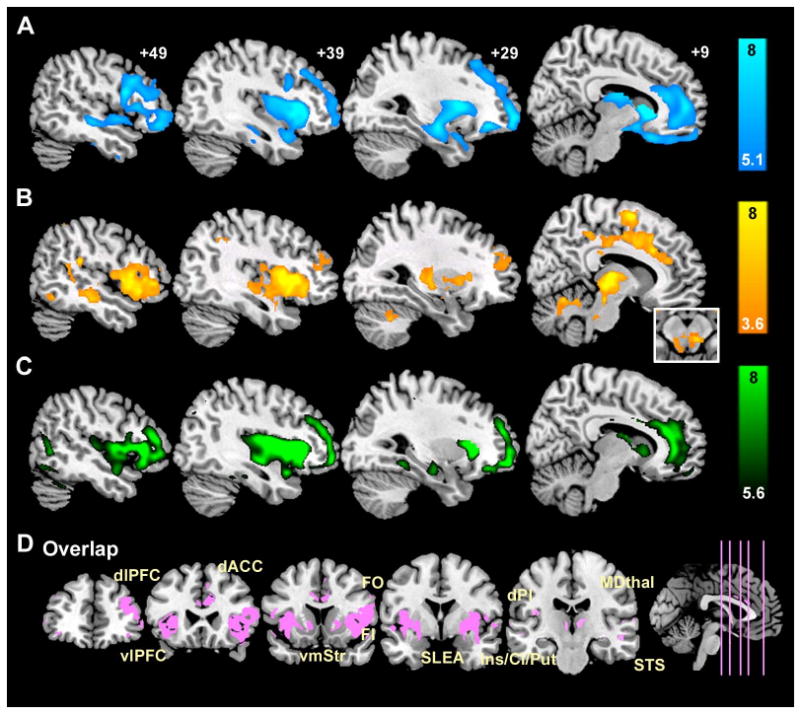
(A) Reduced gray matter volume in bvFTD vs. controls (P < 0.05, whole brain FWE corrected) occurs within regions showing (B) intrinsically correlated BOLD signals in controls (P < 0.001, whole-brain corrected height and extent thresholds) and (C) structural covariance in controls (P < 0.05, whole brain FWE corrected). These distributed spatial maps overlap (D) within a “network” that reflects known primate neuroanatomical connections. AI = anterior insula, dACC = dorsal anterior cingulate cortex, dlPFC = dorsolateral prefrontal cortex, dPI = dorsal posterior insula, FO = frontal operculum, MDthal = mediodorsal thalamus, SLEA = sublenticular extended amygdala, vlPFC = ventrolateral prefrontal cortex, vmStr = ventromedial striatum.
Normal structural covariance patterns mirror intrinsic functional connectivity
Physiological studies have shown that synchronous neuronal firing promotes network-based synaptogenesis (Bi and Poo, 1999; Katz and Shatz, 1996). Therefore, we further questioned whether coherent spontaneous ICN activity might impact normal cortical structure. Specifically, we hypothesized that functionally correlated brain regions would show correlated gray matter volumes across healthy subjects. One previous VBM study, though not designed to assess the relationship between functional connectivity and structure, selected landmark-based cortical and limbic ROIs and found group-level gray matter density correlations between these ROIs and homologous contralateral and functionally related ipsilateral regions (Mechelli et al., 2005). We adapted these methods to study structural covariance patterns arising from disease-vulnerable foci, applying the same seed ROIs (Figure 2A, Table S2) used to probe our intrinsic functional connectivity data. Local ROI mean gray matter intensities extracted from the five seeds provided covariates for five separate whole brain statistical parametric regression analyses in which age and gender were entered as nuisance covariates. These studies revealed striking convergence between healthy intrinsic functional connectivity, derived within subjects (Figures 2B, 3B, 4A-B), and structural covariance, assessed across subjects (Figures 2C, 3C, 4C-D). As a result, our three data streams converged (Figures 5-6). That is, normal ICN and structural covariance patterns mirrored each other and reflected, with high fidelity, those regions that co-degenerate in distinct human neurodegenerative syndromes. BvFTD was chosen to highlight convergence of the three maps in greater detail (Figure 3). ROI functional timeseries from a representative control subject (Figure 4A-B) and related group-level structural correlation plots (Figure 4C-D) further illustrate the brain's shared functional-structural covariance architecture.
Figure 4. Relationship between intrinsic functional connectivity and structural covariance in the healthy human brain.
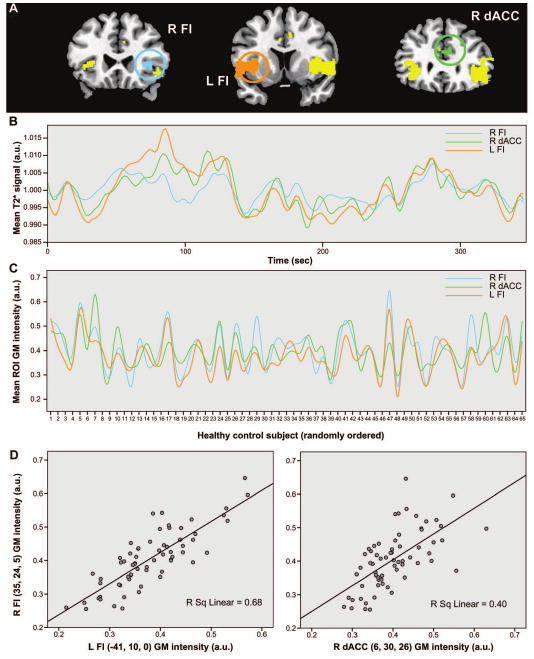
(A) The bvFTD-associated group-level ICA map (parent seed = right FI) was used to extract ROI BOLD signal timeseries from a single representative control subject (B). These timeseries reveal the correlated functional signals arising from the right and left FI and the right dACC, primary neurodegeneration foci in bvFTD. These same ROIs were applied to each of the 65 HC1 subjects to extract and plot local gray matter intensities for each ROI against the subject pool, randomly ordered on the x-axis to illustrate the structural covariance (C). Plots of R FI gray matter intensity against L FI and dACC intensities reveal the strength of within-network gray matter correlations (D). a.u. = arbitrary units.
Figure 5. Quantitative spatial similarity of each ICN and structural covariance map with the five syndromic atrophy maps.
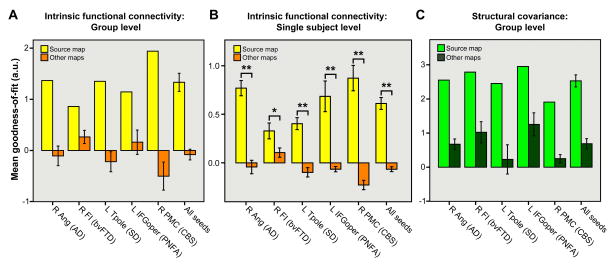
Binary spatial templates derived from the five atrophy maps were used to generate “goodness-of-fit” (GOF) scores that reflect how well the healthy intrinsic functional (A, B) and structural (C) correlation maps fit each syndrome's atrophy pattern. GOF was defined as the difference between the t-score mean within vs. outside each atrophy template, such that each ICN or structural correlation map had one “source” GOF score, for the atrophy map used to derive its seed, and four “other” scores for the four remaining atrophy templates. This procedure revealed higher GOF for source vs. other maps for each seed at the group level (A, C). At the single-subject level (B), all ICNs showed significantly greater GOF to source vs. other atrophy maps. Data are shown as mean +/- s.e.m (where applicable). * P < 0.05. ** P < 0.0005.
Figure 6. Neurodegenerative syndromes target anatomically dissociable brain systems.
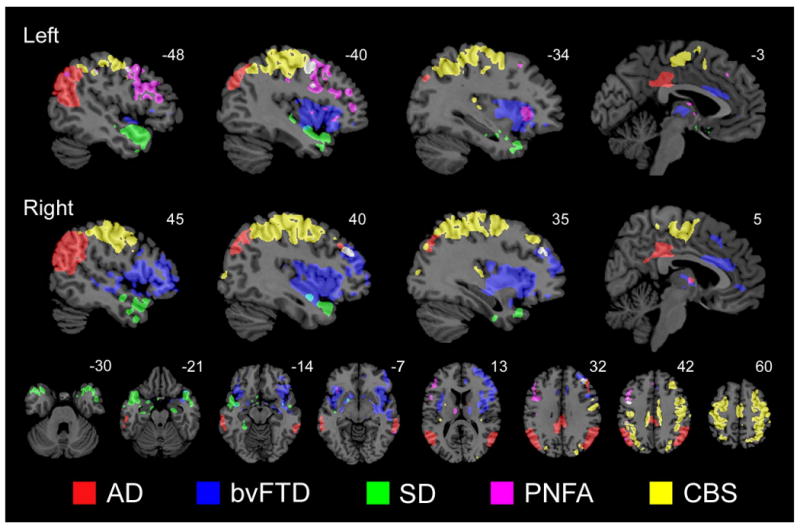
Colored regions highlight voxels found within associated maps of syndromic atrophy (p < 0.0001, uncorrected; patients vs. controls), intrinsic functional connectivity (ICA-derived; p < 0.01, corrected; healthy controls only), and structural covariance (p < 0.0001, uncorrected; healthy controls only). The color code (bottom) refers to the atrophy map used to derive the relevant seed ROI. These results, statistically thresholded to inflate potential overlap across the five 3-map datasets, illustrate the dissociable nature of the targeted brain systems.
Disease-vulnerable networks are dissociable: spatial similarity and overlap analyses
To quantify the spatial similarity between each atrophy pattern and the healthy functional-structural covariance networks, we used the ten control group correlation maps (5 functional, 5 structural) to generate goodness-of-fit (GOF) scores to each syndromic atrophy map. Fit was defined as the difference between the mean t-score of all voxels inside vs. outside each binary spatial atrophy template. These analyses indicated a strong fit between the intrinsic functional and structural covariance maps and their “source” atrophy patterns (Figure 5A, C). Although only a small, single ROI from each source map (Figure 2A) was used to seed the ICN and structural covariance analyses, the resulting healthy networks fit better with their source atrophy maps than with the other four disease patterns.
To confirm our group-level spatial similarity findings, we used each HC2 subject's best-fit ICA components (one for each seed ROI) to generate individual GOF scores to the source and other atrophy maps (Figure 5B). Paired samples t-tests (N = 19, 2-tailed) showed significant source vs. other GOF differences for all five ICNs (right ANG: t = 6.9, p = 0.000002; right FI: t = 2.4, p = 0.03; left Tpole: t = 7.4, p = 0.0000007; left IFG: t = 4.7, p = 0.0002; right PMC: t = 8.6, p = 0.00000009; mean of all seeds: t = 8.8, p = 0.00000006). The least strong (though still significant) source vs. other GOF statistical difference involved the right FI ICN. As highlighted in Figure 3A-C, this ICN and its structural covariance counterpart map showed robust qualitative similarity to the bvFTD atrophy map at the group level. We derived further support for the close atrophy-ICN relationship by comparing each HC2 subject's first and second best-fit ICA components, for each ICN, to the relevant source atrophy maps (see Experimental Procedures). This analysis confirmed a sharp GOF drop-off from the 1st to 2nd best fitting ICA components (paired samples t-tests: right ANG, t = 7.3, p = 0.0000009; right FI, t = 5.3, p = 0.00005; left Tpole, t = 8.6, p = 0.00000008; left IFG, t = 4.7, p = 0.0002; right PMC, t = 5.1; p = 0.00008). By definition, the remaining (unselected) components (3rd best-fit and beyond) for each subject fit the relevant atrophy patterns even less well. Therefore, our ICA and component selection procedures effectively identified the five normal ICNs that correspond best to the five syndromic atrophy patterns.
Finally, to visualize the spatial relationships among the five disease-vulnerable networks, we determined the voxel-wise (whole brain) overlap of each 3-map set (atrophy, intrinsic functional connectivity, and structural covariance associated with each ROI) and plotted the five resulting overlap maps on a shared template. Because we hypothesized that the five systems would prove dissociable, we lowered the statistical threshold for each map used to create the overlaps (see Experimental Procedures), in effect reducing our power to demonstrate spatial divergence among the five networks. Nonetheless, the five overlap maps showed minimal overlap with each other (Figure 6), illustrating the dissociable nature of these targeted brain systems.
Discussion
Our results show that functional and structural network mapping approaches yield robust, convergent, anatomically predictable networks, and that specific neurodegenerative diseases target these patterned brain systems. First, we characterized five early-stage dementia syndromes to isolate five circumscribed atrophy patterns, replicating and extending previous findings (Boccardi et al., 2005; Gorno-Tempini et al., 2004; Josephs et al., 2006; Josephs et al., 2008; Seeley et al., 2008a). We then demonstrated that these spatial disease patterns reflect the healthy brain's intrinsic functional network architecture. Although we and others have noted the concordance between AD-related atrophy and healthy intrinsic functional connectivity (Buckner et al., 2005; Greicius et al., 2004), in this study we confirmed the network degeneration hypothesis across five distinct dementia syndromes. Because these syndromes describe the clinical rather than molecular disease features, and because each syndrome can result from diverse underlying molecular pathologies, our data suggest that numerous disease proteins, including β-amyloid, tau, alpha-synuclein, and TDP-43, have the capacity to misfold and aggregate with specific brain networks.
The ICNs linked here to disease represent canonical findings from the ICN literature. Our AD-affected ICN (right ANG seed) corresponds to the “default mode network” that participates in episodic memory (Buckner et al., 2005) and became known for its task-related deactivations across fMRI studies (Fox et al., 2005; Fransson, 2005; Greicius et al., 2003). The ICN targeted in bvFTD (right FI seed) was first identified with ICA (Beckmann et al., 2005) and later linked to emotional salience processing capacities (Seeley et al., 2007) lost in early bvFTD (Seeley, 2008). SD affects an ICN (left temporal pole seed) that has escaped previous detection in humans but corresponds to a temporal pole-subgenual cingulate-ventral striatum-amygdala network, well-established in non-human primates (Mesulam and Mufson, 1982), that shows progressive atrophy in early-stage SD (Brambati et al., 2007). The PNFA-targeted ICN (left IFG seed) includes the frontal operculum, primary and supplementary motor cortices, and inferior parietal lobule bilaterally, linking the language and motor systems that enable speech fluency. This ICN, often divided into left and right hemispheric systems, has been noted in several previous studies (Beckmann et al., 2005; Damoiseaux et al., 2006; De Luca et al., 2006; van den Heuvel et al., 2008). In PNFA, asymmetric degeneration of this system may reflect its accentuated functional and connectional asymmetry in healthy humans (Stark et al., 2008). In CBS, prominent skeletal and ocular motor abnormalities result from disease within a dorsal sensorimotor association network (right PMC seed) detailed in several ICN studies (De Luca et al., 2006; Fox et al., 2005; Vincent et al., 2008) and elegantly mapped in the macaque using convergent ICN, oculomotor task-based fMRI, and axonal tracer methods (Vincent et al., 2007). ICNs frequently reported (Beckmann et al., 2005; Damoiseaux et al., 2006; De Luca et al., 2006; van den Heuvel et al., 2008) but not studied here include primary and secondary visual networks that may provide substrate for the visual-spatial variant of AD known as the posterior cortical atrophy syndrome (Hof et al., 1997), a primary sensorimotor ICN that may relate to amyotrophic lateral sclerosis (Kassubek et al., 2005), and a lateral frontoparietal executive-control network (Seeley et al., 2007; Vincent et al., 2008) that falters in most neurodegenerative diseases as degeneration spreads beyond the sites of initial injury into widely interconnected supervisory neocortical systems.
Confirmation of the network degeneration hypothesis raises important questions about how specific syndromes will impact ICN strength. Previous studies suggest that AD attenuates connectivity within the right ANG (“default mode”) network (Greicius et al., 2004; Supekar et al., 2008), even during early clinical stages (Sorg et al., 2007). Task-based episodic memory fMRI studies, in contrast, suggest heightened hippocampal activation during prodromal AD (Dickerson et al., 2004), and an AD intrinsic connectivity study revealed focal frontal connectivity enhancements (Supekar et al., 2008). For the non-AD dementia syndromes, no published ICN data are available. Therefore, it remains uncertain whether disease causes network connectivity impairment, upregulation, or effects that vary by disease stage. This issue requires clarification before network connectivity approaches can be used to diagnose dementia or track its progression. Those patients who scale up connectivity within early-affected networks or unaffected compensatory systems may prove more resilient to functional decline. Even more intriguingly, patients with focal connectivity enhancements may develop new or intensified functional capabilities (Seeley et al., 2008b).
Why does neurodegeneration spread throughout specific neural networks? The present study was designed to raise rather than answer this question, but several mechanisms may apply. First, selective neuronal vulnerability within key synaptic convergence zones may disconnect or weaken functional circuits (Hyman et al., 1984), inducing deleterious network-wide compensatory strategies. This notion fits with AD transgenic mouse models, in which aberrant medial temporal circuit excitability disrupts homeostasis and function, leading to progressive degeneration within the circuit (Palop et al., 2006; Palop et al., 2007). Second, retrograde axonal transport deficits may cut off growth factor supply to long-range projection neurons, begetting axonal degeneration, synapse loss, and post-synaptic dendrite retraction (Salehi et al., 2006). Third, as seen in experimental prion disease, misfolded disease proteins may themselves propagate along neural processes, marching throughout local and then long-range circuits via transsynaptic spread (Scott et al., 1992). This account of our data would require that non-prion misfolded disease proteins could trigger misfolding of adjacent same-species proteins, which, in turn, could cascade down processes and between interconnected neurons. Although many aspects of this candidate mechanism remain unexplored, in a recent study pathological tau conformers were shown to induce native (unfolded) tau to adopt a pathological conformation, and this misfolding seeded further conformational changes in adjacent tau molecules (Frost et al., 2008). Finally, each syndrome we studied features sporadic and autosomal dominantly inherited forms. For AD, known susceptibility genes, such as the Apo ε4 allele, increase risk or hasten disease onset. Lifelong possession of a disease-causing mutation (Geschwind et al., 2001) or risk factor allele (Reiman et al., 2004) may destabilize network connectivity and function in young adulthood or even childhood (Shaw et al., 2007), setting the stage for mid- to late-life neurodegeneration. These hypothetical network degeneration mechanisms need not be considered mutually exclusive.
Several recent studies suggest that, in health, functionally correlated brain regions feature defined axonal connections (Greicius et al., 2008; Seeley et al., 2007; Sporns et al., 2005). Our findings demonstrate a less intuitive linkage, revealing that healthy ICNs feature correlated gray matter volume. This finding builds on existing notions of “small worldness” in human cortical gray matter structure (He et al., 2007) by demonstrating a direct, network-based function-structure relationship. Why gray matter function and volume correlate, however, remains unclear. During development, network covariance may emerge (Fair et al., 2008) and remodel itself (Kelly et al., 2008; Lerch et al., 2006) in response to core inherited projection map formation scripts (Ruthazer and Cline, 2004) that interact with the environment to drive individual differences in cognitive, emotional, and motor function (Seeley et al., 2007). Use-related trophic effects, in turn, may continue to reshape the cortical mantle throughout adulthood (Maguire et al., 2000), linking synaptic strength, synaptic density, and neuropil mass within functionally co-active regions.
In summary, neurodegenerative diseases are not diffuse, random, or confluent but instead target specific large-scale distributed networks. In the healthy brain, these networks feature convergent intrinsic functional and structural covariance. To build more comprehensive disease pathogenesis models, neurodegeneration researchers should pursue the interface between disease protein aggregation and selective, network-driven neuronal vulnerability.
Experimental procedures
Subjects
All subjects (or their surrogates) provided informed consent and the procedures were approved by the institutional review boards at UCSF and Stanford University.
Patients: Structural Imaging
Subjects for each patient group were selected from the UCSF Memory & Aging Center (MAC) database. All had undergone a comprehensive neurological, neuropsychological, and functional assessment, and final diagnoses were rendered at a multidisciplinary consensus conference, as detailed previously (Liu et al., 2004). To be considered for inclusion, patients were required to meet published research criteria, which do not include neuroimaging features, for probable AD (McKhann et al., 1984), bvFTD (Neary et al., 1998), SD (Neary et al., 1998), PNFA (Neary et al., 1998), or CBS (criteria developed at the MAC, as previously described (Boxer et al., 2006)) within 90 days of MRI scanning. In addition, the following selection criteria were required: (1) CDR total score ≤ 1, (2) CDR and MMSE completed within 90 days of scanning, (3) absence of comorbid motor neuron disease, and (4) absence of significant vascular or other structural lesions on MRI. We chose to study only these five syndromes because they could be matched for age while preserving adequate sample size. Although potentially relevant to our study aims, patients with the posterior cortical atrophy syndrome, progressive supranuclear palsy, dementia with Lewy bodies, Huntington's disease, amyotrophic lateral sclerosis, and multiple systems atrophy were excluded to constrain the scope of the study and promote matching or because too few subjects with these diagnoses had available research MRI scans. The three patient groups with the most subjects meeting inclusion criteria were bvFTD (n=24), AD (n = 49), and SD (n = 32). Therefore, 24 AD and 24 SD subjects were chosen from their larger pools to match the bvFTD group, as closely as possible, for age, gender, and education (Table S1). PNFA (n = 13) and CBS (n =17) groups were smaller than the others but still comparable to published samples used to illustrate the syndromic regional atrophy patterns (Boxer et al., 2006; Gorno-Tempini et al., 2004; Josephs et al., 2006). These syndromes are known to feature uneven gender distributions (Johnson et al., 2005); therefore, in part because of our strict inclusion criteria (e.g. CDR ≤ 1), all groups could not be matched for gender. Accordingly, gender was added as a nuisance covariate to the neuroimaging analyses.
Importantly, we made a sharp distinction between clinical syndromes and histopathological entities. Therefore, although the terms Alzheimer's disease and corticobasal degeneration are used by pathologists to describe specific histopathologies, in this context we applied the terms Alzheimer's disease and corticobasal syndrome only to suggest clinical, not pathological, features. This approach fits with our scientific objective, which was to determine the relationship between syndromic atrophy patterns and healthy human brain networks. Our overarching hypothesis, indeed, was that large-scale cortical networks provide anatomical scaffoldings that are dismantled to produce clinically recognizable lesion-deficit constellations (syndromes). Applying network imaging to predict neuropathology was not a goal of this study; indeed, most degenerative syndromes can be caused by several underlying pathologies, and all neuropathological entities can give rise to diverse clinical syndromes.
UCSF Controls: Structural Imaging
Healthy controls (HC) were recruited for the structural imaging and neuropsychological analyses from the San Francisco community. These controls, referred to from here forward as HC1, underwent a comprehensive neuropsychological assessment and a neurological exam within 180 days of scanning. HC1 subjects were required to have a CDR total score of 0, an MMSE of 28 or higher, no significant history of neurological disease or structural pathology on MRI, and a consensus diagnosis of cognitively normal; 101 subjects met these criteria. This group was reduced to 65 subjects to match, as closely as possible, the overall patient group for age, gender, and education. No UCSF control subject took neuropsychiatric medications except one, who took buproprion 100 mg daily for a remote history of depression.
Stanford Controls: Functional Imaging
HC subjects for the functional imaging analyses, referred to henceforth as HC2, were recruited from the UCSF Memory and Aging center (as described above) and from the Stanford Medical Center community. The Stanford subjects denied any significant neurologic or psychiatric history and were not taking psychoactive medications. All had MMSE scores of 27/30 or higher.
Image acquisition
Structural Imaging
Structural MRI scans were obtained at UCSF on all patients and HC1 subjects. Images were acquired on a 1.5 Tesla Magneton VISION system (Siemens Inc., Iselin, NJ) using a standard quadrature head coil. A volumetric magnetization prepared rapid gradient echo (MP-RAGE) MRI (TR/TE/inversion time = 10/4/300 msec) sequence was used to obtain a T1-weighted image of the entire brain (15 degree flip angle, coronal orientation perpendicular to the double spin echo sequence, 1.0×1.0 mm2 in-plane resolution of 1.5 mm slab thickness).
Functional Imaging
Functional MRI (fMRI) scanning was performed at Stanford University on all HC2 subjects. Images were acquired on a 3 Tesla GE Signa Excite scanner (GE Medical Systems, Milwaukee, WI) using a standard GE whole head coil. Twenty-eight axial slices (4 mm thick, 1 mm skip) parallel to the plane connecting the anterior and posterior commissures and covering the whole brain were imaged using a T2* weighted gradient echo spiral pulse sequence (repetition time, 2000 ms; echo time, 30 ms; flip angle, 80 degrees and 1 interleave) (Glover and Lai, 1998). The field of view was 200×200mm2, and the matrix size was 64×64, yielding an in-plane spatial resolution of 3.125 mm. To reduce blurring and signal loss arising from field inhomogeneities, an automated high-order shimming method based on spiral acquisitions was used before acquiring functional MRI scans (Kim et al., 2000). All subjects underwent two 6-minute task-free fMRI scans after being instructed only to remain awake with their eyes closed. For two of 17 subjects, one of the two runs was excluded due to technical factors.
Image preprocessing and analysis
Structural Imaging
Voxel-based morphometry (VBM) is a flexible whole brain statistical analysis technique that can be used to assess between-group differences in local brain tissue content or correlations between tissue content and other measures of interest. Before analysis, T1-weighted MR images underwent several preprocessing steps, following an optimized VBM protocol (Good et al., 2001). First, a study-specific template and priors were created to minimize spatial normalization and segmentation errors. This approach helps to identify group differences in patients with neurodegenerative disease (Senjem et al., 2005). All subjects were used to create the template, and custom images for each subject were generated by applying affine and deformation parameters obtained from normalizing the grey matter images, segmented in native space, to the custom template. Voxel values were modulated by multiplying them by the jacobian determinants derived from the spatial normalization step, and images were smoothed with a 12-mm isotropic Gaussian kernel.
To identify gray matter regions significantly atrophied in each syndrome vs. HC1, each group was entered as a condition into a single model, and linear contrasts were applied to derive five syndromic atrophy maps. From these maps, we isolated the most significantly atrophied cortical region, designated as the voxel with the peak t-score (Table S2). We elected to use only cortical seed ROIs because subcortical/limbic nuclei, such as the caudate or amygdala, feature a dense admixture of subnuclear projection fields and outputs with multi-network connectivity. Therefore, after 4-mm fMRI data smoothing, subcortical/limbic connectivity maps become less straightforward to interpret than those derived using cortical seeds, and we sought to treat all five network analyses similarly in this regard. In addition, VBM can overestimate periventricular volume loss due to atrophy-related spatial registration errors, even with the optimized methods employed here (Senjem et al., 2005). This concern further justifies exclusion of subcortical seeds for functional connectivity and structural covariance analyses. For the SD map, we chose the second most atrophied cortical region because the peak region fell within an area of susceptibility artifact on our T2* images (Ojemann et al., 1997). Collectively, the syndromic peak voxels were used to create five 4-mm radius spherical ROIs using MarsbaR v0.41, a region of interest toolbox for SPM (Brett et al., 2002). These five cortical “seeds” were used in all subsequent structural and functional correlation analyses.
Structural correlation analyses were applied only to the 65 HC1 subjects. Adapting previous approaches by Mechelli and colleagues (Mechelli et al., 2005), we extracted each subject's mean grey matter intensity from the five seed ROIs. These values were then entered into separate covariate-only models to identify each voxel, across the whole brain, whose gray matter intensity was significantly correlated with that of each seed across subjects.
For all VBM analyses, age and gender were entered as nuisance covariates and total intracranial volume served as a global correction factor. Preprocessing and analysis was implemented in the SPM5 software package (www.fil.ion.ucl.ac.uk/spm). For group contrast and correlation analyses, voxels were considered significant if they met statistical threshold of p < 0.05, corrected for family wise error (FWE).
Functional imaging
Functional images were realigned, slice-time corrected, normalized, and smoothed with a 4 mm Gaussian kernel. Normalization was carried out by calculating the warping parameters between the mean T2* (spiral in/out) image and the MNI EPI template and applying them to all images in the sequence. Subsequently, the images were resampled at a voxel size of 2mm3.
ROIs derived from the five syndromic atrophy patterns were then used to seed five separate functional connectivity analyses, following previous methods (Seeley et al., 2007). That is, after removing the first eight n frames to allow for stabilization of the magnetic field, the average time series from the first task-free scan was extracted from each ROI by averaging the time series of all voxels within the ROI. Before averaging individual voxel data, scaling and filtering steps were performed across all brain voxels as follows. To minimize the effect of global drift, voxel intensities were scaled by dividing the value of each time point by the mean value of the whole-brain image at that time point. Next, the scaled waveform of each brain voxel was filtered using a bandpass filter (0.0083/s < f < 0.15/s) to reduce the effect of low frequency drift and high-frequency noise (Lowe et al., 1998). The scaling and filtering steps were applied equivalently to all voxels (including those in the ROIs). The resulting time series, representing the average intensity (after scaling and filtering) of all voxels in the ROI, was then used as a covariate of interest in a whole-brain, linear regression, statistical parametric analysis. As a means of controlling for non-neural noise in the ROI time series, we included, as a nuisance covariate, the global average T2* time series. Contrast images corresponding to the ROI time series regressors were derived individually for each subject, and entered into second-level, random-effects analyses (joint height and extent thresholds of p < 0.001 for significant clusters, corrected at the whole brain level) (Poline et al., 1997) to determine the brain areas that showed significant functional connectivity with each seed across HC2 subjects. The resulting group maps were used as spatial templates to select the best-fit independent component from each subject in subsequent ICA analyses.
We used ICA to further refine our intrinsic functional connectivity maps (Seeley et al., 2007). ICA decomposes a timecourse of whole brain volumes (a 4-D image) from a single subject into independent spatiotemporal components. After preprocessing, images were concatenated into 4-D files and entered into FSL 4.0 Melodic ICA software (http://www.fmrib.ox.ac.uk/fsl/index.html). We allowed the program to automatically determine the dimensionality of each data set, including the number of components. Among the 19 control subjects, ICA extracted an average of 36.7 components (range 19-51) from Run 1 and 36.5 components (range 29-52) from Run 2. After high frequency filtering, an average of 23.2 components (range 7-42) remained from Run 1 and 20.7 components (range 12-42) remained for Run 2. Temporal filtering was not applied to the ICA data because ICA has been shown to separate non-neural noise components from components of interest (Beckmann et al., 2005). The components for each subject (from Run 1 or 2) that best fit the ROI-derived group ICN template maps were selected using an automated three step procedure (Seeley et al., 2007). For two of nineteen subjects, the algorithm selected the same component for two of the five spatial templates. For one of these subjects, the right parietal (AD) and left inferior frontal (PNFA) seed-based ROI maps were best fit by the same component, and for the other subject the right FI (bvFTD) and left inferior frontal (PNFA) seed maps best fit the same component. One-sample t-tests were performed on the best-fit component images to derive a group-level ICN map for each seed. Significance was determined using joint expected probability distribution with height and extent thresholds of p < 0.001. The ICA-derived group maps were used in all figures and spatial similarity analyses.
Spatial similarity analyses
The spatial similarity between atrophy maps and their related ICN and structural covariance maps was first quantified by measuring the GOF between each atrophy map, binarized at a p < 0.05 family wise error (FWE) corrected threshold, and the group-level ICA-derived ICNs and VBM-derived structural correlation maps. GOF was defined as the difference between the mean of the t-scores inside vs. outside the binarized atrophy map. Therefore, each group-level correlation map had five GOF scores: one “source” and four “other”. Here, “source” refers to the GOF score from the atrophy map used to derive the correlation map's seed, whereas “other” refers to the four other atrophy maps. We further compared each HC2 subject's five best-fit ICA components (one for each seed ROI) to the five atrophy maps, using the same GOF procedure. “Other” GOF scores were averaged for each subject, and paired samples t-tests were used to compare source vs. mean other map GOF scores. Finally, to assess the specificity of each atrophy-ICN pairing at the single-subject level, we calculated mean source map GOF score for the best-fit ICA components (one each from Runs 1 and 2) and compared these scores to the mean GOF for the 2nd best-fit components (Runs 1 and 2) using paired samples t-tests (two-tailed). Because structural covariance cannot be derived for single subjects, statistical comparisons of structural covariance GOF results were not performed.
To visualize the spatial overlap within related atrophy, functional connectivity, and structural covariance maps, we loaded all three maps as overlays, rendered at less stringent statistical thresholds (atrophy: p < 0.0001, uncorrected; functional connectivity: ICA maps at p < 0.01 joint height and extent thresholds, corrected at the whole brain level; structural covariance: p < 0.0001, uncorrected), and determined the intersection of the three maps, defined as those voxels contained in 3/3 maps at these thresholds. This procedure allowed us to demonstrate the convergence of the three maps within each set, as well as the spatial dissimilarity between the five separate 3-map sets (Figure 6).
Supplementary Material
01
Acknowledgments
We thank M.L. Gorno-Tempini and V. Menon for discussion and our patients and their families for participating in neurodegeneration research. This work was supported by the National Institute of Aging (NIA grants K08 AG027086 to W.W.S.; P01 AG19724 and P50 AG1657303-75271 to B.L.M.), the National Institute of Neurological Disorders and Stroke (NINDS grant K23NS048302 to M.D.G.) and the Larry L. Hillblom Foundation (W.W.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi G, Poo M. Distributed synaptic modification in neural networks induced by patterned stimulation. Nature. 1999;401:792–796. doi: 10.1038/44573. [DOI] [PubMed] [Google Scholar]
- Boccardi M, Sabattoli F, Laakso MP, Testa C, Rossi R, Beltramello A, Soininen H, Frisoni GB. Frontotemporal dementia as a neural system disease. Neurobiol Aging. 2005;26:37–44. doi: 10.1016/j.neurobiolaging.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Boxer AL, Geschwind MD, Belfor N, Gorno-Tempini ML, Schauer GF, Miller BL, Weiner MW, Rosen HJ. Patterns of brain atrophy that differentiate corticobasal degeneration syndrome from progressive supranuclear palsy. Arch Neurol. 2006;63:81–86. doi: 10.1001/archneur.63.1.81. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological staging of Alzheimer-related changes. Acta Neuropathologica. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Brambati SM, Rankin KP, Narvid J, Seeley WW, Dean D, Rosen HJ, Miller BL, Ashburner J, Gorno-Tempini ML. Atrophy progression in semantic dementia with asymmetric temporal involvement: A tensor-based morphometry study. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett M, Anton JL, Valabregue R, Poline JB. Region of interest analysis using an SPM toolbox [abstract]. Paper presented at: 8th International Conference on Functional Mapping of the Human Brain; Sendai, Japan. 2002. [Google Scholar]
- Buckner RL, Snyder AZ, Shannon BJ, LaRossa G, Sachs R, Fotenos AF, Sheline YI, Klunk WE, Mathis CA, Morris JC, Mintun MA. Molecular, structural, and functional characterization of Alzheimer's disease: evidence for a relationship between default activity, amyloid, and memory. J Neurosci. 2005;25:7709–7717. doi: 10.1523/JNEUROSCI.2177-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci U S A. 2006;103:13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M, Beckmann CF, De Stefano N, Matthews PM, Smith SM. fMRI resting state networks define distinct modes of long-distance interactions in the human brain. Neuroimage. 2006;29:1359–1367. doi: 10.1016/j.neuroimage.2005.08.035. [DOI] [PubMed] [Google Scholar]
- Dickerson BC, Salat DH, Bates JF, Atiya M, Killiany RJ, Greve DN, Dale AM, Stern CE, Blacker D, Albert MS, Sperling RA. Medial temporal lobe function and structure in mild cognitive impairment. Ann Neurol. 2004;56:27–35. doi: 10.1002/ana.20163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Cohen AL, Dosenbach NU, Church JA, Miezin FM, Barch DM, Raichle ME, Petersen SE, Schlaggar BL. The maturing architecture of the brain's default network. Proc Natl Acad Sci U S A. 2008;105:4028–4032. doi: 10.1073/pnas.0800376105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8:700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Raichle ME. Intrinsic fluctuations within cortical systems account for intertrial variability in human behavior. Neuron. 2007;56:171–184. doi: 10.1016/j.neuron.2007.08.023. [DOI] [PubMed] [Google Scholar]
- Fransson P. Spontaneous low-frequency BOLD signal fluctuations: an fMRI investigation of the resting-state default mode of brain function hypothesis. Hum Brain Mapp. 2005;26:15–29. doi: 10.1002/hbm.20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson P. How default is the default mode of brain function? Further evidence from intrinsic BOLD signal fluctuations. Neuropsychologia. 2006;44:2836–2845. doi: 10.1016/j.neuropsychologia.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Frost B, Ollesch J, Wille H, Diamond MI. Conformational diversity of wild-type tau fibrils specified by templated conformation change. J Biol Chem. 2008 doi: 10.1074/jbc.M805627200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind D, Robidoux J, Alarcon M, Miller B, Wilhelmsen K, Cummings J, Nasreddine Z. Dementia and neurodevelopmental predisposition: cognitive dysfunction in presymptomatic subjects precedes dementia by decades in frontotemporal dementia. Ann Neurol. 2001;50:741–746. doi: 10.1002/ana.10024. [DOI] [PubMed] [Google Scholar]
- Glover GH, Lai S. Self-navigated spiral fMRI: interleaved versus single-shot. Magn Reson Med. 1998;39:361–368. doi: 10.1002/mrm.1910390305. [DOI] [PubMed] [Google Scholar]
- Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- Gorno-Tempini ML, Dronkers NF, Rankin KP, Ogar JM, Phengrasamy L, Rosen HJ, Johnson JK, Weiner MW, Miller BL. Cognition and anatomy in three variants of primary progressive aphasia. Ann Neurol. 2004;55:335–346. doi: 10.1002/ana.10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveland GA, Williams RS, DiFiglia M. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington's disease. Science. 1985;227:770–773. doi: 10.1126/science.3155875. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci U S A. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer's disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci U S A. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Supekar K, Menon V, Dougherty RF. Resting-State Functional Connectivity Reflects Structural Connectivity in the Default Mode Network. Cereb Cortex. 2008 doi: 10.1093/cercor/bhn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Chen ZJ, Evans AC. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb Cortex. 2007;17:2407–2419. doi: 10.1093/cercor/bhl149. [DOI] [PubMed] [Google Scholar]
- Hesselmann G, Kell CA, Eger E, Kleinschmidt A. Spontaneous local variations in ongoing neural activity bias perceptual decisions. Proc Natl Acad Sci U S A. 2008;105:10984–10989. doi: 10.1073/pnas.0712043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof PR, Vogt BA, Bouras C, Morrison JH. Atypical form of Alzheimer's disease with prominent posterior cortical atrophy: a review of lesion distribution and circuit disconnection in cortical visual pathways. Vision Res. 1997;37:3609–3625. doi: 10.1016/S0042-6989(96)00240-4. [DOI] [PubMed] [Google Scholar]
- Hyman BT, Damasio AR, Van Hoesen GW, Barnes CL. Alzheimer's disease: cell-specific pathology isolates the hippocampal formation. Science. 1984;298:83–95. doi: 10.1126/science.6474172. [DOI] [PubMed] [Google Scholar]
- Johnson JK, Diehl J, Mendez MF, Neuhaus J, Shapira JS, Forman M, Chute DJ, Roberson ED, Pace-Savitsky C, Neumann M, et al. Frontotemporal lobar degeneration: demographic characteristics of 353 patients. Arch Neurol. 2005;62:925–930. doi: 10.1001/archneur.62.6.925. [DOI] [PubMed] [Google Scholar]
- Josephs KA, Duffy JR, Strand EA, Whitwell JL, Layton KF, Parisi JE, Hauser MF, Witte RJ, Boeve BF, Knopman DS, et al. Clinicopathological and imaging correlates of progressive aphasia and apraxia of speech. Brain. 2006;129:1385–1398. doi: 10.1093/brain/awl078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs KA, Whitwell JL, Dickson DW, Boeve BF, Knopman DS, Petersen RC, Parisi JE, Jack CR., Jr Voxel-based morphometry in autopsy proven PSP and CBD. Neurobiol Aging. 2008;29:280–289. doi: 10.1016/j.neurobiolaging.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassubek J, Unrath A, Huppertz HJ, Lule D, Ethofer T, Sperfeld AD, Ludolph AC. Global brain atrophy and corticospinal tract alterations in ALS, as investigated by voxel-based morphometry of 3-D MRI. Amyotroph Lateral Scler Other Motor Neuron Disord. 2005;6:213–220. doi: 10.1080/14660820510038538. [DOI] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- Kelly AM, Di Martino A, Uddin LQ, Shehzad Z, Gee DG, Reiss PT, Margulies DS, Castellanos FX, Milham MP. Development of Anterior Cingulate Functional Connectivity from Late Childhood to Early Adulthood. Cereb Cortex. 2008 doi: 10.1093/cercor/bhn117. [DOI] [PubMed] [Google Scholar]
- Kim DH, Adalsteinsson E, Glover GH, Spielman S. SVD regularization algorithm for improved high-order shimming. Denver: 2000. [Google Scholar]
- Lerch JP, Worsley K, Shaw WP, Greenstein DK, Lenroot RK, Giedd J, Evans AC. Mapping anatomical correlations across cerebral cortex (MACACC) using cortical thickness from MRI. Neuroimage. 2006;31:993–1003. doi: 10.1016/j.neuroimage.2006.01.042. [DOI] [PubMed] [Google Scholar]
- Liu W, Miller BL, Kramer JH, Rankin K, Wyss-Coray C, Gearhart R, Phengrasamy L, Weiner M, Rosen HJ. Behavioral disorders in the frontal and temporal variants of frontotemporal dementia. Neurology. 2004;62:742–748. doi: 10.1212/01.wnl.0000113729.77161.c9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe MJ, Mock BJ, Sorenson JA. Functional connectivity in single and multislice echoplanar imaging using resting-state fluctuations. Neuroimage. 1998;7:119–132. doi: 10.1006/nimg.1997.0315. [DOI] [PubMed] [Google Scholar]
- Maguire EA, Gadian DG, Johnsrude IS, Good CD, Ashburner J, Frackowiak RS, Frith CD. Navigation-related structural change in the hippocampi of taxi drivers. Proc Natl Acad Sci U S A. 2000;97:4398–4403. doi: 10.1073/pnas.070039597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Mechelli A, Friston KJ, Frackowiak RS, Price CJ. Structural covariance in the human cortex. J Neurosci. 2005;25:8303–8310. doi: 10.1523/JNEUROSCI.0357-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesulam MM, Mufson EJ. Insula of the old world monkey. I. Architectonics in the insulo-orbito-temporal component of the paralimbic brain. J Comp Neurol. 1982;212:1–22. doi: 10.1002/cne.902120102. [DOI] [PubMed] [Google Scholar]
- Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, Freedman M, Kertesz A, Robert PH, Albert M, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- Ojemann JG, Akbudak E, Snyder AZ, McKinstry RC, Raichle ME, Conturo TE. Anatomic localization and quantitative analysis of gradient refocused echo-planar fMRI susceptibility artifacts. Neuroimage. 1997;6:156–167. doi: 10.1006/nimg.1997.0289. [DOI] [PubMed] [Google Scholar]
- Palop JJ, Chin J, Mucke L. A network dysfunction perspective on neurodegenerative diseases. Nature. 2006;443:768–773. doi: 10.1038/nature05289. [DOI] [PubMed] [Google Scholar]
- Palop JJ, Chin J, Roberson ED, Wang J, Thwin MT, Bien-Ly N, Yoo J, Ho KO, Yu GQ, Kreitzer A, et al. Aberrant excitatory neuronal activity and compensatory remodeling of inhibitory hippocampal circuits in mouse models of Alzheimer's disease. Neuron. 2007;55:697–711. doi: 10.1016/j.neuron.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poline JB, Worsley KJ, Evans AC, Friston KJ. Combining spatial extent and peak intensity to test for activations in functional imaging. Neuroimage. 1997;5:83–96. doi: 10.1006/nimg.1996.0248. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer's dementia. Proc Natl Acad Sci U S A. 2004;101:284–289. doi: 10.1073/pnas.2635903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthazer ES, Cline HT. Insights into activity-dependent map formation from the retinotectal system: a middle-of-the-brain perspective. J Neurobiol. 2004;59:134–146. doi: 10.1002/neu.10344. [DOI] [PubMed] [Google Scholar]
- Salehi A, Delcroix JD, Belichenko PV, Zhan K, Wu C, Valletta JS, Takimoto-Kimura R, Kleschevnikov AM, Sambamurti K, Chung PP, et al. Increased App expression in a mouse model of Down's syndrome disrupts NGF transport and causes cholinergic neuron degeneration. Neuron. 2006;51:29–42. doi: 10.1016/j.neuron.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Scott JR, Davies D, Fraser H. Scrapie in the central nervous system: neuroanatomical spread of infection and Sinc control of pathogenesis. J Gen Virol. 1992;73(Pt 7):1637–1644. doi: 10.1099/0022-1317-73-7-1637. [DOI] [PubMed] [Google Scholar]
- Seeley WW. Selective functional, regional, and neuronal vulnerability in frontotemporal dementia. Curr Opin Neurol. 2008;21:701–707. doi: 10.1097/WCO.0b013e3283168e2d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Bauer AM, Miller BL, Gorno-Tempini ML, Kramer JH, Weiner M, Rosen HJ. The natural history of temporal variant frontotemporal dementia. Neurology. 2005;64:1384–1390. doi: 10.1212/01.WNL.0000158425.46019.5C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Carlin DA, Allman JM, Macedo MN, Bush C, Miller BL, Dearmond SJ. Early frontotemporal dementia targets neurons unique to apes and humans. Ann Neurol. 2006;60:660–667. doi: 10.1002/ana.21055. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Crawford R, Rascovsky K, Kramer JH, Weiner M, Miller BL, Gorno-Tempini ML. Frontal paralimbic network atrophy in very mild behavioral variant frontotemporal dementia. Arch Neurol. 2008a;65:249–255. doi: 10.1001/archneurol.2007.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Matthews BR, Crawford RK, Gorno-Tempini ML, Foti D, Mackenzie IR, Miller BL. Unravelling Bolero: progressive aphasia, transmodal creativity and the right posterior neocortex. Brain. 2008b;131:39–49. doi: 10.1093/brain/awm270. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer's disease is a synaptic failure. Science. 2002;298:789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- Senjem ML, Gunter JL, Shiung MM, Petersen RC, Jack CR., Jr Comparison of different methodological implementations of voxel-based morphometry in neurodegenerative disease. Neuroimage. 2005;26:600–608. doi: 10.1016/j.neuroimage.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Lerch JP, Pruessner JC, Taylor KN, Rose AB, Greenstein D, Clasen L, Evans A, Rapoport JL, Giedd JN. Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: an observational study. Lancet Neurol. 2007;6:494–500. doi: 10.1016/S1474-4422(07)70106-0. [DOI] [PubMed] [Google Scholar]
- Sorg C, Riedl V, Muhlau M, Calhoun VD, Eichele T, Laer L, Drzezga A, Forstl H, Kurz A, Zimmer C, Wohlschlager AM. Selective changes of resting-state networks in individuals at risk for Alzheimer's disease. Proc Natl Acad Sci U S A. 2007;104:18760–18765. doi: 10.1073/pnas.0708803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kotter R. The human connectome: a structural description of the human brain. PLoS Comput Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark DE, Margulies DS, Shehzad ZE, Reiss P, Kelly AM, Uddin LQ, Gee DG, Roy AK, Banich MT, Castellanos FX, Milham MP. Regional variation in interhemispheric coordination of intrinsic hemodynamic fluctuations. J Neurosci. 2008;28:13754–13764. doi: 10.1523/JNEUROSCI.4544-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar K, Menon V, Rubin D, Musen M, Greicius MD. Network analysis of intrinsic functional brain connectivity in Alzheimer's disease. PLoS Comput Biol. 2008;4:e1000100. doi: 10.1371/journal.pcbi.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel M, Mandl R, Hulshoff Pol H. Normalized cut group clustering of resting-state FMRI data. PLoS ONE. 2008;3:e2001. doi: 10.1371/journal.pone.0002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Kahn I, Snyder AZ, Raichle ME, Buckner RL. Evidence for a frontoparietal control system revealed by intrinsic functional connectivity. J Neurophysiol. 2008;100:3328–3342. doi: 10.1152/jn.90355.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
