Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly (original) (raw)
Abstract
Broad applications of zinc finger nuclease (ZFN) technology—which allows targeted genome editing—in research, medicine, and biotechnology are hampered by the lack of a convenient, rapid, and publicly available method for the synthesis of functional ZFNs. Here we describe an efficient and easy-to-practice modular-assembly method using publicly available zinc fingers to make ZFNs that can modify the DNA sequences of predetermined genomic sites in human cells. We synthesized and tested hundreds of ZFNs to target dozens of different sites in the human CCR5 gene—a co-receptor required for HIV infection—and found that many of these nucleases induced site-specific mutations in the CCR5 sequence. Because human cells that harbor CCR5 null mutations are functional and normal, these ZFNs might be used for (1) knocking out CCR5 to produce T-cells that are resistant to HIV infection in AIDS patients or (2) inserting therapeutic genes at “safe sites” in gene therapy applications.
Zinc finger nucleases (ZFNs) are artificial DNA restriction enzymes that consist of custom-designed DNA-binding zinc finger proteins and the nonspecific nuclease domain derived from FokI endonuclease (Kim et al. 1996). ZFNs can be used for efficient genetic modifications in mammalian, plant, and other higher eukaryotic cells and organisms (Bibikova et al. 2002, 2003; Lloyd et al. 2005; Urnov et al. 2005; Wright et al. 2005; Beumer et al. 2006; Morton et al. 2006; Doyon et al. 2008; Maeder et al. 2008; Meng et al. 2008; Santiago et al. 2008). ZFN technology, indeed, holds great promise for broad applications from cell engineering to plant and animal biotechnology and to gene therapy.
The zinc finger (ZF) moieties of ZFNs consist of three or four tandemly arrayed ZF modules, each of which recognizes 3-bp subsites. ZFNs function as both homo- and heterodimers and, therefore, recognize 18- to 24-bp sequence elements in a genome. Various in vitro and in vivo selection methods can be used to construct ZF arrays (Rebar and Pabo 1994; Greisman and Pabo 1997; Joung et al. 2000; Bae et al. 2003; Hurt et al. 2003; Bae and Kim 2006; Maeder et al. 2008). These selection-based methods may yield effective ZF proteins, but are labor-intensive and time-consuming.
An alternative method for constructing ZF arrays is modular assembly of precharacterized ZFs using standard recombinant DNA technology (Bae et al. 2003; Wright et al. 2006; Cathomen and Joung 2008; Ramirez et al. 2008). Sangamo Biosciences has used its own modular-assembly method to construct, with high efficiency, ZFNs that target two endogenous genomic sites in human cells and one site in Chinese hamster ovary cells (Urnov et al. 2005; Perez et al. 2008; Santiago et al. 2008). However, not all of the amino acid sequences of the ZFs used in these studies were reported, and, therefore, this method currently is not available for use in the academic science community (Scott 2005; Wilson 2008).
Recently, Ramirez et al. (2008) reported unexpected high failure rates of modularly assembled ZFNs. Unlike Sangamo Biosciences, which assembles four-finger ZFNs from its own proprietary archive of two-finger modules, Ramirez et al. (2008) used publicly available one-finger libraries to assemble three-finger ZFNs. Although this method should allow any researcher to assemble ZFNs readily by linking ZF modules using standard recombinant DNA technology and has yielded ZFNs that have successfully modified genomic sequences in Drosophila (Beumer et al. 2006, 2008) and Caenorhabditis elegans (Morton et al. 2006), it is important to note that not a single endogenous genomic site in human or other mammalian cells has been modified thus far with ZFNs constructed via this method. However, our analyses of the data provided by Ramirez et al. (2008) and re-examination of our own data (Bae et al. 2003) cited in Ramirez et al. suggest that success rates for modularly assembled ZFNs that can modify specific genomic sites in mammalian cells may depend on one's choice of ZF modules.
Here, we report on the successful production of genome-modifying ZFNs constructed by modular assembly of carefully chosen ZFs. We assembled hundreds of ZFNs that target dozens of genomic sites in human cells and found that many of these ZFNs show genome editing activity. Our approach provides a rapid, easy-to-practice, and publicly available platform for ZFN design.
Results
ZFN design and synthesis
In order to test our ZFN assembly method, we designed and synthesized ZFNs that target the human chemokine (C-C motif) receptor 5 (CCR5) gene. The CCR5 protein is the major co-receptor used by the human immunodeficiency virus (HIV) to infect target cells and is one of the two human genes that have been successfully edited with ZFNs generated by Sangamo Biosciences (Urnov et al. 2005; Perez et al. 2008). ZFNs that knock out CCR5 may be used for producing immune cells that are resistant to HIV infection, which holds promise for the development of a novel gene therapy approach for the treatment of AIDS. In addition, the CCR5 locus might provide a “safe harbor” for the insertion of therapeutic genes (Camenisch et al. 2008; Cathomen and Joung 2008), because individuals that harbor a naturally occurring homozygous 32-bp deletion in CCR5 (_CCR5Δ_32) are healthy and show no visible phenotypic differences when compared with _CCR5Δ_32 heterozygous or wild-type individuals (Liu et al. 1996).
To assemble our ZFNs, we first chose 54 ZF modules with diverse DNA-binding specificities. Of these 54 ZF modules, 31 were derived from DNA sequences in the human genome, and two were from those in the Drosophila genome (termed “ToolGen modules”). These naturally occurring ZFs have been used repeatedly for the regulation of many genes in bacterial, yeast, and mammalian cells (Bae et al. 2003; Lee et al. 2003, 2004; Park et al. 2003, 2005a,b; Kwon et al. 2006; Yun et al. 2008). The remaining 21 ZF modules were either selected from libraries of ZF variants using phage display (Barbas modules) (Dreier et al. 2000, 2001, 2005; Liu et al. 2002) or produced by site-directed mutagenesis (Sangamo modules) (Liu et al. 2001), and were chosen to supplement the DNA-binding specificities of ToolGen modules.
Next, for the design of multi-finger ZFNs, we used a computer algorithm (available at http://www.toolgen.com/ZFNfinder) to identify potential ZFN target sites in the DNA sequence of the CCR5 coding region. Because ZFNs function as dimers, two ZFN monomers need to be prepared to target a single DNA site. Each of the two monomeric ZFNs that compose a ZFN pair binds to one of the two 9- or 12-bp half-sites that are separated by a 5- or 6-bp spacer sequence. These ZFNs contain either three or four ZF modules, each of which recognizes a 3-bp subsite. For a single half-site, multiple monomeric ZFNs can be designed, which consist of different sets of ZFs with identical or similar DNA-binding specificities. Thus, a single site can be targeted with many combinatorial ZFN pairs. We synthesized 208 ZFN monomers and tested 315 ZFN pairs at 33 genomic sites.
In vitro and in vivo assays of ZFN activity
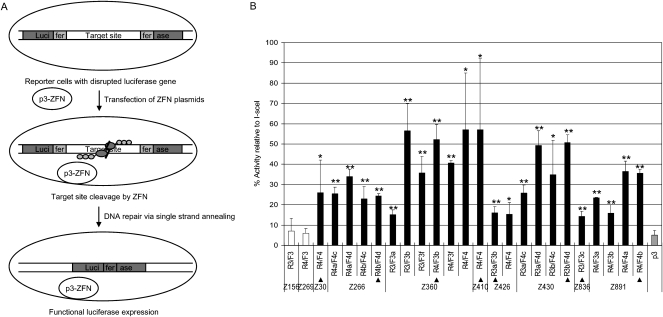
First, ZFNs were prepared using an in vitro transcription and translation (IVTT) system and incubated with a DNA segment that contained the CCR5 coding sequence to assess their DNA restriction activities in vitro. As shown in Supplemental Figure 1, many ZFN pairs (44%) showed efficient (that is, >5%) site-specific cleavage of the target DNA. We then tested whether these ZFNs were able to induce homologous recombination in human cells using a mammalian cell-based single-strand annealing (SSA) system (Chames et al. 2005). Plasmids that encoded ZFNs were transfected into human embryonic kidney cells (HEK293 cells) whose genome contained a stably integrated, partially duplicated firefly luciferase gene that was disrupted by insertion of the CCR5 sequence. Effective ZFNs would generate a double-strand break (DSB) in the CCR5 sequence, which should allow the functional luciferase gene to be restored via SSA. The efficiency of DNA cleavage by the ZFNs can be estimated by measuring luciferase enzyme activity. The meganuclease I-SceI was used as a positive control.
As shown in Figure 1, many ZFN pairs yielded significant luciferase activity in this assay. Out of 315 ZFN pairs, 23 pairs showed 15%–57% luciferase activity, compared with I-SceI. It is possible that ZFN pairs that exhibited <15% activity still would be able to induce a DSB in cells, but we focused on the highly active 23 pairs for further studies. These 23 ZFN dimers did not yield significant luciferase activity when we used targetless reporter cells, whose genome contained the partially duplicated luciferase gene disrupted by a DNA sequence unrelated to CCR5 (data not shown). These results suggest that ZFN-mediated DNA repair is _CCR5_-specific. Collectively, these 23 active ZFN pairs were designed to target eight different sites in the CCR5 gene. All of these 23 pairs had shown strong endonuclease activity in the IVTT assay.
Figure 1.
Assessment of ZFN activities using a cell-based single-strand annealing (SSA) system. (A) Schematic overview of a single-strand annealing system. ZFN expression plasmids are transfected into HEK293 cells whose genome contains an inactive, partially duplicated, and disrupted luciferase gene. If ZFNs cleave target sites in cells, DNA is efficiently repaired by the SSA mechanism, and the functional luciferase gene is restored. (B) Luciferase activities of cells in which various ZFNs are expressed. p3 (gray bar) is the empty plasmid, which was used as a negative control. The target sequence contains the recognition site of I-SceI, which was used as a positive control. The activity of each ZFN pair is reported as the percentage relative to the I-SceI control. ZFN pairs and their constituent monomers are indicated. The ZFN pairs used in further studies are marked with triangles. Means and standard deviations (error bars) from at least three independent experiments are shown. Black and white bars indicate active and inactive ZFNs, respectively. _P_-values were calculated with the Student's _t_-test; (*) P < 0.01; (**) P < 0.001 (p3 control vs. ZFN).
Mutation detection in HEK293 cells treated with ZFNs
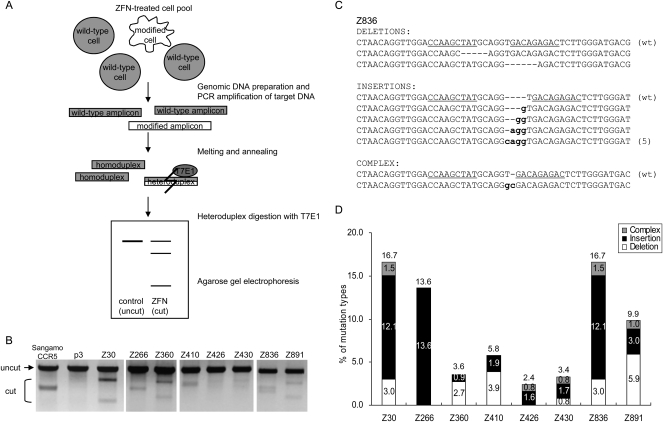
Next, we investigated whether the ZFNs that both displayed endonuclease activity in the IVTT assay and induced luciferase activity in the cell-based assay could modify endogenous target sequences in cells. A DSB induced by ZFNs can be repaired by error-prone nonhomologous end-joining (NHEJ) (Bibikova et al. 2002; Morton et al. 2006). The resulting DNA often contains small insertions or deletions (“indel” mutations) near the DSB site. These indel mutations can be detected in vitro by treating amplified DNA fragments with mismatch-sensitive T7 endonuclease I (T7E1) (Fig. 2A).
Figure 2.
ZFN-mediated genome editing in human cells. (A) Schematic overview of mismatch detection using T7E1 assay. Genomic DNA was purified from cells transfected with plasmids encoding ZFNs. The DNA segments encompassing the sites of ZFN recognition were PCR-amplified, and the DNA amplicons were melted and annealed. If the DNA amplicons contain both wild-type and mutated DNA sequences, heteroduplexes would be formed. T7E1 recognizes and cleaves heteroduplexes, but not homoduplexes. The DNA fragments were assessed by agarose gel electrophoresis; a schematic of an idealized gel result is shown. (B) ZFN-mediated genomic modification revealed by the T7E1 assay. The ZFN pairs are shown at the top of the agarose gels. The expected positions of the resulting DNA bands are indicated by an arrow (uncut) and a bracket (cut) at the left of the gel panels. p3 is the empty plasmid used as a negative control. Sangamo's _CCR5_-targeting ZFN pair (Sangamo CCR5) was included in this assay. (C) DNA sequences of a genomic site targeted by the Z836 ZFN pair. (Underlined) ZFN recognition elements. (Dashes) Deletions; (small letters) inserted bases. In cases in which a mutation was detected more than once, the number of occurrences is shown in parentheses. (wt) Wild type. (D) Types of mutations at various ZFN-targeted sites. The number of deletions, insertions, and complex mutations for each ZFN pair (as shown in C for Z836) were counted, and the percentages of these incidents were plotted.
HEK293 cells were transfected with plasmids that encoded ZFN pairs, and genomic DNA was isolated 72 h later. To assess the genome editing activities of the ZFNs, the DNA segments that encompassed the ZFN target sites were PCR-amplified and then treated with T7E1. As shown in Figure 2B, small fractions of amplified DNA from ZFN-treated cells were cleaved by T7E1. Each ZFN pair gave rise to distinctive cleavage patterns, reflecting the fact that these ZFNs targeted eight different sites (Supplemental Fig. 2). In addition, the sizes of the resulting DNA bands were as expected. Out of 23 ZFN pairs tested in the T7E1 assay, 21 showed detectable DNA bands with expected size (data not shown). We also used the T7E1 assay to test representative ZFN pairs that either did not show significant endonuclease activity in the IVTT assay or did not give rise to significant luciferase activity in the cell-based SSA system, but passed the IVTT test. None of these ZFN pairs induced detectable levels of DNA mutation (data not shown).
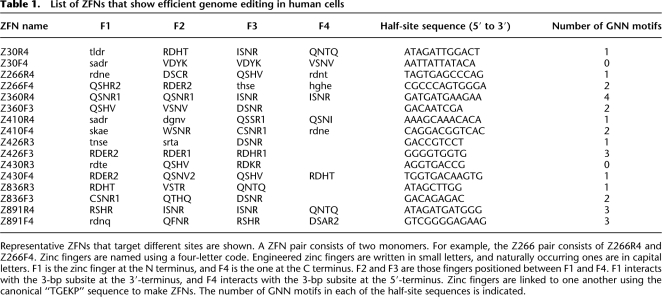
We chose for further study eight representative ZFN pairs, each of which targeted different sites in the CCR5 gene (Table 1). Amplified DNA from cells transfected with each of the eight ZFN pairs was cloned and sequenced. Various deletions and insertions were observed near the sites of DSBs (Fig. 2C,D; Supplemental Fig. 3). These mutagenic patterns are the signature of error-prone NHEJ-associated mutagenesis, and similar patterns have been reported by others (Bibikova et al. 2002; Morton et al. 2006; Santiago et al. 2008).
Table 1.
List of ZFNs that show efficient genome editing in human cells
Eight ZFN pairs that targeted distinct sites showed 2.4%–17% mutagenic rates, which were calculated by dividing the number of mutant clones by the number of total clones analyzed. These high efficiencies of ZFN-induced mutagenesis suggest that clonal mutant cells can be isolated by screening only 10–100 single-cell-derived colonies.
Isolation of clonal cells after ZFN treatment
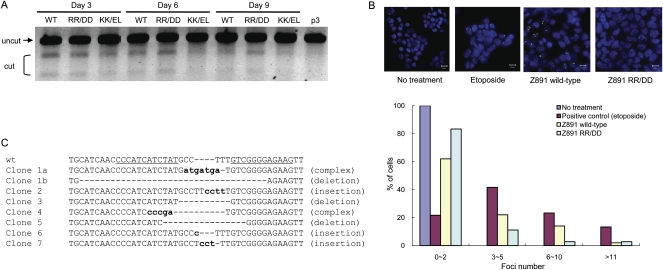
It has been shown by others that certain ZFNs have off-target effects and are cytotoxic when expressed in mammalian cells (Miller et al. 2007; Szczepek et al. 2007). In order to investigate whether mammalian cells that carry ZFN-induced mutations were growth-impaired and thus outgrown by unmodified cells, we performed the T7E1 assay with DNA isolated from cells at 3, 6, and 9 d after ZFN transfection. We chose the Z891 pair for this analysis and, as shown in Figure 3A, the cleaved DNA fragments were reduced at day 6, compared with those at day 3, and were barely detectable at day 9 post-ZFN treatment. These results suggest that ZFN-mediated mutagenesis does, indeed, have cytotoxic effects on cell growth.
Figure 3.
Obligatory heterodimeric ZFNs and isolation of mutant clones. (A) Time-course analysis of wild-type and obligatory heterodimeric ZFNs. The T7E1 mismatch detection assay was performed at various time points with DNA isolated from cells treated with different forms of the FokI nuclease domain. (WT) Wild-type nuclease domain; (RR/DD or KK/EL) obligatory heterodimeric nuclease domains. (B) ZFN-induced DSBs detected by TP53BP1 staining. Both the wild-type and obligatory heterodimeric Z891 ZFN pair were transfected into HEK293T/17 cells, and intracellular TP53BP1 foci were detected by immunofluorescence at day 2 post-transfection. Etoposide (1 μM) was used as a positive control. The distribution of the numbers of TP53BP1 foci is plotted. At least 100 cells were analyzed for each treatment in two independent measurements. (C) DNA sequences of mutant clones. Seven mutant clones were isolated, by limiting dilution, from cells treated with the RR/DD ZFN dimer. The DNA sequences of the target site in these clonal cells are shown. (Dashes) Deletions are indicated with dashes; (small letters) inserted bases. Clones 1a and 1b indicate DNA sequences that resulted from biallelic modifications in a single clone.
Off-target effects of ZFNs are caused largely by the activity of ZFN monomers that form both homo- and heterodimers. These effects could be reduced significantly by using FokI nuclease variants that form heterodimers, but cannot form homodimers (Miller et al. 2007; Szczepek et al. 2007). Therefore, to reduce the cytotoxic effects of ZFNs, we prepared and tested, in the T7E1 assay, two different types of these so-called obligatory heterodimers (shown as “RR/DD” dimers and “KK/EL” dimers in Fig. 3A). The T7E1 assay revealed that, when cells were transfected with the RR/DD ZFN pair but not with the KK/EL pair, the cleaved DNA fragments persisted even at 9 d after ZFN treatment.
In order to confirm that the reduced cytotoxicity of the obligatory heterodimeric ZFN resulted from its enhanced specificity of DNA cleavage in cells, we analyzed the number of DSBs in ZFN-treated cells using an antibody against TP53BP1 (also known as 53BP1), a protein that is recruited to DSBs (Schultz et al. 2000). As shown in Figure 3B, the number of TP53BP1 foci was reduced significantly in cells treated with the RR/DD ZFN pair, when compared with cells treated with the corresponding wild-type ZFN pair.
Seven different mutant clonal HEK293 cells were subsequently isolated using the RR/DD ZFN pair after screening 225 single-cell colonies that had been grown separately in 96-well plates. We examined the DNA sequences of the CCR5 regions in these clonal cells to confirm the ZFN-induced genomic modifications. Because of a trisomic chromosome (Bylund et al. 2004), HEK293 cells carry three copies of the CCR5 gene in their genome. DNA sequence analyses of the genomes from the modified cells revealed that six of the clones showed monoallelic deletions or insertions, and one showed biallelic modifications in the CCR5 gene (Fig. 3C). In the clone that showed biallelic modifications, two different indel sequences were observed as well as one unmodified, wild-type sequence. This high frequency of bialleic modification by ZFN treatment suggests that homozygous knockout cells could be isolated in a single step without the use of selection markers.
Off-target effects of _CCR5_-targeting ZFNs at the homologous CCR2 sites
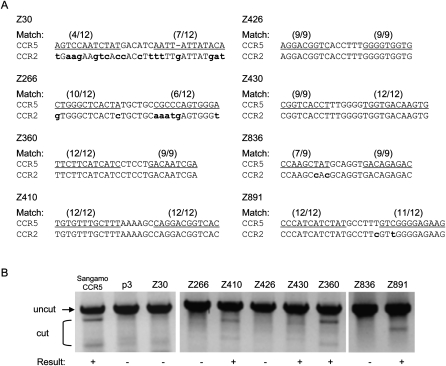
The CCR2 gene is homologous to the CCR5 gene, and many of the CCR5 ZFN recognition elements are conserved in the CCR2 locus. Therefore, we examined whether the eight ZFNs designed for the CCR5 sites could also modify homologous or identical sequences at the CCR2 locus. Three ZFN pairs (Z360, Z410, and Z430) that have identical recognition elements in the CCR2 gene were able to induce efficient genomic modification in the T7E1 assay (Fig. 4). In the CCR2 gene, the recognition element of the Z891 pair consisted of a 12-bp half-site that perfectly matched the corresponding site in the CCR5 gene and a 12-bp half-site that carried a one-base mismatch. As expected, this ZFN pair also showed significant gene editing activity at the CCR2 site. (We also tested Sangamo's _CCR5_-targeting ZFN pair whose recognition element in the CCR2 locus contains one-base mismatches in each of the 12-bp half-sites [Perez et al. 2008] and confirmed that this ZFN shows off-target genomic modification at the homologous CCR2 site [Fig. 4].) In contrast, no detectable ZFN activities were observed with ZFN pairs (Z30, Z266, and Z836) whose recognition elements at the CCR2 locus displayed at least two mismatches.
Figure 4.
Off-target effects of ZFNs at the CCR2 locus. (A) ZFN recognition elements at the CCR5 and CCR2 loci. The ZFN pairs are indicated at the left of the DNA sequences. (Parentheses) The numbers of base matches between the CCR5 and CCR2 loci; (lowercase bold letters) mismatched bases; (underlined) the half-site ZFN recognition elements. (B) T7E1 assay at CCR2 sites. PCR-amplified DNA corresponding to the CCR2 coding region from cells treated with the ZFN pairs (shown at the top of the gel panels) was analyzed. (+) ZFN pairs that gave rise to the modification at the CCR2 sites. p3 is the empty plasmid used as a negative control. Sangamo's _CCR5_-targeting ZFN pair (Sangamo CCR5) was included in this assay.
We also examined whether the seven mutant clonal cells obtained by transfection of HEK293 cells with the Z891 pair induced mutations in the DNA sequence of the highly homologous CCR2 site in addition to that of the intended CCR5 site. Although this pair showed significant gene editing activity at the CCR2 site as well as at the CCR5 site in the T7E1 assay as described above (Fig. 4), the CCR2 locus was not mutated in these clonal cell lines. These results demonstrate that it is possible to isolate clonal cells in which only the intended target site, but not homologous sites, is mutated by screening cells after ZFN treatment.
Summary of ZFN analyses
In order to target various sites in the human CCR5 gene, we synthesized 208 ZFN monomers and tested 315 ZFN pairs for their DNA restriction and genome editing activities. Collectively, these 315 pairs were intended to target 33 sites in the CCR5 gene. Many ZFNs (44%) were able to cleave target DNA efficiently in vitro, but only a fraction of our ZFNs (7.3%) were able to function efficiently in the cell-based SSA system. Most of these ZFNs (21 out of 23) that showed significant activities in the cell-based assay induced efficient genomic modifications at the intended endogenous sites (Supplemental Table 1). These results suggest that the cell-based SSA system, but not the in vitro DNA digestion assay, is a reliable method with which to assess the genome editing potential of ZFNs. These 21 ZFN pairs collectively targeted eight different sites. Thus, out of 33 sites, eight sites were successfully modified by at least one ZFN pair, and the overall success rate of our approach was 24%.
We observed higher success rates with four-finger ZFNs (ZFN pairs that consist of two four-finger ZFN monomers) than with three-finger ZFNs (P < 0.05, Student's _t_-test; Supplemental Table 2). Thus, 26% of potential cleavage sites were targeted successfully with four-finger ZFNs, whereas only 9.1% of potential sites were targeted with three-finger ZFNs. The success rate (12%) of ZFN pairs that consisted of one four-finger ZFN and one three-finger ZFN fell in between the two aforementioned categories.
Although success rates that range from 9.1% to 26% are acceptable, we think that this range is an underestimate of what is possible with our system. First, we chose for subsequent analysis only those ZFNs that showed at least 15% luciferase activity compared to I-SceI in the cell-based SSA system. It is possible that ZFNs that did not pass this test might still be active in genome editing. Second, the T7E1 assay cannot identify the existence of <1% of the modified sequence using gel electrophoresis. ZFNs that allow <1% genomic modification might still be useful for certain applications, but these ZFNs cannot be identified in this assay. Third, the mutagenic NHEJ process often generates large deletions (Morton et al. 2006), which may not be detected by the T7E1 assay.
Module swap analysis
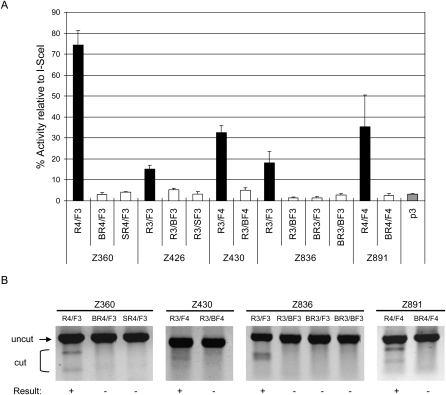
We next investigated why our approach of making ZFNs via modular assembly led to high success rates in genome editing, but other similar approaches had not (Cathomen and Joung 2008; Ramirez et al. 2008). The key difference might reside in the ZF modules. Previously, we reported that naturally occurring ZFs were better building blocks for constructing custom DNA-binding proteins than were the engineered ZFs isolated by phage display or site-directed mutagenesis (Bae et al. 2003). Many of the functional ZFNs that showed efficient genome editing in our study were, indeed, composed exclusively of naturally occurring modules (Table 1). To test whether the module source is a critical determinant in the construction of functional ZFNs, we performed module swap experiments in which our ZFNs made from naturally occurring modules were replaced with those generated from engineered ZFs (i.e., Barbas or Sangamo modules) that displayed identical DNA-binding specificities.
To this end, we prepared eight new ZFN monomers composed exclusively of Barbas or Sangamo modules and used them to replace six different ZFN monomers composed exclusively of ToolGen modules (Supplemental Table 3). Each of these new ZFN monomers was paired with appropriate partner ZFN monomers, and the resulting ZFN pairs were tested in both the SSA and T7E1 assays. As shown in Figure 5, none of the ZFN pairs that consisted of at least one of these newly synthesized ZFN monomers showed any significant activity in the SSA system (P < 0.05, Student's _t_-test). In addition, none of these ZFNs showed any detectable gene-editing activity in the T7E1 assay.
Figure 5.
Module swap analysis. New ZFNs were prepared using engineered ZFs to test whether these ZFNs can functionally replace ZFNs composed exclusively of naturally occurring modules that recognize identical sequences. The names of these ZFNs are indicated by the Z numbers at (A) the bottom of the graph or (B) the top of the gel panels. The ZFN monomers whose names start with “B,” such as “BR4,” are composed exclusively of Barbas modules, and those with “S,” such as “SR4,” are composed exclusively of Sangamo modules. The ZFs that compose these ZFNs are described in Supplemental Table 4. These ZFNs were analyzed using (A) the cell-based SSA system and (B) the T7E1 assay. (A) Luciferase activities of cells in which the various ZFNs were expressed. (Gray bar) p3 is the empty plasmid, which was used as a negative control. The target sequence contains the recognition site of I-SceI, which was used as a positive control. The activity of each ZFN pair is reported as the percentage relative to the I-SceI control. ZFN pairs and their constituent monomers are indicated. Means and standard deviations (error bars) from at least three independent experiments are shown. (B) ZFN-mediated genomic modification revealed by the T7E1 assay. ZFN pairs and their constituent monomers are shown at the top of the agarose gels. (+) ZFN pairs that gave rise to positive gene-editing events.
These results strongly support our previous conclusion that naturally occurring ZFs constitute a more reliable framework for the modular assembly of functional ZF arrays than do engineered ZFs (Bae et al. 2003). It should be noted, however, that Sangamo Biosciences uses two-finger rather than one-finger modules (Urnov et al. 2005). The Barbas group demonstrated that new ZF arrays constructed by grafting only the base-contacting alpha-helixes of appropriate ZFs to a given ZF framework were more effective than those constructed by modular assembly of entire ZF domains (Beerli et al. 1998).
Evaluating ZF modules
Statistical analysis of our 315 ZFN pairs could provide a basis for the design of new ZFNs that can be used for targeted mutagenesis of additional endogenous genes of interest. To this end, we counted the number of occurrences of each ZF in the 23 ZFN pairs that scored positively in the SSA system. We then determined an “activity score” for each ZF by dividing this number by the number of occurrences of the module in all 208 ZFN monomers (Supplemental Table 4). Certain highly active ZFs, for example, “QNTQ” and “RSHR,” had activity scores of 50% and 38%, respectively; these high scores suggest that these modules are reliable when it comes to predicting in vivo site-specific nuclease activity. Other ZFs, such as “QSNK” and “rdae,” which were shown to be inefficient modules in our assays, displayed activity scores of zero. Both of these ZFs were used 15 times each, but none of the ZFNs that contained these modules were active in our assays. The difference in activity scores between the reliable ZFs and the inefficient ZFs was statistically significant (P < 0.05, Student's _t_-test).
We also compared the activity scores of ZFs with identical DNA recognition specificities. Certain ZFs were clearly better than other target-equivalent modules (P < 0.05, Student's _t_-test). For example, both “DSNR” and “HSNK” recognized the same GAC sequence. The use of “DSNR” in the construction of ZFNs yielded many active nucleases (activity score = 35%), while the incorporation of “HSNK” yielded only inactive ZFNs (activity score = 0%).
On the basis of our analysis, we tentatively recommend 37 ZFs (24 ToolGen modules, 1 Sangamo module, and 12 Barbas modules) for use in future gene-editing studies (Supplemental Table 4). These 37 modules all were found at least once in active ZFNs. If we had used only these 37 modules in our experiments, we would have synthesized 78 ZFN monomers (but not 208 monomers) and tested 82 ZFN pairs (but not 315 pairs), and the overall success rate could have been 53%, which is a significant improvement over the current rate (24%). The 37 ZFs collectively recognize 38 out of 64 3-bp subsites. For a given 1-kb random DNA sequence, we predict that there would be 88 [= (38/64)6 × 1000 × 2] potential target sites (that contain either a 5- or 6-base spacer between the half sites) for three-finger ZFN dimers and 31 sites for four-finger ZFN dimers. Assuming 9.1%–26% success rates with our approach (the actual rates might be significantly higher if we use the 37 ZFs, but not the other inefficient modules), ZFN-mediated genome editing would be possible at eight sites, on average, in the 1-kb target sequence. This suggests that most, if not all, eukaryotic genes could be edited with our ZFN assembly method.
Discussion
ZFNs have emerged as important new tools for reverse genetic studies and biotechnology. Despite the broad interest in this technology, a convenient, robust, and easy-to-practice method of producing functional ZFNs currently is not available to the entire research community. The most critical parameter in the use of ZFN technology is how to make effective and specific ZF arrays that selectively bind to predetermined DNA sequences. Target site compositions appear to be especially critical in the design of functional ZFNs. Indeed, most previous studies (Bibikova et al. 2002; Lloyd et al. 2005; Morton et al. 2006) in model organisms used ZFNs that target GNN-repeat sequences, such as 5′-GNNGNNGNN-3′. However, these sequences occur only rarely in a given gene of interest. For example, a three-finger ZFN pair can be designed to target the GNN-repeat DNA sequence, 5′-NNCNNCNNCNNNNNNGNNGNNGNN-3′, which occurs, on average, only once in a 4096-bp (= 46) sequence. GNN-repeat sites for four-finger ZFNs are even more scarce, occurring, on average, only once in a 65,536-bp (= 48) sequence. Therefore, it is likely that such sites do not exist in many genes of interest. In this regard, we note that most of our ZFNs that targeted various sites in CCR5 did not recognize GNN-repeat sequences (Table 1). Among the 16 ZFN monomers that successfully modified the CCR5 sequences, only two (Z426F3 and Z360R4) recognized GNN-repeat sites (Table 1), while two (Z430R3 and Z30F4) recognized half-site elements free of the GNN motif, and 12 recognized sites that consisted of both GNN and non-GNN motifs. The capability of targeting sites other than GNN-repeat sequences greatly expands the utility of ZFN technology.
Recently, Ramirez et al. (2008), after carefully characterizing hundreds of ZF proteins, duly raised concerns about the modular-assembly method for making ZFNs and suggested that more time-consuming and laborious selection systems should be used. At first glance, their conclusion appears in sharp contrast with our results. Using the modular-assembly method, we obtained high success rates both with four-finger (26%) and three-finger ZFN pairs (9.1%).
After an in-depth analysis, we have identified several critical differences between our methods and those of Ramirez et al. (2008). First, Ramirez et al. constructed and characterized three-finger, but not four-finger arrays, whereas we tested both four-finger and three-finger ZFNs. As shown in Supplemental Table 1, our success rates were significantly higher when we used four-finger rather than three-finger ZFNs. Furthermore, all of the active ZFNs reported by Sangamo Biosciences consist of four ZFs. Apparently, four-finger proteins bind to 12-bp target sites more stably and specifically than three-finger proteins bind to 9-bp sites. However, target sites for four-finger ZFN dimers are 24-bp long, and these sites occur much more rarely than do the 18-bp target sites that bind the three-finger ZFN dimers. In this regard, it is encouraging that six out of 16 of our functional ZFN monomers consisted of 3 ZFs (Table 1).
Second, Ramirez et al. (2008) did not mix ZFs from different sources (ToolGen, Sangamo, and Barbas modules) when they generated their ZF arrays. By mixing ZFs from different sources, we obtained nine (out of 16) functional ZFN monomers (Table 1).
Third, most of the ZF arrays characterized in Ramirez et al. were composed exclusively of ZFs isolated by phage display or site-directed mutagenesis (Barbas or Sangamo modules, respectively). In contrast, we used many naturally occurring ZFs (ToolGen modules). Seven out of 16 of our functional ZFN monomers were composed exclusively of naturally occurring ZFs, and two ZFN pairs (out of eight representative pairs) consisted of ZFN monomers composed exclusively of naturally occurring ZFs. All of the other functional ZFN monomers in our study, with the exception of Z426R3, contained at least two naturally occurring ZFs. Module swap experiments confirmed the importance of using naturally occurring ZFs. All of the new ZFNs that we synthesized using engineered ZFs were found to be inefficient and failed to functionally replace ZFNs composed exclusively of naturally occurring modules. Taken together, all of these findings are in line with previous reports that modular assembly with engineered ZFs does not yield functional ZFNs (Maeder et al. 2008; Ramirez et al. 2008).
Why do naturally occurring ZFs fare better than engineered modules in the production of active ZF-containing proteins? We have discussed this issue previously (Bae et al. 2003). Briefly, all of the naturally occurring ZFs we used in this and other studies (Lee et al. 2003, 2004; Park et al. 2003, 2005a,b; Kwon et al. 2005, 2006) had been selected by a stringent cell-based assay. These ZFs may be more suitable for mixing-and-matching than are ZFs selected by other means. We note, however, that the Sangamo and Barbas modules are useful for extending the DNA-binding specificities of ZFNs when used together with naturally occurring modules, and that many of our functional ZFN monomers (nine out of 16) contained at least one of these engineered modules.
Recently, the Joung group (Joung et al. 2000) and members of the Zinc Finger Consortium (www.zincfingers.org) reported a highly efficient _Escherichia coli_-based selection system for producing ZFNs (termed “OPEN ZFNs”) (Maeder et al. 2008). These OPEN ZFNs were shown to successfully target seven of 14 different endogenous DNA sites both in human and plant cells; thus, the success rate in this study was 50%. (Currently, the target sites of OPEN ZFNs are limited to those rich in GNN motifs and free of CNN and ANN motifs.) Importantly, OPEN ZFNs were selected among pools of ZFs with identical DNA-binding specificities, and, unlike modular assembly, this selection process is likely to consider context-dependent effects of neighboring ZFs on DNA binding. However, the OPEN approach is more time-consuming and labor-intensive than is the modular-assembly method, as the OPEN approach requires two rounds of pre-selection, in E. coli, of DNA-binding ZF arrays, which subsequently must be attached to the FokI nuclease domain to make ZFNs.
It is interesting to compare our results with those of Sangamo Biosciences, which reported a _CCR5_-targeting ZFN pair that consists of two four-finger ZFN monomers (Perez et al. 2008). Because we used different sets of ZF modules, our ZFNs targeted eight distinct sites in CCR5 that differ from the one site recognized by Sangamo's ZFN pair (Supplemental Fig. 2). These ZFNs could provide diverse options to researchers interested in ZFN-mediated gene therapy applications. It has been suggested that naturally occurring CCR5Δ32 is not only a loss-of-function mutation, but also a gain-of-function mutation (Chelli and Alizon 2001; Agrawal et al. 2004), and the mutant form may have _trans_-inhibitory effects on the expression of both CCR5 and CXCR4, the two host co-receptors used by HIV during the infection process. Whereas Sangamo's ZFN targets upstream of the Δ32 mutation, two of our ZFNs (Z836 and Z891) target downstream from this mutation, and mutant forms of the CCR5 protein generated using these ZFNs may retain the _trans_-inhibitory effects and make cells more resistant to HIV infection. Interestingly, one of our ZFN pairs cleaves at position +893 in the CCR5 gene, and this ZFN pair could generate CCR5 variants that mimic a naturally occurring mutant form of the CCR5 protein that is observed exclusively in Asians and results from a single base deletion at +893 (Shioda et al. 2001).
The sequence specificity of ZFNs is of particular importance in clinical applications. Sangamo's _CCR5_-targeting ZFN pair showed somewhat efficient genome editing at a homologous site in the CCR2 gene, as well as at the intended site in the CCR5 gene (Perez et al. 2008). Disruption of CCR2 gene function may not itself be deleterious. But it is always desirable to reduce nonspecific effects as much as possible to minimize the potential unwanted side effects of gene therapy. Off-target effects of our ZFNs must also be carefully characterized before we attempt to use these reagents in future clinical applications. We do note, however, that some of our ZFNs did not show any significant off-target effects at related sites in the CCR2 locus. These ZFNs should be more suitable for the targeted insertion of therapeutic genes in gene therapy applications than are those with off-target effects at the CCR2 locus.
In this paper, we report successful genomic modifications at several endogenous sites in human cells using ZFNs constructed by modular assembly. To our knowledge, this report is the first demonstration that modularly assembled ZFNs using publicly available sources can target endogenous sites in mammalian cells. Our method is readily accessible to any researchers interested in using ZFN technology. All the amino acid sequences of ZF modules used in this study are reported here, and most of the plasmids that encode these modules are available from Addgene (http://www.addgene.org/zfc). We believe that our modular-assembly approach provides a highly efficient, rapid, and easy-to-practice platform for the broad application of ZFN technology in research, medicine, and biotechnology.
Methods
Plasmid construction
Zinc finger nuclease construction
The FokI endonuclease domain derived from Flavobacterium okeanokoites genomic DNA was cloned into p3, which is a modified version of the pcDNA3.0 plasmid (Invitrogen). DNA segments that encoded three- or four-finger arrays were assembled as described previously (Bae et al. 2003). The list of ZFNs and their target sequences are shown in Supplemental Table 5. The amino acid sequence of a ZFN pair is shown in Supplemental Figure 4.
Reporter plasmid used in cell-based assays
The 3′ portion of the firefly luciferase gene was amplified from pGL3-control (Promega) using the primers 1 and 2 (Supplemental Table 6), and the amplified product was cloned into the BamHI and XhoI sites of pcDNA5/FRT/TO (Invitrogen). The 5′ portion of the luciferase gene amplified using the primers 3 and 4 was sequentially cloned into the HindIII and BamHI sites of the resulting plasmid. The I-SceI binding site was cloned into the BamHI site of the reporter plasmid using primers 5 and 6. The CCR5 coding sequence amplified using primers 7 and 8 was cloned into the BamHI site of the reporter plasmid.
In vitro digestion assay
The designed ZFNs were transcribed and translated in vitro using the TnT-Quick coupled transcription/translation system (Promega) as described by the manufacturer. Briefly, plasmids (0.5 μg each) that encoded ZFNs were added to TnT-Quick master mix (20 μL) and 1 mM methionine (0.5 μL), and incubated for 90 min at 30°C. The target plasmid containing the CCR5 coding sequence was first digested with the restriction enzyme and made linear. The target plasmid (1 μg) was then digested by incubation with a pair of ZFN IVTT lysates (1 μL each) for 2 h at 37°C in NEBuffer 4 (New England Biolabs). The reaction mixtures were inactivated by heat (65°C) and then centrifuged at 13,000 rpm. The supernatant was used for agarose gel analysis.
Cell culture and establishment of reporter cell line
HEK293T/17 (ATCC, CRL-11268TM) cells and Flp-In T-REx 293 cells (Invitrogen) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 100 units/mL penicillin, 100 μg/mL streptomycin, 0.1 mM nonessential amino acids, and 10% fetal bovine serum (FBS). The reporter plasmid encoding the disrupted luciferase gene was stably integrated into Flp-In T-REx 293 cells according to the manufacturer's instructions. Briefly, the cells were co-transfected with the reporter plasmid and pOG44, the Flp recombinase expression vector, and selected using hygromycin B. A clonal cell line bearing the disrupted luciferase gene was identified and used for the SSA assay.
Cell-based assay using the SSA system
Each pair of ZFN expression plasmids (100 ng each) was transfected into 30,000 reporter cells/well in a 96-well plate format using Lipofectamine 2000 (Invitrogen). After 48 h, the luciferase gene was induced by incubation with doxycycline (1 μg/mL). After 24 h of incubation, the cells were lysed in 20 μL of 1× lysis buffer (Promega), and luciferase activity was determined using 10 μL of luciferase assay reagent (Promega) plus 2 μL of cell lysate.
T7E1 assay
HEK293T/17 cells (3 × 104) pre-cultured in a 96-well plate were transfected with two plasmids encoding a ZFN pair (100 ng each) using Lipofectamine 2000 (Invitrogen). After 72 h of incubation, the genomic DNA was extracted from ZFN-transfected cells using the G-spin Genomic DNA Extraction Kit (iNtRON Biotechnology) as described by the manufacturer. The genomic region encompassing the ZFN target site was amplified, melted, and annealed to form heteroduplex DNA. The primers 9 and 12 were used for the Z30 site; 10 and 12 for the Z266 and Z360 sites; 11 and 12 for the Z410, Z426, and Z430 sites; and 13 and 14 for the Z836 and Z891 sites (Supplemental Table 6). The annealed DNA was treated with 5 units of T7 endonuclease 1 (New England BioLabs) for 15 min at 37°C and then precipitated by addition of 2.5 volumes of ethanol. The precipitated DNA was analyzed by agarose gel electrophoresis.
DSB foci staining
Intranuclear stain for TP53BP1 was performed with HEK293T/17 cells. Slides were prepared by attaching the cells using a Lab-Tek chamber slide with a cover (NUNC) and fixing the cells with 3.7% formaldehyde. The cells were then permeabilized by treatment with 0.2% Triton X-100 in phosphate-buffered saline (PBS) at room temperature (RT) for 10 min. Cells were then incubated with anti-TP53BP1 rabbit polyclonal antibodies (Bethyl Laboratories) in the presence of 5% bovine serum albumin (BSA) to block nonspecific staining, followed by incubation with Alexa Fluor 488-conjugated secondary antibodies (Invitrogen-Molecular Probes). Slides were mounted in the presence of DAPI (Sigma) to counterstain cell nuclei and examined under an immunofluorescence microscope (Carl Zeiss-LSM510).
Note added in proof
Since the submission of this paper, Foley et al. (2009) reported genome editing in zebrafish using OPEN ZFNs.
Footnotes
References
- Agrawal L, Lu X, Qingwen J, VanHorn-Ali Z, Nicolescu IV, McDermott DH, Murphy PM, Alkhatib G. Role for CCR5Δ32 protein in resistance to R5, R5X4, and X4 human immunodeficiency virus type 1 in primary CD4+ cells. J Virol. 2004;78:2277–2287. doi: 10.1128/JVI.78.5.2277-2287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae KH, Kim JS. One-step selection of artificial transcription factors using an in vivo screening system. Mol Cells. 2006;21:376–380. [PubMed] [Google Scholar]
- Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, et al. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat Biotechnol. 2003;21:275–280. doi: 10.1038/nbt796. [DOI] [PubMed] [Google Scholar]
- Beerli RR, Segal DJ, Dreier B, Barbas CF., III Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics. 2006;172:2391–2403. doi: 10.1534/genetics.105.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer KJ, Trautman JK, Bozas A, Liu JL, Rutter J, Gall JG, Carroll D. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc Natl Acad Sci. 2008;105:19821–19826. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- Bylund L, Kytola S, Lui WO, Larsson C, Weber G. Analysis of the cytogenetic stability of the human embryonal kidney cell line 293 by cytogenetic and STR profiling approaches. Cytogenet Genome Res. 2004;106:28–32. doi: 10.1159/000078556. [DOI] [PubMed] [Google Scholar]
- Camenisch TD, Brilliant MH, Segal DJ. Critical parameters for genome editing using zinc finger nucleases. Mini Rev Med Chem. 2008;8:669–676. doi: 10.2174/138955708784567458. [DOI] [PubMed] [Google Scholar]
- Cathomen T, Joung JK. Zinc-finger nucleases: The next generation emerges. Mol Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- Chames P, Epinat JC, Guillier S, Patin A, Lacroix E, Paques F. In vivo selection of engineered homing endonucleases using double-strand break induced homologous recombination. Nucleic Acids Res. 2005;33:e178. doi: 10.1093/nar/gni175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelli M, Alizon M. Determinants of the trans-dominant negative effect of truncated forms of the CCR5 chemokine receptor. J Biol Chem. 2001;276:46975–46982. doi: 10.1074/jbc.M106432200. [DOI] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier B, Segal DJ, Barbas CF., III Insights into the molecular recognition of the 5′-GNN-3′ family of DNA sequences by zinc finger domains. J Mol Biol. 2000;303:489–502. doi: 10.1006/jmbi.2000.4133. [DOI] [PubMed] [Google Scholar]
- Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF., III Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- Dreier B, Fuller RP, Segal DJ, Lund CV, Blancafort P, Huber A, Koksch B, Barbas CF., III Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- Foley JE, Yeh JR, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by oligomerized pool engineering (OPEN) Plos One. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greisman HA, Pabo CO. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- Hurt JA, Thibodeau SA, Hirsh AS, Pabo CO, Joung JK. Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection. Proc Natl Acad Sci. 2003;100:12271–12276. doi: 10.1073/pnas.2135381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Ramm EI, Pabo CO. A bacterial two-hybrid selection system for studying protein–DNA and protein–protein interactions. Proc Natl Acad Sci. 2000;97:7382–7387. doi: 10.1073/pnas.110149297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HS, Shin HC, Kim JS. Suppression of vascular endothelial growth factor expression at the transcriptional and post-transcriptional levels. Nucleic Acids Res. 2005;33:e74. doi: 10.1093/nar/gni068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon RJ, Kim SK, Lee SI, Hwang SJ, Lee GM, Kim JS, Seol W. Artificial transcription factors increase production of recombinant antibodies in Chinese hamster ovary cells. Biotechnol Lett. 2006;28:9–15. doi: 10.1007/s10529-005-4680-7. [DOI] [PubMed] [Google Scholar]
- Lee DK, Park JW, Kim YJ, Kim J, Lee Y, Kim JS. Toward a functional annotation of the human genome using artificial transcription factors. Genome Res. 2003;13:2708–2716. doi: 10.1101/gr.1397903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DK, Kim YH, Kim JS, Seol W. Induction and characterization of taxol-resistance phenotypes with a transiently expressed artificial transcriptional activator library. Nucleic Acids Res. 2004;32:e116. doi: 10.1093/nar/gnh114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, MacDonald ME, Stuhlmann H, Koup RA, Landau NR. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- Liu PQ, Rebar EJ, Zhang L, Liu Q, Jamieson AC, Liang Y, Qi H, Li PX, Chen B, Mendel MC, et al. Regulation of an endogenous locus using a panel of designed zinc finger proteins targeted to accessible chromatin regions. Activation of vascular endothelial growth factor A. J Biol Chem. 2001;276:11323–11334. doi: 10.1074/jbc.M011172200. [DOI] [PubMed] [Google Scholar]
- Liu Q, Xia Z, Zhong X, Case CC. Validated zinc finger protein designs for all 16 GNN DNA triplet targets. J Biol Chem. 2002;277:3850–3856. doi: 10.1074/jbc.M110669200. [DOI] [PubMed] [Google Scholar]
- Lloyd A, Plaisier CL, Carroll D, Drews GN. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc Natl Acad Sci. 2005;102:2232–2237. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Morton J, Davis MW, Jorgensen EM, Carroll D. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc Natl Acad Sci. 2006;103:16370–16375. doi: 10.1073/pnas.0605633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Lee DK, Lee H, Lee Y, Jang YS, Kim YH, Yang HY, Lee SI, Seol W, Kim JS. Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nat Biotechnol. 2003;21:1208–1214. doi: 10.1038/nbt868. [DOI] [PubMed] [Google Scholar]
- Park KS, Jang YS, Lee H, Kim JS. Phenotypic alteration and target gene identification using combinatorial libraries of zinc finger proteins in prokaryotic cells. J Bacteriol. 2005a;187:5496–5499. doi: 10.1128/JB.187.15.5496-5499.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Seol W, Yang HY, Lee SI, Kim SK, Kwon RJ, Kim EJ, Roh YH, Seong BL, Kim JS. Identification and use of zinc finger transcription factors that increase production of recombinant proteins in yeast and mammalian cells. Biotechnol Prog. 2005b;21:664–670. doi: 10.1021/bp049658x. [DOI] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebar EJ, Pabo CO. Zinc finger phage: Affinity selection of fingers with new DNA-binding specificities. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CT. The zinc finger nuclease monopoly. Nat Biotechnol. 2005;23:915–918. doi: 10.1038/nbt0805-915. [DOI] [PubMed] [Google Scholar]
- Shioda T, Nakayama EE, Tanaka Y, Xin X, Liu H, Kawana-Tachikawa A, Kato A, Sakai Y, Nagai Y, Iwamoto A. Naturally occurring deletional mutation in the C-terminal cytoplasmic tail of CCR5 affects surface trafficking of CCR5. J Virol. 2001;75:3462–3468. doi: 10.1128/JVI.75.7.3462-3468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Wilson JH. Knockout punches with a fistful of zinc fingers. Proc Natl Acad Sci. 2008;105:5653–5654. doi: 10.1073/pnas.0802298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DA, Townsend JA, Winfrey RJ, Jr, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF. High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 2005;44:693–705. doi: 10.1111/j.1365-313X.2005.02551.x. [DOI] [PubMed] [Google Scholar]
- Wright DA, Thibodeau-Beganny S, Sander JD, Winfrey RJ, Hirsh AS, Eichtinger M, Fu F, Porteus MH, Dobbs D, Voytas DF, et al. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat Protocols. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- Yun CO, Shin HC, Kim TD, Yoon WH, Kang YA, Kwon HS, Kim SK, Kim JS. Transduction of artificial transcriptional regulatory proteins into human cells. Nucleic Acids Res. 2008;36:e103. doi: 10.1093/nar/gkn398. [DOI] [PMC free article] [PubMed] [Google Scholar]