Tudor domain protein Tdrd6 is required for spermiogenesis, chromatoid body architecture and regulation of miRNA expression (original) (raw)
. Author manuscript; available in PMC: 2010 Apr 28.
Published in final edited form as: Curr Biol. 2009 Apr 2;19(8):630–639. doi: 10.1016/j.cub.2009.02.047
Summary
Background
Chromatoid bodies (CBs) are characteristic spermatid organelles, which were suggested to function in RNA storage and small RNA processing, but whose functions remain largely unknown. CB components include Mili, Miwi, and Tudor-domain proteins such as Tdrd6, whose contribution to CB structure and function is elusive.
Results
We determined gametogenesis stage- and male-specific expression and localization of Tdrd6, identified a C-terminally truncated form as predominant after meiosis I, and demonstrate direct physical interaction of Tdrd6 with the CB components Mili and Miwi. Development from round into elongated spermatids is abrogated in _Tdrd6_−/− mice. Their round spermatids bear “ghost” CBs, whose architecture is greatly disrupted. Mael, Miwi, and Mvh do not localize to the Tdrd6-deficient CBs, but retrotransposons are not significantly activated. However, more than 50 miRNAs are more abundant in _Tdrd6_−/− testes, as are exemplary pre- and pri-miRNAs.
Conclusion
We conclude that Tdrd6 is essential for spermiogenesis, for CB structure, and for proper mature and precursor miRNA expression.
Keywords: Tdrd6, spermiogenesis, chromatoid bodies, miRNA
Introduction
In germ cells of various organisms dense fibrous material accumulates into a cytoplasmic structure, which in the initial stages of meiosis is scattered and has cloud-like appearance, inspiring the term nuage [1]. In Drosophila melanogaster, the nuage is localized at the posterior end of the egg termed germ or pole plasm. The pole plasm gives rise to the polar granule in a cytoplasmic region within the oocyte, which after fertilization has an essential function in the specification of germline lineage and the initiation of embryonic development. A morphologically distinct germ plasm is not readily identifiable in mammalian oocytes or eggs. On the basis of structural features and protein composition, the chromatoid body (CB) in the cytoplasm of spermatogenic cells has been suggested to be the mammalian counterpart of the polar granule. The CB of mammalian germ cells is the most widely studied form of nuage and appears first as fibrous and granulated material in the interstices of mitochondria clusters and in the perinuclear area of pachytene spermatocytes [2–4]. In early studies the origin of the CBs was suggested as dense material appearing in the nucleus or nucleolus of pachytene spermatocytes [4–8], or as intermitochondrial cement [8]. The nuage aggregates diminish in number and density with very few faint granules present in late diplotene and during the first meiotic division. Large dense bodies appear de novo scattered throughout the cytoplasm of newly formed secondary spermatocytes. After meiosis, these CBs condense into one single lobulated, perinuclear granule in round spermatids and remain a distinctive feature in the cytoplasm of post-meiotic spermatids until the nucleus begins to elongate [3]. We refer to the nuage/CB of spermatocytes I as “CB type 1”, and the later CB as “CB type 2”. CBs type 2 migrate via intercellular cytoplasmic bridges [9], are closely associated with the nuclear envelope, and also often reside in close proximity to the Golgi apparatus [7, 10]. Electron microscopy studies showed that ribonucleoprotein material of nucleolar origin translocates to the cytoplasm and contributes to CBs [11].
Molecularly the CB consists mainly of RNA and various, predominantly RNA-binding proteins. Monoclonal anti-DNA antibodies and DNA-specific staining indicate lack of DNA [12]. Some CB components such as snRNPs and hnRNPs originate from the nucleus and the nucleolus, while others are derived from polysomes [13]. In addition, many enzymes that belong to the RNAi machinery were found in the CB suggesting that functionally this organelle may be the counterpart of the P-bodies found in somatic cells[14]. Therefore, the CB may be a storage site for mRNA and may regulate translation through RNAi mechanisms.
Tudor domains are related to plant Agenet, Chromo PWWP and MBT domains, which together form the Tudor domain ‘Royal Family’[15], but the function of this protein domain remains unclear. Several Tudor domain containing proteins exist in mammalian germ cells including Tdrd1, Tdrd4, Tdrd6, Tdrd5 and Tdrd7/Trap[16–20]. In D. melanogaster, the tudor protein localizes to the polar granules, may act downstream of vasa and oskar in assembly of germ plasm, and controls the size and number of polar granules [21]. The tudor domain of the Survival Motor Neuron (SMN) protein binds directly to spliceosomal Sm proteins during spliceosome assembly [22–25]. Direct interaction of the SMN-type tudor domain with other proteins is supported by methylated arginine or lysine residues in the target protein [22, 24–30]. Several tudor domain proteins such as 53BP1 interact with methylated histones [28, 31, 32]. Tudor proteins may also interact with ribonucleic acids [33], but the mode of interactions remains unclear [15].
The only known domains of Tdrd6 are its multiple Tudor domains, which defines it as the closest homolog of Drosophila tudor. Tdrd6 was initially identified as a gene overexpressed in colon cancer cells [34], but in primary cells is specifically expressed in germ cells [19]. Tdrd6 (2134 amino acids) was reported to migrate as an app. 250 kDa protein in SDS gel electrophoresis [19]. By immuno fluorescence (IF) staining of testis sections, Tdrd6 was undetectable in spermatogonia, appeared diffusely in pachytene spermatocytes, and localized to CB in round spermatids [19]. In a mouse mutated in Mvh1098/1098, Tdrd6 aberrantly localizes in cytoplasmic granules, indicating that Mvh/Ddx4 is required for proper localization of Tdrd6 [19].
The function of Tdrd6, however, remained elusive. We used a _Tdrd6_−/− mouse model to define an essential function for Tdrd6 in spermiogenesis, particularly in CB formation.
Results
Differential pattern of Tdrd6 expression in the male germline
We characterized the temporal pattern of Tdrd6 expression in male gonads by Northern analysis of total RNA from juvenile and adult mouse testis (Fig. 1A). In mice, spermatogenesis is initiated on day 3 pp and progresses as a synchronous wave during the first weeks of life (e.g. type B spermatogonia appear at day 8 pp; elongated spermatids appear after 20 days pp) [35]. A weak hybridization signal was detected as a single band in the testis of 16-days old mice, which coincides with the mid-pachytene stage of primary spermatocytes. The signal increases at 18 days post-partum (dpp) and remains elevated with the appearance of round spermatids at 22 dpp and in mature testis. There is no indication for alternative splice products from this and another study[19]. Co-expression of Tdrd6 with Dmc1, Sycp3 and Mouse vasa homolog/DEAD box protein 4 (Mvh/Ddx4) confirmed prophase I onset of Tdrd6 gene expression in testis, which is not seen in the embryonic ovary or in the somatic Sertoli cells (Suppl. fig. 1A). The Tdrd6 expression data were also confirmed by RT-PCR of testis RNA samples of an extended age-range (Suppl. fig. 1B). Tdrd6 expression was not detected in the testis of _Smc1β_−/− mice (Fig. 1A), which are devoid of germ cells beyond the early pachytene stage [36], defining the onset of Tdrd6 expression as past stage IV.
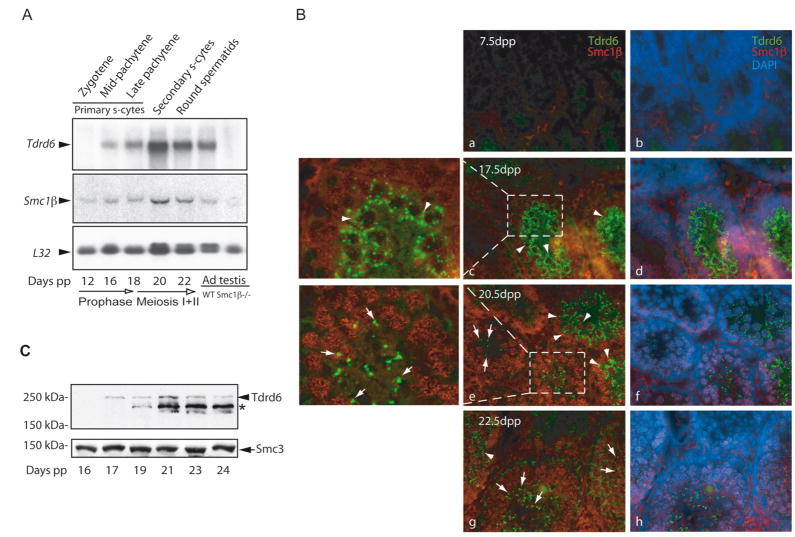
Figure 1. Kinetics of Tdrd6 expression in testis.
(A) Northern analysis of total RNA (5 μg/lane) from juvenile mice of ascending age and adult wild-type and Smc1β−/− testes. The L32 gene encoding a ribosomal protein was used as a loading control. The mouse age and the phase of meiosis are indicated at the bottom. The latest differentiated germ cell type corresponding to the mouse age is shown above each lane. (B) Immunofluorescence staining of frozen testicular sections from 7.5 (a, b), 17.5 (c plus area enlarged, d), 20.5 (e plus area enlarged, f) and 22.5 days old mice (g, h). The Tdrd6 stained in green first appears in the multilobular cytoplasmic CBs of primary spermatocytes (arrowheads) and then re-appears as a single perinuclear dot in spermatids (arrows). DAPI (blue) marks the nuclei and Smc1β stained in red shows the synapsed meiotic chromosomes in primary spermatocytes. Non-specific green staining appears in the center and non-specific red staining at the basal lamina of tubules from 7.5 dpp old mice. (C) Western analysis of Tdrd6 protein expression in testis extracts from juvenile mice. An arrow indicates the 250-kDa Tdrd6 reported previously [19]. A star marks the position of an abundant smaller product of the Tdrd6 gene (approx. 230 kDa) also recognized by α-265. Cohesin protein Smc3 was used as loading control.
To analyze the presence of Tdrd6 in situ, we immuno-stained frozen testis sections collected from mice ages 7.5, 17.5, 20.5 and 22.5 dpp with rabbit antibodies raised against a peptide representing amino acids 265–373 of Tdrd6 (α-265). As expected, testes from day 7.5 pp mice were negative for Tdrd6. At later time points we observed specific signals that varied by cell type. Double staining with antibodies against Smc1β [37] showed that Tdrd6 appears first in primary spermatocytes (day 17.5 pp; late pachytene) as multiple fine filamentous cytoplasmic granules (Fig. 1B–c, d). These appear to surround the nuclei. In post-meiotic round spermatids found at day 20.5 and 22.5 dpp the Tdrd6 signal appeared as bright distinct perinuclear dots (Fig. 1B–e, f, g, h). This staining pattern is in concert with the localization of Tdrd6 to CBs reported previously [19]. Tdrd6 was not detected in sections from adult ovaries or kidney (data not shown).
The same antibodies were used to probe immunoblots of total testis extracts from mice aged 16–24 dpp (Fig. 1C). While in the extract from day 17 pp the expected 250-kDa Tdrd6 [19] protein was recognized, extracts from older mice display a strong band of 230 kDa and a weak band of 250 kDa. Mass spectrometric analysis of the two immuno-precipitated proteins confirmed that both are products of the Tdrd6 gene.
C-terminal processing of Tdrd6 occurs in stage-specific manner
To identify the origin of the shorter Tdrd6 polypeptide and investigate in detail the expression and localization of the two Tdrd6 forms in testis we raised rabbit polyclonal antibodies against fragments corresponding to the N-terminal (α-Nterm) or C-terminal (α-Cterm) ends of Tdrd6 (amino acids 3-130 and 2008-2134, respectively). Cyto-spun germ cells were co-stained with these antibodies conjugated to Alexa-488 or Alexa-555. Staining with α-Nterm was equally strong in the fine cytoplasmic filaments of primary spermatocytes as well as the larger perinuclear granules in secondary spermatocytes and spermatids (Fig. 2A–a). The α-Cterm antibody yielded bright Tdrd6 signals in pachytene and diplotene spermatocytes marked by the synaptonemal complex protein Sycp3 present on meiotic chromosomes (Fig. 2A–b, c), but produced weak signals in CBs of secondary spermatocytes and spermatids (Fig. 2A–b, c, d). This difference is clearly visible after merging the green and red fluorescence, which resulted in yellow spots in primary spermatocytes, while the red spots (α-Nterm) in secondary spermatocytes and spermatids remain unaffected (Fig. 2A–c, d).
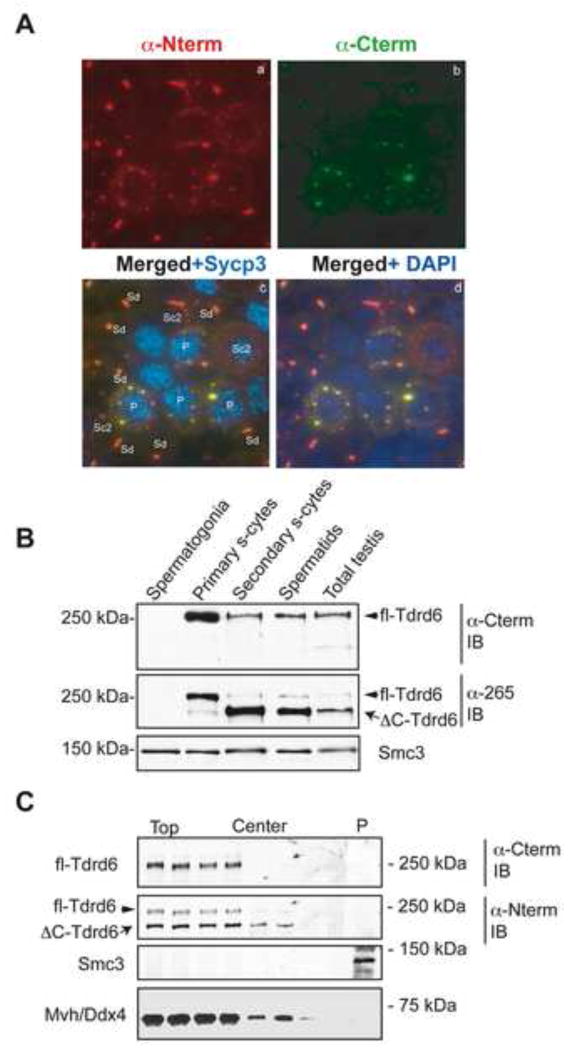
Figure 2. Removal of the C-terminal end of Tdrd6 at the meiosis I meiosis II transition.
(A) Immunofluorescence staining of sorted cytospun germ cells from adult testis with anti-Tdrd6 antibodies raised against the N-terminus (a) or C-terminus (b). Images were merged to visualize the relative intensity of staining with each antibody in different germ cell types (c, d). Primary spermatocytes are labeled by Sycp3 on synapsed chromosomes in meiosis I (blue in c) and nuclei in all cells are labeled by DAPI (blue in d). (B) Differential distribution of the two Tdrd6 forms. Adult testis cell suspension was used for sorting and isolation of the main germ cell subtypes. Total extracts from the sorted populations were probed with either α–Cterm or α-265 antibodies. Cohesin Smc3 was used as loading control. (C) Centrifugation of nuclei and cytoplasm components from total testis sucrose-containing homogenate on a sucrose cushion. Fractions were collected from the top to near the bottom of the centrifugation tube. The pellet containing nuclei was resuspended and pelleted again through the same type of cushion (P). Fractions and the extracted nuclear pellet were then immunoblotted and probed with the indicated antibodies.
Using a modification of a flow-cytometry method reported previously [38] to isolate specific testis populations we analyzed extracts from such populations by immunoblotting (Fig. 2B). The α-Cterm antibodies recognize a single band of 250-kDa (Fig. 2B, top panel), the predominant form in primary spermatocytes. α-Nterm (not shown) or α-265 recognize two bands. The lower (230-kDa) band is present only in secondary spermatocytes and spermatids (Fig. 2B, middle panel). This indicates that the C-terminus of Tdrd6 is removed during the transition from meiosis I to meiosis II and we designated the 250-kDa protein as full-length Tdrd6 (fl-Tdrd6) and the 230-kDa protein as C-terminal deleted (ΔC-Tdrd6). Analysis of crude cytoplasmic and nuclear extract from individual, purified germ cell populations confirmed this pattern and revealed the association of Tdrd6 as a component of nuclei-associated CBs (Suppl. fig. 2).
To address whether Tdrd6 is stably associated with nuclei as the perinuclear localization of CBs may suggest, we subjected testis sucrose-containing homogenates to centrifugation through a 2 M sucrose cushion according to [39] (Fig. 2C). There is neither fl-Tdrd6 nor ΔC-Tdrd6 detectable in the pelleted nuclei. The fl-Tdrd6 appears to accumulate preferentially at the top of the sucrose solution, while the ΔC-Tdrd6 migrated towards the center fractions.
CB component Mvh/Ddx4 associates with Tdrd6
The germ cell-specific ATP-dependent RNA helicase Mvh is essential for male gametogenesis, associates with perinuclear RNA-protein complexes, resides in the CBs [40], and is required for proper localization of Tdrd6 to the intermitochondrial cement and CBs [19]. Tdrd6 was found to form a complex with Tdrd1 and Tdrd7/TRAP. However, mislocalization of these three proteins to non-overlapping cellular loci in the Mvh mutant mouse suggested that Tdrd6 may localize independently of Tdrd1 and Tdrd7 [19]. We examined the localization of Tdrd6 and Mvh in sorted, cytospun germ cell populations by double staining with α-Cterm and α-Ddx4/Mvh antibodies (Fig. 3A). As expected, prior to Tdrd6 expression, Mvh was present in the filamentous nuage of primary spermatocytes (Fig. 3A–a, b, c and d). In primary spermatocytes Mvh and Tdrd6 signals partially co-localize, but completely co-localize in secondary spermatocytes and spermatids (Fig. 3A – e, f, g, h). Physical interaction of Mvh with either full-length or truncated Tdrd6 was tested by immunoprecipitation of complexes from total testis extract with either α-Cterm, α-265, or preimmune antibodies (Fig. 3B). Immunoblots with α-Ddx4/Mvh antibodies revealed that both full-length and ΔC-Tdrd6 co-precipitate with Mvh. Vice versa, α-Ddx4/Mvh immunoprecipitation yielded Tdrd6 in the precipitates. Fl-Tdrd6 and ΔC-Tdrd6 may have additional binding partners, which we sought to determine by mass spectrometry. Samples were prepared from total testis extract from wild-type and – for early meiocytes – from _Smc1β_−/− mice, and immunoprecipitated with α-Cterm, α-265, or purified antibodies from pre-immune rabbits as background subtraction control. Analysis was done by mass spectrometry. Mvh was observed as a major component of both, α-Cterm and α-265 Tdrd6 co-precipitates. Other proteins important for piRNA metabolism such as Miwi and Mili were also represented.
Figure 3. Tdrd6 interacts with Mvh.
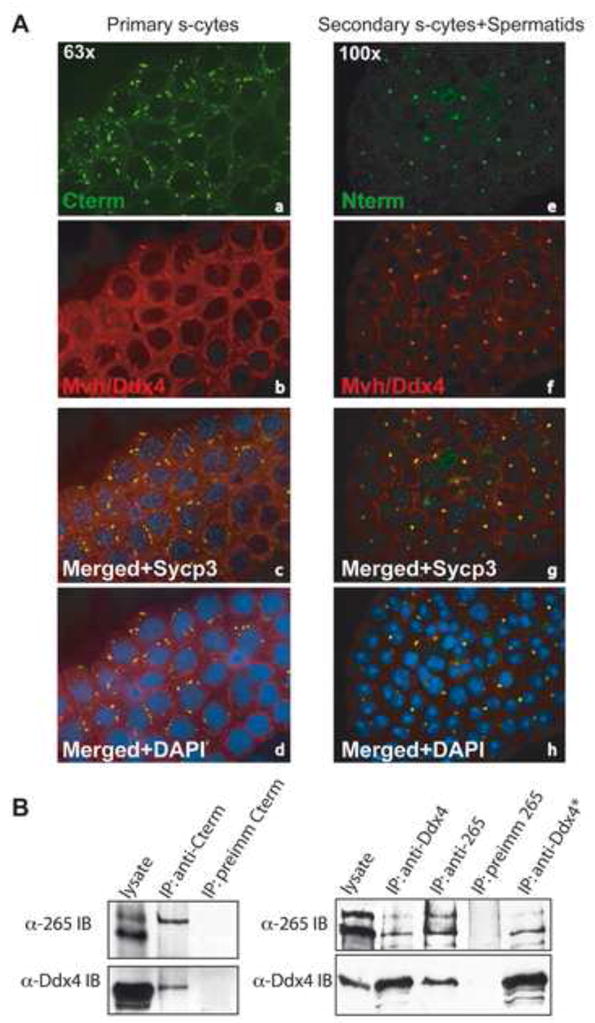
(A) Immunolabeling of cyto-spun primary spermatocytes (a–d) or secondary spermatocytes and spermatids (e–h) with α-Cterm or α-Nterm (green) and α-Ddx4/Mvh antibodies. (B) Immunoprecipitation with α-Cterm (left panel), or α-Ddx4/Mvh and α-256 antibodies (right panel). Precipitates were separated by SDS-PAGE and probed with either α-Ddx4/Mvh or α-Tdrd6 antibodies. Purified preimmune sera were used as control; * indicates highly stringent washing conditions of the bead-bound material using SDS-containing RIPA buffer.
Full-length and ΔC-Tdrd6 bind to Piwi family poteins
In addition to Mvh, Tdrd6 may associate with other proteins, which take part in RNA processing and localize to the CB. The two Piwi domain-containing proteins, Miwi and Mili form a complex and interact directly with Mvh [41], and were major components of α-Cterm immunoprecipitates. To assess the cytoplasmic localization of Miwi and for comparison RNF17 relative to Tdrd6, we co-stained sorted germ cell populations with α-Hiwi (predicted to cross-react with mouse Miwi) or α-RNF17 and α-Tdrd6 antibodies. Miwi and Tdrd6 were both found in the CBs of all germ cells examined, while RNF17 associated with a different component of the germ cell nuage that is distinct from the CBs and did not overlap with the Tdrd6 loci as previously reported [17] (Suppl. fig. 3).
Considering the co-precipitation of Mili and Miwi with Tdrd6 we used in vitro transcribed and translated, tagged proteins to test for potential direct interactions. A screen for candidate proteolytic activity yielded a single caspase I recognition sequence located approximately 20 kDa from the C-terminus of Tdrd6 (Fig. 4A). Accordingly, we designed the HA-ΔC-Tdrd6 by deleting the C-terminus up to the caspase I site. The HAΔC-Tdrd6 shows the same electrophoretic mobility as the native 230-kDa protein. Myc-tagged Miwi or Mili were mixed in equal ratio with HA-tagged fl-Tdrd6 or ΔC-Tdrd6 and incubated with α-HA antibodies. As shown in Fig. 4 α-myc antibodies recognize the tagged Mili and Miwi associated with both forms of Tdrd6. Compared to fl-Tdrd6 the amount of Mili-myc bound to ΔC-Tdrd6 was significantly reduced (Fig. 4B-left panel). Both forms of Tdrd6 pulled down about the same amount of Miwi-myc (Fig. 4B – right panel). To confirm these data, we immunoprecipitated Miwi with α-256 antibody from testis lysates, and vice versa precipitated Tdrd6 with α-Hiwi/Miwi antibody (Fig. 4C).
Figure 4. Association of Tdrd6 with Mili and Miwi.
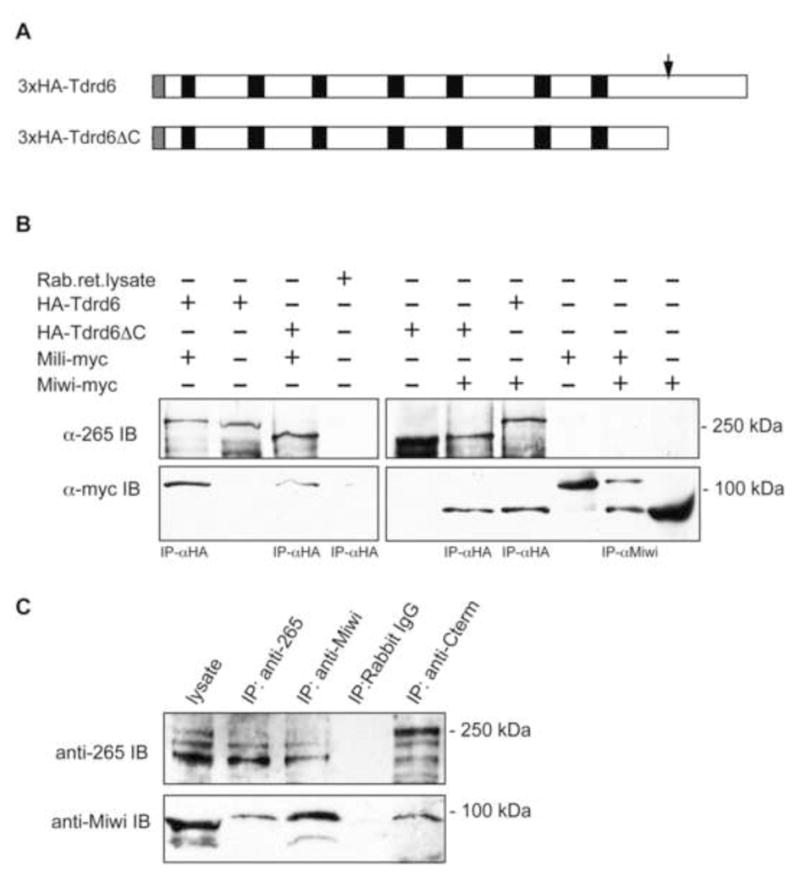
(A) Schematic structure of the truncated Tdrd6 protein. Black boxes show the tudor domains, grey box depicts the 3xHA tag. The red box indicates the predicted Caspase I cleavage site. (B) Co-immunoprecipitation of Mili and Miwi with Tdrd6. The proteins were expressed in rabbit reticulocyte lysate system from plasmids containing HA-tagged full-length and C-terminally deleted Tdrd6 cDNA or myc-tagged MILI or MIWI cDNA [41]. The lysates were immunoprecipitated as indicated and detected with the anti-Tdrd6 or anti-Myc antibodies. (C) Immunoprecipitation from testis lysates using α-265, α-Cterm, or α-Miwi antibodies or rabbit IgG for control. The resulting blots were probed with α-Nterm or α-Miwi.
Spermatogenesis in Tdrd6−/− mice is blocked at the round spermatid stage
To investigate the function of Tdrd6 in vivo, we generated _Tdrd6-_deficient mice by homologous recombination (Suppl. methods and Suppl. fig. 4). Exon 1 representing 95 % of the Tdrd6 gene was replaced with the hCD4 gene lacking the cytoplasmic tail responsible for intracellular signaling [42]. We determined that the Tdrd6 protein is present in Tdrd6+/− and absent from _Tdrd6_−/− testes. Expression of hCD4 is driven by the endogenous Tdrd6 promoter and therefore, Tdrd6+/− derivatives of this strain may serve as a very useful tool to identify, sort and investigate separate post-pachytene germ cell populations (sorting of specific populations see Fig. S4D).
_Tdrd6_−/− mice are viable and show apparently normal development. Interbreeding of Tdrd6+/− mice produces offspring of normal litter size, and yield the Mendelian ratios of Tdrd6+/+, Tdrd6+/− and _Tdrd6_−/− mice. While the female _Tdrd6_−/− mice are fertile, the males are sterile. _Tdrd6_−/− testes of 20 days old males are the same size as those of WT littermates, which suggests normal development of spermatogonia, primary and secondary spermatocytes. However, at 6 weeks of age, the _Tdrd6_−/− males present with up to 25 % smaller testes than those of control littermates. Histological analysis of adult testes revealed that sperm are completely absent from the epididymes of _Tdrd6_−/− mice and that elongated spermatids are almost completely lacking (Fig. 5A). By FACS we compared the ratio of specific germ cell populations from 10-week-old Tdrd6+/− and _Tdrd6_−/− mice. There is a large reduction in the number of elongated spermatids from 11.6 % in Tdrd6+/− to 1.4 % in the _Tdrd6_−/− testis (Fig. 5B). In addition, we observed higher numbers of spermatogenic cells of earlier stages including primary spermatocytes, secondary spermatocytes and round spermatids.
Figure 5. Defective spermatogenesis in _Tdrd6_−/− mice.
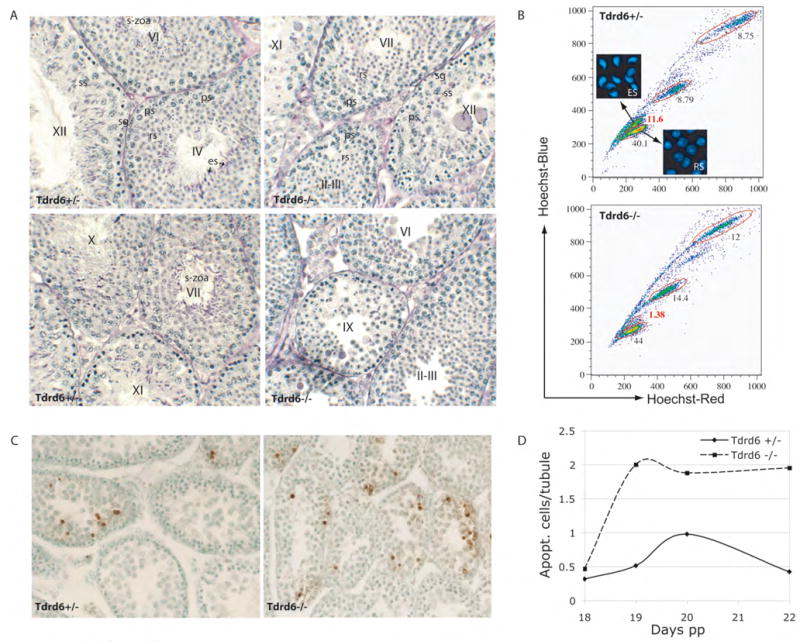
(A) Testis sections from adult heterozygous (+/−) and homozygous mutant (−/−) littermates were stained with periodic acid-Schiff (PAS) reagent. Seminiferous tubule stages are indicated by roman numbers in the center. Examples of cell types labeled in the center with abbreviations as follows: sg – spermatogonium, ps – primary spermatocytes, ss-secondary spermatocytes, rs round spermatid, es – elongated spermatid and s-zoa – spermatozoa. (B) Comparison of the distribution of germ cell populations in 10-week-old heterozygous (+/−) and homozygous mutant (−/−) mice. Very few elongated spermatids are found in tubules of _Tdrd6_−/− mouse. (C) Apoptosis in Tdrd6+/− and _Tdrd6_−/− testes. Testes from 20-day old heterozygous and homozygous mutant littermates were stained using the TUNEL technique. (D) Quantification of apoptosis in the testis of juvenile mice.
TUNEL labeling was used to determine the timing of abnormal cell death in the first wave of spermatogenesis and the subsequent loss of elongated spermatids in adulthood (Fig. 5C). TUNEL-positive cells were rarely detected in _Tdrd6_−/− testes on day 14 (data not shown), which coincides with the appearance of pachytene spermatocytes, and remained scarce until day 18 pp. Thereafter, we observed an average 2-fold increase in the number of apoptotic cells in the _Tdrd6_−/− testis compared to Tdrd6+/− testis (Fig. 5D).
CB components mis-localize and CB architecture is distorted in Tdrd6−/− spermatids
We then addressed if the lack of Tdrd6 affects interacting proteins. Immunoblot analysis for Mvh, Mili and Miwi showed that these proteins are still present in the _Tdrd6_−/− testis (Suppl. fig. 5). However, immunostaining of testicular cryosections from homozygous mutant mice shows a severely disrupted localization pattern for both Mvh and Miwi (Fig. 6). There is diffuse cytoplasmic staining instead of the typical “single-dot” appearance of CBs in round spermatids. Similar diffuse localization was observed for Mael, another CB component associating with Mvh [43], but not for RNF17, which resides in a different compartment [18] (not shown). Mvh appeared in faint ring-like structures resembling the disruption of the CBs seen after actinomycin D treatment [44–46]. Thus, localization of CB components and likely CB architecture itself depend on Tdrd6.
Figure 6. Mis-localization of CB proteins in _Tdrd6_−/− spermatdis.
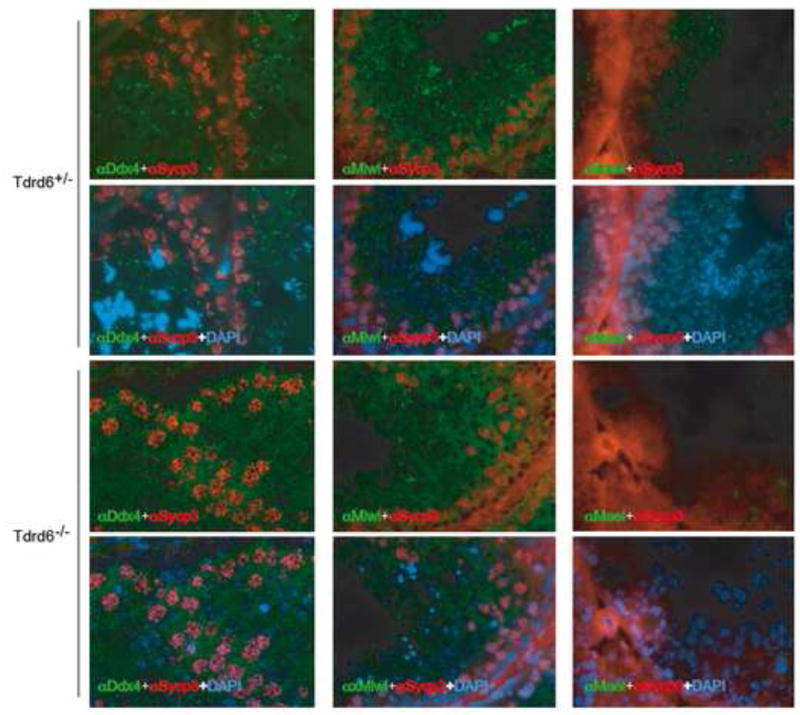
Testicular frozen sections from heterozygous or homozygous null mice were immunostained with either α-Ddx4/Mvh, α-Hiwi/Miwi or α-Mael antibodies (green). For all three proteins the typical pattern of CB staining seen in the heterozygous testis is disrupted in the _Tdrd6_−/−. Sycp3 (red) was used to visualize synaptonemal complexes on chromosomes and nuclei were stained with DAPI (blue).
To further define a possible CB deficiency in _Tdrd6_−/− mice we subjected individual germ cells and tubules to high-resolution morphological analysis by transmission electron microscopy. CBs were observed at similar frequencies in round spermatids of the same stage of Tdrd6+/− and _Tdrd6_−/− mice. In _Tdrd6_−/− spermatocytes and round spermatids, subcellular structures such as synaptonemal complex and acrosomes do not differ from control mice. The most striking difference in round spermatids of _Tdrd6_−/− mice is the disrupted appearance of the CBs (Fig. 7). Whereas the CBs in Tdrd6+/− littermates consist of dense, amorphous material, they appear in _Tdrd6_−/− mice as diffuse structures. The CBs look disrupted, less condense and incorrectly assembled. In _Tdrd6_−/− stage I seminiferous tubules only very few elongated spermatids are visible, and in stage V seminiferous tubules the elongated spermatids are completely absent (Suppl. Fig. 6). Therefore, we conclude that Tdrd6 is essential for the correct architecture of the CBs in round spermatids, which require Tdrd6 to differentiate into elongated spermatids.
Figure 7. Electron microscopy of the chromatoid body.
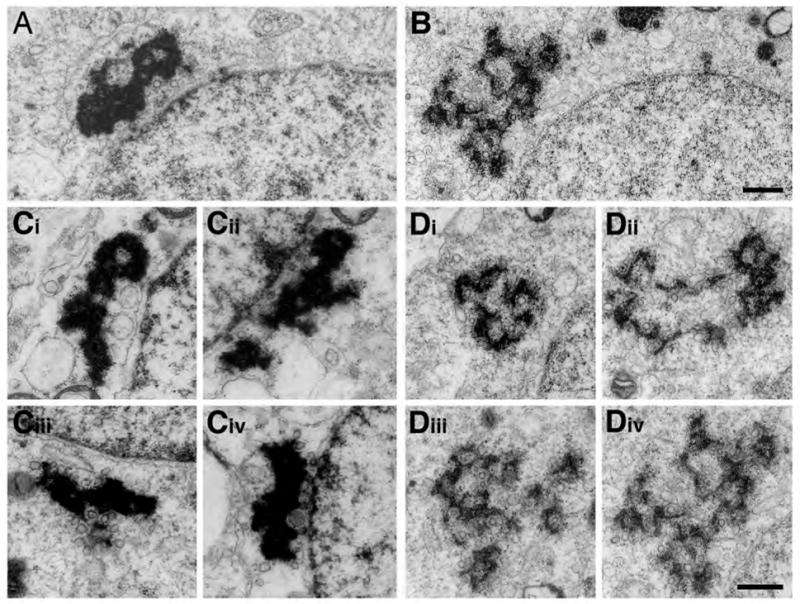
A and C show the fully condensed chromatoid bodies near the nuclear membrane in stage I round spermatids in Tdrd6+/− mice; whereas, B and D show the dispersed chromatoid bodies in round spermatids of the same stage in _Tdrd6_−/− mice (bar 500 nm).
Altered miRNA expression profiles in Tdrd6−/− testis
RNA and RNA processing enzymes reside in CBs, including proteins involved in the miRNA pathway. The abnormal architecture of the CB in _Tdrd6_−/− spermatids and the mislocalization of Mvh and Miwi may affect the regulation of miRNAs. We analyzed RNA isolated from 18 days old Tdrd6+/− and _Tdrd6_−/− testis on miRNA expression arrays. Three pairs of 18-day old littermates were used. Of the app. 793 miRNAs known or predicted for mice (http://www.tarmir.rgcb.res.in/disspc.php), 54 miRNA were found up-regulated (>1.5-fold), but only 5 are down-regulated (< 0.7-fold) (Suppl. Table 1). For several of these miRNAs, the data were validated by quantitative real-time PCR [47], including miRNAs that are known to be expressed in wild-type testis (e.g. 29B, 29C, 34A, 34B, 101B, 463), and those, which are apparently not expressed in adult wild-type testis (e.g. 489, 497). All are up-regulated in _Tdrd6_−/− testis. To evaluate whether upregulation of such miRNAs occurs on the level of precursor or mature miRNAs, the pri- and pre-forms along with the mature forms of three miRNAs were analyzed using primers for real-time PCR, which are specific for each form (Suppl. Fig. 7). Significant upregulation of the primary miRNA transcript and of the pre-miRNA was observed.
Piwi-domain containing proteins Mili and Miwi2 as well as Mael are known to regulate retrotransposon activation [45, 46, 48]. In the respective “knockout” mice activity of transposons like Line-1 (L1) or intracisternal A-particle (IAP) is strongly increased. Using qRT-PCR and CpG methylation-diagnostic Southern blotting (HpaII versus MspI digests), we examined L1 and IAP transcription in Tdrd6−/− testis. No significant up-regulation of retrotransposon activity was observed (data not shown).
Discussion
CBs may serve as a storage site for proteins and RNAs during spermatid differentiation. RNA processing enzymes, including Mvh/Ddx4 and Ddx25 [49], and components of the microRNA pathway like Dicer, Miwi and Mili, localize to CBs, indicating a role in small RNA processing. Another common feature of germ-cell nuage and P-bodies is the presence of tudor domain containing proteins like Tdrd1 [16], Tdrd4, Tdrd5 [20], Tdrd6 and Tdrd7/TRAP [19]. Unlike Tdrd1 and Tdrd5 [20], which are expressed in male gonads during embryonic development, Tdrd6 first appears later in pachytene beyond stage IV. In primary spermatocytes Tdrd6 exists as a single polypeptide, which during the transition from meiosis I to meiosis II undergoes proteolytic cleavage near the C-terminus by an unknown protease. Using _Tdrd6_−/− mice we determined if fl-Tdrd6 has an essential role in meiosis I or if ΔC-Tdrd6 is a critical protein after meiosis I. Since germ cell differentiation in _Tdrd6_−/− mice arrests at step 12–13, Tdrd6 – and thus ΔC-Tdrd6 – acts primarily in post-meiotic germ cells. This does not exclude non-essential, subtle or redundant function(s) of fl-Tdrd6 during meiosis I. Fl-Tdrd6 may alternatively serve as an inactive precursor.
In direct protein interaction experiments, both, fl-Tdrd6 and ΔC-Tdrd6 interact with Mili and independently with Miwi and thus both forms may affect Mili and Miwi localization or function. The stage of arrest in germ cell development in the _Tdrd6_−/− testis is similar to that in the _Miwi_−/− testis [50], but _Mili_−/− spermatogenesis arrests in mid-pachytene [51], suggesting a critical CB-independent function for Mili. In contrast, Miwi and Tdrd6 become essential at the stage of formation of CBs since spermiogenesis in _Tdrd6_−/− and _Miwi_−/− mice arrest between step 8 and 14, with no elongated spermatids formed thereafter [50]. The CBs were not analyzed in detail in Miwi_−/− mice, but spermatogenesis shows multiple impairments already in spermatocytes I and II and possibly in spermatogonia, clearly differing from Tdrd6_−/−, where no such deficiencies were seen. Our interaction data imply that of those proteins Tdrd6 appears the most CB-specific and may serve as a key subunit of the Mvh-(Mili)-Miwi CB complexes.
While generating _Tdrd6_−/− mice, we introduced a non-functional truncated version of hCD4 into the Tdrd6 locus. FACS analysis confirmed stage-specific cell surface expression of hCD4, starting in mid meiosis I and lasting into the spermatid stage. Since hCD4 can be used to sort cells, the Tdrd6+/− mice may serve as a new and rare tool in preparing post-pachytene male germ cells.
The most striking intracellular phenotype of _Tdrd6_−/− mice is their highly aberrant architecture of CBs in round spermatids. _Tdrd6_−/− CBs appear as diffuse, disrupted and less condensed structures. Their total diameter does not seem much affected, and their outer borders appear similar to that of wt CBs. However, their interior is largely missing, and only a “ghost” structure remains, expected to be significantly impaired in function. Localization of Miwi, Mvh and Mael to CBs depends on Tdrd6, and their absence most likely contributes significantly to the ghost-like phenotype of _Tdrd6_−/− CBs.
Emergence of defective CBs and a block of development of round into elongated spermatids strongly correlate in _Tdrd6_−/− mice. Whether the architectural deficiency of the CBs itself or an associated functional failure causes the developmental block, or whether another CB-independent function of Tdrd6 – as unlikely as it may be – causes the block, cannot yet be determined. We hypothesized that one of the most likely functions of the CBs, i.e. maintenance or processing of small RNAs, is affected by Tdrd6 deficiency. This was confirmed by showing altered miRNA expression in _Tdrd6_−/− testis. Many more miRNAs are up-regulated than down-regulated, which may indicate less turnover of miRNAs, and thus accumulation of these transcripts, or may be consequence of altered miRNA gene transcription. Indeed, we observed increased levels of pre- and pri-forms of the three miRNAs, which were analyzed in this regard. This suggests an unexpected direct or indirect control of Tdrd6 on miRNA precursor transcription and/or half-life. Since we did not find Tdrd6 in purified nuclei, an indirect involvement of Tdrd6 in regulating transcription or half-life appears most likely. Several families of miRNAs are aberrantly regulated, including the miR-29, miR-34, miR-463 and miR-489 families. The predicted targets of these miRNAs frequently include other RNA processing enzymes such as Dicer, DNA modifying enzymes such as DNA methyl transferases, and generally include many testis-specific RNAs (Bartel’s lab TargetScan program [52]). The miR-463 targets also Tdrd6 itself, besides many other RNAs, and the absence of Tdrd6 RNA may cause accumulation of this miRNA or its precursor. There is no correlation of aberrantly regulated miRNAs with CpG islands or miRNA transcriptional start sites (Suppl. Table 1). There is no strong preference for a certain chromosome, but relatively many of these miRNAs cluster on the X chromosome, which, however, is rich in miRNA clusters (19 % of known mouse testis miRNAs; [53]).
Contrasting mice deficient in Miwi2, Mili or Mael, there is no or only very weak activation of retrotransposons in _Tdrd6_−/− testes, indicating that Tdrd6 may represent a new class of CB proteins, specifically involved in CB formation and small RNA transcription and processing. This also indicates that despite malformed CBs and despite mislocalization of Miwi, Mvh, and Mael, protection from retrotransposon activation is still functional.
Conclusions
The characterization of mice deficient in the CB component Tdrd6 revealed unexpected findings with respect to the relationship between CB structure, retrotransposon activation and miRNA expression. Tdrd6 as a major CB component is necessary for CB formation including incorporation of several other CB proteins. A functional and properly structured CB is not required for protection from retrotransposon activation. Notably, more than 50 miRNAs are present at increased concentrations. This may be due to increased half-life of mature miRNAs caused by the non-functional CB, but it may also be caused by increased expression of pri-miRNAs as indicated by real-time PCR analysis. Since not only pri-miRNA, but also pre-miRNA and mature miRNAs accumulate in Tdrd6−/− spermatocytes, a function of Tdrd6 solely in processing of pri-miRNAs is unlikely. Rather, we suggest Tdrd6 to play an unexpected, probably indirect role in a nuclear process, i.e. in transcription.
Experimental Procedures
Antibody generation
Three Tdrd6 cDNA fragments (amino acid residues 52-171 (Tdrd6-Nterm), 265-373 (Tdrd6-265) and 2058-2182 (Tdrd6-Cterm)) were subcloned into the Escherichia coli expression vector pGEX-4T-3 (Invitrogen). The GST-tagged peptides were expressed and purified on glutathione agarose (Sigma). The Tdrd6 C-term was soluble, the Tdrd6-Nterm and Tdrd6-265 peptides were partially soluble under native conditions. A mixture of native and denatured proteins of the latter two was used for immunization of rabbits and polyclonal antibodies were generated. The obtained sera were tested by IB and specific antibodies purified on Tdrd6-Cterm-Sepharose resin, or Protein A-Sepharose Fast-flow (Invitrogen) for Tdrd6-Nterm and Tdrd6-mid. The resulting antibodies were designated as α-Nterm, α-265 (Tdrd6-265) and α-Cterm.
Protein extracts, immunobloting, immunoprecipitation
Cytoplasmic and nuclear extracts from tissue or sorted cells were prepared as described previously [37, 54]. For immunoblot analysis we used the following antibodies: rabbit polyclonal anti-Ddx4/Mvh (1 μg/ml, Abcam), rabbit polyclonal anti-PiwiL2 (1 μg/ml, Abcam), rabbit polyclonal anti-SMC3 (1 μg/ml [37]), rabbit polyclonal anti-RNF17 (kind gift from Jeremy Wang, UPenn), rabbit polyclonal anti-Mael (Abcam), rabbit-polyclonal anti-Hiwi (1 μg/ml) and anti-Sp1 (1 μg/ml, Bethyl Labs). For immunoprecipitation testes from wt or _Smc1β_−/− adult mice were Dounce-homogenized, lysed in RIPA buffer for 20 min on ice and then centrifuged to remove cellular debris. Cell lysates, diluted 1:5 in PBS, 0.1 % Tween-20, were incubated with either purified α-Cterm or α-265 antibodies coupled to Protein A-agarose for 16 h at 4°C. Rabbit antibodies purified from pre-immune serum were used as a control. The beads were then washed twice with incubation buffer and once with PBS. The retained proteins were resolved by SDS-PAGE and the gel stained with Coomassie Blue R. Polypeptides were excised from the gel and identified by mass spectrometry (MPI-CBG, Dresden, Germany).
Immunofluorescence
Immunofluorescence labeling of dried-down preparations and frozen sections of mouse testis was performed as described before [55]. For staining of individual germ cell types single-cell suspensions were prepared from testes as previously described [38] with minor modifications. Suspensions were sorted on Influx cell sorter (CYTOPEIA, Seattle, WA) with UV laser excitation wavelength 355nm and emission filters UV-Blue 460/50 and UV-Red 692/40 using Spigot 5.3.8 software. Appropriate numbers of each sorted cell population were spot-spun on slides using Cytospin 3 (ThermoShandon, USA). The following antibodies were used: rabbit polyconal anti-Tdrd6 N- and C-term, rabbit-polyclonal anti-Mvh (Abcam), rabbit polyclonal anti-PiwiL2 (Abcam), mouse monoclonal anti-SYCP3 [55]. For double immunostaining rabbit polyclonal antibodies were labeled using the Zenon Rabbit IgG Labeling Kits (Molecular probes, Invitrogen). Secondary antibodies used were Alexa-555-labeled donkey anti-mouse IgG (1:800, Molecular Probes, Invitrogen), Alexa-488-labeled goat anti-rabbit, (1:800, Molecular Probes, Invitrogen) and Cy5-labeled goat anti-mouse IgG (Jackson Immunochemicals), biotin-labeled donkey anti-mouse IgG (1:500, Dianova, Jackson Lab), Fluorescein isothiocyanate (FITC)-labeled ExtrAvidin® (1:1000, Sigma), Cy3-labeled goat anti-rabbit, (1:500, Dianova Jackson Lab). Nuclei or sorted cells were stained with 5 μg/ml Hoechst 33242 dye (Sigma) and nuclei of testis sections were visualized with DAPI.
Electron microscopy
Seminiferous tubules were dissected from mouse testis, fixed in 0.1 M phosphate buffer (pH 7.4) containing 2 % glutaraldehyde for 1.5 h and post-fixed in 1% osmium tetroxide in water for 1 h. Samples were dehydrated through a graded series of ethanol and thin-layer embedded in Epon/Araldite as described [56]. Ultrathin sections (70 nm) were stained using 2% uranyl acetate in 70% methanol followed by lead citrate and imaged in a TECNAI 12 transmission electron microscope (FEI) operated at 100 kV.
MiRNA expression analyses
Total testis RNA was isolated from 18 days old mice using Trizol reagent (InVitrogen) according to the manufacturer’s recommendations. MiRNA expression analyses was carried out using the miRXplore™ Standard Service (Miltenyi Biotech GmbH). Data were normalized by applying the 50th percentile of background intensity values and by performing the Lowess normalization to correct for dye bias. Then, the ratio of sample versus control was calculated for each spot, and the mean of the ratios of 4 spots calculated. Data were validated by quantitative RT-PCR according to [47]. Briefly, reverse transcription on total testis RNA was performed with an oligonucleotide (RT6-miR-x) that hybridizes to the 3′ end of the respective miRNA. For subsequent real-time qRT-PCR (QuantiTect SYBR Green PCR Kit, Qiagen)) another miRNA-specific 5′ primer as well as two pairs of universal primers for control (actin, GAPDH) were used. For primer sequences refer to supplemental table S1. Precursors of miRNAs were analyzed according to [57]. The results of real-time qRT-PCR assays presented are averages of three independent RNA preparations of which each sample was analyzed in triplicates. The ratio of these miRNA data to the combined average of the actin and GAPDH normalization controls are shown. Statistical analyses were performed using One-way ANOVA followed by Bonferroni post tests (GraphPad Prism software).
In Vitro Transcription and Translation
The TNT Quick coupled transcription/translation system from Promega was used for the in vitro expression of recombinant Mili-myc, Miwi-myc and HA-Tdrd6 as per the manufacturer’s instructions. Total testis RNA was used to generate cDNA by reverse transcription. Fragments of the Tdrd6 cDNA were amplified with high-fidelity Pfx polymerase, sequenced and assembled in vector pcDNA3.1-3xHA-Tdrd6. In brief, 1 μl (1.0 μg) of supercoiled plasmid carrying the recombinant gene of interest under the T7 promoter was used in a 25 μl TNT reaction containing 20 μl TNT Quick Master Mix, 1.5 μl cold amino acids, 1mM methionine, 1.0 μl RNasein and 1.5 μl RNase-free water. The reaction was incubated at 30 °C for 90 min and aliqots of the products separated on a 7.5 % denaturing polyacrylamide gel, followed by immunoblot analysis.
Additional information on Experimental Procedures can be found in the Supplemental Information.
Supplementary Material
01
Suppl. Fig. 1 Analysis of Tdrd6 RNA expression. (A) Comparison of Tdrd6 expression and other germ cell-specific genes in male and female gonads. The label at the bottom of each lane indicates the tissue or cell type used for total RNA isolation and the age of the mouse. E15.5, E18.5 and E19.5 designate embryos at 15.5, 18.5 and19.5 days post coitum (dpc), respectively. Neonat - newborn female; Primary s-cytes - sorted primary spermatocytes (B) Time-course of Tdrd6 mRNA expression in juvenile and adult testis assayed by RT-PCR. Actin was used as loading control.
Suppl. Fig. 2 Immunoblot analysis of Tdrd6 protein expression in cytoplasmic or nuclear extract. Spermatocyte and spermatid populations from adult testis were separated by FASC sorting. Tdrd6 expression in 10 μg cytoplasmic and 5 μg nuclear extract (including CBs, which co-sediment with nuclei) was analysed with α-265 antibody.
Suppl. Fig. 3 Tdrd6 co-localizes with Miwi and not with RNF17 loci. Cytospun primary spermatocytes or secondary spermatocytes and spermatids were co-stained with either α-Hiwi/Miwi or α-RNF17 (green) and α-Cterm antibodies (red). Synaptonemal complexes on chromosomes are marked with α-Sycp3 (blue) and nuclei are stained with DAPI.
Suppl. Fig. 4 Targeted disruption of the_Tdrd6_ gene and generation of Tdrd6 null/hCD4 knock-in allele. (A) Schematic representation of the wild-type allele, the targeting vector and the mutated allele. The numbered white boxes (1–5) denote the five coding exons of the gene. The targeting vector includes the PGK_puro_ gene (puromycin resistance) for positive selection and the PGK-DTA (diphteria toxin) gene for negative selection as well as the hCD4 gene in frame with the Tdrd6 5′ UTR and ATG (start) codon. (B) Southern blot analysis of representative offspring from heterozygous mating. The wild-type allele produces a 6.6 kb _Bgl_II product, while the disrupted allele gives rise to a 4.25 kb band with a 1.4 kb 5′-end hybridization probe. (C) Western blot analysis of the Tdrd6 in testes of the same offspring using antibodies against the N-terminus of Tdrd6. Lysates (10 μg of protein per lane) of the testes or kidney were loaded on the gel. The +/+, +/− and −/− indicate wildtype, heterozygous and homozygous mutant testes or kidney, respectively. Sp1 was used as loading control. (D) FACS analysis of anti-hCD4 stained total testis cell suspension and sorted germ cell subpopulations from Tdrd6+/− mice. Kidney cell suspension and human peripheral blood monocytes (hPBMC), which normally express CD4, were used as controls.
Suppl. Fig. 5 Immunoblot of nuclear extracts from testis and kidney from_Tdrd6_+/+, Tdrd6+/− and Tdrd6_−/_− mice. Extracts were probed with specific antibodies for chromatoid body markers Tdrd6, PiwiL2/Mili, Mvh, Mael, Hiwi/Miwi, and for control Rnf17 and Sp1.
Suppl. Fig. 6 Electron microscopy of seminiferous tubules. In stage I seminiferous tubules of Tdrd6+/− (A) and _Tdrd6_−/− mice (B) elongated spermatids are apparent. Due to a block in spermiogenesis, stage V seminiferous tubules of _Tdrd6_−/− mice (D) lack elongated spermatids, while they are present in Tdrd6+/− mice (C) (bar 10 μm).
Suppl. Fig. 7 Differentially expressed precursor and mature miRNAs in testes of Tdrd6−_/_− mice. Analysis was performed with total testis RNA from 18 days old Tdrd6−/− and Tdrd6+/− mice. Precursors (Pri- and pre-miRNAs) were analyzed for miR-29b, miR-29c and miR-34a. Mature miRNAs were analyzed using primer for miR-29b, miR-29c, miR-34a, miR-34c, miR-322 and miR-342. The values for the Tdrd6+/− control were set to 1.0 and the respective increase or decrease in Tdrd6−/− mice are represented as mean +/− SEM. (*** P value < 0.001, ** P value 0.001 to 0.01, * P value 0.01 to 0.05, ns not significant P value > 0.05)
Suppl. Table 1: PCR primer used in this study.
Suppl. Table 2: Differentially expressed miRNAs in testes of 18 days old Tdrd6−**_/_**− mice. Shown are the miRNA names and GeneIDs, the fold change observed on microarrays, the genomic localtion, the miRNA gene cluster, features such as known transcriptional start sites or CpG islands, and the location in intragenic or intergenic regions.
Acknowledgments
We thank Drs. Ekaterina Revenkova and Attila Toth for critical reading of the manuscript. We thank Dr. S. Kuramochi-Miyagawa for the kind gift of myc-tagged Miwi and Mili. This work was in part supported by the NIH/NIGMS (R01-062517), by the DFG (JE150/4-1), and the Deutsche Krebshilfe.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Andre J, Rouiller C. The ultrastructure of the vitelline body in the oocyte of the spider Tegenaria parietina. J Biophys Biochem Cytol. 1957;3:977–984. doi: 10.1083/jcb.3.6.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fawcett DW, Eddy EM, Phillips DM. Observations on the fine structure and relationships of the chromatoid body in mammalian spermatogenesis. Biology of reproduction. 1970;2:129–153. doi: 10.1095/biolreprod2.1.129. [DOI] [PubMed] [Google Scholar]
- 3.Russell L, Frank B. Ultrastructural characterization of nuage in spermatocytes of the rat testis. The Anatomical record. 1978;190:79–97. doi: 10.1002/ar.1091900108. [DOI] [PubMed] [Google Scholar]
- 4.Soderstrom KO, Parvinen M. Transport of material between the nucleus, the chromatoid body and the Golgi complex in the early spermatids of the rat. Cell and tissue research. 1976;168:335–342. doi: 10.1007/BF00215311. [DOI] [PubMed] [Google Scholar]
- 5.Comings DE, Okada TA. The chromatoid body in mouse spermatogenesis: evidence that it may be formed by the extrusion of nucleolar components. Journal of ultrastructure research. 1972;39:15–23. doi: 10.1016/s0022-5320(72)80003-0. [DOI] [PubMed] [Google Scholar]
- 6.Soderstrom KO. Formation of chromatoid body during rat spermatogenesis. Zeitschrift fur mikroskopisch-anatomische Forschung. 1978;92:417–430. [PubMed] [Google Scholar]
- 7.Parvinen M, Parvinen LM. Active movements of the chromatoid body. A possible transport mechanism for haploid gene products. The Journal of cell biology. 1979;80:621–628. doi: 10.1083/jcb.80.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eddy EM. Cytochemical observations on the chromatoid body of the male germ cells. Biology of reproduction. 1970;2:114–128. doi: 10.1095/biolreprod2.1.114. [DOI] [PubMed] [Google Scholar]
- 9.Ventela S, Toppari J, Parvinen M. Intercellular organelle traffic through cytoplasmic bridges in early spermatids of the rat: mechanisms of haploid gene product sharing. Molecular biology of the cell. 2003;14:2768–2780. doi: 10.1091/mbc.E02-10-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parvinen M, Salo J, Toivonen M, Nevalainen O, Soini E, Pelliniemi LJ. Computer analysis of living cells: movements of the chromatoid body in early spermatids compared with its ultrastructure in snap-frozen preparations. Histochemistry and cell biology. 1997;108:77–81. doi: 10.1007/s004180050148. [DOI] [PubMed] [Google Scholar]
- 11.Peruquetti RL, Assis IM, Taboga SR, de Azeredo-Oliveira MT. Meiotic nucleolar cycle and chromatoid body formation during the rat (Rattus novergicus) and mouse (Mus musculus) spermiogenesis. Micron. 2007 doi: 10.1016/j.micron.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Biggiogera M, Fakan S, Leser G, Martin TE, Gordon J. Immunoelectron microscopical visualization of ribonucleoproteins in the chromatoid body of mouse spermatids. Molecular reproduction and development. 1990;26:150–158. doi: 10.1002/mrd.1080260209. [DOI] [PubMed] [Google Scholar]
- 13.Figueroa J, Burzio LO. Polysome-like structures in the chromatoid body of rat spermatids. Cell and tissue research. 1998;291:575–579. doi: 10.1007/s004410051027. [DOI] [PubMed] [Google Scholar]
- 14.Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci U S A. 2006;103:2647–2652. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 16.Chuma S, Hiyoshi M, Yamamoto A, Hosokawa M, Takamune K, Nakatsuji N. Mouse Tudor Repeat-1 (MTR-1) is a novel component of chromatoid bodies/nuages in male germ cells and forms a complex with snRNPs. Mech Dev. 2003;120:979–990. doi: 10.1016/s0925-4773(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 17.Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan J, Goodheart M, Chuma S, Nakatsuji N, Page DC, Wang PJ. RNF17, a component of the mammalian germ cell nuage, is essential for spermiogenesis. Development (Cambridge, England) 2005;132:4029–4039. doi: 10.1242/dev.02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hosokawa M, Shoji M, Kitamura K, Tanaka T, Noce T, Chuma S, Nakatsuji N. Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: domain composition, intracellular localization, and function in male germ cells in mice. Developmental biology. 2007;301:38–52. doi: 10.1016/j.ydbio.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 20.Smith JM, Bowles J, Wilson M, Teasdale RD, Koopman P. Expression of the tudor-related gene Tdrd5 during development of the male germline in mice. Gene Expr Patterns. 2004;4:701–705. doi: 10.1016/j.modgep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Thomson T, Lasko P. Tudor and its domains: germ cell formation from a Tudor perspective. Cell Res. 2005;15:281–291. doi: 10.1038/sj.cr.7290297. [DOI] [PubMed] [Google Scholar]
- 22.Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buhler D, Raker V, Luhrmann R, Fischer U. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum Mol Genet. 1999;8:2351–2357. doi: 10.1093/hmg/8.13.2351. [DOI] [PubMed] [Google Scholar]
- 24.Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, Sattler M. SMN tudor domain structure and its interaction with the Sm proteins. Nat Struct Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 25.Sprangers R, Groves MR, Sinning I, Sattler M. High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. J Mol Biol. 2003;327:507–520. doi: 10.1016/s0022-2836(03)00148-7. [DOI] [PubMed] [Google Scholar]
- 26.Cote J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J Biol Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- 27.Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 28.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anne J, Mechler BM. Valois, a component of the nuage and pole plasm, is involved in assembly of these structures, and binds to Tudor and the methyltransferase Capsuleen. Development. 2005;132:2167–2177. doi: 10.1242/dev.01809. [DOI] [PubMed] [Google Scholar]
- 30.Ramos A, Hollingworth D, Adinolfi S, Castets M, Kelly G, Frenkiel TA, Bardoni B, Pastore A. The structure of the N-terminal domain of the fragile X mental retardation protein: a platform for protein-protein interaction. Structure. 2006;14:21–31. doi: 10.1016/j.str.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 31.Huyen Y, Zgheib O, DiTullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 32.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 33.Ponting CP. Tudor domains in proteins that interact with RNA. Trends Biochem Sci. 1997;22:51–52. doi: 10.1016/s0968-0004(96)30049-2. [DOI] [PubMed] [Google Scholar]
- 34.Scanlan MJ, Welt S, Gordon CM, Chen YT, Gure AO, Stockert E, Jungbluth AA, Ritter G, Jager D, Jager E, et al. Cancer-related serological recognition of human colon cancer: identification of potential diagnostic and immunotherapeutic targets. Cancer Res. 2002;62:4041–4047. [PubMed] [Google Scholar]
- 35.Bellve AR, Millette CF, Bhatnagar YM, O’Brien DA. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J Histochem Cytochem. 1977;25:480–494. doi: 10.1177/25.7.893996. [DOI] [PubMed] [Google Scholar]
- 36.Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R. Cohesin SMC1[beta] is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol. 2004;6:555–562. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- 37.Revenkova E, Eijpe M, Heyting C, Gross B, Jessberger R. Novel Meiosis-Specific Isoform of Mammalian SMC1. Mol Cell Biol. 2001;21:6984–6998. doi: 10.1128/MCB.21.20.6984-6998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bastos H, Lassalle B, Chicheportiche A, Riou L, Testart J, Allemand I, Fouchet P. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A. 2005;65:40–49. doi: 10.1002/cyto.a.20129. [DOI] [PubMed] [Google Scholar]
- 39.Gorski K, Carneiro M, Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986;47:767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- 40.Noce T, Okamoto-Ito S, Tsunekawa N. Vasa homolog genes in mammalian germ cell development. Cell structure and function. 2001;26:131–136. doi: 10.1247/csf.26.131. [DOI] [PubMed] [Google Scholar]
- 41.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 42.Bedinger P, Moriarty A, von Borstel RC, 2nd, Donovan NJ, Steimer KS, Littman DR. Internalization of the human immunodeficiency virus does not require the cytoplasmic domain of CD4. Nature. 1988;334:162–165. doi: 10.1038/334162a0. [DOI] [PubMed] [Google Scholar]
- 43.Costa Y, Speed RM, Gautier P, Semple CA, Maratou K, Turner JM, Cooke HJ. Mouse MAELSTROM: the link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Human molecular genetics. 2006;15:2324–2334. doi: 10.1093/hmg/ddl158. [DOI] [PubMed] [Google Scholar]
- 44.Parvinen LM, Jokelainen P, Parvinen M. Chromatoid body and haploid gene activity: actinomycin D induced morphological alterations. Hereditas. 1978;88:75–80. doi: 10.1111/j.1601-5223.1978.tb01605.x. [DOI] [PubMed] [Google Scholar]
- 45.Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 46.Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharbati-Tehrani S, Kutz-Lohroff B, Bergbauer R, Scholven J, Einspanier R. miR-Q: a novel quantitative RT-PCR approach for the expression profiling of small RNA molecules such as miRNAs in a complex sample. BMC Mol Biol. 2008;9:34. doi: 10.1186/1471-2199-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 49.Tsai-Morris CH, Sheng Y, Lee E, Lei KJ, Dufau ML. Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is essential for spermatid development and completion of spermatogenesis. Proc Natl Acad Sci U S A. 2004;101:6373–6378. doi: 10.1073/pnas.0401855101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng W, Lin H. miwi, a Murine Homolog of piwi, Encodes a Cytoplasmic Protein Essential for Spermatogenesis. Developmental Cell. 2002;2:819. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 51.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 52.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ro S, Park C, Sanders KM, McCarrey JR, Yan W. Cloning and expression profiling of testis-expressed microRNAs. Dev Biol. 2007;311:592–602. doi: 10.1016/j.ydbio.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol. 2004;6:555–562. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- 56.Muller-Reichert T, Hohenberg H, O’Toole ET, McDonald K. Cryoimmobilization and three-dimensional visualization of C. elegans ultrastructure. J Microsc. 2003;212:71–80. doi: 10.1046/j.1365-2818.2003.01250.x. [DOI] [PubMed] [Google Scholar]
- 57.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
Suppl. Fig. 1 Analysis of Tdrd6 RNA expression. (A) Comparison of Tdrd6 expression and other germ cell-specific genes in male and female gonads. The label at the bottom of each lane indicates the tissue or cell type used for total RNA isolation and the age of the mouse. E15.5, E18.5 and E19.5 designate embryos at 15.5, 18.5 and19.5 days post coitum (dpc), respectively. Neonat - newborn female; Primary s-cytes - sorted primary spermatocytes (B) Time-course of Tdrd6 mRNA expression in juvenile and adult testis assayed by RT-PCR. Actin was used as loading control.
Suppl. Fig. 2 Immunoblot analysis of Tdrd6 protein expression in cytoplasmic or nuclear extract. Spermatocyte and spermatid populations from adult testis were separated by FASC sorting. Tdrd6 expression in 10 μg cytoplasmic and 5 μg nuclear extract (including CBs, which co-sediment with nuclei) was analysed with α-265 antibody.
Suppl. Fig. 3 Tdrd6 co-localizes with Miwi and not with RNF17 loci. Cytospun primary spermatocytes or secondary spermatocytes and spermatids were co-stained with either α-Hiwi/Miwi or α-RNF17 (green) and α-Cterm antibodies (red). Synaptonemal complexes on chromosomes are marked with α-Sycp3 (blue) and nuclei are stained with DAPI.
Suppl. Fig. 4 Targeted disruption of the_Tdrd6_ gene and generation of Tdrd6 null/hCD4 knock-in allele. (A) Schematic representation of the wild-type allele, the targeting vector and the mutated allele. The numbered white boxes (1–5) denote the five coding exons of the gene. The targeting vector includes the PGK_puro_ gene (puromycin resistance) for positive selection and the PGK-DTA (diphteria toxin) gene for negative selection as well as the hCD4 gene in frame with the Tdrd6 5′ UTR and ATG (start) codon. (B) Southern blot analysis of representative offspring from heterozygous mating. The wild-type allele produces a 6.6 kb _Bgl_II product, while the disrupted allele gives rise to a 4.25 kb band with a 1.4 kb 5′-end hybridization probe. (C) Western blot analysis of the Tdrd6 in testes of the same offspring using antibodies against the N-terminus of Tdrd6. Lysates (10 μg of protein per lane) of the testes or kidney were loaded on the gel. The +/+, +/− and −/− indicate wildtype, heterozygous and homozygous mutant testes or kidney, respectively. Sp1 was used as loading control. (D) FACS analysis of anti-hCD4 stained total testis cell suspension and sorted germ cell subpopulations from Tdrd6+/− mice. Kidney cell suspension and human peripheral blood monocytes (hPBMC), which normally express CD4, were used as controls.
Suppl. Fig. 5 Immunoblot of nuclear extracts from testis and kidney from_Tdrd6_+/+, Tdrd6+/− and Tdrd6_−/_− mice. Extracts were probed with specific antibodies for chromatoid body markers Tdrd6, PiwiL2/Mili, Mvh, Mael, Hiwi/Miwi, and for control Rnf17 and Sp1.
Suppl. Fig. 6 Electron microscopy of seminiferous tubules. In stage I seminiferous tubules of Tdrd6+/− (A) and _Tdrd6_−/− mice (B) elongated spermatids are apparent. Due to a block in spermiogenesis, stage V seminiferous tubules of _Tdrd6_−/− mice (D) lack elongated spermatids, while they are present in Tdrd6+/− mice (C) (bar 10 μm).
Suppl. Fig. 7 Differentially expressed precursor and mature miRNAs in testes of Tdrd6−_/_− mice. Analysis was performed with total testis RNA from 18 days old Tdrd6−/− and Tdrd6+/− mice. Precursors (Pri- and pre-miRNAs) were analyzed for miR-29b, miR-29c and miR-34a. Mature miRNAs were analyzed using primer for miR-29b, miR-29c, miR-34a, miR-34c, miR-322 and miR-342. The values for the Tdrd6+/− control were set to 1.0 and the respective increase or decrease in Tdrd6−/− mice are represented as mean +/− SEM. (*** P value < 0.001, ** P value 0.001 to 0.01, * P value 0.01 to 0.05, ns not significant P value > 0.05)
Suppl. Table 1: PCR primer used in this study.
Suppl. Table 2: Differentially expressed miRNAs in testes of 18 days old Tdrd6−**_/_**− mice. Shown are the miRNA names and GeneIDs, the fold change observed on microarrays, the genomic localtion, the miRNA gene cluster, features such as known transcriptional start sites or CpG islands, and the location in intragenic or intergenic regions.