Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger (original) (raw)
. Author manuscript; available in PMC: 2009 Dec 11.
Published in final edited form as: Nature. 2009 Jun 11;459(7248):847–851. doi: 10.1038/nature08036
Abstract
Histone H3 Lys4 methylation (H3K4me) was proposed as a critical component in regulating the gene expression, epigenetic states, and cellular identities1. The biological meaning of H3K4me is interpreted via conserved modules including _p_lant _h_omeo_d_omain (PHD) fingers that recognize varied H3K4me states1,2. The dysregulation of PHD finger has been implicated in a variety of human diseases including cancers and immune or neurological disorders3. Here we report that fusing an H3K4-trimethylation (H3K4me3)-binding PHD finger, such as the C-terminal PHD finger of JARID1A or PHF23 (JARID1APHD3, PHF23PHD), to a common fusion partner nucleoporin-98 (NUP98) as identified in human leukemias4,5, generated potent oncoproteins that arrested hematopoietic differentiation and induced acute myeloid leukemia (AML). In these processes, a PHD finger that specifically recognizes H3K4me3/2 marks was essential for leukemogenesis. Mutations in PHD fingers that abrogated H3K4me3-binding also abolished leukemic transformation. NUP98-PHD fusion prevented the differentiation-associated removal of H3K4me3 at many loci encoding lineage-specific transcription factors (Hox(s), Gata3, Meis1, Eya1, Pbx1), and enforced their active gene transcription. Mechanistically, NUP98-PHD fusions act as ‘chromatin boundary factors’, dominating over polycomb-mediated gene silencing to ‘lock’ developmentally crucial loci into an active chromatin state (H3K4me3 with induced histone acetylation), a state that defined leukemia stem cells. Collectively, our studies represent the first report wherein the deregulation of PHD finger, ‘effector’ of specific histone modification, perturbs the epigenetic dynamics on developmentally critical loci, catastrophizes cellular fate decision-making, and even causes oncogenesis during development.
Recent studies have showed that an H3K4me3-binding PHD finger in the NURF, ING2 or TFIID complex helps to recruit and/or stabilize these effectors and associated factors onto appropriate target promoters during transcriptional regulation1,6-10; An unmodified H3K4 (H3K4me0)-engaging PHD finger in the DNMT3L or LSD1-complex connects activities of DNA methylation or H3K4 demethylation to repressive chromatin11,12. Interestingly, germ-line mutation in the PHD finger of RAG2 abrogates its recognition of H3K4me3 and causes immunodeficiency13; Mutations in the PHD finger of ING1 have been implicated in cancers3,8,14. However, evidence supporting a causal role for PHD finger mutation and inappropriate interpretation of histone modification in oncogenesis is still elusive.
In clinically reported AML patients4,5, chromosomal translocation fuses the C-terminal PHD finger of JARID1A (also known as RBP2/KDM5A) or PHF23, together with nuclear localization signals, to NUP98, a common leukemia fusion partner that harbors transactivation activities15-17 (Supplementary Fig.1). Notably, the JARID1APHD3 motif is excluded from an alternatively spliced isoform of JARID1A and the corresponding NUP98-JARID1A fusion (hereafter referred to as NJS), while it is retained in the longer fusion isoform (hereafter referred to as NJL; Fig.1a). We asked whether JARID1APHD3 as a putative chromatin-‘reading’ module is involved in hematopoietic malignancies. To test this, we examined leukemogenic potential of both fusion isoforms using hematopoeitic progenitor transformation assay18 (Supplementary Fig.2a). While bone marrow-derived hematopoietic stem/progenitor cells transduced with empty retrovirus or retrovirus encoding NJS proliferated transiently and differentiated into mature cells, those transduced with NJL proliferated indefinitely as undifferentiated progenitors (Fig.1b-c). NJL-transduced marrow cells proliferated in a cell-autonomous manner, exhibited typical myeloblast morphology (Fig.1d) and expressed early myeloid progenitor antigens (c-Kit+/Cd11b+/Cd34+/Gr-1-/Cd19-/B220-/low; Fig.1e and Supplementary Fig. 2b). The arres of myeloid differentiation by NJL indicated that it would induce leukemia in vivo. Indeed, all of 12 mice transplanted with bone marrow progenitors transduced with NJL died of AML in an average of 69 days, whereas those reconstituted with vector- or NJS-transduced progenitors remained healthy after one year (Fig.1f). NJL-induced leukemia exhibited a myeloid phenotype (Supplementary Fig.2c-d), and typically presented with an enlarged spleen, packed progenitors in bone marrow, and massive increase in peripheral white blood cells (Supplementary Table 1; Fig.1g-h). Taken together, NJL represents a potent leukemia oncogene in both cellular and animal models.
Figure 1. The PHD finger-containing NUP98-JARID1A fusion isoform (NJL), but not that lacking the PHD finger (NJS), confers leukomogenic potentials to hematopoietic stem/progenitor cells.
a, NUP98-JARID1A and NUP98-PHF23 structure (see Supplementary Fig.1 for details). b, Immunoblot of hematopoietic cells transduced with empty vector (lanes 1-2) or that encoding FLAG-tagged NJS (lanes 3-4) or NJL (lanes 5-6). c, Proliferation kinetics of lineage-negative hematopoietic cells after transduction of empty vector, NJL or NJS. Data are presented as mean ±s.d. of 6 experiments. d, Wright-Giemsa staining (insert, microscopy image) and e, FACS of NJL-transformed cells. f, Leukemia kinetics in mice (12 each group) after transplantation of bone marrow transduced with vector, NJL or NLS. g, Hematoxylin-Eosin staining of spleen section and h, Wright-Giemsa staining of bone marrow from NJL-induced AML mice. Scale bar, 20μM.
The fact that NJS failed to induce leukemia indicated that the PHD finger is required for leukemogenesis. Indeed, deletion of JARID1APHD3, but not JARID1A sequences prior to or following it, abolished NJL-mediated transformation of hematopoietic cells (Supplementary Fig.2f-h). We next asked whether JARID1APHD3 recognizes histone methylation. First, only histone H3 associated with recombinant JARID1APHD3 using total histone extracts (Supplementary Fig.3a). When a mini-library of H3 peptides harboring either unmodified, mono-, di- or tri-methylated K4, K9, K27, K36 or K79 were screened in biotinylated peptide pull-down, JARID1APHD3 only interacted with those containing H3K4me3/2 (Fig.2a; Supplementary Fig.3b). Such specificity was further confirmed by immunostaining and co-immunoprecipitation using Flag-NJL stable expression cells— NJL exhibited a speckled nuclear staining pattern and significantly co-localized with H3K4me3, but not H3K9me3 (Supplementary Fig.4); The vast majority of NJL were bound to mononuclesomes containing H3K4me3, but not H3K27me3 (Supplementary Fig.3c). Calorimetry-based measurements revealed a dissociation constant (Kd) of ∼0.75 μM for JARID1APHD3 binding to H3K4me3, with reduced affinities to H3K4me2/1/0 (Supplementary Fig.3d).
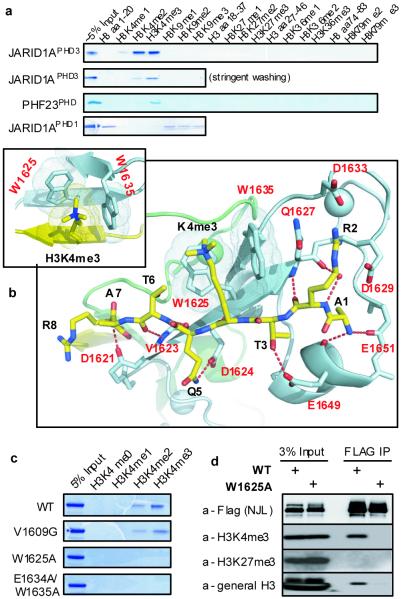
Figure 2. JARID1APHD3, an essential motif for NJL-mediated leukemia, specifically recognizes H3K4me3/2 marks.
a, Capability of JARID1APHD3, PHF23PHD and JARID1APHD1 (the first PHD finger of JARID1A, Supplementary Fig.1) to interact with H3 peptides harboring different Kme in peptide pull-down assay. JARID1APHD1 interacted with H3K4me0 as BHC80PHD11. b, Crystal structure of JARID1APHD3 (cyan) complexed with H3K4me3 peptide (yellow) and a close-up view of the H3K4me3-binding channel (inset) formed by two orthogonally aligned Trp residues. The residue of JARID1APHD3 and H3 is shown in red and black, respectively. c, Capability of wildtype or mutant JARID1APHD3 to bind to H3K4me3/2. d, CoIP showing that NJL containing the wildtype, but not mutant (W1625A) PHD finger, associated with H3K4me3 or H3 in transiently trasfected 293 cells.
We determined the structure of JARID1APHD3:H3K4me3 complexes using X-ray crystallographic and NMR spectroscopic techniques. Both analyses revealed that the JARID1APHD3-H3K4me3 interaction was established via (i) anti-parallel β-sheet pairing between the H3 backbone and a β-sheet of JARID1APHD3, (ii) a hydrophobic cleft formed by two Trp residues (W1625, W1635) that anchor the H3K4me3 side chain, and (iii) positioning of H3R2 in an acidic pocket (Q1627/D1629/D1633) (Fig.2b; Supplementary Fig. 5b,6c). H3K4me3 is stacked between the indole rings of two orthogonally aligned Trp residues with intermolecular contacts showed in Fig.2b and Supplementary Fig.5b,6d. The X-ray (a domain-swapped dimer of one molecule and a crystallographic symmetry-related molecule) and solution NMR (monomer) analyses are summarized in Supplementary Fig.5 (statistics in Supplementary Table 2) and Supplementary Fig.6-7 (statistics in Supplementary Table 3), respectively. Comparison between JARID1APHD3 structures in the free and H3K4me3-bound state (Supplementary Fig.6a-b) revealed no overall conformational changes. Residues W1625 and W1635 are evolutionarily conserved among JARID1 homologues (Supplementary Fig.8a). Mutations targeting these Trp residues disrupted the H3K4me3-binding in vitro (Fig.2c) and in cells (Fig.2d). Such a two-sided H3K4me3-binding tryptophan channel is a varied form of the H3K4me3-engaging pocket involving 3-4 hydrophobic residues found in the PHD finger of BPTF7, ING28, Yng119 or RAG213 (Supplementary Fig.8b-d). Yet, it exhibited a stronger H3K4me3-binding affinity (Kd=0.75μM). Collectively, the PHD finger, an essential motif of NUP98-JARID1A, uniquely recognizes H3K4me3/2 using an aromatic engaging channel.
To gain insight into mechanisms of NJL-induced AML, we used microarray analyses to compare the transcriptome of NJL-transformed progenitors and control cells— committed myeloid progenitors generated as described before18. Strikingly, a significant portion of genes upregulated in NJL-transformed progenitors were those either targeted by polycomb proteins20,21 or exhibiting ‘bivalent domain pattern’22 in stem cells, many of which encode developmentally critical transcription factors (Hoxa5/a7/a9/a10, Gata3, Meis1, Eya1, Pbx1; Supplementary Table 4). Such upregulation was further confirmed by RT-PCR using vector- versus NJL-transduced marrow cells (Supplementary Fig. 9a-c). Other Hox-A genes (a1, a2, a11, a13) were not expressed in NJL-transformed progenitors. We detected a similar target specificity for Hox-A genes using chromatin immunoprecipitation (ChIP)— NJL directly bound to the promoters of Hoxa6-a10, but not distal Hoxa1-a3 or Hoxa11-a13 (Fig.3a and Supplementary Fig.9d; green); NJL-binding specificity among Hox clusters was correlated to H3K4me3— H3K4me3 was abundant in Hoxa6-a10, while low/absent in Hoxa1-a4 or Hoxa11-a13 (Fig.3b). Enforced expression of Hox and Meis1 has been shown sufficient to induce AML23. This indicated that NJL blocks hematopoietic differentiation and induces AML by enforcing the transcription of these genes.
Figure 3. NUP98-JARID1A enforced high H3K4me3 and active transcription associated with developmentally critical loci such as Hox.
a, ChIP for NJL- or Ezh2-binding to _A_-cluster Hox promoters in committed myeloid progenitor line18 (cell 1) or in hematopoietic stem/progenitor cells three weeks after transduction of control vector (cell 2) or 3xFlag-tagged NJL (cell 3-4). b, ChIP of H3K4me3, H3K27me3 and general H3 among _Hox_-A gene cluster in hematopoietic progenitors three weeks after transduction of vector or NJL. c, Hoxa9/a10 expression in hematopoietic stem/progenitor cells after days of in vitro cultivation (day 0, 4, 8, 12), macrophages (mϕ), NIH-3T3 fibroblasts or NJL-transformed progenitors. d, α-Hoxa9 blot in marrow progenitors 10 days after transduction of MLL-ENL, empty vector, NJS or NJL. e, ChIP for Hoxa9/a10 promoter-associated H3K4me3 in hematopoietic stem/progenitor cells after days of in vitro culture, mϕ and marrow progenitors 20 days post transduction of vector or NJL. n=3, error bar indicates s.d; *, P<0.01; **, P<0.001; ***, P <10-4.
It has been reported that the A-cluster Hox gene expression is high in hematopoietic stem cells (HSC) and early progenitors, followed by down-regulation and shut-off during terminal differentiation24. Our ex vivo hematopoietic stem/progenitor cell system recapitulated such dynamics— coincident to the silencing of HSC marker and activation of differentiation marker (Supplementary Fig.9f), Hoxa9/a10 were down regulated >10- or 60-fold respectively in 8 days of culture (Fig.3c); Concurrent loss of _Hoxa9/a10_-associated H3K4me3 was observed in these cells (Fig.3e). Strikingly, NJL persistently enforced high levels of Hoxa9/a10 expression and _Hoxa9/a10_-associated H3K4me3 in marrow cells, whereas Hoxa9/a10 was silenced ten days after transduction of vector or NJS in similarly maintained cells (Fig.3c-e). To rigorously test the role of H3K4me3 recognition during leukemogenesis, we mutated the H3K4me3-engaging residues. NJL harboring mutation on the residueW1625 or W1635 failed to bind to H3K4me3 or H3 (Fig. 2d), failed to bind to the Hoxa9 promoter that exhibited high H3K4me3 in 293 cells (Fig.4a; Supplementary Fig.9i), failed to enforce the Hoxa9 expression (Fig.4b) or _Hoxa9_-associated H3K4me3 in hematopoietic progenitors (Fig.4c), and failed to transform the hematopoietic cells (Fig.4d), whereas the irrelevant mutation (V1609G) did not affect these activities (Supplementary Fig.10e). To assess whether NJL-induced phenotype was unique to JARID1APHD3, we investigated another similar de novo translocation, NUP98-PHF23 (Fig.1a)5, and also swapped JARID1APHD3 with other PHD fingers reported before. PHF23PHD specifically engaged H3K4me3/2 as predicted1 (Fig.2a); NUP98-PHF23 robustly enforced _Hoxa9_-associated H3K4me3 and transformed hematopoietic progenitors (Fig.4c,e; Supplementary Fig.10). Strikingly, swapping JARID1APHD3 with another H3K4me3/2-binding PHD finger from ING28 or even S. cerevisiae Yng119 also succeeded in the transformation, whereas replacing it with an H3K4me0-binding PHD finger, either BHC80PHD11 or JARID1APHD1 (Fig.2a), abolished the transformation (Fig.4c,e). Therefore, engaging H3K4me3/2 by NUP98-PHD fusion causes leukemia by enforcing an active state on developmentally critical loci.
Figure 4. The H3K4me3/2 engagement by NUP98-JARID1A perturbs the epigenetic state of developmentally critical loci during hematopoiesis.
a, Impact of mutations on the Flag-NJL binding to HOXA9 in 293 cells. b, Immunoblot of hematopoietic progenitors ten days post transduction of vector, wildtype or mutant NJL. Phospho-c-Kit, marker of mast cells. c, ChIP for Hoxa9 promoter-associated NUP98-fusion proteins (3xFlag-tagged) and H3K4me3 in marrow progenitors 10 days after transduction. d, Transforming capacities after introducing mutation to NJL or e, those by NUP98-PHF23 or after replacing JARID1APHD3 with another PHD finger that engages either H3K4me3/2 or H3K4me0. Total progenitor number was counted at day 1, 10, 25 and 40. f, ChIP for SUZ12 and g, MLL2 binding to Hoxa9/a11 and h, _Hoxa9_-associated H3 acetylation in marrow progenitors 15 days after transduction of vector or NJL. Error bar indicates s.d (n=3); *, P<0.05; **, P<0.005; ***, P<10-4; *****, P<10-6. i, A scheme that NUP98-PHD fusion acts as “boundary factor” and prevents the spreading of polycomb factors from Hoxa13/a11 to Hoxa9, thus inhibiting H3K4me3 removal and H3K27me3 addition during hematopoiesis.
Because the H3K4me3 recognition cannot provide DNA sequence specificity and yet NJL-upregulated genes were enriched with polycomb-targeted 20,21 or ‘bivalent domain’ genes22 in stem cells (e.g., Hox(s), Gata3, Meis1; Supplementary Table 4), we asked whether such specificity is due to their dynamically regulated characteristics. Towards this end, we examined the effect of NJL on two distinct gene classes— developmentally critical genes, and housekeeping genes that exhibit constitutive H3K4me3 (Supplementary Fig.11a, top panel). Interestingly, although NJL bound to housekeeping genes, it had little affect on their expression during cell differentiation (Supplementary Fig.11a, middle and bottom panels). Thus, NJL tends to affect the developmentally critical loci specifically during hematopoeisis. We next pursued the possibility that NJL interferes with activities of polycomb proteins at these developmentally critical loci. Using ChIP, we found that, while Ezh2 or Suz12 was spread throughout Hox-A clusters in vehicle-infected marrow progenitors that underwent differentiation, these polycomb factors were restricted within Hoxa11-a13 in NJL-infected progenitors (Fig.3a,4f, red). In the NJL-transduced cells, H3K27me3 was also only detected at _Hoxa13-a11_— the differentiation-associated spreading of H3K27me3 was inhibited at a region from Hoxa10 to Hoxa1 (Fig.3b). The spreading of polycomb factors from distal Hox loci (a13-a11) seemed to be blocked at Hoxa10-a9 by NJL that were bound there (Fig.3a; Supplementary Fig.9d). Similar result was also found at Meis1 (Supplementary Fig.9e). Consistent to previous reports15,16, the recruitment of p300 and dramatic elevation of H3 acetylation (H3K27ac by >2,000 fold) were observed on Hoxa9 in NJL-transduced cells (Fig.4h; Supplementary Fig.11b). Collectively, NUP98-PHD fusion dominated over the spreading of polycomb and enforced an H3K4me3/acetylated histone state at developmentally critical loci, an epigenetic state that defines leukemia stem cells.
In summary, we have demonstrated for the first time that fusing an H3K4me3-engaging PHD finger (plus nuclear localization signal) to a common partner NUP98 is sufficient to induce leukemia. We showed that NUP98-PHD fusion prevented the silencing of critical loci encoding master transcription factors (Hox(s), Gata3, Mesi1, Pbx1) during hematopoietic differentiation. NUP98 fusion partners can be grouped into two major groups, DNA-binding homoedomain and chromatin-associated factors including PHD fingers (JARID1A, PHF23)17. Although the existence of additional unknown ligand is possible for PHD fingers in the latter group (as H3K4 site cannot be mutated in mammals), the most straightforward interpretation of our findings is that binding H3K4me3/2 marks is responsible for leukemia described here. In support, a genetic interaction was demonstrated in yeast between H3K4 and the Yng1 PHD finger25, a module that imparted similar oncogenic properties when swapping into our assays (Fig.4e). Several PHD fingers exist in NSD1, another NUP98-fusion partner16, however, none contains critical H3K4me3-engaging residues1. Thus, our report represents the first example wherein inappropriate interpretation of histone modification can actively induce a deregulation of developmentally critical loci, perturb cellular/epigenetic identities, and even induce oncogenesis. NUP98-PHD fusion coordinates acts of H3K4me3/2 and histone acetylation, mimicking mechanisms utilized by evolutionarily conserved ING(s)-complexes for robust gene activation19,26 (Supplementary Fig.12). H3K4me3 bound by NUP98-PHD may serve as ‘seed’ of propagation mediated by WDR5-MLL2/3 complexes1,27 that is also coupled with UTX/Jmjd3-mediated H3K27 demethylation28,29, as we detected high levels of WDR5, RBBP5, and MLL2 on Hoxa9 in NJL-transduced marrow cells (Fig.4g; Supplementary Fig.11c-d). We suggest that NUP98-PHD acts as ‘boundary factors’, using the PHD finger to protect H3K4me3 from JARID1(s)-mediated demethylation29 and also inducing H3K27ac to block H3K27me addition (Fig.4i). In support, we observed a ‘bivalent domain’ feature22 at Hoxa11-a10, the junction region of two antagonizing mechanisms (Fig.3b). Loss-of-function mutation of RAG2PHD in immunodeficiency and gain-of-function mutation involving PHD fingers in malignancies described here indicate a new type of diseases that arise from ‘misinterpreting’ the ‘histone code’3,30. With ∼200 PHD fingers in human genome and some intimately associated to diseases3, we expect similar ‘mis-reading’ mechanisms responsible for some unstudied diseases. These pathologies together with those caused by ‘mis-writing’ or ‘mis-erasing’29 histone modification, underscore the significance in investigating the biological readout of histone marks.
METHODS SUMMARY
Hematopoietic cell transformation assays
Protocols for the culture of primary hematopoietic stem/progenitor cells were previously described18. Briefly, 100,000 lineage-negative bone marrow stem/progenitor cells were subjected to retroviral infection, followed by kinetics analyses of proliferation versus differentiation in ex vivo culture system as described before18.
Peptide pull-down assay
Pull-down using biotinylated histone peptide and recombinant protein was performed as described6,11. After binding, peptide-Avidin beads were washed extensively in solution containing 50mM Tris pH 7.5, 150mM NaCl (250mM as stringent washing), 0.05% NP-40, 0.3mg/ml BSA and 1mM DTT.
Supplementary Material
1
2
Acknowledgement
We thank L. Baker, P. Chi, A. Ruthenberg and A. Xiao for critical reading and help in manuscript preparation, C. Hughes for antibodies, and other Allis lab member for their advice. We are extremely grateful to Drs. M. Kamps and S. Rafii for shared plasmids and expert advice. Thanks to E. Coutavas, W. Chen, E. Ezhkova, and the Bob Roeder lab for help on cell and animal work. G.G.W. is supported by a Leukemia & Lymphoma Society Fellow award and a Choh-Hao Li Memorial Fund Scholar award; H.L.D. is supported by a predoctoral Boehringer Ingelheim Foundation fellowship; This research was supported by the NIH grant and funds of Rockefeller University to C.D.A, funds of Abby Rockefeller Mauze Trust and the Dewitt Wallace and Maloris Foundations to D.J.P, U.S. Department of Defense CDMRP grant to J.L., and a joint Starr Foundation Cancer Consortium grant.
Appendix
METHODS
Plasmid construction and retroviral expression system
The NUP98-JARID1A fusion cDNA4 was generated by ligating NUP98 sequences encoding amino acids 1-514 to those encoding amino acids 1489-1690 of JARID1A transcript variant 1 (NCBI accession No. NM_001042603) or amino acids 1489-1641 of JARID1A transcript variant 2 (NCBI accession No. NM_005056), producing two fusion isoforms (NJL or NJS) respectively. The same method was used to generate NUP98-PHF235. The fusion cDNA with an N-terminal 3xFLAG was cloned into MSCV retroviral expression vector (Clontech). JARID1A (RBP2), PHF23 and BHC80 cDNAs were purchased from Open Biosystems. NUP98 plasmids were kindly provided by Dr. J.M. van Deursen, Hoxa9 by Dr. M.P. Kamps, MLL-ENL by Dr. R.K. Slany, Yng1 by Dr. S.D. Taverna, CBX7 by Dr. E. Bernstein, and ING2 by Dr. Z. Tang. Mutations were generated by site-directed mutagenesis, and all used plasmids were confirmed by sequencing.
Purification and culture of primary hematopoietic cells
Bone marrow cells harvested from femur and tibia of balb/C or b/6 mice were subject to lineage-negative (Lin-) enrichment using Hematopoietic Progenitor Enrichment Kit (StemCell Technologies or Miltenyi Biotec) to remove cells expressing differentiation antigens as described before16. ∼400,000 of Lin- enriched hematopoietic progenitors were obtained per mouse with ∼10% c-Kit+Lin-Sca1+ HSCs. Before retroviral infection, Lin--enriched hematopoietic progenitors were stimulated in OptiMEM base medium (Invitrogen, cat#31985) complemented with 10% of FBS (Invitrogen, cat#16000-044), 1% of antibiotics, 50μM of β-mercaptoethanol and a cytokine cocktail containing SCF (supernatant of SCF-producer cells), 5ng/mL FLT3 ligand (Sigma), 5ng/mL IL3 and IL6 (Miltenyi) for 2-3 days as described18,31. After retroviral infection and selection (1μg/mL puromycin), marrow cells were plated in the same medium with SCF as the sole cytokine. Cell splitting and replating to fresh medium were performed every 3-4 days to keep cell number <2 million per well (6- or 12-well plate). Cell morphology was examined by Wright-Giemsa staining. Macrophages were obtained by culture of marrow cells in M-CSF (Miltenyi) for 1-2 weeks as described32; Immortalized cell lines that mimic committed neutrophil-macrophage progenitors were generated as described before18,31,33.
Murine bone marrow transplantation leukemogenic assay
Leukemogenic potentials of oncogenes were evaluated in sublethally irradiated syngeneic mice after tail vein injection with 100,000 of bone marrow-derived Lin- cells that were infected with retrovirus encoding the fusion gene as described18. Mice exhibiting leukemic phenotype were subjected to pathological analyses.
Recombinant protein production and GST pull-down
JARID1APHD3 (amino acids 1601-1660) GST-fusion proteins were produced using a previously described protocol19. GST pull-down using total histone extracts was performed as described with modification9. Briefly, ∼2μg GST-fusion protein bound to glutathione beads (Amersham) were incubated with 10μg of calf thymus histone extracts (Worthington) in a binding buffer containing 50mM Tris-HCl pH 7.5, 0.5 M NaCl, 0.5% NP-40, 0.2mM EDTA, 1mM DTT and protease inhibitor cocktail (Roche) at 4°C for 4 h.
Native co-immunoprecipitation (CoIP)
Mononucleosomes-containing fractions were prepared as described before6. Briefly, intact nuclei were subject to limited micrococcal nuclease (MNase) digestion so that the major form of released chromatin is mononucleosome. After the removal of insoluble fraction by centrifugation, supernatant containing mononucleosomes was then incubated with FLAG or HA-agarose beads (Sigma), or with Dynal magnetic beads (Invitrogen) coupled with αH3K4me3 (Abcam) or control antibodies. After extensive washing, precipitated proteins were subject to immunoblot.
Isothermal titration calorimetry (ITC) measurements
Calorimetric experiments were conducted at 25.0°C with an MicroCal iTC200 instrument (Northampton, MA) as described7. Recombinant JARID1APHD3 protein and H31-15K4me peptides were dialyzed overnight against 25mM Tris-HCl pH7.5, 50mM KCl, and 2mM β-mercatoethanol. Protein concentration was determined by absorbance spectroscopy (Tyr ε280=1,420 M-1cm-1; Trp ε280=5,600 M-1cm-1; Cys ε280=125 M-1 cm-1). H31-15K4me peptides were quantified by the absorbance of an added C-terminal Tyr with ε280=1,280 M-1cm-1 for peptide. Acquired calorimetric titration data were analyzed using software Origin7.0 (MicroCal, LLC ITC 200) based on a 1:1 binding stoichiometry.
Antibodies and immunoblot
Antibodies used were α-FLAG (Sigma; M2), α-HA (Covance, MMS101), α-Hoxa9 (Upstate, 07-178), α-Pbx1 (Santa Cruz, sc889), α-phosph-c-Kit (Cell signaling) and α-Tubulin (Sigma).
ChIP analysis
ChIP analysis was performed using Upstate ChIP kit and a protocol described before34. 1∼2 million cells per ChIP were used for histones, and 2∼3 million for others. Antibodies and amount used were α-Flag (Sigma M2, 1-3μg), α-HA (Covance MMS101, 1-3μg), α-H3K4me3 (Upstate 07-473, 1μL; Abcam 8580, 0.5μg), α-H3K27me3 (Upstate 07-449, 0.5μg), α-acetyl-H3 (Upstate 06-599, 1μg), α-general H3 (Abcam 1791, 0.5μg), α-acetyl-H3K9 (Upstate 06-942, 1μg), α-acetyl-H3K27 (Abcam 4729, 1μg), α-Ezh2 (Cell signaling 4905, 4-5ul), α-Suz12 (Upstate 07-379, 2ul), α-MLL2 (Bethyl A300-113A, 4μg), α-WDR5 (Upstate 07-706, 2μg), α-RBBP5 (Bethyl A300-109A, 3μg; a gift of Dr. Christina Hughes) and α-p300 (Santa Cruz, N15/C20, 10μg). The same amount of nonspecific IgG (Upstate) was used as antibody control, and a silenced intragenic locus, Chr8Int, as locus control for H3K4me3 or activator binding as described21. The promoter sequence was acquired from UCSC genomic browser (http://genome.ucsc.edu). ChIP primers were shown in Supplementary Table 5. ChIP signals were represented in the percentage (%) of signals from total chromatin used, and fold of enrichment calculated by normalizing against signals of nonspecific IgG.
Microarray analysis
Total RNA was extracted and the transcript expression quantified using Affymetrix GeneChip Mouse arrays as described18. RNA hybridization, scanning and signal quantification were performed by RU Gemonic Resources Center. Hybridization signals were retrieved and normalized, followed by differential expression analysis and statistical analysis using GeneSpring Analysis Platform GX 7.0 (Agilent Technologies).
RT-PCR analysis
Reverse transcription of RNA was performed using the random hexamer and Invitrogen Superscript III kit. Usually the PCR amplicon (size ∼90-200bp) is designed to span over large intron regions. Exon-intron information was obtained from UCSC genomic browser. Quantitative PCR was performed in triplicate using SYBR green master mix reagent (Applied Biosystem) on a Stratagene Mx3005P QPCR system. Primer information is shown in Supplementary Table 5.
Flow cytometry (FACS)
Cells were blocked with BD FcBlock (2.4G2) and stained on ice with fluro-conjugated antibodies (1:1,000 dilution of Cd117FITC, Sca-IPE:CY7, Cd34APC, Cd34FITC, Cd11bAPC, Gr-1PE, Cd19PE or B220PE, BD Biosciences) and analyzed on BD FACSCalibor cytometer. Data was collected and analyzed using CellQuestPro and FlowJo software.
Immunofluorescence microscopy
Suspension cultured hematopoietic cells were attached to cover slips treated with 0.01% (w/v) poly-lysine, followed by 15-minute fixation in 4% of parafomaldehyde and 10-min solubilization in PBS, 0.2% Triton-X100 and 0.2% NP-40. After a 30-min block in PBS, 2.5% BSA and 10% normal goat serum, cells were stained with primary antibodies (M2 α-FLAG [1:1,000∼2,000 dilution of 1mg/mL], rabbit α-H3K4me3 [Upstate 07-473 or Abcam 8580, 1:2,000] or rabbit α-H3K9me3 antibodies [Upstate 07-442, 1:1,000]) followed by washing and staining with fluorescent-labeled secondary antibodies. After washing, fluorescent signal was visualized and analyzed with a DeltaVision Image Restoration Microscope and a Cofocal Microscope (Applied Precision/Olympus). Deconvolution microscopy image analysis was performed to reassign the out-of-focus blurred light to its origin35, and subcellular co-localization analysis was carried out from stacks of deconvolved images using ImageJ (Rasband WS, et al. NIH, Bethesda, USA; http://rsb.info.nih.gov/ij/) and the plugin JACoP36. Confocal microscopy analysis was performed as previously described37. Co-immunostaining statistics was analyzed using Pearson’s Coefficient of Correlation method. Image acquisition, processing and analyses were performed with the expert help from RU Bio-Imaging Center, and detailed protocols are available upon request.
Statistics
All results are presented as the mean and standard deviation (s.d). Statistical analyses were performed using Student’s t-test.
Protein preparation for structural studies
The gene fragment encoding JARID1APHD3 was fused to the C-terminus of a His(6x)-SUMO tag in a modified pRSFDuet-1 vector (Novagen), with a ubiquitin-like-protease (ULP) cleavage site located at the linker region. The bacterial expressed protein was purified using a Ni-NTA affinity column, followed by ULP cleavage, separation of JARID1APHD3 from His(6x)-SUMO via a second Ni-NTA chromatography step, and gel filtration. The JARIPD1APHD3-H3K4me3 complex was obtained by mixing JARIPD1APHD3 protein with an equal molar amount of H31-9K4me3 peptides (H3 amino acids 1-9, with Lys4 tri-methylated), then purified by gel filtration, and concentrated by ultrafiltration.
Crystal growth
The crystals of JARIPD1APHD3-H3K4me3 complexes were obtained by equilibrating a reservoir consisting of 20% (w/v) poly(ethylene glycol) monomethyl ether 2000, 10mM nickel (II) chloride hexahydrate, and 0.1 M Tris (pH 8.5) with a hanging drop consisting of 1 _μ_L of the reservoir solution and 1 _μ_L of a 27 mg/mL protein solution in 10 mM Tris pH 8.0, 0.1 mM ZnCl2, 5mM DTT and 50 mM NaCl (Crystal Screen 2 kit, Hampton Research). A mixture of the well solution with 10% (v/v) glycerol was used as cryoprotectant.
Data collection and structure determination
An anomalous diffraction data set for JARID1APHD3-H31-9K4me3 complex was collected at the zinc anomalous peak wavelength (1.28215 Å) at beamline NE-CAT 24ID-C, Advanced Photon Source, Chicago. The data set was indexed, integrated and merged to 2.2 Å using the program HKL2000. The crystal belongs to I41 space group and contains one molecule per asymmetric unit. Heavy-atom search, SAD phasing and model building were performed with the PHENIX38 software package. Three zinc atoms were unambiguously identified for SAD phasing, and ∼90% residues of the protein-peptide complex were successfully built into the initial model. The PHENIX-model was further manually rebuilt using COOT39 and refined using REFMAC540 in successive cycles. The final refined structure has Rwork and Rfree values of 0.200 and 0.234, respectively (Supplementary Table 2). One molecule forms a domain-swapped dimer with a crystallographic symmetry-related molecule (Supplementary Fig. 5a). The swapped segment spans the first 14 residues from the N-terminus.
Using a crystal of the complex following pH optimization of crystallization conditions, we were able to collect one 1.9 Å data set at wavelength 0.97949 Å at the same beamline. The crystal belongs to the same crystal form as the previous one. We solved the high-resolution structure by molecular replacement using PHASER41 with the above 2.2 Å model after removing all the water molecules and some flexible residues. Structure refinement was done using CNSsolve42, cycled with manual model building in COOT. Prior to the refinement, the same Rfree set of reflections were transferred from the low-resolution data using the program Freerflag in CCP4 suite43 for effective cross validation. For both data sets, ∼10% reflections were selected in a ‘random’ mode throughout the resolution range. After resetting the overall B-factor to 20 Å2 and rigid body refinement, simulated annealing starting at 5,000 K was performed to reduce model bias before extensive B-factor and positional refinement. The final model contains full length JARID1APHD3 (1609-1659) with one additional serine at the N-terminus from the expression vector, histone H31-8K4me3, three zinc ions and thirty-two water molecules. The JARID1APHD3-H3K4me3 complex in the crystal shows that one molecule forms a domain-swapped dimmer with a crystallographically symmetry-related molecule (Supplementary Fig. 5a). Two zinc ions are integral to the folding of the PHD finger, while the third zinc ion locates at the interface between two domain-swapped dimers, thereby mediating crystal packing (Supplementary Fig. 5c). The Rwork and Rfree of the final structure are 0.208 and 0.225 respectively (Supplementary Table 2).
Isotopic labeling, NMR data collection and structure determination
Samples used for NMR chemical shift assignments, 15N relaxation measurements, and structure determination contained 0.2-0.5 mM uniformly-[15N]- or [13C, 15N]-labeled JARID1APHD3 in the free state and in complex with unlabeled H31-9K4me3 peptide dissolved in NMR buffer (20 mM Na-phosphate, 1 mM ZnCl2, 5 mM DTT, 90% H2O/10% D2O) at pH 7.0. The sample used for measurements of 15N-1H residual dipolar couplings (RDCs) contained 0.2 mM JARID1APHD3 aligned in 12 mg/ml of bacteriophage Pf1 (Alsa, Riga, Latvia), 10 mM MOPS, 200 mM NaCl, pH 7.0.
All NMR spectra were collected at the New York Structural Biology Center (NYSBC) using 800 MHz Bruker NMR spectrometers equipped with 1H, 15N, 13C triple-resonance cryogenic probes. Unless indicated otherwise, the sample temperature was controlled at 20°C. A suite of 3D heteronuclear NMR experiments, including HNCACB, CBCA(CO)NH, HNCO, HBHA(CO)NH, and HCCH-TOCSY were acquired for sequential backbone and non-aromatic side chain assignments of JARIPD1APHD3 both in the free state and in complex with H31-9K4me3 peptide in solution. 2D NOESY (_τ_mix = 100 ms), 3D 15N-edited NOESY-HSQC (_τ_mix = 100 ms), 3D aromatic 13C-edited NOESY-HSQC (_τ_mix = 100 ms) and 3D aliphatic 13C-edited NOESY-HSQC (_τ_mix = 100 ms) data sets were acquired and used for additional assignments (side chain amide and aromatic groups) and distance constraints. To selectively observe the NOEs between JARIPD1APHD3 and H31-9K4me3 peptide, a [13C,15N]-filtered,13C-edited NOESY (_τ_mix = 120 ms) spectrum44 of uniformly [15N,13C]-labeled JARIPD1APHD3 bound to unlabeled H31-9K4me3 peptide was recorded. One-bond N-H RDCs were determined by using the IPAP 15N-HSQC sequence at 25°C45. Standard pulse sequences46 were used for measurements of the 15N relaxation rates (_R_1, _R_2) of JARIPD1APHD3 at 25°C. The spectra were processed and analyzed, respectively, with the NMRPipe47 and Sparky (http://www.cgl.ucsf.edu/home/sparky) software. The solution structures of JARIPD1APHD3 both in the free state and in complex with H31-9K4me3 peptide were first calculated using the CYANA program48. Interproton distance constraints were derived from 2D NOESY, 3D 15N-edited NOESY-HSQC and 3D 13C-edited NOESY-HSQC spectra. Backbone ϕ and ψ angles were derived from TALOS-based analysis of backbone chemical shifts49. A number of hydrogen bonds derived from chemical shift analysis and from observed NOEs characteristic for _α_-helices and _β_-sheets, were added in the final rounds of structure refinement. Of the 100 final structures calculated by CYANA, 20 structures with the lowest target functions were chosen for further refinement using the Xplor-NIH program50, in which 1_D_NH RDC restraints, physical force field terms and explicit solvent terms51 were added to the calculation. The final structures were validated by Procheck-NMR52, and the statistics for the 20 final structures are listed in Supplementary Table 3.
Monomeric state of JARID1APHD3 in free and H31-9K4me3-bound states in solution
The oligomeric states of JARID1APHD3 (mol. wt. 5.8 kD) and JARID1APHD3-H31-9K4me3 complex (mol. wt. 6.8 kD) were first evaluated by comparing their elution volumes on a Superdex G75 16/60 column, with the calibration line derived from a number of molecular standards. As shown in Supplementary Fig. 7a, the elution volumes of both free JARID1APHD3 and JARID1APHD3-H31-9K4me3 complex are comparable with those expected for their monomeric states, but considerably larger that those expected for their dimeric states. This suggests that JARID1APHD3 is monomeric in solution, for both the free and H31-9K4me3 states. Furthermore, the rotational correlation times of JARID1APHD3 in the free and H31-9K4me3-bound states were estimated as 3.5 ns and 4.5 ns, respectively, based on an analysis of 15N R2/R1 relaxation time ratios (Supplementary Fig. 7b) using the quadratic diffusion program53. These values are consistent with isotropic tumbling values of a monomeric protein of their respective sizes, further supporting that both free and complexed JARID1APHD3 exist as monomers in solution.
Thus, although JARID1APHD3-H3K4me3 complex exhibits a domain-swapped dimer in the crystal, gel filtration and NMR relaxation measurements (Supplementary Figure 7) clearly showed such a complex to be monomeric in solution. Hence the domain-swapped dimerization observed in the crystal is likely to be a characteristic feature of the crystalline state, originating perhaps in packing interactions.
- 31.Wang GG, et al. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat Methods. 2006;3:287–93. doi: 10.1038/nmeth865. [DOI] [PubMed] [Google Scholar]
- 32.Sawka-Verhelle D, et al. PE-1/METS, an antiproliferative Ets repressor factor, is induced by CREB-1/CREM-1 during macrophage differentiation. J Biol Chem. 2004;279:17772–84. doi: 10.1074/jbc.M311991200. [DOI] [PubMed] [Google Scholar]
- 33.Calvo KR, Sykes DB, Pasillas M, Kamps MP. Hoxa9 immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced meis expression. Mol Cell Biol. 2000;20:3274–85. doi: 10.1128/mcb.20.9.3274-3285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okada Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–78. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 35.Wallace W, Schaefer LH, Swedlow JR. A workingperson’s guide to deconvolution in light microscopy. Biotechniques. 2001;31:1076–8. 1080, 1082. doi: 10.2144/01315bi01. passim. [DOI] [PubMed] [Google Scholar]
- 36.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–32. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 37.Fischle W, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–22. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 41.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–33. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 43.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 44.Zwahlen C, et al. Methods for Measurement of Intermolecular NOEs by Multinuclear NMR Spectroscopy: Application to a Bacteriophage gamma N-Peptide/boxB RNA Complex. J. Am. Chem. Soc. 1997;119:6711–6721. [Google Scholar]
- 45.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson. 1998;131:373–8. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 46.Farrow NA, et al. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 47.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 48.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–98. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 49.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 50.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 51.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 52.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 53.Lee LK, Rance M, Chazin WJ, Palmer AG., 3rd Rotational diffusion anisotropy of proteins from simultaneous analysis of 15N and 13C alpha nuclear spin relaxation. J Biomol NMR. 1997;9:287–98. doi: 10.1023/a:1018631009583. [DOI] [PubMed] [Google Scholar]
Footnotes
References
- 1.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 2.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker LA, Allis CD, Wang GG. PHD fingers in human diseases: Disorders arising from misinterpreting epigenetic marks. Mutat Res. 2008;647:3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Zutven LJ, et al. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer. 2006;45:437–46. doi: 10.1002/gcc.20308. [DOI] [PubMed] [Google Scholar]
- 5.Reader JC, Meekins JS, Gojo I, Ning Y. A novel NUP98-PHF23 fusion resulting from a cryptic translocation t(11;17)(p15;p13) in acute myeloid leukemia. Leukemia. 2007;21:842–4. doi: 10.1038/sj.leu.2404579. [DOI] [PubMed] [Google Scholar]
- 6.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 7.Li H, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–5. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pena PV, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–3. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–9. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vermeulen M, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 11.Lan F, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–22. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–7. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matthews AG, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–10. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong W, Suzuki K, Russell M, Riabowol K. Function of the ING family of PHD proteins in cancer. Int J Biochem Cell Biol. 2005;37:1054–65. doi: 10.1016/j.biocel.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Kasper LH, et al. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19:764–76. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–12. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 17.Moore MA, et al. NUP98 dysregulation in myeloid leukemogenesis. Ann N Y Acad Sci. 2007;1106:114–42. doi: 10.1196/annals.1392.019. [DOI] [PubMed] [Google Scholar]
- 18.Wang GG, Pasillas MP, Kamps MP. Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood. 2005;106:254–64. doi: 10.1182/blood-2004-12-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taverna SD, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–96. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 22.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroon E, et al. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. Embo J. 1998;17:3714–25. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pineault N, Helgason CD, Lawrence HJ, Humphries RK. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp Hematol. 2002;30:49–57. doi: 10.1016/s0301-472x(01)00757-3. [DOI] [PubMed] [Google Scholar]
- 25.Martin DG, et al. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol. 2006;26:7871–9. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyon Y, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–72. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 28.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–50. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 29.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–40. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2