Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution (original) (raw)
. Author manuscript; available in PMC: 2009 Jul 25.
Summary
The biological causes of selective pressures on coding-sequence evolution remain controversial, despite the surprising consistency of covariation between common measures of evolutionary change (substitution rates) and gene expression (mRNA levels, codon usage) across taxa. We carry out a unified analysis which reveals these conserved patterns in E. coli, yeast, worm, fly, mouse, and human, and suggests that all trends stem largely from a unified underlying selective pressure. In metazoans, these trends are strongest in tissues composed of neurons, whose structure and lifetime confer extreme sensitivity to protein misfolding. We propose, and demonstrate using a molecular-level evolutionary simulation, that selection against toxicity of misfolded proteins generated by ribosome errors suffices to create all the observed covariation. The mechanistic model of molecular evolution which emerges yields testable biochemical predictions, calls into question use of nonsynonymous-to-synonymous substitution ratios (Ka/Ks) to detect functional selection, and suggests how mistranslation may contribute to neurodegenerative disease.
Introduction
Evolutionary cell biologists seek to understand how natural selection shapes cellular features and processes. Recent work has revealed the molecular basis of organism-level adaptations by drawing upon phylogenetic information to infer, recreate, and functionally interrogate the sequences of evolving proteins (Dean and Thornton, 2007), such as receptors bound by distinct ligands (Bridgham et al., 2006), or enzymes with distinct coenzyme preferences (Zhu et al., 2005).
But if gene sequences reveal the lineage-specific fits and starts of adaptive evolution, they must also bear signs of cell-biological constraints common to all organisms, such as the biophysical challenges of producing folded polypeptides in a crowded intracellular milieu. Selection for proper protein folding and function causes coding sequences to accumulate nonsynonymous (amino-acid-altering) substitutions at a slower rate than the synonymous (amino-acid-preserving) substitution rate. Differing evolutionary rates between coding sequences in the same lineage hint at further constraints: for example, histones evolve slower than hormones (Wilson et al., 1977). Because residues directly involved in protein function tend not to tolerate substitutions (Anfinsen, 1959; Guo et al., 2004; Wilson et al., 1977; Zuckerkandl, 1976), it has long been hypothesized that slow-evolving proteins have more sites committed to function (Zuckerkandl, 1976) or are more functionally important (Wilson et al., 1977). Plausible alternatives exist; genes may evolve faster or slower for reasons unrelated to their functions, such as regional variation in mutation rates.
Indeed, genome-scale data has revealed that measures of functional importance, such as essentiality or the number of protein-protein interactions, are surprisingly weak correlates of evolutionary rate (Pál et al., 2006). Concurrently, a striking and apparently universal link between gene regulation and coding sequence evolution has emerged: genes with high mRNA expression levels encode slow-evolving proteins, from bacteria (Drummond et al., 2006; Rocha and Danchin, 2004), yeast (Drummond et al., 2005; Pál et al., 2001) and algae (Popescu et al., 2006), to nematodes (Krylov et al., 2003), plants (Ingvarsson, 2007; Wright et al., 2004), fruit flies (Lemos et al., 2005), mice, and humans (Subramanian and Kumar, 2004). Among duplicated yeast genes, the higher-expressed gene generally evolves slower (Drummond et al., 2005) while often showing minimal functional differences from its paralog: the faster-evolving, lower-expression pyruvate kinase 2 can completely substitute for the enzymatic activity of the slower-evolving, higher-expressed pyruvate kinase 1 (Boles et al., 1997).
Evolutionary effects linked strongly to mRNA level but not to function have been argued to indicate selection on translation, including adaptations to combat ribosomal infidelity (Drummond et al., 2005). Missense errors in translation occur at rates of one per 103–104 codons (Kramer and Farabaugh, 2007; Ogle and Ramakrishnan, 2005; Parker, 1989); at an error rate of 5×10–4, 18% of proteins expressed from an average length (~400-codon) gene contain at least one missense substitution. Roughly ~10–50% of random substitutions disrupt protein function (Guo et al., 2004; Markiewicz et al., 1994), and most loss-of-function mutations appear to be loss-of-folding mutations (Pakula and Sauer, 1989), also shown by large-scale folding and functional assays (Bloom et al., 2006). Finally, misfolded proteins possess generic cytotoxicity (Bucciantini et al., 2002): destabilized proteins expose natively buried hydrophobic residues which seek nonpolar surface area and find it in other destabilized proteins, causing protein-protein aggregation. At cell membranes, protein-membrane aggregation disrupts membrane integrity (Kourie and Henry, 2002; Stefani and Dobson, 2003) and with it crucial ionic balances (e.g. Ca2+) required for viability (Stefani, 2007). The expectation that ribosomal infidelity generates cytotoxic species suggests a general selective pressure for genetic adaptations that reduce these costs.
Longstanding efforts to understand connections between genetic change and organismal biology have uncovered a dense set of interrelationships between the nonsynonymous evolutionary rate (dN), the synonymous rate (dS), and other variables (see Supplementary Box S1). (dN and dS are sometimes called Ka and Ks.). The ratio dN/dS measures the strength of selection, assuming synonymous changes have no effect on fitness. However, synonymous changes are weakly selected against in most organisms, including humans (Chamary et al., 2006; Yang and Nielsen, 2008). In some organisms, selection favors efficiently translated (so-called “optimal”) codons in highly expressed genes (Ikemura, 1985), a bias measured most simply as the fraction of optimal codons (Fop) per gene. Codon preference slows the rate of synonymous change, dS (Ikemura, 1985). Notably, a positive dN–dS correlation was detected early on in mammals (Li et al., 1985) and bacteria (Sharp and Li, 1987) but remains unexplained, as have a negative correlation between Fop and dN (Marais et al., 2004; Sharp and Li, 1987) and, recently, a positive relationship between dN/dS and dS in mammals (Wyckoff et al., 2005). Whereas an analysis in yeast suggested that variation in dN and dS reflect a common determinant (Drummond et al., 2006), analyses of a wider array of organisms have concluded that the rates of evolution vary for different reasons in microbes and multicellular eukaryotes (Choi et al., 2007; Koonin and Wolf, 2006; Liao et al., 2006). Whether these divergent conclusions reflect primarily biological, methodological, or interpretive differences has remained unclear.
Here, we pursue a cell-biological understanding of these long-studied evolutionary patterns. We first carry out a comprehensive, methodologically unified evolutionary analysis across a set of six distantly related model organisms. The results confirm multiple conserved genome-wide signatures of selection, including several unexplained and novel observations, and firmly establish selection for translational accuracy in mammals. We introduce a model in which selection against cytotoxic protein misfolding produces all these conserved features. To test the model’s ability to generate the observed evolutionary patterns, we create and analyze a large-scale simulation which incorporates known biological constraints on translation and protein folding into an evolutionary framework. We repeat all analyses on these in silico evolved genomes and show that all conserved patterns emerge. Finally, we examine the simulation results at the molecular level to determine what molecular adaptations arise under selection against misfolding in this simplified system. These analyses yield predictions for future experimental studies.
Results
Organisms show a consistent correlation structure
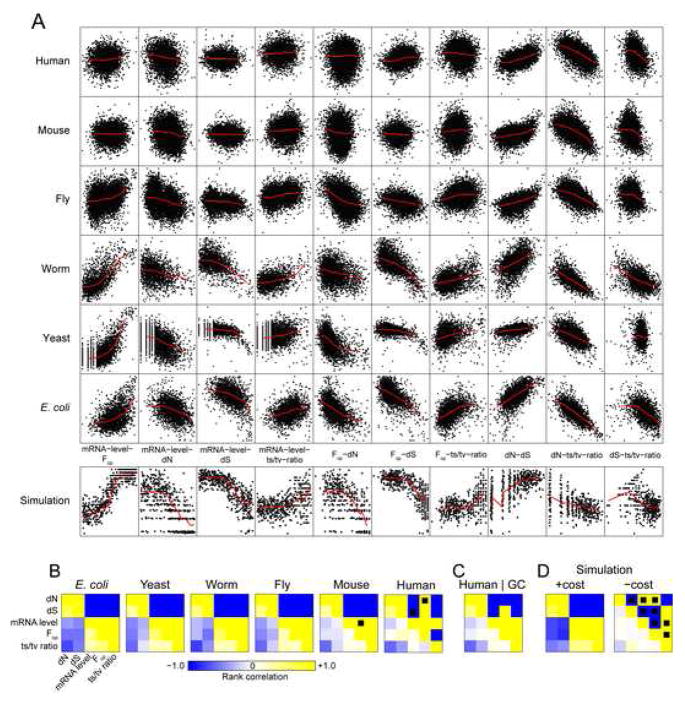
To systematically examine patterns of coding sequence evolution, we assembled measures of commonly studied variables (dN, dS, microarray-quantified mRNA expression level, codon bias measured by the fraction of optimal codons Fop [controlled for guanine+cytosine (GC) content in mammals; see Supplementary Experimental Procedures]), and the less-well-studied transition/transversion (ts/tv) ratio, for each of six model organisms: a gram-negative bacterium (E. coli), baker’s yeast (S. cerevisiae), a nematode worm (C. elegans), fruit fly (D. melanogaster), mouse (M. musculus), and human (H. sapiens), each with orthologous genes in a distinct species (see Experimental Procedures). As shown in Figure 1A, the data display remarkably consistent patterns of covariation despite considerable variability between organisms. Spearman rank correlation matrices show distinct similarities revealed clearly by the signs of the relationships (Figure 1B and Supplementary Table S1). All matrices show a block-like structure, with all signs absolutely conserved from E. coli to mouse. Deviations in human are consistent with known hypermutation at 5-methyl-cytosine in the primate lineage (Supplementary Results) and largely disappear after controlling for intronic GC content (Figure 1C), a reporter for such hypermutation. Weakening of correlations in higher organisms is limited to those involving mRNA level and Fop, suggesting reduced precision or accuracy in these measurements relative rather than weaker selection.
Figure 1.
Covariation of gene expression levels, patterns of codon usage, and rates of gene evolution are conserved across a bacterium, yeast, worm, fly, mouse, and human. A, All pairwise correlations between nonsynonymous and synonymous evolutionary rates (dN and dS), mRNA expression level, fraction of optimal codons (Fop), and transition-transversion ratio reveal conserved patterns of genome evolution across widely diverged taxa. Red lines show lowess-smoothed data. B, Correlation matrices and signs display a block structure. Correlation strengths (lower left of matrix) and signs (upper right of matrix) for each organism are shown; those with P > 0.05 after false-discovery-rate correction for multiple testing are shown overlaid with a black square. C, Human correlations controlled for intronic guanine and cytosine content recapitulate the structure conserved in other organisms, with the exception of a positive Fop–dS correlation. D, Similar correlations arise in a large-scale simulation involving selection against costs of mistranslation-induced protein misfolding (left), but not in the same simulation when mistranslation-induced misfolding imposes no cost (right).
The block-like correlation structure evident in Figure 1 suggests that the correlations reflect a unified underlying selective force. Principal component analysis (PCA) of each organism’s correlation matrix confirmed that a single underlying component explains 36–60% of the variance in all five analyzed variables (Figure 2A) and is conserved across species (Figure 2B, red box).
Figure 2.
Principal component analysis (PCA) of the correlation matrices in Figure 1 reveals a simple structure. A, The percentage of variance in all five variables explained by each of the first three principal components (of five; the remaining two are omitted for clarity). The dotted line indicates 20% of the variance, the cutoff for a component to have any meaningful explanatory value. B, Cluster analysis of the first three principal components reveals a tight cluster which contains the dominant principal components from all organisms and the misfolding-cost simulation (red box), but which excludes the dominant principal component from the no-cost simulation (green box).
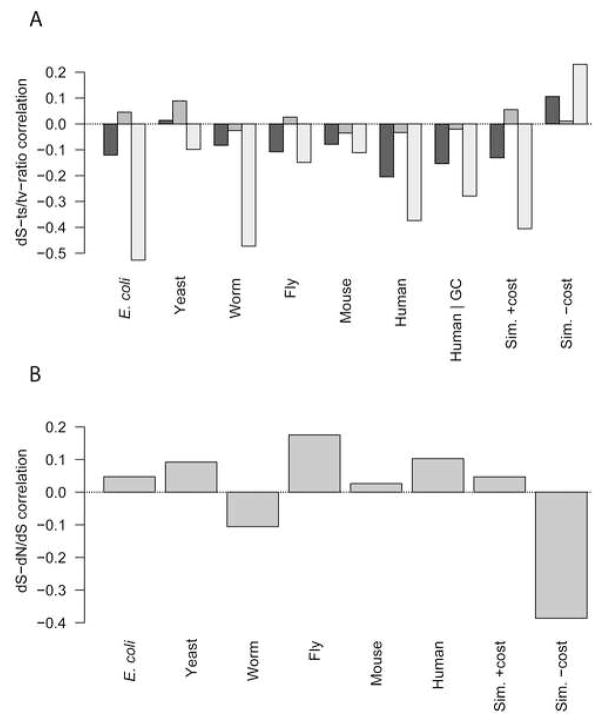
Correlations involving the transition/transversion ratio, an important measure of sequence change (Wakeley, 1996) whose variation between genes is little-studied, are both consistent and surprising. For example, because most substitutions occur in the third codon position, and most transition mutations (C↔T and A↔G)in the third position are synonymous, the ts/tv ratio and dS should positively correlate. Instead, the relationship is negative in all organisms. We consider explanations for these unexpected results later.
Selection for translational accuracy affects all organisms
Selection on translational efficiency in mammals is viewed as weak and contentious (Chamary et al., 2006) or absent (dos Reis et al., 2004). Using Akashi’s test (Akashi, 1994), an elegant and sensitive test for selection on translational accuracy that was previously only applied to fly (Akashi, 1994) and E. coli (Stoletzki and Eyre-Walker, 2007), we demonstrate that selection on translational accuracy affects mammals as well. The test quantifies the association between optimal codons and conserved residues, under the assumption that selection favors accurate codons at sites where substitutions are most harmful. The test’s comparison of only those codons within a gene that encode instances of the same amino acid renders it immune to biases arising from between-gene differences in expression level, evolutionary rate, amino acid composition, mRNA stability, and local nucleotide content.
In every organism including human and mouse, we found that optimal codons significantly associated with conserved sites (Table 1), implying that selection has positioned optimal codons to reduce the consequences of translation errors. We then computed the Akashi association score for all possible alternative sets of synonymous optimal codons. As Table 1 shows, the previously identified optimal codon set scored significantly higher than alternative sets in all organisms (distributions are shown in Supplementary Figure S1). These results suggest that both the set of optimal codons (i.e., the abundant tRNAs (Rocha, 2004)) and codon positions within genes interact to enhance translational accuracy. In yeast, worm, and fly, likelihood of finding optimal codons at conserved sites grew markedly stronger in the top 10% highest-expressed genes (Table 1). Mouse and human showed little change, as did E. coli; the latter finding may be rationalized by strong selection for translational speed in high-expression genes, which optimizes all codons and mutes the signature of accuracy selection.
Table 1.
Translational accuracy selection quantified by the intragenic association between “optimal” codons and conserved sites.
| All genes | Top 10% highest expression | |||||
|---|---|---|---|---|---|---|
| Organism | Z | Odds ratio | P(better codon set)a | Z | Odds ratio | P(better codon set) a |
| E. coli | 12.62*** | 1.108 | 0.0062 | 2.69** | 1.105 | 0.0650 |
| Yeast | 22.08*** | 1.125 | 0.0283 | 8.87*** | 1.308 | 0.0004 |
| Worm | 25.54*** | 1.135 | 4.58×10−6 | 7.66*** | 1.351 | 3.90×10−5 |
| Fly | 65.72*** | 1.362 | 2.72×10−7 | 17.72*** | 1.430 | 1.00×10−6 |
| Mouse | 35.37*** | 1.180 | 0.0049 | 5.86*** | 1.144 | 0.0268 |
| Human | 31.97*** | 1.160 | 0.0015 | 6.20*** | 1.171 | 0.0059 |
| Simulation | ||||||
| +cost | 4.07*** | 1.437 | 0.0255 | 0.82c | 3.000c | 0.1319c |
| −cost | −1.59 | 0.894 | 0.8277 | −1.01 | 0.807 | 0.8514 |
| +noiseb | 4.07*** | 1.437 | 0.0255 | 0.54 | 1.180 | 0.3718 |
Hypothesis: protein misfolding costs impose a major evolutionary constraint
The extraordinary lineage-spanning consistency of covariation in evolutionary rates, codon choice, and gene expression, within and between genes (Table 1 and Figure 1), demands a unified explanation. That the covariation structure suggests a dominant underlying factor or cost (Figure 2) motivates the search for the identity of that cost. We propose that adaptations to reduce the cellular burden imposed by protein misfolding creates all these covariation patterns. Furthermore, we suggest that selection against mistranslation-induced misfolding is necessary and sufficient to create them.
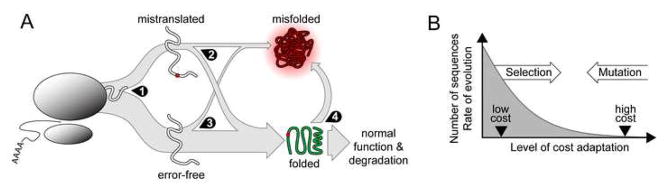
Misfolding costs can be reduced by four main adaptations (Figure 3A): increasing the proportion of properly translated proteins (increased translational accuracy), decreasing the proportion of proteins which misfold or unfold due to mistranslation (increased translational robustness), and decreasing the proportion of properly translated proteins which either fail to attain, or prematurely lose, their native structure (decreased stochastic misfolding and unfolding).
Figure 3.
The misfolding hypothesis. A, Outcomes of translation. Most proteins exit the ribosome (left) with no errors (bottom), but a substantial proportion contain at least one error (top). The probability of misfolding after correct translation is lower than after erroneous translation (center). Some proteins attain native state but then improperly unfold (right). Natural selection can act at four points: at 1), to reduce the frequency of translation errors in certain proteins; at 2), to reduce the proportion of error-containing proteins which misfold; at 3), to reduce the number of error-free proteins which misfold; and at 4), to reduce the number of proteins (with or without errors) which improperly unfold. B, Adaptations to higher misfolding costs constrain sequence evolution because adapted sequences are rare. So long as evolutionarily viable gene sequences are substantially more adapted than random sequences and adaptation levels are roughly bell-shaped in distribution, the number of alternative sequences (possible alleles) compatible with higher levels of adaptation (accuracy, robustness, etc.) declines rapidly. Adaptation to increasing misfolding costs therefore leads to increasing evolutionary constraint and slower sequence evolution.
For transcriptionally regulated genes, translation frequency increases with expression level, and, absent selection, so will the number of costly misfolded proteins generated by mistranslation or by stochastic misfolding (Figure 3A). Thus, high-expression genes impose steeper costs and should disproportionately display adaptations implied by Figure 3A. Assuming highly adapted alleles are rare, fewer mutations will lead to viable alternative alleles (Figure 3B), so costly, well-adapted genes will reject a higher proportion of mutations and evolve slowly.
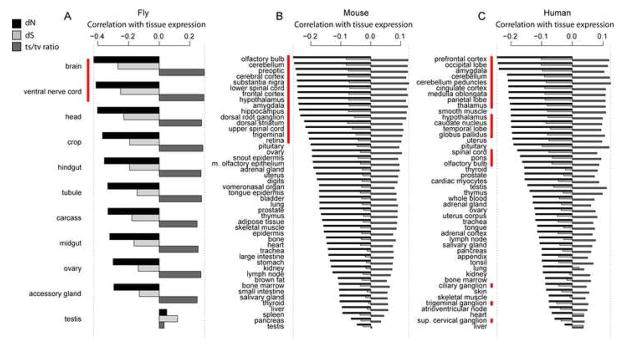
Neuronal tissues appear particularly sensitive to protein misfolding. Neurodegenerative diseases disproportionately involve protein misfolding and aggregation (Soto, 2003). The elaborately ramified structures and extraordinary cell length of many neurons confer a particularly high surface-area-to-volume ratio, increasing the likelihood of disruptive protein-membrane interactions (Kourie and Henry, 2002). Limited neuronal turnover makes cell loss more probable, and more likely permanent, under sustained chronic stress induced by misfolding. Malfunctioning of broadly expressed proteins involved in translation and protein folding manifests specifically neurotoxic effects in mouse (Lee et al., 2006; Zhao et al., 2005). Under our hypothesis, the differing sensitivity of certain cell types to misfolding will cause all the trends we identify in Figure 1 to be amplified in tissues comprised of sensitive cell types and muted in less-sensitive tissues.
Tissue expression modulates coding sequence evolution
To assess the prediction that evolutionary patterns should vary systematically across tissues, we computed correlations between dN, dS, or ts/tv ratio and tissue-specific mRNA levels in fly, mouse, and human, uncovering several strikingly consistent patterns (Figure 4). In all three organisms, expression in neural tissues shows the strongest correlation with dN, dS, and ts/tv ratio; in mouse and fly, all neural tissues show a stronger correlation of tissue-expression with dN than non-neural tissues. Moreover, the degrees to which each evolutionary variable correlates with tissue expression changes in a correlated way (minimum r = 0.78, P < 0.01 in all cases), as expected if all correlations arise from an underlying cost which varies by tissue. Both trends persist after restricting the analysis to those genes with below-median tissue specificity of expression (Supplementary Figure S2).
Figure 4.
Correlations of per-tissue mRNA levels with dN, dS, and ts/tv ratio for fly (A), mouse (B), and human (C, controlled for intronic guanine+cytosine content) vary systematically across tissues. Tissues composed primarily of neurons are indicated with a red bar. Dotted lines indicated the minimum and maximum correlation strengths.
Codon bias measured by Fop correlates with tissue expression similarly to dN, dS, and ts/tv ratio in fly, but not in mouse or human (not shown), likely due to tissue-specific codon usage (Dittmar et al., 2006).
A large-scale evolutionary simulation reproduces conserved patterns
The above results suggest that selection against misfolding underlies broad patterns of sequence evolution. To test our narrower hypothesis, that selection against mistranslation-induced protein misfolding suffices to create all these patterns, we turned to a large-scale computer simulation.
We created a population of 1,000 simulated organisms with 37.5kb genomes consisting of 500 coding nucleotide sequences (genes) expressed at different levels and translated, with occasional codon-dependent translation errors, into computationally foldable model proteins (Supplementary Figure S3). Translationally optimal codons were designated arbitrarily and were translated several-fold more accurately than their synonyms (Supplementary Table S3). The population evolved subject to mutation, drift, and selection, with fitness depending only on stable folding of the wild-type protein and the number of misfolded proteins generated by mistranslation. Proteins that failed to fold into the structure encoded by the native amino-acid sequence with a free energy of unfolding above 5 kcal/mol were designated misfolded. We ran simulations from identical initial conditions with and without a cost imposed by mistranslation-induced misfolding.
Figure 1A (bottom) shows the covariation of expression level, dN, dS, Fop, and ts/tv ratio in the simulation for comparison with real organisms. As Figure 1D shows, the simulated genes display all the consensus correlations found from E. coli to mouse, including the surprising negative correlation of ts/tv ratio with dS (r = −0.42***). When misfolding costs were eliminated, all but two correlations vanished (Figure 1D), and as expected in the absence of synonymous-site selection, the ts/tv-ratio–dS relationship turned positive (r = 0.24***). We hypothesized that the novel and counterintuitive negative association of transition/transversion ratio with dS was mediated by synonymous codon choice. This hypothesis predicts that the negative ts/tv-ratio–dS correlation will be strongest at third-codon-position sites, where transitions mostly lead to synonymous substitutions; weaker at first-codon-position sites, where only a small fraction of transitions are synonymous; and weakest at second-codon-position sites, where all transitions are nonsynonymous. Figure S5A confirms these predictions all organisms, and in the simulation, but only when misfolding costs are applied. These results recall the recent report of a positive dS–dN/dS correlation in mouse and human, claimed to be inconsistent with present models of molecular evolution (Wyckoff et al., 2005). We find that this correlation exists in all organisms but worm, is reproduced in our simulation only in the presence of misfolding costs (Figure S5B), and is expected when dN and dS are not independent (Supplementary Results), as when both are constrained by adaptations to combat misfolding.
Tests for translational accuracy (Table 1) and the results of principal component analysis on the simulation’s correlation matrix (Figure 2) matched the organismal results, but only when misfolding imposed a fitness cost. To examine whether inaccuracy or imprecision of mRNA measurements and codon bias in indicating translational frequency and accuracy could produce the weaker results in higher organisms (Figure 2), we added noise to expression level and Fop, performed PCA on this noisy simulation, and found close qualitative agreement with mouse and human (Figure 2).
In the previous simulations, all genes were essential, suppressing any potential evolutionary variation due to differing importance of genes. We then asked whether the covariation patterns could be explained by functional importance, as measured by the fitness effect of deletion or “dispensability” (Wall et al., 2005). We performed simulations under a fitness function in which each molecule expressed from a gene contributed equally to fitness, but the total contribution of that gene to fitness—the cost of losing all molecules, i.e. the dispensability—was given by the fitness defect upon gene deletion. To ensure the proper relationship between mRNA level and dispensability, values were sampled from the joint distribution observed in yeast. All correlations with mRNA level weakened substantially; the strong link between mRNA-level and dN, and the correlation between mRNA level and ts/tv-ratio, both found in all real organisms, became insignificant (Supplementary Figure S4). To ensure that the alternate fitness function, not the altered distribution of expression levels, was responsible for the failure to match biological observations, we evolved the new sampled genome under the original fitness function, where each misfolded molecule imposed equal fitness costs, and obtained results similar to the original ones (Supplementary Figure S4).
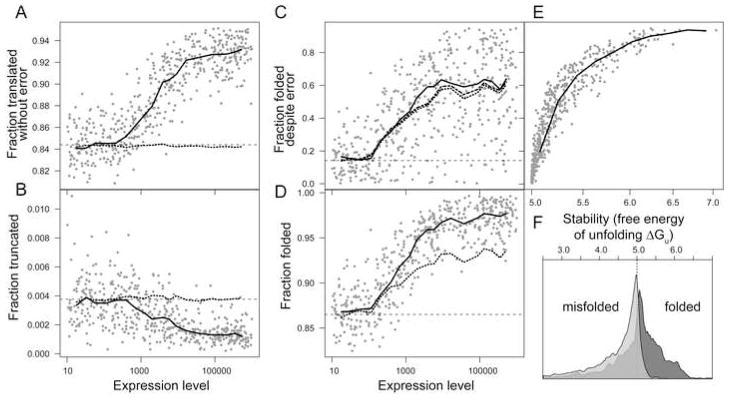
By tracking the fates of 10,000 proteins translated from each evolved gene, we dissected the precise adaptations that occurred as a function of expression level, the only non-random independent variable in the simulation. Figure 6A–D shows that translational accuracy, translational robustness (the propensity of proteins to fold properly despite mistranslation), production of full-length polypeptides, and overall propensity to fold properly all increase with expression level relative to the baseline observed in the no-cost simulation.
Figure 6.
Outcomes of translation in the simulation vary with expression level, and can be attributed to selection on the amino-acid sequence or the nucleotide sequence. A, Fraction of accurately translated polypeptides. B, Fraction of mistranslated polypeptides that fold properly, a measure of translational robustness. C, Fraction of truncated polypeptides. D, Fraction of folded proteins. Lines show the sliding-window medians for translation outcomes generated by genes evolved with a cost for misfolding (solid), by ensembles of genes with randomly chosen codons encoding the same protein sequences (dotted), and by genes evolved under no cost (dashed baseline). In C, a dash-dot line shows the proportion of mistranslated but full-length (not truncated) polypeptides that fold properly. E, Translational robustness is tightly linked to increases in thermodynamic stability (free energy of unfolding). F, Stability distributions of mistranslated proteins (containing at least one error) translated from the 100 lowest-expression (light gray) and 100 highest-expression (dark gray) genes. Arrows indicate the median free energy of each subpopulation. A free energy of unfolding of 5 kcal/mol (dotted line) is the minimum to be considered stably folded in the model.
After recoding each gene with randomly chosen synonymous codons, the proportion of accurately translated proteins regressed to the mean of the no-cost simulation (Figure 6A), tying differences in translational accuracy to synonymous codon usage. By contrast, the fraction of tolerated errors was dominated by adaptation at the protein level (Figure 6B). As predicted (Drummond et al., 2005), translationally robust proteins achieved error tolerance through increased stability as measured by the free energy of unfolding (Figure 6E), and fewer translation errors resulted in destabilization past the threshold for stable folding (Figure 6F). Note that relative to the average protein, high-expression proteins tolerate more substitutions at the ribosome (Figure 6B) but tolerate fewer substitutions over evolutionary time (Figure 1A, bottom). Genes evolve slowly because most mutations are deleterious. Most mutations to a highly expressed gene encoding a translationally robust protein yield sequences encoding other folded proteins, but not other robust proteins. Because loss of translational robustness elevates mistranslation-induced misfolding and is therefore deleterious, such mutations will fail to rise to fixation, and the gene will accumulate few changes over evolutionary time (Wilke and Drummond, 2006).
Discussion
Our analyses reveal conserved patterns of covariation linking common measures of evolutionary change and gene expression across the genomes of E. coli, yeast, worm, fly, mouse, and human. Principal component analysis suggests that all these pairwise correlations are manifestations of a unified underlying selective pressure. We propose, and demonstrate using a molecular-level simulation, that selection against protein misfolding, particularly misfolding induced by missense errors at the ribosome, suffices to create all the observed patterns. Because we focus on patterns of covariation, our results do not imply that all variation in molecular evolution can be explained by protein misfolding, or that protein function plays a negligible role.
What is the nature of the fitness cost imposed by misfolding? Misfolded proteins manifest fitness costs unrelated to their function; such costs may involve direct toxicity, such as disruption of membrane integrity or inappropriate interactions with other cellular components (Stefani and Dobson, 2003); indirect toxicity, such as heightened sensitivity to stresses such as heat shock; or other indirect efficiency burdens, such as the energy expended in synthesizing, detecting, and degrading misfolded proteins, or the squandering of ribosomal capacity on useless products (Stoebel et al., 2008). Although our results cannot rule out any of these costs, previous analyses have shown little to no role for amino-acid cost or polypeptide length, the major correlates of synthesis and degradation cost, in shaping evolutionary rate variation (Drummond et al., 2006; Rocha and Danchin, 2004). Under theories—often informed by results of microbial competition assays (Stoebel et al., 2008)—which link fitness costs to ribosomal throughput or translational efficiency, the strengthening of evolutionary effects with expression in neurons has no clear explanation, since many neurons do not divide at all and it seems unlikely that neuronal translational capacity limits animal reproduction in a way analogous to microbes. By emphasizing neuronal cell loss, with its clear consequences for the animal (Lee et al., 2006), the hypothesis that cells are sensitive to cytotoxic misfolded proteins accounts for these trends in a way which also applies to microbial growth.
Misfolding also steals a potentially functional molecule from the cell. If the loss of functional molecules imposes the dominant fitness cost, essential genes determined by systematic deletion studies should evolve much more slowly than dispensable genes, because deletions result in the loss of all functional molecules. Instead, in yeast, essential proteins evolve 16% slower than all proteins, compared to a ~10-fold difference linked to gene expression (mRNA level, codon usage, and protein abundance) (Drummond et al., 2006). When our simulation was modified to model functional importance as measured by yeast deletion studies (Wall et al., 2005; Warringer et al., 2003), the strong mRNA-level–dN correlation vanished (Supplementary Figure S4). In this alternate model, highly expressed proteins evolve slowly only as a side-effect of the relationship between dispensability and mRNA level, a weak correlation in yeast (r = −0.18***). In general, functional arguments to explain why low-expression proteins evolve rapidly imply that low-expression proteins have little functional importance. Cell-biological considerations undermine this notion: many essential gene products (such as those involved in DNA replication) have intrinsically low-copy-number targets (such as replication forks) and are therefore weakly expressed. The misfolding hypothesis resolves the conflict by decoupling evolutionary rate from importance. However, while our results suggest that loss of functional molecules is unlikely to be the dominant evolutionary cost uncovered here, function doubtless applies some constraint. The nature of the cellular cost imposed by misfolding remains unresolved in studies of misfolding diseases after substantial study, and obtaining unambiguous evidence to resolve this question in an evolutionary context will likely require similar sustained efforts, particularly experimental measurements of the magnitude and mechanistic basis of growth-rate costs arising from chronic, low-frequency protein misfolding.
Influences of the distribution of expression across tissues
Substantial attention has been devoted to the dependence of evolutionary rate, particularly dN, on the breadth, tissue specificity, and aggregate level of expression in multicellular organisms (Duret and Mouchiroud, 2000; Liao et al., 2006; Pál et al., 2006; Parmley et al., 2007; Wright et al., 2004). Our results suggest that such summaries of expression patterns, although predictive of evolutionary rates (Supplementary Table S4), obscure key biological differences between tissues. Recent studies show that mutations in broadly expressed genes involved in translation and protein folding produce brain-specific phenotypes (Lee et al., 2006; Zhao et al., 2005), suggesting that neural tissues suffer disproportionately given systemic misfolding. Our findings hint that such sensitivity shapes sequence change over evolutionary time: mRNA levels in animal neural tissues consistently predict evolutionary rates better than levels in other tissues (Figure 4), even when tissue-specific genes are discarded.
Previous studies have identified brain-expressed genes as slow-evolving (Wang et al., 2007; Zhang and Li, 2004); that slow evolution accompanies expression in neural tissues outside the brain (e.g. spinal cord, or ventral nerve cord in fly) implicates a constraint operating on neurons rather than on the brain per se. That dS and ts/tv ratio relationships parallel dN across tissues demands an explanation beyond functional selection or complex regulation. Neuronal sensitivity to mistranslation-induced misfolding (Lee et al., 2006) predicts all these observations.
Experimental predictions
Our hypothesis makes a number of experimentally testable predictions. Given a pair of genes of similar structure where the first is expressed more highly and evolves more slowly than its partner, the first encoded protein is predicted to misfold less often than the second. In yeast, duplicated genes provide ample such pairs (Drummond et al., 2005), including many constitutive and glucose-repressed enzyme isoforms (e.g. pyruvate kinases 1 and 2). Similarly, the higher-expressed, slower-evolving duplicate gene is predicted to encode a protein with higher thermodynamic stability than its paralog.
We predict that misfolded proteins will disproportionately contain translation errors when compared to folded proteins. If instead stochastic misfolding of error-free polypeptides dominates protein misfolding, this prediction would be falsified. We further predict that putatively translationally robust (higher-expressed, slower-evolving) genes should more often generate folded proteins after ribosomal errors—and DNA point mutations which simulate those errors—than their more fragile counterparts.
In formulating our hypotheses, we have drawn heavily on studies of disease-related protein misfolding. On virtually any dimension (population of the unfolded state, rate of premature unfolding, failure to export, aggregation propensity, and so on), proteins containing translational missense errors are generally expected to show pathological behavior more often than error-free proteins—indeed, this is why genetic missense mutations are thought to cause disease.
Decreased translational fidelity arising from mutations in translation-related genes such as alanyl-tRNA synthetase (Lee et al., 2006) has been argued to contribute to heritable disease (Bacher and Schimmel, 2007), but is likely to yield widespread misfolding, inconsistent with some misfolding diseases in which specific proteins unrelated to translation or protein folding carry mutations. Our hypothesis emphasizes that proteins differ in their tolerance to translation errors that occur given normal fidelity, and that lower tolerance to translation errors of certain proteins and associated mutants provides a novel mechanism to explain their propensity to misfold pathologically. Such a mechanism is most likely to operate when 1) the disease arises despite proper folding of the majority of disease-associated protein molecules, 2) disease symptoms are associated with toxic protein misfolding and cell stress; 3) disease severity increases with the degree of mutation-induced destabilization of the protein, 4) many missense mutations across the gene cause disease; and 5) the protein is highly expressed in the affected tissues. SOD1-linked familial and sporadic forms of amyotrophic lateral sclerosis (ALS) (Gruzman et al., 2007) and central nervous system amyloidosis (TTR-CNSA) linked to kinetically stable, thermodynamically unstable variants of transthyretin (Sekijima et al., 2005) meet all of these criteria.
Mistranslation-induced misfolding offers a largely unexplored mechanism for generating pathological misfolding of a subset of protein molecules in the absence of genetic mutations, a potentially valuable avenue of inquiry given the large fraction of sporadic ALS cases. Our hypothesis, which holds that mistranslation is a contributor but not the sole cause, makes the following testable predictions: misfolded, aggregated SOD1 and TTR molecules, particularly those in early-forming toxic oligomers, will contain significantly more missense errors than the soluble species, and aggregation and premature degradation, predictors of disease onset and severity, will be reduced under conditions of elevated translational fidelity.
Broader consequences
Our results suggest a substantial rethinking of several widely credited hypotheses in molecular evolution. First, most variation in evolutionary rates appears to be attributable to regulatory rather than functional differences, and translation, often ignored, may play a central role. Second, synonymous changes are not silent in any of the organisms studied, and as a direct consequence, the common practice of using dN/dS (equivalently, Ka/Ks) as a measure of protein divergence controlled for neutral divergence should be reconsidered. If, as our results strongly suggest, both dN and dS reflect translational selection in most organisms, then analyses of brain evolution which ascribe evolutionary significance to increases in this ratio (Dorus et al., 2004) should be revisited. Finally, in place of common claims that sequence evolution in multicellular organisms differs in qualitative ways from that in microbes, our results rather suggest a continuum. C. elegans is a behaving multicellular eukaryotic animal with multiple tissues, and yet on the dimensions analyzed in the present work, its genes evolve almost indistinguishably from those of E. coli. If infidelity of the evolutionary ancient translational apparatus indeed governs sequence change, the model-organism paradigm may be exploited with unusual confidence in efforts to uncover the relevant cell biology.
Experimental Procedures
Genome and transcriptome data and evolutionary rate measurements
Detailed methods, data sources, and gene count data are listed in Supplementary Experimental Procedures. Briefly, coding DNA sequences were built from coding exon sequences which were extracted from chromosomal DNA sequences. Protein alignments of translated orthologous gene sequences were generated with MUSCLE 3.6 (Edgar, 2004) and used to align nucleotide sequences. A single cDNA per gene was randomly chosen from each gene that showed evidence of alternative splicing. Only cDNAs with 80% alignment to their ortholog, dS < 2 except as noted, and at least 30 codons were retained. Whole-organism and per-tissue mRNA-level data were downloaded and, for human, mouse, and fly, combined using a geometric mean across tissues (detailed procedures in Supplementary Materials). Evolutionary rates and transition/transversion rate ratios were computed by maximum likelihood with PAML (Yang, 2006; Yang, 1997) using a physical-sites definition (Bierne and Eyre-Walker, 2003) operating on codons (codeml program) with the F3×4 codon frequency model, one dN/dS ratio per branch (model 0), and an arbitrary seed ts/tv rate ratio of 3.4. Ts/tv ratios were computed by counting transitions and transversions separating orthologous sequences, adding 1 to each (Laplace estimation), and taking their ratio. Distributions of dN and dS for all organisms are shown in Supplementary Figure S5. The fraction of optimal codons, Fop, was calculated as described (Duret and Mouchiroud, 1999) using published optimal codons except in mouse (cf. Supplementary Table S2).
Translational accuracy selection was first tested exactly as described using Akashi’s test (Akashi, 1994). Sites with the same amino acid at the aligned position in the orthologous gene (or for the simulation, in all ancestral proteins on the line of descent) were designated conserved. In a second test, significance of the optimal–conserved association, randomized over the choice of optimal codon set, was assessed by computing the odds ratio for all possible alternate optimal codon sets which preserve the number of optimal codons per synonymous family in the naturally occurring set.
Simulation protocol
500 in silico nucleotide sequences of length L = 75 encoding model polypeptides that folded into a specific native structure with a stability (free energy of unfolding) of at least 5 kcal/mol were found and folded as described (Wilke, 2004). Evolution of each gene at a rate of μ = 10−6 mutations/site/generation proceeded in parallel in a population of 1,000 genes. Parallel evolution, employed to make large-scale evolution tractable, rests on the assumptions of no linkage, independent mutations (plausible because Ne μL = 0.075 < 1), and fitness as defined below. Nucleotide mutat\ions were equally likely (e.g., no mutational transition bias). To simulate regulated expression, polypeptides translated from a gene were folded until a target number of folded proteins, the gene’s expression level x, was obtained. The translation error spectrum was implemented as described (Freeland and Hurst, 1998). Translation at codons identified as optimal for yeast (Sharp and Cowe, 1991) proceeded with five-fold higher accuracy on average than at non-optimal codons and roughly 15% of low-expression polypeptides contained an error, consistent with biological expectations (Drummond et al., 2005). Misfolding resulted from mistranslation leading to truncation, adoption of a non-native conformation, or native-state stability of less than 5 kcal/mol. Toxicity of misfolded proteins was assumed to derive from exposure of interaction-prone buried residues (Bucciantini et al., 2002), and aggregation was not modeled explicitly. The likelihood of reproduction was proportional to organism fitness (Wright-Fisher sampling), which was in turn determined by the number of toxic misfolded proteins m according to fitness(m) = e_−_cm (see Supplementary Results) where c is a positive constant and m is the amount of misfolded protein. If f is the fraction of folded proteins translated, then m = x(1_−_f)/f. We chose c = 0.0001; altering c is equivalent to raising or lowering all expression levels. Supplementary Figure S6 assesses the fitness disadvantage of codon changes as a function of expression level for perspective. An alternate fitness function incorporating protein importance or dispensability was also used in which fitness = es(1_−_f), where s is the additive growth-rate effect of gene deletion (i.e., f = 0), s = ln(deletion-strain growth rate/max growth rate) (see Supplementary Experimental Procedures). Source code is available upon request.
Supplementary Material
01
Figure 5.
Unexpected correlations between dS and other variables can be explained by selection against mistranslation-induced misfolding. A, An unexpected negative correlation between dS and transition/transversion ratio is strongest at third-position sites and weakest at second-codon-position sites in all six organisms, and the simulation under conditions where mistranslation-induced protein misfolding imposes a cost. For each organism, from left to right, the Spearman rank correlation between whole-gene dS and the transition/transversion ratio only for substitutions occurring in the first, second, or third codon positions in each gene are shown. B, An unexpected positive correlation arises between dS and dN/dS in most organisms and in the simulation when mistranslation-induced misfolding is costly; Spearman rank dS–dN/dS correlations in each organism and the simulations are shown.
Acknowledgments
We are grateful to F. H. Arnold for invaluable guidance and support in developing this work. We thank C. Adami for support; S. Eddy for tRNA gene count data; J. Cuff and B. Baer for computational assistance; B. Stern, E. O’Shea, A. Murray, M. Laub, J. Plotkin, A. Kieu, and members of the FAS Center for Systems Biology for helpful discussions. This work was supported by grants from the National Institutes of Health (C.O.W., and an NIH Center grant to the FAS Center for Systems Biology). D.A.D. conceived of the study and performed the analyses, and D.A.D. and C.O.W. designed the research, constructed the simulation, and wrote the paper. The authors declare no conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfinsen C. The Molecular Basis of Evolution. New York: John Wiley & Sons, Inc; 1959. [Google Scholar]
- Bacher JM, Schimmel P. An editing-defective aminoacyl-tRNA synthetase is mutagenic in aging bacteria via the SOS response. Proc Natl Acad Sci U S A. 2007;104:1907–1912. doi: 10.1073/pnas.0610835104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N, Eyre-Walker A. The problem of counting sites in the estimation of the synonymous and nonsynonymous substitution rates: implications for the correlation between the synonymous substitution rate and codon usage bias. Genetics. 2003;165:1587–1597. doi: 10.1093/genetics/165.3.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD, Labthavikul S, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc Natl Acad Sci U S A. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles E, Schulte F, Miosga T, Freidel K, Schluter E, Zimmermann FK, Hollenberg CP, Heinisch JJ. Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is catalytically insensitive to fructose-1,6-bisphosphate. J Bacteriol. 1997;179:2987–2993. doi: 10.1128/jb.179.9.2987-2993.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006;312:97–101. doi: 10.1681/01.asn.0000926836.46869.e5. [DOI] [PubMed] [Google Scholar]
- Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- Choi JK, Kim SC, Seo J, Kim S, Bhak J. Impact of transcriptional properties on essentiality and evolutionary rate. Genetics. 2007;175:199–206. doi: 10.1534/genetics.106.066027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: the functional synthesis. Nat Rev Genet. 2007;8:675–688. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS genetics. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, Mahowald M, Wyckoff GJ, Malcom CM, Lahn BT. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci USA. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Raval A, Wilke CO. A single determinant dominates the rate of yeast protein evolution. Mol Biol Evol. 2006;23:327–337. doi: 10.1093/molbev/msj038. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci U S A. 1999;96:4482–4487. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17:68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeland SJ, Hurst LD. The genetic code is one in a million. J Mol Evol. 1998;47:238–248. doi: 10.1007/pl00006381. [DOI] [PubMed] [Google Scholar]
- Gruzman A, Wood WL, Alpert E, Prasad MD, Miller RG, Rothstein JD, Bowser R, Hamilton R, Wood TD, Cleveland DW, et al. Common molecular signature in SOD1 for both sporadic and familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2007;104:12524–12529. doi: 10.1073/pnas.0705044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HH, Choe J, Loeb LA. Protein tolerance to random amino acid change. Proc Natl Acad Sci U S A. 2004;101:9205–9210. doi: 10.1073/pnas.0403255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Ingvarsson PK. Gene Expression and Protein Length Influence Codon Usage and Rates of Sequence Evolution in Populus tremula. Mol Biol Evol. 2007;24:836–844. doi: 10.1093/molbev/msl212. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI. Evolutionary systems biology: links between gene evolution and function. Curr Opin Biotechnol. 2006;17:481–487. doi: 10.1016/j.copbio.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Kourie JI, Henry CL. Ion channel formation and membrane-linked pathologies of misfolded hydrophobic proteins: the role of dangerous unchaperoned molecules. Clinical and experimental pharmacology & physiology. 2002;29:741–753. doi: 10.1046/j.1440-1681.2002.03737.x. [DOI] [PubMed] [Google Scholar]
- Kramer EB, Farabaugh PJ. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. Rna. 2007;13:87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov DM, Wolf YI, Rogozin IB, Koonin EV. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13:2229–2235. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- Lemos B, Bettencourt BR, Meiklejohn CD, Hartl DL. Evolution of Proteins and Gene Expression Levels are Coupled in Drosophila and are Independently Associated with mRNA Abundance, Protein Length, and Number of Protein-Protein Interactions. Mol Biol Evol. 2005 doi: 10.1093/molbev/msi122. [DOI] [PubMed] [Google Scholar]
- Li WH, Wu CI, Luo CC. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- Liao BY, Scott NM, Zhang J. Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Mol Biol Evol. 2006;23:2072–2080. doi: 10.1093/molbev/msl076. [DOI] [PubMed] [Google Scholar]
- Marais G, Domazet-Loso T, Tautz D, Charlesworth B. Correlated evolution of synonymous and nonsynonymous sites in Drosophila. J Mol Evol. 2004;59:771–779. doi: 10.1007/s00239-004-2671-2. [DOI] [PubMed] [Google Scholar]
- Markiewicz P, Kleina LG, Cruz C, Ehret S, Miller JH. Genetic studies of the lac repressor. XIV. Analysis of 4000 altered Escherichia coli lac repressors reveals essential and non-essential residues, as well as “spacers” which do not require a specific sequence. J Mol Biol. 1994;240:421–433. doi: 10.1006/jmbi.1994.1458. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Pakula AA, Sauer RT. Genetic analysis of protein stability and function. Annu Rev Genet. 1989;23:289–310. doi: 10.1146/annurev.ge.23.120189.001445. [DOI] [PubMed] [Google Scholar]
- Pál C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C, Papp B, Lercher MJ. An integrated view of protein evolution. Nat Rev Genet. 2006;7:337–348. doi: 10.1038/nrg1838. [DOI] [PubMed] [Google Scholar]
- Parker J. Errors and alternatives in reading the universal genetic code. Microbiol Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmley JL, Urrutia AO, Potrzebowski L, Kaessmann H, Hurst LD. Splicing and the evolution of proteins in mammals. PLoS Biol. 2007;5:e14. doi: 10.1371/journal.pbio.0050014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu CE, Borza T, Bielawski JP, Lee RW. Evolutionary rates and expression level in Chlamydomonas. Genetics. 2006;172:1567–1576. doi: 10.1534/genetics.105.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP. Codon usage bias from tRNA’s point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 2004;14:2279–2286. doi: 10.1101/gr.2896904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP, Danchin A. An analysis of determinants of amino acids substitution rates in bacterial proteins. Mol Biol Evol. 2004;21:108–116. doi: 10.1093/molbev/msh004. [DOI] [PubMed] [Google Scholar]
- Sekijima Y, Wiseman RL, Matteson J, Hammarstrom P, Miller SR, Sawkar AR, Balch WE, Kelly JW. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121:73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Cowe E. Synonymous codon usage in Saccharomyces cerevisiae. Yeast. 1991;7:657–678. doi: 10.1002/yea.320070702. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol. 1987;4:222–230. doi: 10.1093/oxfordjournals.molbev.a040443. [DOI] [PubMed] [Google Scholar]
- Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nature reviews. 2003;4:49–60. doi: 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- Stefani M. Generic cell dysfunction in neurodegenerative disorders: role of surfaces in early protein misfolding, aggregation, and aggregate cytotoxicity. Neuroscientist. 2007;13:519–531. doi: 10.1177/1073858407303428. [DOI] [PubMed] [Google Scholar]
- Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- Stoebel DM, Dean AM, Dykhuizen DE. The Cost of Expression of Escherichia coli lac Operon Proteins Is in the Process, Not in the Products. Genetics. 2008;178:1653–1660. doi: 10.1534/genetics.107.085399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletzki N, Eyre-Walker A. Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol Biol Evol. 2007;24:374–381. doi: 10.1093/molbev/msl166. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics. 2004;168:373–381. doi: 10.1534/genetics.104.028944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J. The excess of transitions among nucleotide substitutions: new methods of estimating transition bias underscore its significance. Trends Ecol Evol. 1996;11:158–162. doi: 10.1016/0169-5347(96)10009-4. [DOI] [PubMed] [Google Scholar]
- Wall DP, Hirsh AE, Fraser HB, Kumm J, Giaever G, Eisen MB, Feldman MW. Functional genomic analysis of the rates of protein evolution. Proc Natl Acad Sci U S A. 2005;102:5483–5488. doi: 10.1073/pnas.0501761102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Chien HC, Osada N, Hashimoto K, Sugano S, Gojobori T, Chou CK, Tsai SF, Wu CI, Shen CK. Rate of evolution in brain-expressed genes in humans and other primates. PLoS Biol. 2007;5:e13. doi: 10.1371/journal.pbio.0050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warringer J, Ericson E, Fernandez L, Nerman O, Blomberg A. High-resolution yeast phenomics resolves different physiological features in the saline response. Proc Natl Acad Sci U S A. 2003;100:15724–15729. doi: 10.1073/pnas.2435976100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke CO. Molecular clock in neutral protein evolution. BMC Genet. 2004;5:25. doi: 10.1186/1471-2156-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke CO, Drummond DA. Population genetics of translational robustness. Genetics. 2006;173:473–481. doi: 10.1534/genetics.105.051300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Carlson SS, White TJ. Biochemical evolution. Annu Rev Biochem. 1977;46:573–639. doi: 10.1146/annurev.bi.46.070177.003041. [DOI] [PubMed] [Google Scholar]
- Wright SI, Yau CB, Looseley M, Meyers BC. Effects of Gene Expression on Molecular Evolution in Arabidopsis thaliana and Arabidopsis lyrata. Mol Biol Evol. 2004;21:1719–1726. doi: 10.1093/molbev/msh191. [DOI] [PubMed] [Google Scholar]
- Wyckoff GJ, Malcom CM, Vallender EJ, Lahn BT. A highly unexpected strong correlation between fixation probability of nonsynonymous mutations and mutation rate. Trends Genet. 2005;21:381–385. doi: 10.1016/j.tig.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Yang Z. Computational Molecular Evolution. Oxford, UK: Oxford University Press; 2006. [Google Scholar]
- Yang Z, Nielsen R. Mutation-selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol Biol Evol. 2008;25:568–579. doi: 10.1093/molbev/msm284. [DOI] [PubMed] [Google Scholar]
- Yang ZH. PAML: a program package for phylogenetic analysis by maximum likelihood. Computer Applications in the Biosciences. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li WH. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol Biol Evol. 2004;21:236–239. doi: 10.1093/molbev/msh010. [DOI] [PubMed] [Google Scholar]
- Zhao L, Longo-Guess C, Harris BS, Lee JW, Ackerman SL. Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat Genet. 2005;37:974–979. doi: 10.1038/ng1620. [DOI] [PubMed] [Google Scholar]
- Zhu G, Golding GB, Dean AM. The selective cause of an ancient adaptation. Science. 2005;307:1279–1282. doi: 10.1126/science.1106974. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E. Evolutionary Processes and Evolutionary Noise at the Molecular Level I. Functional Density in Proteins. J Mol Evol. 1976;7:167–183. doi: 10.1007/BF01731487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01