MicroRNA-155 Suppresses Activation-Induced Cytidine Deaminase-Mediated Myc-Igh Translocation (original) (raw)
. Author manuscript; available in PMC: 2009 Jul 21.
SUMMARY
MicroRNAs (miRNAs) are small noncoding RNAs that regulate vast networks of genes that share miRNA target sequences. To examine the physiologic effects of an individual miRNA-mRNA interaction in vivo, we generated mice that carry a mutation in the putative microRNA-155 (miR-155) binding site in the 3′-untranslated region of activation-induced cytidine deaminase (AID), designated Aicda155 mice. AID is required for immunoglobulin gene diversification in B lymphocytes, but it also promotes chromosomal translocations. Aicda155 caused an increase in steady-state Aicda mRNA and protein amounts by increasing the half-life of the mRNA, resulting in a high degree of Myc-Igh translocations. A similar but more pronounced translocation phenotype was also found in miR-155-deficient mice. Our experiments indicate that miR-155 can act as a tumor suppressor by reducing potentially oncogenic translocations generated by AID.
INTRODUCTION
Antibody genes are assembled in developing B cells by RAG1-RAG2 recombinase-mediated site-specific recombination of immunoglobulin (Ig) variable (V), diversity (D), and joining (J) gene segments. Although V(D)J recombination produces a diverse repertoire of IgM antibodies, high-affinity IgG antibody responses require further Ig diversification by somatic hypermutation (SHM) and class-switch recombination (CSR) in antigen-activated B cells (Klein and Dalla-Favera, 2008; Meffre et al., 2000; Rajewsky, 1996). These reactions are both initiated by activation-induced cytidine deaminase (AID), which deaminates cytosine residues and introduces U:G mismatches in DNA (Di Noia and Neuberger, 2007; Muramatsu et al., 2007). AID activity is primarily restricted to Ig genes but can also produce off-target lesions in non-Ig sites such as oncogenes (Liu et al., 2008), and the double-strand breaks it creates can be substrates for translocation (Dorsett et al., 2007; Ramiro et al., 2004). Therefore, the regulation of this enzyme is essential to maintain genomic integrity.
MicroRNAs (miRNAs) are a class of ~20–23 nt noncoding RNAs produced from genes encoding RNAs with a hairpin secondary structure (Bartel, 2004; Filipowicz et al., 2008; Meister and Tuschl, 2004). The hairpin RNAs are processed to produce mature single-stranded small RNAs (the microRNA) that are then incorporated into protein complexes termed, miRNPs, that contain one of several different Argonaute proteins (Filipowicz et al., 2008; Meister and Tuschl, 2004). miRNA binding to complementary sequences, usually located in the 3′-untranslated regions (3′ UTRs) of messenger RNAs (mRNAs), can regulate expression of large groups of genes by a variety of mechanisms including cleavage, degradation, cell-cycle control, translational inhibition, and mRNA transport (Bartel, 2004; Filipowicz et al., 2008; Leung and Sharp, 2007; Meister and Tuschl, 2004).
miR-155 is an oncogenic miRNA product of the MIRN155 gene in humans or Bic in mice and is deregulated in a number of different cancers, most of which are of B cell origin (Costinean et al., 2006; Eis et al., 2005; Fulci et al., 2007; Gironella et al., 2007; Iorio et al., 2005; Jay et al., 2007; Kluiver et al., 2005, 2006, 2007; Lagos-Quintana et al., 2002; Landgraf et al., 2007; Lawrie et al., 2007; Lee et al., 2007; Marton et al., 2008; Nikiforova et al., 2008; Roldo et al., 2006; Tam, 2001; Volinia et al., 2006). Overexpression of miR-155 in transgenic mice leads to preleukemic pre-B cell proliferation in bone marrow and spleen, followed by high-grade B cell neoplasms (Costinean et al., 2006). Conversely, the mature form of miR-155 is absent in primary cases of Burkitt’s lymphoma (Kluiver et al., 2006, 2007; Landgraf et al., 2007). Although the precise role of miR-155 in promoting lymphomagenesis has not been determined, this miRNA is an important immune modulator whose expression is induced along with AID in activated leukocytes and germinal center B cells (Haasch et al., 2002; Taganov et al., 2006; Tam, 2001; Thai et al., 2007; van den Berg et al., 2003). miR-155 deficiency in mice leads to the deregulated expression of hundreds of mRNAs, some of which are direct targets of miR-155, resulting in abnormalities in the germinal center reaction and antibody responses in vivo (Rodriguez et al., 2007; Thai et al., 2007; Vigorito et al., 2007).
Aicda encodes AID and contains a single miR-155 binding site in its 3′ UTR (John et al., 2004; Lewis et al., 2005). It is among the genes deregulated in miR-155-deficient B cells, where its expression is increased 1.6-fold as measured by gene array (Vigorito et al., 2007). However, this small change in mRNA expression was not confirmed by a direct measurement, and whether this is a direct or indirect effect of miR-155 deletion has not been determined. With a few exceptions, miRNA control in living organisms has been studied by overexpression and deletion experiments (Choi et al., 2007; Mayr et al., 2007). These approaches result in simultaneous effects on many genes that share miRNA target sequences, complicating interpretation because indirect regulators are themselves miRNA targets.
To determine whether miR-155 regulates AID expression directly, and to investigate the potential significance of that regulation in Burkitt’s lymphoma-associated Myc-Igh translocation, we produced a mouse strain that carries a mutation in the putative miR-155 target site in the 3′ UTR of the gene encoding AID (Aicda155 mice) and compared them to miR-155-deficient mice.
RESULTS
Aicda155 Mice
Like Aicda, miR-155 appears to have emerged in evolution in bony fish and the 3′ UTR of Aicda contains a candidate miR-155 binding site that is conserved between fish and humans, suggesting that the two may have coevolved (Figures S1A–S1C available online) (Barreto et al., 2005; Conticello et al., 2005, 2007; Lewis et al., 2003, 2005; Wakae et al., 2006). To examine the effects of miR-155 on AID expression directly, we replaced the conserved miR-155 target sequence (seed-match nucleotides 1–8) in the 3′ UTR of Aicda with a G- and C-rich nucleotide sequence that does not match the seed sequence of any known miRNAs (Figures S1A and S2). The mutation was confirmed by sequencing of the Aicda mRNA from the mutant mice (not shown). Aicda155/+ mice were bred to C57Bl/6 _Aicda_−/− mice to produce Aicda155/− mice that were born at normal frequencies (not shown). When compared to wild-type or Aicda+/− controls by flow cytometry, Aicda155/− showed normal B cell development in the bone marrow and normal numbers of peripheral B cells in spleen (Figure S3A). In addition, the amount of serum-immunoglobulin isotypes was normal as measured by enzyme-linked immunoassays (Figure S3B).
Aicda155 and miR-155-Deficient Mice Express Higher Amounts of AID Protein
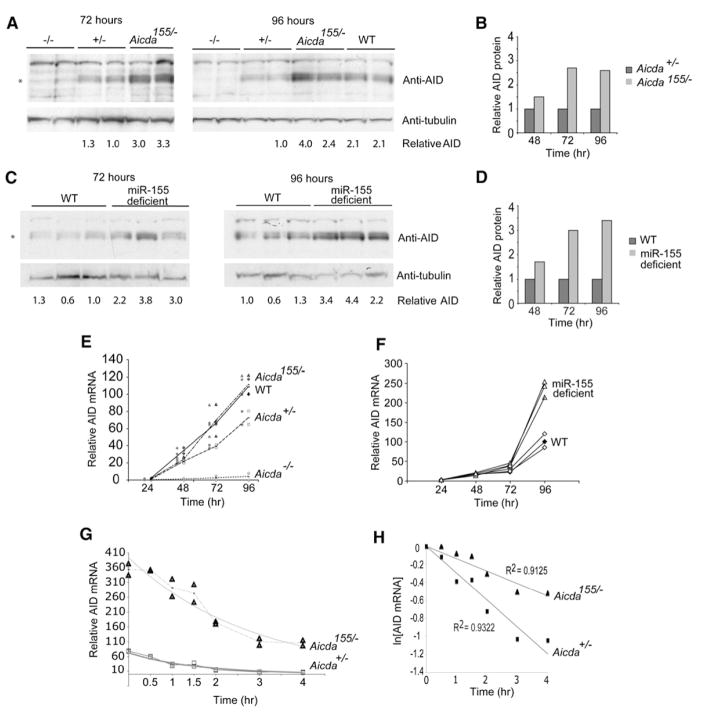
To determine whether the Aicda155 mutation altered AID protein expression, we stimulated B cells with lipopolysacharide (LPS) and interleukin 4 (IL-4) in vitro to induce AID and miR-155, and we measured AID protein expression over 4 days (Figures 1A and 1B). In control B cells, AID protein expression was initially detected 2 days after stimulation and increased on days 3 and 4 in culture (Figures 1A and 1B, and data not shown). Aicda155/− showed a similar expression pattern, but in all cases the amounts were 2- to 3-fold higher than in Aicda+/− controls, as determined by immunoblot (Figures 1A and 1B). Similar effects were also found in miR-155-deficient B cells (Figures 1C and 1D). We conclude that miR-155 regulates the amount of AID protein in stimulated B cells.
Figure 1. Aicda155 Mice.
(A) Immunoblots for AID-protein and tubulin loading controls on extracts of B cells after stimulation with LPS and IL-4 for the indicated times in hours for wild-type (+/+), Aicda155/−, Aicda+/−, and _Aicda_−/− mice. The blots show samples prepared from two separate mice for each genotype. Numbers indicate relative amounts of AID determined by AID/tubulin ratio within each blot measured by densitometry. * indicates the position of AID protein.
(B) Combined data for immunoblot analysis for AID protein from five separate mice. Numbers indicate average relative AID protein expression of Aicda155/− versus Aicda+/− for each time point.
(C) As in (A) but for matched wild-type (WT) and miR-155-deficient B cells.
(D) As in (B) but for miR-155-deficient versus WT B cells (n = 3 independent mice).
(E) Quantitative PCR analysis for Aicda mRNA from wild-type (WT), Aicda155/−, Aicda+/−, and _Aicda_−/− B cells after stimulation with LPS and IL-4 for 1, 2, 3, or 4 days. Lines show means from two separate mice, each indicated as a separate symbol (WT = diamond, Aicda155/− = triangle, Aicda+/− = square, and _Aicda_−/− = circle).
(F) Quantitative PCR analysis for Aicda mRNA from matched wild-type (WT) and miR-155-deficient mice after stimulation with LPS and IL-4 for 1, 2, 3, or 4 days. Data from three separate mice are shown. WT = diamonds, miR-155−/− = triangle.
(G) Quantitative PCR analysis, as in (E), for Aicda mRNA from Aicda155/− or Aicda+/− B cells stimulated with LPS and IL-4 for 3 days, after treatment with Actinomycin-D for the indicated time. Graph represents triplicate Aicda q-PCR for two mice normalized to GAPDH. Aicda155/− = triangles, Aicda+/− = squares.
(H) Linear regression analysis of the data in (G) shown as ln[RNA] versus time of Actinomycin-D treatment. Aicda155/− = triangles, Aicda+/− = squares.
miR-155 Destabilizes Aicda mRNA
Consistent with elevated amounts of AID protein, the corresponding mRNA was elevated in Aicda155/− when compared to Aicda+/− controls beginning 2 days after stimulation with LPS and IL-4 (Figure 1E). Similar results were found with miR-155-deficient B cells, which show a 2.5-fold increase in Aicda mRNA after 4 days (Figure 1F). Increased Aicda mRNA expression suggests that miR-155 regulates the expression of this gene by altering messenger stability. To determine whether miR-155 regulates Aicda mRNA stability, we stimulated B cells with LPS and IL-4, blocked transcription with Actinomycin-D, and measured the decay of Aicda mRNA (Figures 1G and 1H). We found that the half-life of Aicda transcripts was increased from 1.05 hr in control to 1.94 hr in the mutant as determined by linear regression analysis of two mice assayed in triplicate, indicating that miR-155 regulates the level of Aicda mRNA by increasing its turnover (Figures 1G and 1H).
Class Switching in Aicda155 and miR-155-Deficient Mice
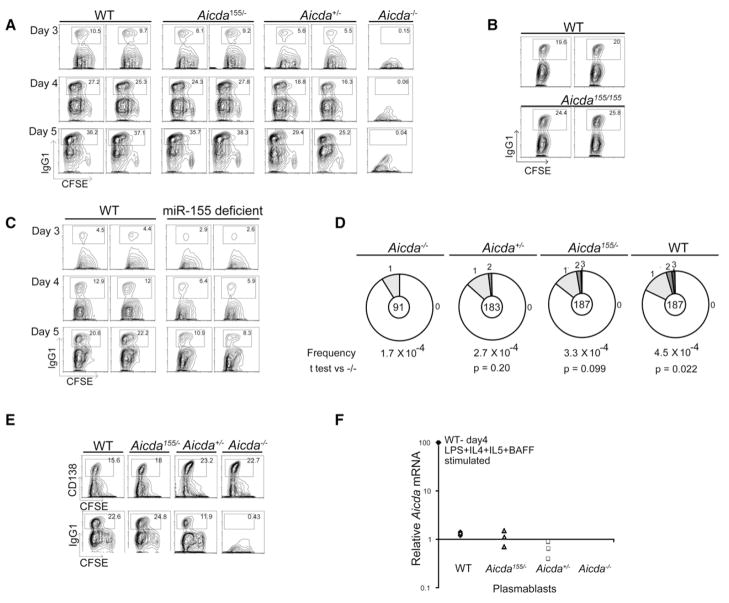
To examine the effects of Aicda155 on class-switch recombination, we labeled B cells with 5-(6)-carboxyfluorescein diacetate succinimidyl diester (CFSE), a reporter dye for cell division, and stimulated them with LPS and IL-4. Cell-surface IgG1 expression was monitored by flow cytometry over a time course of 4 days in culture. Although cell division was normal, class switching was enhanced in Aicda155/− when compared to Aicda+/− B cells and was similar to Aicda+/+ controls (Figure 2A). A similar increase in switching was seen in Aicda155/155 when compared to Aicda+/+ (Figure 2B). This effect was most pronounced early in the culture period, when the number of IgG1-expressing Aicda+/+ and Aicda155/− cells was nearly double that of Aicda+/− controls (Figure 2A). In contrast, miR-155-deficient B cells showed a subnormal amount of class switching despite increased AID expression (Figures 1C, 1D, and 2C) (Rodriguez et al., 2007; Thai et al., 2007; Vigorito et al., 2007)). We conclude that a 2- to 3-fold increase in AID protein expression leads to increased class switching in vitro in Aicda155/− but not miR-155-deficient B cells.
Figure 2. Class-Switch Recombination by Aicda155 and miR-155-Deficient B Cells.
(A) Contour plots show IgG1 expression and CFSE dye dilution by B cells from pairs of wild-type (WT), Aicda155/−, Aicda+/−, and _Aicda_−/− mice after stimulation with LPS and IL-4 for 3, 4, or 5 days. Numbers indicate percentage of cells in the gate.
(B) Contour plots show IgG1 expression by B cells from matched pairs of wild-type (WT) and Aicda155/155 mice after stimulation with LPS and IL-4 assayed after 4 days. Numbers indicate percentage of cells in the gate.
(C) As in (A) but for pairs of matched wild-type (WT) and miR-155-deficient mice after stimulation with LPS and IL-4 for 3, 4, or 5 days.
(D) Mutations in the 5′ of the Sμ region in wild-type (WT), Aicda155/−, Aicda+/−, and _Aicda_−/− B cells stimulated to undergo CSR in vitro with LPS and IL-4 and sorted for five cell divisions and IgM expression. Segment sizes in the pie charts are proportional to the number of sequences carrying the number of mutations indicated in the periphery of the charts. The frequency of mutations per base pair sequenced and the total number of independent sequences analyzed are indicated underneath and in the center of each chart, respectively. The total number of mutations per number of bases analyzed was as follows: WT, 44/97,240 bp; Aicda155/−, 33/97,240 bp; Aicda+/−, 27/95160 bp; and _Aicda_− −/−, 8/47320 bp.
(E) Contour plots show CD138 expression and CFSE dye dilution by B cells from pairs of wild-type (WT), Aicda155/−, Aicda+/−, and _Aicda_−/− mice after stimulation with LPS and IL-4, IL-5, and BAFF for 5 days. Numbers indicate percentage of cells in the gate.
(F) Q-PCR analysis for Aicda mRNA from purified CD138 expressing plasmablasts from cultures of wild-type (WT), Aicda155/−, Aicda+/−, and _Aicda_−/− B cells after stimulation as in (E). Data are relative to B cells stimulated with LPS and IL-4 for 4 days (100%) and represent three separate mice, each indicated as a separate symbol.
Somatic Hypermutation in Aicda155 Mice
Class switching is associated with AID-induced somatic mutations in the 5′ of the switch μ (Sμ) region (Petersen et al., 2001). To determine whether Aicda155 also alters the production of AID-mediated lesions in the Igh switch regions, we measured the mutations that occur 5′ of the switch μ region in LPS- and IL-4-stimulated B cells that had undergone five cell divisions (Petersen et al., 2001). The small increase in mutations in Aicda155/− B cells 5′ of the switch μ region was not statistically significant but corresponded to the increase in class switching at the same time point (Figure 2D).
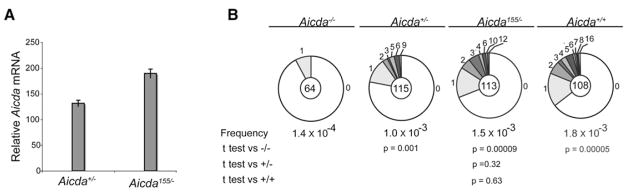
AID expression is also necessary to induce somatic hypermutation of immunoglobulin genes (Muramatsu et al., 2000; Revy et al., 2000; Yoshikawa et al., 2002). To examine the effects of Aicda155 on somatic mutation in vivo, we immunized mice and purified germinal-center B cells, which actively mutate their Ig genes. Like LPS- and IL-4-activated B cells, Aicda155/− germinal-center B cells contained higher amounts of Aicda mRNA than controls (Figure 3A). Similar to 5′ of the switch μ region, we found a statistically insignificant effect on somatic hypermutation of the noncoding DNA region 3′ of IgJH4, which cannot be selected for or against during the germinal-center reaction (Jolly et al., 1997) (Figures 2D and 3B). Although Bcl6 is also mutated during the germinal-center reaction (Liu et al., 2008), we found no increase in mutation at this locus in Aicda155/− germinal-center B cells (Figure S4). In conclusion, neither the miR-155 deficiency (Thai et al., 2007; Vigorito et al., 2007) nor Aicda155 mutation significantly increases somatic hypermutation despite elevated AID expression, and therefore this process is probably regulated by additional mechanisms.
Figure 3. Aicda mRNA Expression and Somatic Hypermutation in Germinal-Center B Cells from Aicda155 Mice.
(A) Quantitative PCR analysis for Aicda mRNA pooled from germinal-center B cells purified from three immunized Aicda155/− and Aicda+/− mice. The relative value obtained with wild-type B cells stimulated with LPS and IL-4 for 4 days is set to 100. Error bars represent standard deviation.
(B) Mutation analysis of the JH4 intron (Jolly et al., 1997) cloned from purified germinal-center B cells (CD19+, Fas+, GL-7+) obtained from the lymph nodes of immunized wild-type (WT), Aicda155/−, Aicda+/−, and _Aicda_−/− mice. Pie charts and statistics are as in Figure 2D. The total number of mutations per number of bases analyzed was as follows: WT, 107/58,644 bp; Aicda155/−, 89/61,359 bp; Aicda+/−, 65/62,445 bp; and _Aicda_−/−, 5/34,752 bp.
Aicda155 mRNA and Protein Do Not Persist in Plasmablasts
To determine whether Aicda155 would result in persistence of Aicda mRNA in plasmablasts, where it is not normally transcribed, we cultured B cells under conditions where they undergo class switching and develop into CD138 plasmablasts (Horikawa and Takatsu, 2006) (Figure 2E). As in LPS and IL-4 cultures, Aicda155 enhanced switching to IgG1 but did not alter plasmablast development (Figure 2E). Furthermore, the amount of Aicda mRNA expressed in plasmablasts was two orders of magnitude less than in B cells stimulated with LPS and IL-4 (Figure 2F). Thus, Aicda155 increases AID expression in developing B cells, yet it does not extend AID expression into the plasmablast stage.
Myc-Igh Translocations in Aicda155 and miR-155-Deficient B Cells
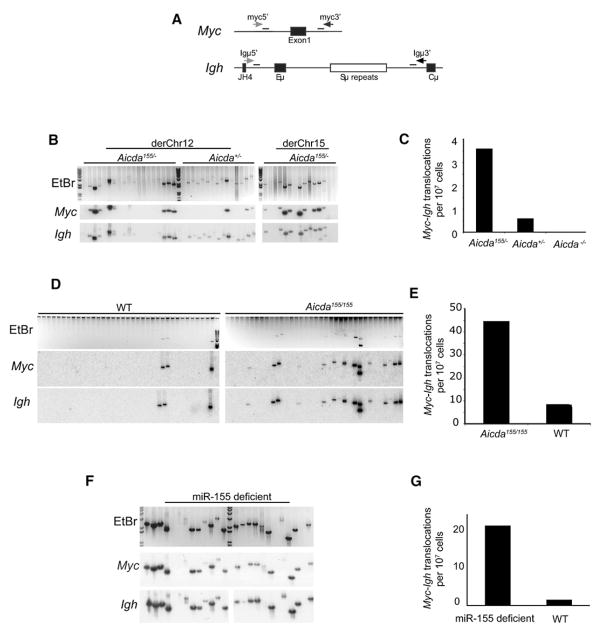
In addition to class-switch recombination and somatic mutation, AID induces potentially oncogenic reciprocal chromosome translocation between Igh and Myc (Myc-Igh) (Dorsett et al., 2007; Ramiro et al., 2004, 2006). To examine the effect of Aicda155 on these translocations, we assayed stimulated B cells for aberrant juxtaposition of the chromosomes that carry Myc and Igh (Potter, 2003; Ramiro et al., 2004) (Figure 4A). In contrast to the modest effects on somatic mutation, Aicda155/− B cells showed a 3- to 6-fold increase in translocation frequency compared to Aicda+/− controls (Figures 4B and 4C, and data not shown). A similar increase in translocation was seen in Aicda155/155 compared to wild-type controls (Figure 4D). Therefore, the mechanisms that restrict somatic mutation in cells expressing elevated amounts of AID do not limit translocation.
Figure 4. Igh to Myc Translocations in Stimulated B Cells from Aicda155 Mice.
(A) Schematic representation of PCR assay for Myc-Igh translocations. Primers used to detect derivative chromosome 12 (derChr12) and derivative chromosome 15 (derChr15) translocations are represented as horizontal black and gray arrows, respectively. Internal oligonucleotide probes used in Southern-blot experiments are shown as horizontal black bars.
(B) Myc-Igh translocations. B cells from Aicda155/− and Aicda+/− (_Aicda_−/− mice not shown) were stimulated with LPS and IL4 for 4 days. Ethidium-bromide agarose gels (EtBr, upper gels) and Southern blots of candidate translocations with Igh or Myc oligonucleotide probes collected from representative samples (three independent spleens 6 × 107 cells per genotype) are shown.
(C) Total number of translocations obtained from Aicda155/− (p = 0.000139 versus Aicda+/−), Aicda+/−, and _Aicda_−/− B cell. p value was determined with a two-tailed Fisher’s exact test.
(D) Myc-Igh translocations. B cells from Aicda155/155 and Aicda+/+ were stimulated with LPS and IL4 for 4 days. Amplification products were separated on agarose gels and stained with ethidium bromide (EtBr, upper gels). Southern blots of the EtBr stained gels were probed for Igh and Myc (three independent spleens, 100,000 cell per lane, 3.6 × 106 cells per genotype).
(E) Total number of translocations obtained from Aicda155/155 (p = 0.00879 versus Aicda+/+) and Aicda+/+ B cell. p value was determined with a two-tailed Fisher’s exact test.
(F) _Myc_-Igh translocations assayed as in (B) except B cells were from miR-155-deficient and WT mice (five separate spleens assayed and representative samples shown, 2 × 107 cells assayed per genotype).
(G) As in (C) except shows total number of translocations obtained from miR-155-deficient and WT mice. p = 0.0003 with a two-tailed Fisher’s exact test.
In addition to Aicda, miR-155 is predicted to target numerous different genes that play a role in the maintenance of genomic integrity (John et al., 2004; Lewis et al., 2005). To determine whether deregulation of miR-155 might further increase the frequency of AID-dependent Myc-Igh translocations, we assayed miR-155-deficient B cells. We found a ~15-fold enhanced translocation frequency in the absence of miR-155 compared to matched controls (Figures 4F and 4G). Thus, miR-155 expression in activated B cells suppresses Myc-Igh translocations.
DISCUSSION
AID initiates Ig gene diversification in activated B cells by deaminating cytosine to produce U:G mismatches in DNA (Di Noia and Neuberger, 2007; Muramatsu et al., 2007). These lesions are processed by ubiquitous DNA-repair pathways and error-prone polymerases to produce somatic mutations or double-strand DNA breaks, which are obligate intermediates in immunoglobulin class-switch recombination (Di Noia and Neuberger, 2007; Muramatsu et al., 2007). However, in addition to diversifying immune responses, AID can damage the genome by mutating oncogenes and creating substrates for chromosomal translocations (Dorsett et al., 2007; Klein and Dalla-Favera, 2008; Perez-Duran et al., 2007; Ramiro et al., 2004). Indeed, AID is essential for translocation between Myc and Igh, and the frequency of these rare off-target events is directly related to the level of AID expressed (Dorsett et al., 2007; Ramiro et al., 2006; Ramiro et al., 2004). For example, Myc-Igh translocations are rare events in B cells expressing physiological levels of AID (1 in 3 × 107 cells) and increase in frequency to (1 in 2.5 × 104) in B cells overexpressing AID (Ramiro et al., 2006). It is therefore essential that AID expression is tightly regulated in vivo.
Under physiologic circumstances, AID expression is restricted to activated B cells (Gonda et al., 2003; Sayegh et al., 2003), and its concentration in the nucleus is limited by active export (McBride et al., 2004). Furthermore, posttranslational modification by phosphorylation at position S38 is required for interaction between AID and replication protein A for optimal AID activity on transcribed DNA (Basu et al., 2005; Chatterji et al., 2007; McBride et al., 2006; Pasqualucci et al., 2006). The results presented here and in an accompanying paper in this issue of Immunity (Teng et al., 2008) define an additional mechanism for regulating AID expression in vivo. Aicda mRNA half-life is decreased by a single miR-155 binding site within the 3′ UTR. Our results also highlight phenotypic differences between the miRNA binding-site mutation in Aicda and the deletion of miR-155. Both mutations result in 2- to 3-fold-higher levels of _Aicd_a mRNA and protein in activated B cells. In contrast, Aicda155 is associated with increased class-switch recombination, whereas mice with a deficiency in miR-155 show an unexpected decrease in this reaction (Thai et al., 2007). Nevertheless, both mutations result in a substantial increase in Myc-Igh translocations. In the case of miR-155-deficient B cells, translocation frequency is similar to that found in ataxia-telangiectasia-mutated kinase (ATM)-deficient B cells, which also display reduced amounts of class-switch recombination (Ramiro et al., 2006; Reina-San-Martin et al., 2004). The disproportionate increase in Myc-Igh translocations underlines the importance of AID regulation and suggests that these events are not limited by the same factors that restrict the degree of SHM and CSR.
Overexpression of Myc in B cells in transgenic mice results in B cell lymphomas (Adams et al., 1985; Schmidt et al., 1988). However, transgenic overexpression of AID does not (Muto et al., 2006), and it is therefore not surprising that we have not found lymphomas in Aicda155 or miR-155-deficient mice after 3 months and 1 yr of observation, respectively.
miRNAs regulate expression of large groups of genes by a variety of mechanisms including cleavage, stability, cell-cycle control, translation inhibition, and mRNA transport (Bartel, 2004; Filipowicz et al., 2008; Leung and Sharp, 2007; Meister and Tuschl, 2004). It is therefore not surprising that they have been implicated in malignant transformation (Esquela-Kerscher and Slack, 2006; Hammond, 2007; Thomson et al., 2006). By mutating the 3′ UTR in Aicda, we have isolated the effects of miR-155 on a single mRNA and determined that it destabilizes the message, which in turn reduces the amount of AID protein and the frequency of Myc-Igh translocations. Throughout evolution, miR-155 and its binding site in the 3′ UTR of Aicda have been conserved. Therefore, we speculate that the emergence of miR-155 and AID in vertebrates and the conservation of their interaction through evolution are related to minimizing chromosome translocations.
miR-155 mutant mice showed higher levels of translocation than Aicda155, indicating that miR-155 may target additional mRNAs that cooperate to prevent Myc-Igh translocation. These translocations are the key transforming events in Burkitt’s lymphoma (Klein and Dalla-Favera, 2008) and are known to be deficient in miR-155 as a result of defects in processing of the gene encoding miR-155 (Kluiver et al., 2006; Kluiver et al., 2007; Landgraf et al., 2007). Thus, our findings suggest a molecular rationale for the development of human Burkitt’s lymphoma, in that absence of miR-155 predisposes activated B cells to Myc-Igh translocations by increasing expression of AID and yet-to-be-identified proteins that contribute to maintaining genomic integrity (Di Noia and Neuberger, 2007; Kluiver et al., 2007; Kuppers and Dalla-Favera, 2001).
EXPERIMENTAL PROCEDURES
Mice
Aicda nucleotides AGCATTAA, located in the Aicda 3′ UTR, 468 bp downstream of the stop codon, were replaced with GCGCGCGC by gene targeting (Figure S1A). The long arm of the targeting vector was 6.9 kb long and introduced a LoxP site within the intron between Aicda exons 4 and 5 (Figure S2). The short arm was a 1.5 kb fragment extending downstream of the 3′ UTR. A LoxP-flanked neomycin-resistance gene was used for positive selection, and a diphtheria toxin gene was used for negative selection (Yagi et al., 1990). The targeting construct was linearized and transfected into C57Bl/6 embryonic stem cells (ESCs). ESC clones were screened and seven positive clones were injected into C57Bl/6 blastocysts, and one produced chimeric mice that transmitted the mutation. The genotype was confirmed by amplifying the mutation with a primer external to the targeting construct and proximal to the end of the short arm. The resulting amplification product was verified by sequencing and digesting with AsceI, resulting in a digested wild-type allele amplification product and nondigested Aicda155 product. The identity of the resulting Aicda transcript was confirmed by reverse transcription of total RNA from activated B cells followed by amplification of the Aicda transcript from the 5′ UTR to the 3′ UTR at the end of the short arm and then by sequencing. To produce Aicda155/− mice, we crossed heterozygous Aicda155/+ mice to _Aicda_−/− C57Bl/6 mice and used littermate Aicda+/− as controls. miR-155-deficient mice were previously described (Thai et al., 2007). All mice were maintained under specific pathogen-free conditions, and experiments were performed under Rockefeller University IACUC approved protocols.
Lymphocyte Cultures and Translocation Assays
Resting B lymphocytes were isolated from the spleen with CD43 microbeads (Miltenyi Biotech), cultured in RPMI supplemented with L-glutamine, sodium pyruvate, 50 μM 2-Mercaptoethanol, and 10% FBS (GIBCO-BRL), and where indicated cells were labeled with CFDA-SE (5 μM, Molecular Probes). The B cells were stimulated with LPS (25 μg/ml) and IL-4 (5 ng/ml, Sigma) alone or together with IL-5 (15 ng/ml, PharMingen) and BAFF (10 ng/ml, R&D systems) for production of plasmablasts (Horikawa and Takatsu, 2006). For the plasma cell experiment, CD138+ B cells were sorted, and FACS was done on day 5. Transloaction assays were exactly as previously described (Dorsett et al., 2007; Ramiro et al., 2006). In brief, PCR of wild-type, _Aicda_−/−, miR-155 deficient, Aicda155/−, or Aicda155/155 was performed with total DNA prepared from day 3 or 4 LPS + IL4 cultures. Data shown for Aicda155/− and Aicda155/155 were from day 4 cultures without dead cell removal prior to DNA preparation. Data for miR-155-deficient mice were from day 3 cultures with dead cell removal prior to DNA preparation. Approximate cell number for each sample was determined by DNA quantitation on an ethidium-bromide-stained agarose gel (500 ng DNA = ~100,000 cells). Subsequent amplification of Myc to Igh switch-region translocations was done as previously described. Amplification products were verified with Southern blots by probing for Myc and Igh as previously described (Ramiro et al., 2004). Bands that probe for both Myc and Igh represent Myc to Igh chromosomal translocations.
Flow Cytometry
Single-cell suspensions from bone marrow, spleen, or lymph node were incubated with biotin-conjugated monoclonal antibodies anti-CD43, anti-IgM, anti-B220, anti-CD95, anti-GL7, anti-CD19, anti-FAS, or anti-IgG1 and then stained with streptavidin conjugated to FITC, PE, or APC (BD Biosciences). Germinal-center cells were CD19+, Fas+, and GL-7+ cells sorted from lymph nodes 14 days after immunization. Data were collected with a FACSCalibur and analyzed with CellQuest and FloJo software. Cell sorting was on a FACSAria and FACSVantage.
Immunizations and ELISA
Age- and sex-matched 8- to 12-week-old mice were immunized by footpad injection with 50 μg of alum precipitated NP21-CGG (both from Biosearch Technologies). To measure serum-antibody amounts, we used goat anti-mouse Ig (H+L) for capture and horseradish peroxidase (HRP)-conjugated goat anti-mouse isotype-specific antibodies (Southern Biotechnology) for detection. Values were calculated by comparison with mouse immunoglobulin standards (Southern Biotechnology). Serial dilutions were performed for each sample, and readings were taken within the linear range for each sample and adjusted for dilution. Results reflect relative absorbance for each sample compared with the standard control. All plates were developed with a Peroxidase Substrate Kit (Bio-Rad), and absorbance was measured at 415 nm.
Immunoblotting
AID antibody was an affinity-purified polyclonal antibody against the carboxyl terminus (EVDDLRDAFRMLGF) of AID and was previously described (McBride et al., 2004). For immunoblot assays, cells were lysed in 20 mM Tris (pH 8.0), 200 mM NaCl, 1% NP-40, 0.5% Deoxycholate, 0.1% SDS, 1 mM DTT, 0.5 mM EDTA, 1 mM PMSF, Protease inhibitor cocktail (Sigma). Fifty micrograms of protein was immunoblotted with the AID antibody or tubulin antibody (abcam). Band densities were quantified with ImageJ software, and relative AID amount is a comparison of AID/tubulin ratios within each gel.
Quantitative PCR
Total RNA was isolated from sorted or LPS- and IL4-activated cells with TRIzol reagent (Life Tecchnologies) according to the manufacturer’s instructions. The first-strand cDNA synthesis was performed with 200 ng of total RNA primed with random primers via the RT reaction protocol provided by the manufacturer (Invitrogen). qPCR was performed with Brilliant SYBR Green QPCR master mix (Stratagene) containing 500 nM primers via standard amplification procedure. All samples were analyzed in triplicate and normalized to GAPDH levels, and the result expressed as _n_-fold induction compared to wild-type day 4 control. Primers for Aicda were forward 5′-GAAAGTCACGCTGGAGACC G-3′ and reverse 5′-TCTCATGCCGTCGCTTGG-3′, and primers for GAPDH were forward 5′-TGAAGCAGGCATCTGAGGG-3′ and reverse 5′-CGAAGGTGGAAGAGTGGGAG-3′. To determine the half-life of _Aicda_155/− mRNA, we treated B cells cultured with LPS + IL-4 for 72 hr when Actinomycin D was added to the culture medium at 10 μg/ml final concentration for 0.5, 1, 1.5, 2, 3, or 4 hr. First-strand cDNA synthesis was performed with 200 ng of total RNA primed with random primers (Invitrogen). Triplicate Aicda q-PCR were preformed for each time point for each of two mice and normalized to GAPDH.
Calculation of Translocation p Values and Aicda RNA Half-Life
p value was calculated with a two-tailed Fisher’s exact test. Comparison between genotypes of the number of translocations per number of PCR reactions was used to do the calculations. To determine the Aicda RNA half-life, we used an exponential regression model of the data generated by Excel. That exponential decay models the data correctly is supported by high R2 values (0.93 for Aicda155/− and 0.94 for Aicda+/−) in the regression and the finding that the log plot of the normalized data is linear. The half-life was determined from the slope of the resulting lines.
Mutation Analysis
Genomic DNA from sorted CD19+Fas+GL7+ germinal-center cells of NP-KLH-immunized mice was PCR amplified in 50 μl with PfuTurbo (Stratagene) for 30 cycles from 10,000–100,000 sorted cell equivalents in four independent reactions that were pooled for cloning experiments. For 5′ Sμ, the primers and PCR conditions have been described (Jolly et al., 1997; Reina-San-Martin et al., 2003). The JH4 intron was amplified with 5′-GGAATTCGCCTGACATCTGAGGACTCTGC-3′ and 5′-CTGGACTTTCGGTTTGGTG-3′ for 14 cycles at 94 °C (30 s), 55°C (30 s), and 72°C (90 s) and then with 5′-GGTCAAGGAACCTCAGTCA-3′ and 5′-TCTCTAGACAGCAACTAC-3′ for 21 cycles at 94°C (30 s), 55°C (30 s), and 72°C (30 s). Statistical significance was determined by a two-tailed t test assuming unequal variance. Bcl6 primers were p369 5′-CTTTCTTGGTTGGAGTCGAGG-3′ and p370 5′-CGGGCTTGAGGTCATTTCTC-3′, as previously described in (Muto et al., 2006). PCR reactions were performed in triplicates, the products were pooled, and bands at the expected size were gel extracted and cloned with TOPO-TA (Invitrogen). Bacterial colonies were sequenced by Biotic Solutions and analyzed with CodonCode Aligner software (CodonCode Coporation). Only good-quality sequence was considered, as determined by inspection of the chromatograms.
Supplementary Material
figures
Acknowledgments
We thank D. Dorsett, M. Dorsett, D. Scheinker for suggestions; K. Velinzon and T. Shengelia for cell sorting; and D. Bosque for animal husbandry. This work was supported by the Schering Foundation (T.A.S.), the Leukemia and Lymphoma Society (D.F.R.), and grants from National Institutes of Health AI064345 (to K.R.) and AI06231 (to M.C.N.). M.C.N is a Howard Hughes Medical Institute investigator.
Footnotes
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Barreto VM, Pan-Hammarstrom Q, Zhao Y, Hammarstrom L, Misulovin Z, Nussenzweig MC. AID from bony fish catalyzes class switch recombination. J Exp Med. 2005;202:733–738. doi: 10.1084/jem.20051378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Basu U, Chaudhuri J, Alpert C, Dutt S, Ranganath S, Li G, Schrum JP, Manis JP, Alt FW. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–511. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- Chatterji M, Unniraman S, McBride KM, Schatz DG. Role of activation-induced deaminase protein kinase A phosphorylation sites in Ig gene conversion and somatic hypermutation. J Immunol. 2007;179:5274–5280. doi: 10.4049/jimmunol.179.8.5274. [DOI] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Dorsett Y, Robbiani DF, Jankovic M, Reina-San-Martin B, Eisenreich TR, Nussenzweig MC. A role for AID in chromosome translocations between c-myc and the IgH variable region. J Exp Med. 2007;204:2225–2232. doi: 10.1084/jem.20070884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, Castellano L, Magrelli A, Citarella F, Messina M, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang KT, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda H, Sugai M, Nambu Y, Katakai T, Agata Y, Mori KJ, Yokota Y, Shimizu A. The balance between Pax5 and Id2 activities is the key to AID gene expression. J Exp Med. 2003;198:1427–1437. doi: 10.1084/jem.20030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasch D, Chen YW, Reilly RM, Chiou XG, Koterski S, Smith ML, Kroeger P, McWeeny K, Halbert DN, Mollison KW, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol. 2002;217:78–86. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- Hammond SM. MicroRNAs as tumor suppressors. Nat Genet. 2007;39:582–583. doi: 10.1038/ng0507-582. [DOI] [PubMed] [Google Scholar]
- Horikawa K, Takatsu K. Interleukin-5 regulates genes involved in B-cell terminal maturation. Immunology. 2006;118:497–508. doi: 10.1111/j.1365-2567.2006.02382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Jay C, Nemunaitis J, Chen P, Fulgham P, Tong AW. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 2007;26:293–300. doi: 10.1089/dna.2006.0554. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly CJ, Klix N, Neuberger MS. Rapid methods for the analysis of immunoglobulin gene hypermutation: application to transgenic and gene targeted mice. Nucleic Acids Res. 1997;25:1913–1919. doi: 10.1093/nar/25.10.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- Kluiver J, Haralambieva E, de Jong D, Blokzijl T, Jacobs S, Kroesen BJ, Poppema S, van den Berg A. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer. 2006;45:147–153. doi: 10.1002/gcc.20273. [DOI] [PubMed] [Google Scholar]
- Kluiver J, van den Berg A, de Jong D, Blokzijl T, Harms G, Bouwman E, Jacobs S, Poppema S, Kroesen BJ. Regulation of pri-micro-RNA BIC transcription and processing in Burkitt lymphoma. Oncogene. 2007;26:3769–3776. doi: 10.1038/sj.onc.1210147. [DOI] [PubMed] [Google Scholar]
- Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie CH, Soneji S, Marafioti T, Cooper CD, Palazzo S, Paterson JC, Cattan H, Enver T, Mager R, Boultwood J, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–1161. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. microRNAs: a safeguard against turmoil? Cell. 2007;130:581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih Ih, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- Marton S, Garcia MR, Robello C, Persson H, Trajtenberg F, Pritsch O, Rovira C, Naya H, Dighiero G, Cayota A. Small RNAs analysis in CLL reveals a deregulation of miRNA expression and novel miRNA candidates of putative relevance in CLL pathogenesis. Leukemia. 2008;22:330–338. doi: 10.1038/sj.leu.2405022. [DOI] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J Exp Med. 2004;199:1235–1244. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KM, Gazumyan A, Woo EM, Barreto VM, Robbiani DF, Chait BT, Nussenzweig MC. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc Natl Acad Sci USA. 2006;103:8798–8803. doi: 10.1073/pnas.0603272103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffre E, Casellas R, Nussenzweig MC. Antibody regulation of B cell development. Nat Immunol. 2000;1:379–385. doi: 10.1038/80816. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Nagaoka H, Shinkura R, Begum NA, Honjo T. Discovery of activation-induced cytidine deaminase, the engraver of antibody memory. Adv Immunol. 2007;94:1–36. doi: 10.1016/S0065-2776(06)94001-2. [DOI] [PubMed] [Google Scholar]
- Muto T, Okazaki IM, Yamada S, Tanaka Y, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. Negative regulation of activation-induced cytidine deaminase in B cells. Proc Natl Acad Sci USA. 2006;103:2752–2757. doi: 10.1073/pnas.0510970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: Biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008 doi: 10.1210/jc.2007-2696. in press. Published online February 12, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Kitaura Y, Gu H, Dalla-Favera R. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc Natl Acad Sci USA. 2006;103:395–400. doi: 10.1073/pnas.0509969103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Duran P, de Yebenes VG, Ramiro AR. Oncogenic events triggered by AID, the adverse effect of antibody diversification. Carcinogenesis. 2007;28:2427–2433. doi: 10.1093/carcin/bgm201. [DOI] [PubMed] [Google Scholar]
- Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter M. Neoplastic development in plasma cells. Immunol Rev. 2003;194:177–195. doi: 10.1034/j.1600-065x.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B, Difilippantonio S, Hanitsch L, Masilamani RF, Nussenzweig A, Nussenzweig MC. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J Exp Med. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B, Chen HT, Nussenzweig A, Nussenzweig MC. ATM is required for efficient recombination between immunoglobulin switch regions. J Exp Med. 2004;200:1103–1110. doi: 10.1084/jem.20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, Croce CM. Micro-RNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- Sayegh CE, Quong MW, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- Schmidt EV, Pattengale PK, Weir L, Leder P. Transgenic mice bearing the human c-myc gene activated by an immunoglobulin enhancer: A pre-B-cell lymphoma model. Proc Natl Acad Sci USA. 1988;85:6047–6051. doi: 10.1073/pnas.85.16.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam W. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene. 2001;274:157–167. doi: 10.1016/s0378-1119(01)00612-6. [DOI] [PubMed] [Google Scholar]
- Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasilious FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg A, Kroesen BJ, Kooistra K, de Jong D, Briggs J, Blokzijl T, Jacobs S, Kluiver J, Diepstra A, Maggio E, Poppema S. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer. 2003;37:20–28. doi: 10.1002/gcc.10186. [DOI] [PubMed] [Google Scholar]
- Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, et al. microRNA-155 Regulates the Generation of Immunoglobulin Class-Switched Plasma Cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakae K, Magor BG, Saunders H, Nagaoka H, Kawamura A, Kinoshita K, Honjo T, Muramatsu M. Evolution of class switch recombination function in fish activation-induced cytidine deaminase, AID. Int Immunol. 2006;18:41–47. doi: 10.1093/intimm/dxh347. [DOI] [PubMed] [Google Scholar]
- Yagi T, Ikawa Y, Yoshida K, Shigetani Y, Takeda N, Mabuchi I, Yamamoto T, Aizawa S. Homologous recombination at c-fyn locus of mouse embryonic stem cells with use of diptheria toxin A-fragment gene in negative selection. Proc Natl Acad Sci USA. 1990;87:9918–9922. doi: 10.1073/pnas.87.24.9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa K, Okazaki IM, Eto T, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 2002;296:2033–2036. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
figures