L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism (original) (raw)
Abstract
Long Interspersed Element 1 (L1) is a retrotransposon that comprises ∼17% of the human genome. Despite its abundance in mammalian genomes, relatively little is understood about L1 retrotransposition in vivo. To study the timing and tissue specificity of retrotransposition, we created transgenic mouse and rat models containing human or mouse L1 elements controlled by their endogenous promoters. Here, we demonstrate abundant L1 RNA in both germ cells and embryos. However, the integration events usually occur in embryogenesis rather than in germ cells and are not heritable. We further demonstrate L1 RNA in preimplantation embryos lacking the L1 transgene and L1 somatic retrotransposition events in blastocysts and adults lacking the transgene. Together, these data indicate that L1 RNA transcribed in male or female germ cells can be carried over through fertilization and integrate during embryogenesis, an interesting example of heritability of RNA independent of its encoding DNA. Thus, L1 creates somatic mosaicism during mammalian development, suggesting a role for L1 in carcinogenesis and other disease.
Keywords: Retrotransposon, Line-1, somatic mosaicism, RNA carryover
L1s (Long Interspersed Element 1s) are important genome modifiers, altering mammalian genomes in many ways, both constructively and destructively (Kazazian 2004). L1s have contributed to >35% of the mass of the human genome (Lander et al. 2001), not only by creating L1 insertion (retrotransposition), but also by indirect replication of short interspersed elements (SINEs) and processed pseudogenes. The full-length human L1 retrotransposon is ∼6 kb and encodes two proteins necessary for its mobilization by a “copy and paste” mechanism (Moran et al. 1996). The parent element is transcribed into RNA, and the bicistronic mRNA is exported from the nucleus to the cytoplasm where it is translated into ORF1p, a nucleic acid chaperone (Hohjoh and Singer 1997; Martin and Bushman 2001), and ORF2p, containing endonuclease and reverse transcriptase activity (Mathias et al. 1991; Feng et al. 1996). The L1 RNA forms a ribonucleoprotein (RNP) complex with its proteins (Kulpa and Moran 2006), and reverse transcription and integration occur in a single step on genomic DNA, a process called target primed reverse transcription (Luan et al. 1993).
Despite the tremendous impact of L1 on the human genome, much of the process of L1 retrotransposition in vivo remains unexplored. The several hundred thousand copies of L1 in the human genome demonstrate that they have accumulated over time. If L1s are still accumulating in the genome, they must either retrotranspose directly in germ cells during gametogenesis or early enough in embryonic development to be incorporated into germ cells. Somatic retrotransposition events that do not get incorporated into germ cells are not heritable and will not accumulate in the genome. In addition, L1 RNA and proteins have been found predominantly in germ cells and infrequently in differentiated tissues (Branciforte and Martin 1994; Trelogan and Martin 1995). Therefore, the prevailing view has been that the bulk of L1 retrotransposition occurs in germ cells. Approximately 20 human de novo L1 retrotransposition insertions have been characterized to date. However, since in all cases there is no definitive evidence as to when the retrotransposition event occurred, many or all of these de novo L1 retrotransposition insertions may have occurred early in embryogenesis. Studies of human L1 elements in transgenic mice have demonstrated direct germline retrotransposition only when the L1 transgene was driven by a heterologous promoter (Ostertag et al. 2002).
Thus, in order to investigate when and where most L1 retrotransposition occurs, we created mouse and rat transgenic models with native human or mouse L1 elements driven by their endogenous promoters, and also analyzed a previous transgenic mouse model in which the human L1 was driven by its endogenous promoter (Muotri et al. 2005). Our transgenes comprised a human or mouse L1 tagged in its 3′ untranslated region (UTR) with a retrotransposition cassette and followed by an SV40 poly(A) signal (Supplemental Fig. S1).
Some new transgenic mouse lines contain the L1LRE3 element, a highly active human L1 (Brouha et al. 2002, 2003). Previous transgenic mouse models have demonstrated significant variability in retrotransposition activity (Ostertag et al. 2002; Prak et al. 2003; Muotri et al. 2005; An et al. 2006; Babushok et al. 2006), possibly the result of methylation of the transgene. To study retrotransposition independent of position effects in these lines, we added two copies of a chicken β-globin insulator sequence (West et al. 2002) at each end of the transgene (L1LRE3 mouse; three lines).
The previously created mouse line (Muotri et al. 2005) is similar to those described here, but contains the human L1RP element (another highly active human L1) followed by an EGFP (enhanced green fluorescent protein) retrotransposition cassette (Ostertag et al. 2000) and does not contain insulators. These mice had exhibited L1 retrotransposition in neuronal precursor cells, but retrotransposition was not studied in sperm or sperm precursor cells, or in early development (L1RP mouse; one line).
We also made the following transgenic animals as controls: (1) a transgenic rat model using human L1RP without insulators to control for possible species-specific effects of the host organism (L1RP rat; eight lines), and (2) a mouse model of an active mouse L1, GF21, with a retrotransposition cassette to control for possible species-specific effects of the L1 element (L1 GF21 mouse; two lines).
Results
L1 retrotransposition events in transgenic animals
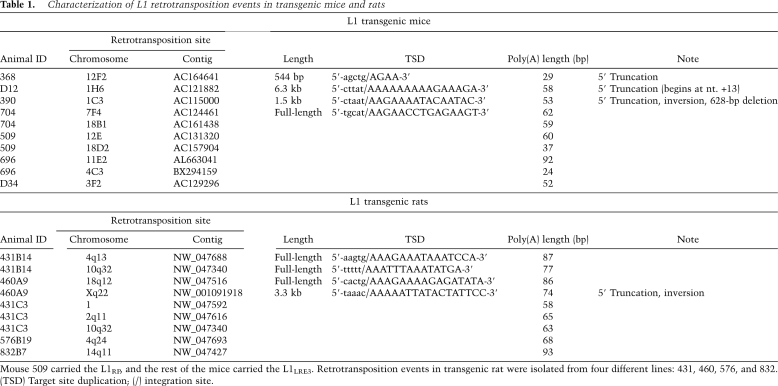
A retrotransposition cassette contains an intron that can only be removed during a retrotransposition event. An intron-flanking PCR distinguishes the L1 transgene from a retrotransposition event (Supplemental Fig. S1). All mice and rats were genotyped for the presence or absence of the transgene by at least two different PCR assays and/or Southern blot. A total of 19 L1 retrotransposition events from the mouse and rat models were cloned and sequenced using thermal asymmetric interlaced PCR (Babushok et al. 2006), and showed all features typical of authentic retrotransposition (Table 1).
Table 1.
Characterization of L1 retrotransposition events in transgenic mice and rats
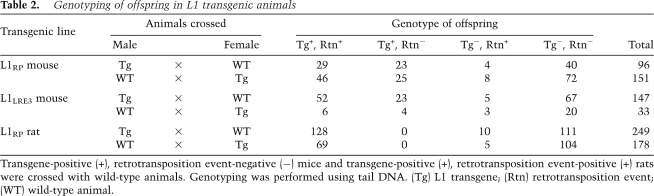
Offspring of human L1 transgenic animals exhibited high retrotransposition activity. In mice, L1 retrotransposition events were seen in >60% of offspring inheriting the transgene and in 9% of offspring lacking the transgene (Table 2). Transgenic rats exhibited greater retrotransposition activity than mice. Human L1 retrotransposition events were identified in all 197 rats inheriting the transgene as well as 6% of rats lacking the transgene. In total, we found 35 animals (both mice and rats) that were transgene-negative, retrotransposition event-positive. These retrotransposition events were clearly not false positives, as they were detected in the same animal multiple times with different tail DNA preps and different PCR primers.
Table 2.
Genotyping of offspring in L1 transgenic animals
L1 RNA in germ cells is heritable and integrates into the genome during development
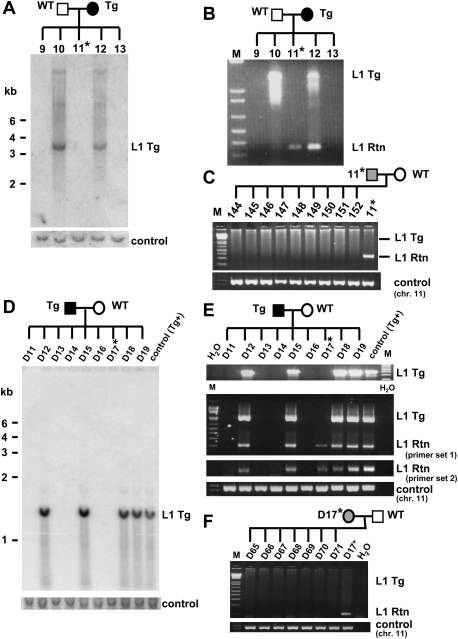
Retrotransposition events in animals lacking the transgene might be explained by germline retrotransposition events in the parent prior to the end of meiosis I, followed by segregation from the donor transgene, as demonstrated previously (Ostertag et al. 2002). In that scenario, retrotransposition events would be present in all cells of the offspring, and would be inherited in 50% of their offspring. Among the 35 transgene-negative, retrotransposition event-positive animals, we bred 10 different mice and rats (five offspring of transgenic males and wild-type females and five offspring of transgenic females and wild-type males). Instead of the expected 50% transmission rate, we found that none of 170 offspring of these matings (72 mice and 98 rats) inherited the L1 retrotransposition event (see Fig. 1 for data on two of these noninherited events). These results suggest that L1 RNA transcribed before the end of spermatogenesis and oogenesis can be carried over in both male and female germ cells and integrated into the genome during embryogenesis.
Figure 1.
L1 retrotransposition caused by L1 RNA carried over through meiosis, fertilization, and embryogenesis in the L1RP mouse (A_–_C) and in the L1LRE3 mouse (D_–_F). (A) Southern blot analysis on tail DNA isolated from offspring of an L1RP transgenic female mouse. A 1.4-kb DNA probe generated from the retrotransposition cassette of the L1RP transgene, which was expected to hybridize to both the transgene and retrotransposition insertion, exhibited only transgene bands in spite of the presence of retrotransposon amplicons by PCR. The membrane was rehybridized with an unrelated DNA probe generated from mouse chromosome 11 as a DNA loading control. (B) Genotyping PCR on tail DNA indicates an L1 retrotransposition event in a mouse lacking the transgene (mouse 11). (C) Mouse 11 (transgene-negative, retrotransposition event-positive) was bred with a wild-type mouse, and its offspring were genotyped. No offspring of this mouse inherited the retrotransposition insertion, indicating mosaicism of the L1 retrotransposition event in mouse 11. A control PCR on mouse chromosome 11 was performed to confirm the amount and quality of DNA. (D_–_F) Similar data to those in A–C are shown for the offspring of an L1LRE3 transgenic male mouse using tail DNA. The transgenic male mouse was bred with a wild-type female mouse, and its offspring were genotyped by Southern blot using a 503-bp probe generated from the L1 3′UTR and SV40 poly(A) signal sequence of the L1LRE3 transgene (D), and by PCR (E). Two independent PCR primer sets were used to confirm the presence of retrotransposition events. (F) The single offspring (D17) that had a retrotransposition event while lacking the L1 transgene was bred with a wild-type mouse. As shown in C, none of its offspring inherited the retrotransposition event. Asterisk denotes a transgene-negative, retrotransposition event-positive mouse. (Tg) transgene; (Rtn) retrotransposition event; (WT) wild-type animal; (M) 1-kb plus DNA Ladder (Invitrogen).
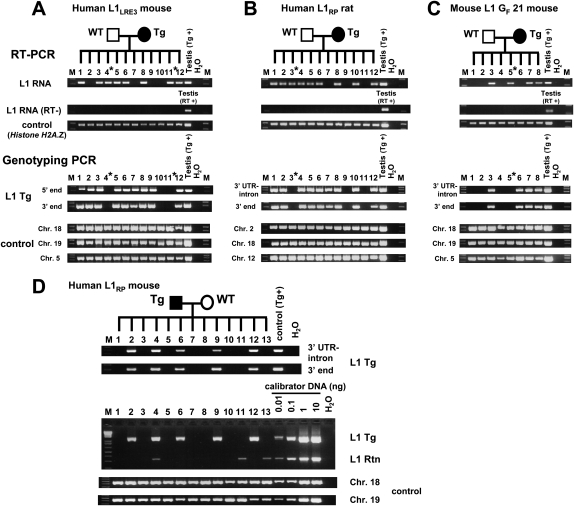
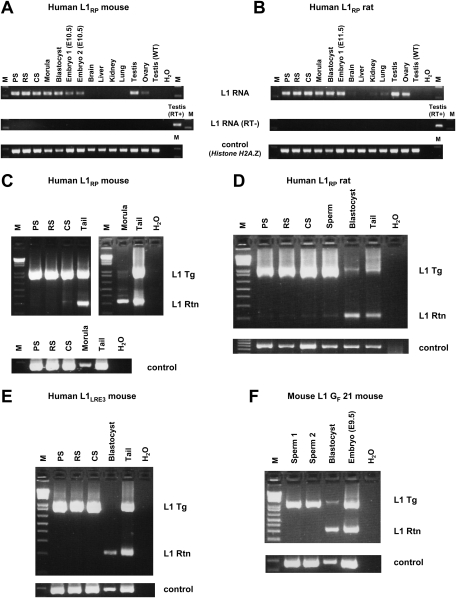
In order to provide further evidence of RNA carryover from germ cells, we analyzed the RNA and DNA from single morulae and early blastocysts obtained from matings between transgenic animals and wild-type animals. Among 12 mouse morulae of a single litter, there were three that did not inherit the L1 transgene from the mother. Two of the three had readily detectable L1 RNA (Fig. 2A). Similarly, among 12 rat blastocysts from a single litter, three did not inherit the L1 transgene, and one of the three had L1 RNA (Fig. 2B), indicating that, in both mice and rats, L1 RNA can be carried over from germ cells to the next generation. Although carried-over L1 RNA appeared quite stable and was present at least until the preimplantation embryo stage, we failed to detect it in embryonic day 10.5 (E10.5) transgene-negative embryos (data not shown).
Figure 2.
Single preimplantation embryos lacking the transgene contain L1 RNA (A–C) and L1 retrotransposition events (D). RT–PCR analysis and genotyping PCR on offspring of an L1LRE3 mouse (A), an L1RP rat (B), and an L1 GF 21 mouse (C). RNA isolated from single morulae or blastocysts was subjected to RT–PCR to detect L1 RNA from the L1 transgene. For genotyping, DNA from each embryo was subjected to genotyping PCR. To exclude the possibility of a false negative genotype for the transgene, each embryo was genotyped by two independent primer sets for the L1 transgene, and three independent genomic loci were amplified as controls. In A_–_C, an asterisk denotes a transgene-negative, L1 RNA-positive preimplantation embryo. (D) Retrotransposition events in individual late blastocysts. Single blastocysts of the L1RP mouse were subjected to genotyping PCR. For semiquantification, mouse DNA that carries one retrotransposition event per diploid genome (Ostertag et al. 2002) was used as a calibrator DNA. (Middle) The amount of DNA of each blastocyst used in the intron-flanking PCR was presumed to be 0.1–0.5 ng. Thus, retrotransposition events appear to be present in much less than one copy per cell. (RT) Reverse transcriptase; (Tg) transgene; (Rtn) retrotransposition event; (WT) wild-type animal; (M) 1-kb plus DNA Ladder (Invitrogen).
To investigate whether L1 RNA carryover was due to non-species-specific transgenes, we created transgenic mouse lines using a native mouse L1 element. Mouse L1s are generally homologous to human L1s but differ significantly in their 5′UTR and the 5′ half of ORF1. The mouse L1 transgene consisted of a highly active mouse L1, GF21 (Goodier et al. 2001), driven by its own promoter and tagged with the retrotransposition cassette (L1 GF21 mouse). As in human L1 experiments, mouse L1 RNA was observed in one of four transgene-negative blastocysts (Fig. 2C).
Furthermore, upon PCR genotyping of individual late blastocysts from a human L1 transgenic male mated with a wild-type female, we found retrotransposition events in both transgene-positive and transgene-negative embryos (Fig. 2D). To estimate the copy number of retrotransposition events in these embryos, mouse DNA with a germline retrotransposition event (one retrotransposition insertion per diploid genome) created previously (Ostertag et al. 2002) served as a calibrator. Comparing genotyping PCRs of single blastocysts with those of serial dilutions of the calibrator DNA demonstrated that the retrotransposition events in individual blastocysts were present in much less than one copy per diploid genome regardless of whether the blastocyst carried the transgene or not, indicating somatic L1 retrotransposition events in these embryos. In addition, since L1 retrotransposition events in single blastocysts lacking the transgene were derived from a cross in which the male carried the transgene, these data further support L1 RNA carryover from male germ cells into the embryo.
Most L1 retrotransposition events occur during development, creating nonheritable de novo L1 insertions
To investigate more precisely when L1 retrotransposition occurs, we compared L1 expression and retrotransposition in various developmental stages of transgenic animals, as well as adult tissues. We tested pachytene spermatocytes (PS), round spermatids (RS), condensing spermatids (CS), and sperm from transgene-positive mice and rats. We also assayed transgene-positive preimplantation embryos (morulae and blastocysts), E10.5–E11.5 embryos, and adult somatic tissues. We found L1 RNA in all spermatogenic cell fractions, morulae, blastocysts, E10.5–E11.5 embryos, and adult testes and ovaries (Fig. 3A,B). Faint amplicons were also identified in other adult tissues, such as lung.
Figure 3.
L1 transcripts and retrotransposition events in various developmental stages and adult tissues. (A,B) RT–PCR on transgenic L1RP mouse (A) and L1RP rat (B) spermatogenic cell fractions, preimplantation embryos (morulae and blastocysts), E10.5–E11.5 embryos, and adult tissues. Only a head portion of E10.5–E11.5 embryos was subjected to RT–PCR in order to eliminate contamination of germ cells in the embryonic developmental stages. Testis from wild-type adult animals was used as a negative control for L1 RNA from the transgene. Histone H2A.Z gene was used as an endogenous control. (C–F) Genotyping PCR on L1RP mouse (C), L1RP rat (D), L1LRE3 mouse (E), and L1 GF 21 mouse (F) spermatogenic cell fractions and pooled preimplantation embryos (L1RP mouse line, 10 morulae; L1RP rat line, 25 blastocysts; L1LRE3 mouse line, nine blastocysts; L1 GF 21 mouse line, 12 blastocysts). Spermatogenic cell fractions were prepared from transgene-positive, retrotransposition event-negative mice (C,E,F) and transgene-positive, retrotransposition event-positive rats (D). Nested PCR was performed on each sample, which was optimized to amplify small products preferentially. In D, similar amounts of DNA (5 ng) from rat spermatogenic cell fractions, pooled blastocysts, and tail were subjected to PCR. Genomic DNA of the Actb region was amplified to confirm the amount of DNA. (M) 1-kb plus DNA Ladder (Invitrogen); (RT) reverse transcriptase; (Tg) transgene; (Rtn) retrotransposition event; (WT) wild type.
Genomic DNA of spermatogenic cell fractions and pooled preimplantation embryos were subjected to the intron-flanking PCR to identify L1 retrotransposition events (Fig. 3C–F). To our surprise, L1 retrotransposition events were rarely detectable in spermatogenic cell fractions of L1RP mice, and only a faint retrotransposition insertion amplicon was observed in the CS fraction (Fig. 3C). In contrast, L1 retrotransposition was much more frequent in preimplantation embryos than in spermatogenic fractions, even though far less embryonic DNA was used in PCR. Similar results were obtained in all human L1 transgenic models, including the L1RP transgenic rat lines (Fig. 3D) and the L1LRE3 mouse lines (Fig. 3E). To rule out a species-specific effect on L1 retrotransposition, we also studied the timing of retrotransposition of an active mouse L1 transgene and found that it was similar to that of the human L1 transgenes. Intron-flanking PCR revealed a much greater frequency of mouse L1 retrotransposition in preimplantation embryos than in sperm (Fig. 3F). Thus, for all transgenic lines, a relative post-transcriptional block to retrotransposition present in germ cells is alleviated after fertilization.
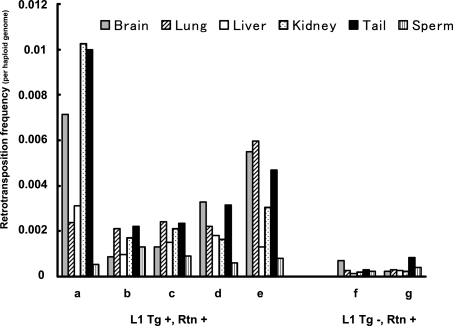
The great majority of de novo L1 retrotransposition events observed in this study integrated during embryogenesis. To estimate the frequency of developmental retrotransposition, we performed quantitative PCR (qPCR) on several adult tissues (brain, lung, liver, kidney, and tail) and sperm DNA of human L1 transgene-positive, retrotransposition event-positive mice (Fig. 4). Animals displayed an unequal tissue distribution of retrotransposition frequency, with higher frequencies of retrotransposition in somatic tissues than in sperm. Retrotransposition events were identified in one out of 50 to one out of 500 somatic cells and in about one out of 1000 sperm. To estimate the frequency of human L1 retrotransposition from RNA carryover, we performed qPCR on adult tissues and sperm DNA of transgene-negative, retrotransposition event-positive mice. The retrotransposition frequencies were lower than those of transgene-positive mice without significant differences between somatic tissues and sperm. Retrotransposition from RNA carryover was estimated to be about one in 1000 cells (about one in 2000 genomes) in both somatic tissues and sperm (Fig. 4).
Figure 4.
L1 retrotransposition frequency in the L1RP mouse. DNA derived from brain, lung, liver, kidney, tail, and sperm of transgene-positive, retrotransposition event-positive mice (A–E) and transgene-negative, retrotransposition event-positive mice (F,G) was analyzed by qPCR. Genomic DNA of each tissue was subjected to the intron-flanking PCR reactions and internal control PCR. Both PCR reactions were normalized to those of the calibrator DNA. The calibrator DNA was obtained from a mouse with a germline L1 retrotransposition event identified in our previous studies (Ostertag et al. 2002), which had 0.5 retrotransposition insertions per haploid genome. (Tg) L1 transgene; (Rtn) retrotransposition event.
Discussion
Previously, L1 retrotransposition was thought to occur predominantly in germ cells (Ostertag and Kazazian 2001; Bourc'his and Bestor 2004). Here, we demonstrate that retrotransposition occurring directly in germ cells is uncommon. Most human and mouse L1 retrotransposition events occur in embryonic development; only a fraction of these events get into the germline and are heritable. These findings in transgenic animals suggest that the majority of L1 retrotranspositions in human beings occur in early embryonic development. Recently, retrotransposition of a transfected L1 has been demonstrated in human embryonic stem cells (Garcia-Perez et al. 2007). In addition, an L1 retrotransposition event during early human embryogenesis leading to somatic and germinal mosaicism was reported (van den Hurk et al. 2007). In fact, all known human retrotransposition events could have occurred early in human development. In one case, we reported a retrotransposition event that we believed had occurred prior to the end of maternal meiosis I (Brouha et al. 2002). On the basis of the present study, the alternative of RNA carryover from the mother followed by retrotransposition in the embryo is just as likely.
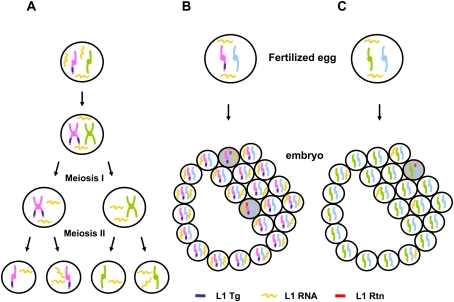
In our study, human and mouse L1 RNA was carried over through both male and female germ cells to the next generation. Thus, L1 RNA in this context is quite stable, with a half-life likely >24 h. We speculate that the previously described L1 RNP (Martin 1991) protects L1 RNA from degradation. Interestingly, male transgenic animals give rise to a similar percentage of transgene-negative, insertion-positive offspring as female transgenics (Table 2). Although it has been widely accepted that there is little, if any, RNA in mature sperm, a recent study in mice has demonstrated strong evidence of non-Mendelian inheritance of RNA molecules through both genders (Rassoulzadegan et al. 2006). The meiotically stable and heritable RNA can cause a phenotypic change in the absence of the inducing allele. L1 RNA is similarly transmitted to the next generation. Moreover, L1 RNA that is inherited independently of its parental allele can modify DNA sequence by creating a de novo L1 retrotransposition event in the genome of the offspring. The mechanism of L1 RNA carryover remains unclear. L1 RNA that is assembled into its RNP complex might be sufficiently stable to complete this unusual scenario (Fig. 5A). Sequestration of the L1RNP in a cellular organelle as seen with the I-factor RNA of Drosophila melanogaster is another possibility (Chambeyron et al. 2008).
Figure 5.
L1 transcripts and L1 retrotransposition in germ cells and early development. (A) Abundant L1 RNA is present in both developing germ cells and meiotic cells. Some L1 RNA segregates from its encoding DNA. However, L1 retrotransposition events are rare in these cells. (B) L1 transgene-positive gametes can be fertilized, carrying L1 RNA into the fertilized egg. L1 RNA transcribed either during germ cell development or during embryogenesis can retrotranspose into the genome. (C) L1 transgene-negative gametes (both sperm and egg) can carryover L1 RNA that has segregated from its encoding DNA into the fertilized egg. This L1 RNA can be the source of infrequent L1 retrotransposition events during embryogenesis. Retrotransposition events in scenario B occur more frequently than those in scenario C (see Fig. 4). (Tg) L1 transgene; (Rtn) retrotransposition event. Shaded cells represent retrotransposition-positive cells.
L1 RNA was easily identified in spermatogenic cells and individual preimplantation embryos, as reported previously (Packer et al. 1993; Branciforte and Martin 1994; Ostertag et al. 2002; Peaston et al. 2004). Furthermore, L1 RNA was also present in post-implantation embryos and a low amount of L1 RNA was present in adult tissues, suggesting a potential role of L1 in human development. In spite of abundant L1 RNA in spermatogenic cells, most actual L1 integration occurs after fertilization and may continue throughout development, resulting in highly unequal tissue distributions of L1 retrotransposon insertions. The retrotransposition frequency in somatic tissues of transgene-positive mice was roughly 10 times greater than the retrotransposition frequency in somatic tissues of transgene-negative mice. These data indicate that most somatic L1 retrotransposition events are caused by L1 RNA that is transcribed after fertilization, creating mosaicism in both somatic and germline tissues (Fig. 5B,C).
More interestingly, the L1 retrotransposition frequency in the sperm of transgene-positive mice was generally lower than that of other somatic tissues. Embryonic germ cells are believed to carry abundant L1 RNA because of L1 demethylation in the fetal testes (Kuramochi-Miyagawa et al. 2008), while L1s in somatic tissues are kept continuously silenced. In fact, abundant L1 RNA was found predominantly in spermatogenic cell fractions. These results indicate that germ cells might have a post-transcriptional defense mechanism that prevents L1 integration into the genome. They also indicate that de novo L1 retrotransposition events can occur even after the time of establishment of germ cells (E7.25), or even in adult tissues, which cannot be transmitted to the next generation. These nonheritable L1 retrotransposition events in somatic tissues may play a significant role in creating genomic diversity within an individual.
It is of great interest that most human L1 retrotransposition events likely occur in development rather than in the germline, using L1 RNAs transcribed in the embryo and in developing germ cells. These data suggest a role for somatic L1 retrotransposition events in human development with potential effects on learning and behavior, as previously suggested (Muotri et al. 2005), carcinogenesis (Miki et al. 1992), and other conditions with somatic, nonheritable mutations in their etiology. That L1 RNA can alter the DNA of a subsequent generation is another example of the noncanonical role RNA can play in determining the human phenotype.
Materials and methods
All primer sequences are listed in Supplemental Table 1. Animal studies were performed in compliance with Institutional Animal Care and Use Committee guidelines of the University of Pennsylvania.
Generation of transgenic animals
A DNA fragment containing the γ-globin intron (INT) was cloned into the BstZ17I and XmaI sites in the L1 3′UTR of pJCC5-L1LRE3 (Ostertag et al. 2000; Brouha et al. 2002) to create pJCC5-L1LRE3-INT. We cloned L1LRE3-INT as a NotI/blunted ApaI fragment into the multiple cloning site of pRJD099 cut with NotI/blunted SfiI to create pRJD099-L1LRE3-INT-SV40 poly(A) signal. To create an insulator backbone, two copies of the chicken β-globin insulator (INS) were isolated from pJC13-1 (a gift of Gary Felsenfeld) as a SalI/KpnI fragment and cloned into pBluescript KS− (Stratagene) to create pBS-2xINS. Another two copies of the insulator were cloned into pBS-2xINS as a SpeI/BamHI and a BamHI/EcoRI fragment to create pBS-4xINS. We cloned the L1LRE3-INT-SV40 poly(A) signal as a blunted NotI/SalI fragment into the multiple cloning site of pBS-4xINS cut with EcoRV/SalI to create pBS-2xINS-L1LRE3-INT-SV40 poly(A) signal-2xINS (L1LRE3 transgene). The full-length mouse L1 element, GF21, was liberated as a NotI/blunted XhoI fragment (Goodier et al. 2001). The GF21 fragment was swapped into pLRE3-EGFP (Brouha et al. 2002) using the NotI and BstZ17I sites to create the GF21 element tagged with the EGFP retrotranspositon cassette but lacking insulators (pGF21-EGFP; mouse L1 GF21 transgene). The transgene-containing plasmid was tested in a cell culture assay of retrotransposition to confirm its activity. The transgene was prepared for injection by cutting it from its backbone plasmid with BssHII or NotI/SalI digestion followed by purification of the transgene-containing fragment using Elutip column (Schleicher & Schuell). The transgenes were microinjected into fertilized C57BL/6J or B6D2F1 oocytes. The transgenic mice were maintained by crossing with wild-type C57BL/6J mice. The transgenic rat was created using the L1RP tagged with a markerless retrotransposition cassette (Babushok et al. 2006). This transgene was microinjected into fertilized Sprague-Dawley oocytes.
Genotyping PCR
Genotyping PCR was performed on 100–500 ng of DNA using an intron-flanking PCR as described (Ostertag et al. 2002; Muotri et al. 2005; Babushok et al. 2006). PCR products were purified using QIAquick PCR purification (Qiagen) and subjected to second-round PCR as necessary. Nested PCR was optimized to amplify small products preferentially. Cycling conditions of the second PCR were initial denaturation for 1 min at 95°C, followed by a two-step profile: denaturation for 30 sec at 95°C and annealing extension for 15 sec at 68°C for 40 cycles. Three different spermatogenic germ cell fractions of PS, RS, and CS were purified from transgene-positive, retrotransposition event-negative mice and transgene-positive, retrotransposition event-positive rats as described previously (Romrell et al. 1976; Ostertag et al. 2002). DNA was purified from pooled preimplantation embryos after proteinase K treatment and was subjected to PCR. When both DNA and RNA were required from single preimplantation embryos, RNA was purified using magnetic beads (Dynal Biotech) and then DNA was purified from the unbound fraction of the beads using the Wizard DNA clean-up system (Promega). The purified DNA was subjected to the genotyping PCR directly (L1RP rat and L1 GF 21 mouse) or was subjected to whole-genome amplification (WGA) using the PEPLI-g kit (Qiagen) prior to the genotyping PCR (L1LRE3 mouse). Three independent WGA reactions on each embryo were combined and subjected to genotyping PCR.
Southern blot
Genomic DNA isolates from mouse tails of the L1RP line mice were digested with PvuII or SalI/EcoRV, and those from the L1LRE3 line animals were digested with PvuII or BglII. Digested samples were separated by electrophoresis on 0.8% agarose gels and transferred to Hybond-N+ membranes (Amersham Bioscience) using alkaline transfer. Membranes were prehybridized for 12 h at 65°C in hybridization buffer (10% polyethylene glycol, 7% SDS, 200 μg/mL denatured sonicated salmon sperm DNA). Blots were probed with a 1.4-kb probe consisting of the CMV promoter and EGFP sequence liberated from pEGFP-N1 as a NotI/AseI fragment (Clontech) or with a 503-base-pair (bp) probe consisting of partial L1 3′UTR and SV40 poly(A) signal sequence liberated from the L1LRE3 transgene as a XmaI/SalI fragment. Each probe was radiolabeled with 32P by random priming, and membranes were hybridized overnight at 65°C. Membranes were then washed and exposed to X-ray film. Each membrane was stripped and hybridized with a 759-bp probe generated from an unrelated region on chromosome 11 or a 532-bp probe generated from the Actb gene region as a DNA loading control. Southern blot for the L1RP rat lines was performed as described (Babushok et al. 2006).
RT–PCR
RNA was purified using either magnetic beads (Dynal Biotech) from individual preimplantation embryos or the RNeasy kit (Qiagen) from other tissues and was reverse transcribed with oligo(dT) primers using SuperScript III (Invitrogen) to synthesize cDNA according to the manufacturer's protocol. L1 transcript was amplified using a primer set that was specific to the L1 transgene. Histone H2A.Z was used as an internal control.
qPCR
Real-time qPCR was performed with LightCycler FastStart DNA Masterplus SYBR Green I (Roche). Genomic DNA of the L1RP mouse was subjected to PCR. L1 retrotransposition events were amplified by the intron-flanking PCR. An unrelated genomic DNA around the Actb gene region was amplified as an internal control. We used calibrator DNA to normalize retrotransposition frequency. The calibrator DNA was derived from a mouse with a germline retrotransposition insertion in our previous studies (Ostertag et al. 2002). The calibrator DNA carried 0.5 L1 retrotransposition events per haploid genome. The intron-flanking PCR and the internal control PCR of each tissue were normalized to those of the calibrator DNA. Each DNA sample was subjected to qPCR in duplicate. qPCR experiments were performed at least twice for each sample. Each reaction was analyzed by melting curve and/or gel electrophoresis to confirm that nonspecific products or the L1 transgene was not amplified. Cycling conditions were initial denaturation for 10 min at 95°C, followed by a three-step profile: denaturation for 10 sec at 95°C, annealing for 15 sec at 60°C, and extension for 3 sec at 72°C.
Acknowledgments
We thank J. Goodier and M. Bartolomei for critical reading of the manuscript. We thank Lu Zheng and Tanya Merdiushev for technical assistance. We thank F. Gage and A. Muotri for a gift of the L1RP mouse transgenic line. We thank the Transgenic Core of the University of Pennsylvania and the Rat Resource and Research Center of the University of Missouri for generation of transgenic mouse lines and transgenic rat lines, respectively. H.H.K. and E.M.O. are supported by grants from the NIH.
Footnotes
References
- An W, Han JS, Wheelan SJ, Davis ES, Coombes CE, Ye P, Triplett C, Boeke JD. Active retrotransposition by a synthetic L1 element in mice. Proc Natl Acad Sci. 2006;103:18662–18667. doi: 10.1073/pnas.0605300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babushok DV, Ostertag EM, Courtney CE, Choi JM, Kazazian HH., Jr L1 integration in a transgenic mouse model. Genome Res. 2006;16:240–250. doi: 10.1101/gr.4571606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: Implications for transposition. Mol Cell Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Meischl C, Ostertag E, de Boer M, Zhang Y, Neijens H, Roos D, Kazazian HH., Jr Evidence consistent with human L1 retrotransposition in maternal meiosis I. Am J Hum Genet. 2002;71:327–336. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Popkova A, Payen-Groschene G, Brun C, Laouini D, Pelisson A, Bucheton A. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci. 2008;105:14964–14969. doi: 10.1073/pnas.0805943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez JL, Marchetto MC, Muotri AR, Coufal NG, Gage FH, O'Shea KS, Moran JV. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Du K, Kazazian HH., Jr A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohjoh H, Singer MF. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol Cell Biol. 1991;11:4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH., Jr Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, DeBerardinis RJ, Goodier JL, Zhang Y, Yang N, Gerton GL, Kazazian HH., Jr A mouse model of human L1 retrotransposition. Nat Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- Packer AI, Manova K, Bachvarova RF. A discrete LINE-1 transcript in mouse blastocysts. Dev Biol. 1993;157:281–283. doi: 10.1006/dbio.1993.1133. [DOI] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Prak ET, Dodson AW, Farkash EA, Kazazian HH., Jr Tracking an embryonic L1 retrotransposition event. Proc Natl Acad Sci. 2003;100:1832–1837. doi: 10.1073/pnas.0337627100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Romrell LJ, Bellve AR, Fawcett DW. Separation of mouse spermatogenic cells by sedimentation velocity. A morphological characterization. Dev Biol. 1976;49:119–131. doi: 10.1016/0012-1606(76)90262-1. [DOI] [PubMed] [Google Scholar]
- Trelogan SA, Martin SL. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc Natl Acad Sci. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hurk JA, Meij IC, Del Carmen Seleme M, Kano H, Nikopoulos K, Hoefsloot LH, Sistermans EA, de Wijs IJ, Mukhopadhyay A, Plomp AS, et al. L1 retrotransposition can occur early in human embryonic development. Hum Mol Genet. 2007;16:1587–1592. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- West AG, Gaszner M, Felsenfeld G. Insulators: Many functions, many mechanisms. Genes & Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]