Imaging individual microRNAs in single mammalian cells in situ (original) (raw)
Abstract
MicroRNAs (miRNAs) are potent negative regulators of gene expression that have been implicated in most major cellular processes. Despite rapid advances in our understanding of miRNA biogenesis and mechanism, many fundamental questions still remain regarding miRNA function and their influence on cell cycle control. Considering recent reports on the impact of cell-to-cell fluctuations in gene expression on phenotypic diversity, it is likely that looking at the average miRNA expression of cell populations could result in the loss of important information connecting miRNA expression and cell function. Currently, however, there are no efficient techniques to quantify miRNA expression at the single-cell level. Here, a method is described for the detection of individual miRNA molecules in cancer cells using fluorescence in situ hybridization. The method combines the unique recognition properties of locked nucleic acid probes with enzyme-labeled fluorescence. Using this approach, individual miRNAs are identified as bright, photostable fluorescent spots. In this study, miR-15a was quantified in MDA-MB-231 and HeLa cells, while miR-155 was quantified in MCF-7 cells. The dynamic range was found to span over three orders of magnitude and the average miRNA copy number per cell was within 17.5% of measurements acquired by quantitative RT-PCR.
INTRODUCTION
MicroRNAs (miRNAs) are an abundant class of short noncoding RNAs, ∼18–25 nt in length, that act as potent negative regulators of gene expression. To date, there have been over 600 miRNAs identified in humans (http://www.sanger.ac.uk/Software/Rfam/mirna/) (1) and they are predicted to influence the regulation of over one-third of all human genes (2). MicroRNAs have been implicated in most major cellular processes including proliferation, apoptosis, developmental timing, hematopoiesis and organogenesis (3). Therefore, it is not surprising that miRNAs have also already been linked to a number of diseases such as cancer (4–10), neurological diseases (11), viral diseases (12) and metabolic diseases (13). In general, microRNAs function as post-transcriptional regulators of gene expression by either triggering mRNA cleavage or repressing translation (14); however, evidence that miRNAs direct the localization of mRNAs to P-bodies introduces a third possibility, the sequestration of mRNA from translational machinery (15).
Despite the rapid advances in our understanding of miRNA biogenesis and mechanism, many fundamental questions still remain regarding miRNA function and their influence on central signaling pathways and cell cycle control. Considering recent reports on the complex stochastic nature of gene expression in mammalian cells and the impact of these fluctuations on phenotypic diversity, it is likely that looking at the average miRNA expression of cell populations could result in the loss of important information connecting miRNA expression and cell function. Therefore, it is predicted that insight into the physiologic function of miRNA will require miRNA abundance to be quantified at the single-cell level.
Currently, numerous technique are available for studying miRNA expression levels, including northern blot, microarrays, RNA-primed array-based Klenow enzyme assay (RAKE) and mirVana miRNA labeling and detection kits (Ambion). At least one assay that has been reported in the literature even exhibits single molecule sensitivity (16); however, in general, these techniques all require the lysis of a population of cells and thus do not allow cell-to-cell variations in miRNA expression to be elucidated. Microdissection techniques could potentially be combined with the aforementioned miRNA detection strategies, or single-cell PCR could be used to allow for single-cell analysis; however, these methods are inefficient, laborious and often result in poor sample quality and erroneous conclusions. An alternative option is to perform fluorescence in situ hybridization (FISH). With the recent introduction of locked nucleic acid (LNA) oligonucleotides as hybridization probes, miRNA-FISH has become a powerful technique for imaging the spatial localization of miRNA at the tissue, cellular and even subcellular level (17–21). LNA probes exhibit a remarkable affinity and specificity against RNA targets, allowing for the discrimination of even single-base mismatches (22–25). Unfortunately, miRNA-FISH generally cannot be used to provide accurate quantitative measures of miRNA expression, but rather is typically limited to providing a qualitative assessment of miRNA localization patterns and tissue distribution.
Recently, several quantitative FISH strategies have been developed that allow single messenger RNA (mRNA) to be visualized. Specifically, target mRNA transcripts are hybridized with a small number of heavily labeled probes (e.g. five oligonucleotide probes, each labeled with five fluorophores) or a larger number of singly labeled probes (e.g. 96 oligonucleotide probes) (26–28). The large number of fluorescent labels generates a bright fluorescent spot that indicates the location of each mRNA. These bright spots can then be counted to determine the number of mRNA transcripts in single cells. Unfortunately, it is not possible to extend this approach to miRNA, since their short length cannot accommodate the hybridization of more than one to two probes.
Here, we describe an alternative quantitative FISH approach that does allow individual miRNAs to be visualized within single cells. Our strategy combines LNA-based FISH with enzyme-labeled fluorescence (ELF) signal amplification (29). ELF is a process whereby cleavage of a pro-luminescent substrate by phosphatase yields a brilliant, yellow-green fluorescent product at the site of enzymatic activity. The ELF precipitate is not only photostable compared to commonly used fluorophores, but also results in labeling that is up to 40 times brighter than signals achieved with probes directly labeled with fluorophores (29). To validate the specificity of our methodology we performed mRNA-FISH using both multiple-labeled probes and our LNA-ELF system simultaneously. As expected, these two approaches exhibited a high degree of correlation. Subsequently, we tested our ability to use the LNA-ELF approach to visualize individual miRNAs in single cells. Various cell lines with a wide range of target miRNA expression were studied to establish sensitivity and dynamic range. Results were compared with quantitative RT-PCR to assess accuracy.
MATERIALS AND METHODS
Plasmid construction
A plasmid containing the Firefly luciferase open reading frame followed by 24 MS2 binding sites was constructed using standard cloning techniques. Specifically, the Luciferase coding sequence was amplified from pGL3 basic (Promega) by PCR using the following primers, forward: 5′-GATCAAGCTTATGGAAGACGCCAAAAACATAAAG-3′ and reverse: 5′GATCGGATCCTTACACGGCGATCTTTCCGCCCTTCTT-3′. The underlined regions correspond to HindIII and BamHI restriction sites, which were to introduce the PCR product into the host vector, pcDNA 3.1(+) (Invitrogen, Carlsbad, California, United States). The 24 MS binding sequence was cut from plasmid pSL MS2 24 (a generous gift from Dr. Robert Singer's lab, Albert Einstein College of Medicine) with the restriction enzymes BamHI and XhoI and ligated into the pcDNA-luciferase construct. The new plasmid, pcDNA-Luc-MS2-24, was amplified in stbl2 cells (Invitrogen) and purified using the Qiagen Maxiprep system. The sequence of the final plasmid was confirmed by DNA sequencing.
Cell culture
Human cervical carcinoma HeLa cells and human breast cancer MCF-7 cells (ATCC) were cultured in Dulbecco's MEM (DMEM, high glucose, with L-glutamine and phenol red) supplied with 0.1 mM nonessential amino acids (NEAA) and 10% fetal bovine serum (FBS). MDA-MB-231 human breast cancer cells (ATCC) were cultured in L-15 medium supplied with 2 mM glutamine and 15% FBS. HeLa and MCF-7 cells were maintained at 37°C with 5% CO2 and MDA-MB-231 cells were maintained at 37°C with 0% CO2.
Creation of stable cell lines
HeLa cells were plated in 24-well plates at a density of 8 × 104 cells in antibiotics-free medium. After 24 h (∼90–95% confluency), the cells were transfected with pcDNA-Luc-MS2-24 using Lipofectamine 2000 (Invitrogen). Specifically, 0.8 μg of plasmid DNA was diluted into 50 μl of OPTI-MEM I Reduced Serum medium. Lipofectamine 2000 reagent (1.5 μl) was also diluted into 50 μl OPTI-MEM medium and both samples were incubated for 5 min at room temperature. The diluted lipofectamine was then combined with the plasmid and the mixture was incubated at room temperature for 20 min to allow DNA-Lipofectamine complexes to form. Immediately before adding the DNA-Lipofectamine complexes to the cells, the growth medium on the HeLa cells was replaced with 250 μl of medium without serum. Cells were incubated at 37°C in a humidified 5% CO2 atmosphere. After 24 h, the cells were passaged at a 1:8 ratio into fresh growth medium containing 750 μg/ml G418 selection agent. Single colonies were picked after 3–4 days. Positive colonies were maintained in 200 μg/ml G418 under normal growth conditions, as described above. All stable cells were screened for luciferase activity using One Step Luciferase Assay kit for detection of firefly luciferase (Promega) according to the manufacturer's instructions. Furthermore, mRNA levels were determined via quantitative reverse transcriptional (RT)-PCR as described below.
FISH probes
An oligonucleotide probe that was complementary to the MS2 binding domain was purchased from Integrated DNA Technologies. Each probe was ordered with two Cy3 dyes (Amersham Biosciences) as follows: 5′-/5Cy3/CTGCAGACATGGGTGATCCTCAT/iCy3/GTTTTCTAG-3′ (IDT). An LNA probe complementary to the luciferase coding region was purchased from Exiqon and was ordered with digoxigenin at its 3′-end, 5′-TCCGCAAACACAACTCCTCCGC/3dig/-3′ (Exiqon). The LNA-binding domain on the luciferase transcript was within 100 bases of the first MS2 binding domain so that the two probes were in close proximity. The probes used in miRNA FISH were also purchased from Exiqon with a digoxigenin at the 3′-end. The specific probes utilized were 5′-CACAAACCATTATGTGCTGCTA-3′ for miR-15a and 5′-CCCCTATCACGATTAGCATTAA-3′ for miR-155.
Multiple probe FISH and LNA-ELF-FISH
Cells were seeded into multi-chambered coverglass slides (Lab-Tek, Nalge Nunc, Rochester, New York, United States) and incubated under normal growth conditions overnight, reaching 50–70% confluency. The cells then fixed with 4% formaldehyde for 30 min at room temperature, washed three times with 1× PBS, and permeabilized at 4°C in 70% ethanol overnight. Hybridization with the LNA probe (10 nM) was carried out at 20–22°C below the melting temperature of the probe for 1 hour after incubation in prehybridization buffer (25% formamide, 0.05 M EDTA, 4× SSC, 10% dextran sulfate, 1× Denhardt's solution, 0.5 mg/ml Escherichia coli tRNA and 0.5 mg/ml RVC) for 2 h at 60°C. The optimal level of formamide used during hybridization and washing for maximal signal-to-background was empirically determined to be 25%. After three stringent washes in 4× SSC, 2× SSC and 1× SSC, the cells were incubated in prehybridization buffer again and probed with the MS2 probe at a concentration of 50 nM at 37°C overnight. After three stringent washes, as described above, the cell samples were subject to ELF using the ELF 97 mRNA In Situ Hybridization Kit (Molecular Probes, Inc, Eugene, OR, USA), according to the manufacturer's instructions. Briefly, the cells were incubated in blocking buffer from the ELF 97 mRNA In Situ Hybridization Kit for 1 h at room temperature. Then, 2 μg/ml anti-DIG antibody (Jackson ImmunoResearch) in blocking buffer was added to the cells and incubated at room temperature for 1 h. After three washes in 1× wash buffer, signals were amplified in ELF 97 phosphatase substrate working solution for 10–15 min. For signal preservation, the cell samples were quickly washed with 1× wash buffer and postfixed by incubating the slides in post-fixation solution (2% formaldehyde, 20 mg/ml BSA in 1× PBS) for 30 min at room temperature. The slides were then counterstained in 1 μg/ml Hoechst 33342 and mounted in mounting solution.
Control experiments were conducted using the identical procedure as described above except only a single hybridization step was performed, either with the LNA probes (i.e. LNA-ELF-FISH) or with the MS2 probes (multiple probe FISH). ELF was also only performed as deemed necessary.
LNA-ELF-FISH with EDC treatment
After the cells had been fixed in 4% paraformaldehyde for 30 min at room temperature and permeabilized in 70% ethanol at 4°C overnight, cells were rehydrated and washed three times with 1× PBS. To remove residual phosphate from the PBS washes, slides were incubated twice for 10 min in a freshly prepared solution containing 0.13 M 1-methylimidazole, 300 mM NaCl, pH 8.0 adjusted with HCl. Then 0.16M l-ethyl-3-(3–dimethylaminopropyl) carbodiimide (EDC) (Sigma) was added to the cells and incubated for 1 h at 25°C. The slides were washed in 0.2% (w/v) glycine/TBS and then washed twice in TBS for prehybridization. Subsequent LNA hybridization and ELF signal amplifications steps were carried out as described above.
Conventional LNA-FISH
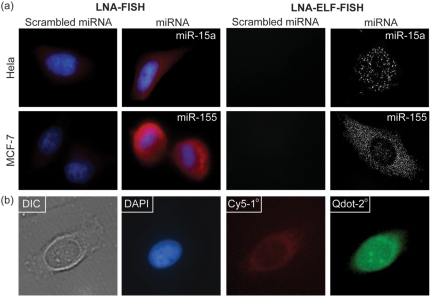
Conventional LNA-FISH (i.e. no ELF signal amplification) was conducted similar to the procedure described above. However, after incubating the cells in the blocking buffer at room temperature for 1 h, 2 μg/ml anti-DIG antibody (Jackson ImmunoResearch) in blocking buffer was added to the cells and incubated in the cold room overnight. After three washes in 1× wash buffer, Texas red-labeled secondary antibody was added and incubated at room temperature for 3 h. The slides were then counterstained in 0.1 μg/ml DAPI (Invitrogen) and mounted in mounting solution.
An analogous experiment was conducted using 2 μg/ml Cy5-labeled anti-DIG antibody (Jackson ImmunoResearch) as the primary antibody and 0.02 μM Qdot® 565 goat F(ab′)2 antimouse IgG conjugate (H+L) as the secondary antibody (Invitrogen).
Image acquisition and analysis
Following in situ hybridization, cells were imaged using an Olympus IX81 motorized inverted fluorescence microscope equipped with a back-illuminated EMCCD camera (Andor), an X-cite 120 excitation source (EXFO) and Sutter excitation and filter wheels. A UPLN 60× oil-immersion objective, N.A. 0.9, was used for all imaging experiments. IPLab acquisition software was used to acquire the 2D and 3D images. Briefly, after randomly selecting cells in a field, a 3D stack viewed image was taken with 0.3 μm increments in the _z_-direction and a total of 35 sections. After 3D deconvolution of the images in IPLab using AutoQuant plug-in software, a 2D image was constructed in IPLab using a maximum-intensity merged image. Images were then opened in ImageJ and processed using the following commands: (I) Process -> Sharpen, (II) Image -> type -> 8-bit and (III) Process -> binary -> make binary. The total number of isolated signals was then counted in ImageJ using the particle analysis counter program (Analyze -> analyze particles).
To evaluate the co-localization between ELF signals and the signal elicited by the binding of multiple Cy3 labeled probes to the same target, images of Cy3 fluorescence and the ELF signal were acquired using IPLab. Only 2D images of the focal plane were acquired due to the rapid photobleaching of Cy3. All 2D images were deconvolved in IPLab using AutoQuant plug-in software. Images were then processed in ImageJ as described above. After overlapping the corresponding Cy3 and ELF images, the number of col-ocalized and nonco-localized signals was then counted manually.
Quantitative RT-PCR of mRNA
Stable HeLa cell lines expressing Luc-MS2 were analyzed using a TaqMan gene expression Assay (Applied Biosystems, Foster City, CA, USA) under conditions defined by the supplier. Luc-MS2 DNA standards were cut from the plasmid pcDNA Luc-MS2 24 with restriction enzymes BamHI/XhoI. After gel purification, Luc-MS2 24 DNA concentration was measured and calculated. After serial dilution, Luc-MS2 was used as a DNA standard for the real time PCR. Total mRNA was isolated from 106 cells using the mirVana miRNA Isolation Kit (Ambion Inc., Austin, TX, USA) according to the manufacturer's instructions. Total mRNA was then reverse-transcribed by using Superscript First-Strand Synthesis Kit for RT-PCR (Invitrogen) according to supplier's instruction. cDNA was quantified by real-time PCR on the ABI Prism 7300 Sequence Detection System (Applied Biosystems). Each sample was run in triplicates and each PCR experiment included three non-template control wells. Each cDNA generated was amplified by quantitative PCR by using sequence-specific primers and probe designed by the supplier (Applied Biosystems). The 20-μl PCR included 10 μl of 2× TaqMan Fast Universal PCR Master Mix (No AmpErase UNG), 1 μl of each 20× TaqMan Gene Expression Assay Mix, 0.2 μl 1 U/μl SuperTaq(Ambion) and 2 μl of reverse transcription (RT) product. The reactions were run in a fast mode as follows: 95°C for 20 s, followed by 40 cycles of 95°C for 1 s and 60°C for 20 s.
Quantitative RT-PCR of miRNA
Total RNA was isolated from 106 cells using the mirVana miRNA Isolation Kit (Ambion Inc., Austin, TX, USA) according to the manufacturer's instructions. RNA concentrations were determined using a Cary100 UV-Vis spectrophotometer (Varian). Reverse transcription was performed on 0.5 μg total RNA samples using mirVana qRT-PCR miRNA detection kit (Ambion Inc) as suggested in the manual-2 μj _mir_Vana 5× RT Buffer, 1 μl 1 × _mir_Vana RT Primer, 25 ng RNA in 1-μl Nuclease-free Water, 0.4 μl ArrayScript Enzyme Mix, and enough nuclease-free water to bring the final volume up to 10 μl. Samples were incubated for 30 min at 37°C and then for 10 min at 95°C.
Subsequently, real-time PCR was performed according to the manufacturer's instructions. RNA oligonucleotides with the same sequence as endogenous miRNA (miRNA-15a and miR-155) were ordered from Integrated DNA Technologies and used as standards for miRNA quantification. 1× SYBR Green I (10 000× in stock; Molecular Probes) was added to the miRNA detection kit Master mix for each miRNA sample including the standards for real-time quantification. 50× ROX was also added, as an internal control. Following denaturation at 95°C for 3 min, 35 cycles of PCR were performed using the following protocol: 95°C for 15 s, 60°C for 30 s. All PCR experiments were performed on an ABI Prism 7300 Sequence Detection System (Applied Biosystems).
RESULTS
Detection of individual mRNA transcripts using multiple-labeled probes and LNA-ELF-FISH simultaneously
Recently, it was reported that individual mRNA transcripts could be detected in situ by hybridizing target mRNA with multiple oligonucleotide probes with one or more fluorescent labels (26–28). The high local concentration of fluorescent labels associated with each mRNA transcript creates a bright fluorescent spot that can be readily visualized by fluorescence microscopy. In this study, we used this multiple-probe approach (MP-FISH) to confirm that individual RNA transcripts could also be detected with single LNA probes, when combined with ELF (LNA-ELF-FISH). Specifically, human cervical carcinoma HeLa cells were engineered to constitutively express luciferase mRNA with 24 MS2 binding sites in the 3′-untranslated region. To obtain single molecule sensitivity using MP-FISH, we hybridized each MS2 binding site with a complementary oligonucleotide probe labeled at its 5′ and 3′-ends with a Cy3 fluorescent dye. Therefore, collectively there were 48 Cy3 dyes per mRNA transcript. Simultaneously, the luciferase mRNA transcripts were also hybridized with digoxigenin (dig)-labeled LNA probes within ∼100 bp of the first MS2 repeat. The LNA probes were subsequently labeled with anti-dig-alkaline phosphatase conjugates and ELF signal amplification was performed.
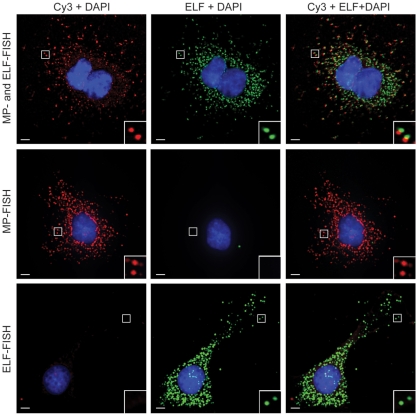
As shown in the top panel of Figure 1, the bright fluorescent spots in the Cy3 image and the ELF image are highly co-localized. The signal elicited by the hybridization of multiple probes to the target RNA was generally not as bright as the ELF signal, particularly in the perinuclear region of most cells, but signal intensity could be improved with the use of more hybridization probes (28). To confirm that the signals in the ELF image did not arise from spectral bleed-through of Cy3 fluorescence into the ELF channel and vice versa, MP FISH and LNA-ELF-FISH were also conducted on separate cell samples (Figure 1, middle and lower panels). No spectral bleed-through was observed in these experiments. Interestingly, the Cy3 signals appeared brighter when MP-FISH was performed independently of LNA-ELF-FISH.
Figure 1.
Simultaneous detection of individual mRNA molecules using MP-FISH and LNA-ELF-FISH. HeLa cells were engineered to constitutively express luciferase mRNA with 24 MS2 binding repeats in the 3′-untranslated region. Each MS2 site was hybridized by an oligonucleotide probe labeled at its 5′- and 3′-end with Cy3. In addition, the coding region of the luciferase RNA was labeled with a single dig-labeled LNA probe. The LNA probes were subsequently labeled with anti-dig-alkaline phosphatase conjugates and ELF signal amplification was performed. Two-dimensional, deconvolved images of the Cy3 fluorescence, ELF signal and a merged image are shown (top panel). Analogous studies were performed using just Cy3 probes (middle panel) or just LNA probes + ELF amplification (bottom panel). Scale bar, 5 μm.
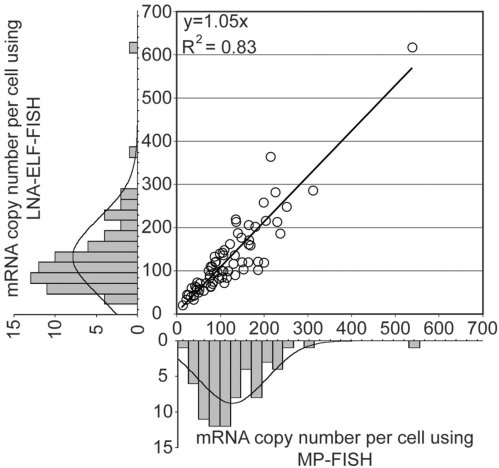
To assess the specificity of LNA-ELF-FISH, the total number of RNAs (i.e. distinct fluorescent spots) within single cells were counted for 72 cells in both the Cy3 and ELF images. Graphical analysis indicated a linear correlation between the two methods with a slope of 1.05 and _R_2 value of 0.83 (Figure 2). Both methods were also in close agreement with quantitative RT-PCR data. Specifically, the average RNA copy number per cell as determined by quantitative RT-PCR was 168 ± 15.2, whereas the average RNA copy number per cells as determined by LNA-ELF-FISH and MP-FISH was 126.6 ± 10.7 (SE) and 118.8 ± 9.6 (SE), respectively. A manual transcript-by-transcript comparison between the LNA-ELF and MP-FISH approaches, in six different cells, revealed that 78% of the fluorescent signals in the ELF images co-localized with fluorescent signals in the Cy3 images and 86% of the fluorescent signals in the Cy3 images co-localized with fluorescent signals in the ELF images (Table 1).
Figure 2.
Quantitative analysis of mRNA copy number in single cells as determined by MP-FISH and LNA-ELF-FISH. Luciferase-MS2 transcripts were fluorescently labeled in HeLa cells by performing MP-FISH and LNA-ELF-FISH, simultaneously. IPLab acquisition software was used to acquire 3D images of both the MP signal (i.e. Cy3) and the ELF signal in 72 randomly selected cells. After 3D deconvolution of the images in IPLab using AutoQuant plug-in software, a 2D image was constructed using a maximum-intensity merged image. The total number of isolated signals was then counted in ImageJ using the particle analysis counter program. The marginal histograms show the distributions of mRNA copy numbers across the population of selected cells as determined by MP-FISH (bottom) and LNA-ELF-FISH (left), respectively.
Table 1.
Analysis of Luc-MS2 mRNA detection in single cells using MP-FISH and ELF-FISH
| Cell Number | Total # of mRNA detected: | # of mRNA observed with only one technique: | # of co-localized signals | % of signals colocalized (ELF/MP) | ||
|---|---|---|---|---|---|---|
| ELF | MP | ELF only | MP only | |||
| 1 | 204 | 188 | 41 | 25 | 163 | 80/87 |
| 2 | 139 | 127 | 36 | 24 | 103 | 74/81 |
| 3 | 46 | 33 | 13 | 0 | 33 | 71/100 |
| 4 | 244 | 216 | 48 | 20 | 196 | 80/91 |
| 5 | 133 | 123 | 38 | 28 | 95 | 71/77 |
| 6 | 96 | 95 | 12 | 11 | 84 | 88/88 |
| Total | 862 | 782 | 188 | 108 | 674 | 78/86 |
Visualization and quantification of miRNA using LNA-ELF-FISH
To investigate whether LNA-ELF-FISH could be extended to the study of individual miRNAs in single cells and to explore the dynamic range of this approach, MDA-MB-231 human breast cancer cells and HeLa cells were stained with a probe for miR-15a. In addition, MCF-7 human breast cancer cells were stained with a probe for miR-155. Fluorescent images revealed that individual bright fluorescent spots, which presumably correspond to individual miRNAs, were dispersed relatively uniformly throughout the cytoplasm of the cells, with a smaller number of miRNA generally localized in the nucleus (Figure 3). Surprisingly absent from the fluorescent images was any clear indication of P-bodies (15). It is not clear whether this was due to an inability to bind and/or detect multiple miRNA within a single P-body, the absence of these specific miRNAs from P-bodies, or another cause.
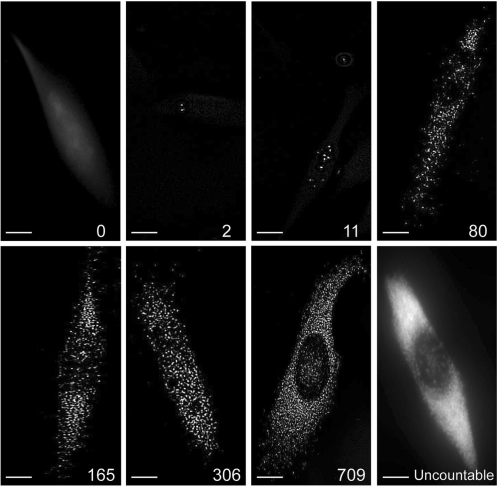
Figure 3.
Fluorescent images of individual miRNAs in single mammalian cells. MDA-MB-231 human breast cancer cells were stained with an LNA probe for miR-15a followed by ELF signal amplification. IPLab acquisition software was used to acquire 3D images of individual cells. After 3D deconvolution of the images, a 2D image was constructed using a maximum-intensity merged image. Representative images illustrating the sensitivity and the dynamic range of the LNA-ELF-FISH approach are shown. The total number of miRNAs identified in each cell is shown in the lower right corner of each panel. The number of miRNA in the cell on the lower right could not be counted directly, due to the inability to distinguish individual fluorescent spots. Scale bar, 5 μm.
Quantification of the individual bright fluorescent spots in single cells revealed that LNA-ELF-FISH could be used to directly quantify anywhere from 0 to ∼1000 copies of miRNA per cell (Figure 3). Therefore, the dynamic range spans over three orders of magnitude. Once the number of miRNAs exceeded ∼1000 copies, it became increasingly difficult to discern individual fluorescent spots and at very high miRNA copy numbers (>3000), the entire cell would exhibit a nearly uniform fluorescent signal. Nonetheless, an estimate of miRNA copy number could still be obtained in these highly fluorescent cells by first drawing a linear correlation between miRNA copy number and total cellular fluorescence in cells where individual miRNA could be discerned. The equation describing this correlation could then be used to calculate the copy numbers in cells with high miRNA expression based on their total fluorescence intensity.
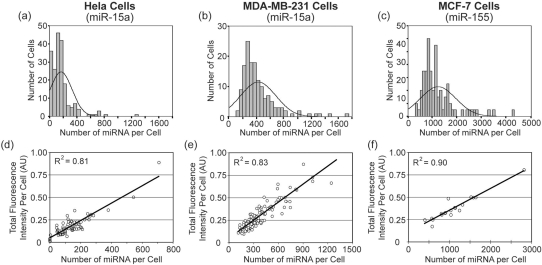
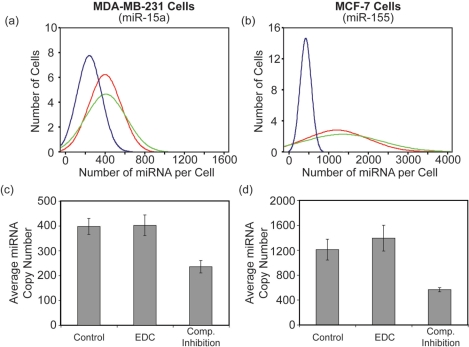
Histograms showing the per cell distribution of miR-15 in MDA-MB-231 and HeLa cells and miR-155 in MCF-7 cells are provided in Figure 4. The respective correlations between total cellular fluorescence and number of miRNA molecules per cell are also shown. Interestingly, a very large variation in the number of miRNAs per cell was observed. Further, for each cell line the distribution appeared to be slightly skewed with a small number of cells exhibiting miRNA copy numbers that were much higher than the mean. Similar observations have been reported for mRNA expression (27). This provides preliminary evidence that miRNAs may also be synthesized in short intense bursts of transcription; however, the increased stability of miRNA likely buffers the fluctuation in copy number compared with mRNA.
Figure 4.
Quantitative analysis of miRNA in three cancer cell lines. The total number of miR-15a molecules in (a) HeLa cells (n = 198) and (b) MDA-MB-231 cells (n = 148) was determined following LNA-ELF-FISH. Similarly, the total number of miR-155 in (c) MCF-7 cells (n = 84) was also quantified. Since individual miRNA could not be discerned in some cells with high miRNA copy number, an estimate was obtained by first drawing a linear correlation between miRNA copy number and total cellular fluorescence. The correlations for (d) HeLa, (e) MDA-MB-231 and (f) MCF-7 cells are shown. The equation describing each correlation was used to calculate the miRNA copy numbers in cells with high expression, based on their fluorescence intensity.
The accuracy of LNA-ELF-FISH was assessed by comparing the mean number of miRNA copies per cell with values obtained by quantitative RT-PCR (Table 2). Assuming quantitative RT-PCR provided the true miRNA copy number per cell, the efficiency of detection with LNA-ELF-FISH was 101.4% for miR-15 in HeLa cells, 82.5% for miR-15 in MDA-MB-231 cells and 108.1% for miR-155 in MCF-7 cells. The close agreement between these two methods suggests that LNA-ELF-FISH can be used to provide accurate quantitative measures of miRNA expression. Of course, LNA-ELF-FISH has the additional advantages of being able to provide information on miRNA localization as well as information on the stochastic distribution of miRNA expression across a population of cells.
Table 2.
Mean number of miRNA/cell detected by LNA-ELF-FISH and qRT-PCR
| miR-15a (HeLa) | miR-15a (MDA-MB-231) | miR-155 (MCF-7) | |
|---|---|---|---|
| LNA-ELF-FISH | 145 ± 19 (SE) | 402 ± 19 (SE) | 1194 ± 99 (SE) |
| qRT-PCR | 143 ± 10 (SE) | 487 ± 16 (SE) | 1105 ± 139 (SE) |
To establish that quantitative RT-PCR itself provides an accurate measure of the miRNA copy number, synthetic miRNA was exposed to the same isolation and amplification procedures as endogenous RNA prior to quantification by RT-PCR. This value was then compared with the amount of starting material. It was found that >88% of the miRNAs could be recovered and detected, suggesting that the quantitative RT-PCR measurements were indeed reflective of the true miRNA copy number; although it should be noted that this control experiment does not account for the effect of cell lysis or interference from cellular biomolecules on the efficiency of miRNA isolation.
While LNA-ELF-FISH measurements of miRNA copy number agreed well with quantitative RT-PCR, one concern was that many of the bright fluorescent spots, which presumably corresponded to miRNA, were actually artifacts of nonspecific LNA or antibody binding. To directly assess whether LNA-ELF-FISH suffered from an abundance of non-specific interactions, we performed two control experiments. The first control experiment simply involved performing LNA-ELF-FISH with a scrambled LNA probe. In general, when a scrambled LNA probe was used we never observed more than one to two bright fluorescent spots per cell and most cells did not contain any bright fluorescent spots (data not shown). These findings suggest that there is essentially no nonspecific or off-target binding. In a second control experiment, competitive inhibition studies were performed. Specifically, LNA-ELF-FISH was conducted using an equal concentration of dig-labeled LNA probe and unlabeled LNA probe, where both probes were designed to target the same miRNA sequence. It was hypothesized that if LNA binding were specific, a 50:50 mixture of labeled and unlabeled probes would lead to a ∼50% reduction in miRNA copy number (i.e. number of bright fluorescent spots) since both probes would be competing for the same binding site. Conversely, if binding of the LNA probes were nonspecific, binding of the unlabeled probe would not prevent binding of the labeled probe and the miRNA copy number would remain unchanged. Histograms showing the per cell distribution of miR-15a in MDA-MB-231 cells and miR-155 in MCF-7 cells, with and without inhibitor, are provided in Figure 5. The presence of the unlabeled LNA probe reduced the mean miRNA copy number from 398 ± 32 (SE) to 236 ± 25 (SE) in MDA-MB-231 cells and from 1209 ± 167 (SE) to 562 ± 35 (SE) in MCF-7 cells. The approximate 50% reduction in mean miRNA copy number, when equal concentrations of labeled and unlabeled LNA probes were used, suggests that nonspecific binding of LNA probes did not have a significant impact on LNA-ELF-FISH measurements.
Figure 5.
Cell-to-cell variation and mean miRNA copy number in cells exposed to various LNA-ELF-FISH conditions. Histograms showing the distribution of (a) miR-15a copy number in MDA-MB-231 cells and (b) miR-155 copy number in MCF-7 cells were plotted following the implementation of several different LNA-ELF-FISH protocols. Specifically, LNA-ELF-FISH was performed using either conventional formaldehyde fixation (red curves) or formalydehyde fixation followed by EDC fixation (green curves). In addition, competitive inhibition studies were performed by incubating formaldehyde-fixed cells with an equal concentration (10 nM) of dig-labeled LNA probe and unlabeled LNA probe (blue curves). The effect of EDC fixation and competitive inhibition on the average number of (c) miR-15a copies in MDA-MB-231 cells and (d) miR-155 copies in MCF-7 cells are shown.
Recently, it has been reported that a substantial amount of miRNA could be lost during in situ hybridization procedures when conventional formaldehyde fixation is performed (30). It was suggested that an additional fixation step with EDC should be conducted to minimize this loss. EDC immobilizes the miRNA by reacting with the 5′-phosphate and coupling it to amino groups in the protein matrix to form stable linkages. To determine the impact of EDC fixation on miRNA quantification, LNA-ELF-FISH was performed with and without EDC fixation. Histograms showing the per cell distribution of miR-15 in MDA-MB-231 cells and miR-155 in MCF-7 cells, with and without EDC fixation, are provided in Figure 5. The addition of EDC increased the mean miRNA copy number from 398 ± 32 (SE) to 402 ± 41 (SE) in MDA-MB-231 cells and from 1209 ± 167 (SE) to 1394 ± 208 (SE) in MCF-7 cells. The marginal increase in miRNA copy number suggests that there was not a significant loss in miRNA in the absence of EDC fixation; however, this could be highly dependent on the experimental parameters, such as sample type (cell culture versus tissue) and the specific miRNA target. Therefore, it is recommended that EDC be used as a precautionary measure.
To highlight the advantages of LNA-ELF-FISH over conventional in situ hybridization techniques, LNA-FISH experiments were also carried out utilizing (i) Texas red-labeled secondary antibodies, (ii) dig-labeled LNA probes combined with Cy5-labeled anti-dig antibodies and (iii) quantum dot labeled secondary antibodies. Although it was found that these approaches could report on the relative level of miRNA expression, the sensitivity was dramatically lower than LNA-ELF-FISH and images often exhibited a high background due to autofluorescence (Figure 6). When quantum dots were applied there was also a loss of specificity, due to a high degree of nonspecific binding.
Figure 6.
Comparison of LNA-ELF-FISH with alternative miRNA-FISH techniques. (a) miRNA FISH was performed using either Texas red-labeled secondary antibodies (left two columns) or dig-labeled LNA probes followed by ELF amplification (right two columns). FISH experiments were performed in both HeLa cells and MCF-7 cells. (b) miRNA FISH was also performed using dig-labeled LNA probes combined with Cy5-labeled anti-dig antibodies and quantum dot labeled secondary antibodies. LNA-ELF-FISH was the only technique that allowed individual miRNAs to be readily visualized.
DISCUSSION
Here we described a highly sensitive and specific method for miRNA detection at the single molecule level in individual cells. Specifically, by combining LNA hybridization probes with ELF signal amplification, single miRNAs could be visualized and counted to yield quantitative information on miRNA expression. The dynamic range of this approach spanned more than three orders of magnitude (i.e. 1 to ∼1000 miRNAs per cell) directly and through the construction of standardization curves could also yield quantitative measurements on cells with higher miRNA copy numbers. Comparisons with MP-FISH on mRNA also revealed that spatial information was retained with LNA-ELF-FISH. Although MP-FISH techniques have previously been reported for the single molecule detection of mRNA, this is the first technique that allows miRNA to be quantified in single cells. Overall, LNA-ELF-FISH is extremely simple and yields reproducible data. Further, in contrast to RT-PCR, no cell lysis, miRNA purification or sample enrichment steps are required and spatial information is retained.
Although the short length of miRNAs prevents MP-FISH approaches from being used for the visualization and quantification of miRNA, we have shown that LNA-ELF-FISH can be applied to both miRNA and mRNA. When used for mRNA detection this approach offers both advantages and disadvantages compared with MP-FISH. One important advantage is the need for only a single hybridization probe. This provides a large savings in cost and eliminates the need to identify a large number of probes with similar melting temperatures. Additional advantages of LNA-ELF-FISH include the long stokes shift and high photostability of the fluorescent precipitate. The long stokes shift results in low autofluorescence and the high photostability allows for repeated imaging. Moreover, the fluorescent precipitate is extremely bright and thus only short exposure times are needed (i.e. ∼10 ms). In contrast, MP-FISH utilizes commercial fluorophores and thus the fluorescent signal was highly susceptible to photobleaching and was much fainter than the ELF signal, often requiring exposure times in excess of 1 s. Surprisingly, even when labeling single mRNA transcripts with 48 fluorescent labels it was still often difficult to differentiate single fluorescent spots from autofluorescence. Of course, image contrast could be improved if a larger number of probes are utilized (28).
Despite the disadvantages of MP-FISH with regard to sensitivity, if enough probes are used to yield high image contrast, this method is likely to offer improved specificity compared with LNA-ELF-FISH. Specifically, since the co-localization of many fluorescent probes is necessary to generate a detectable fluorescent spot, it is unlikely that this would occur by any means other than the specific accumulation of multiple hybridization probes to target mRNA. Alternatively, in a situation where only a fraction of the hybridization probes were bound the target mRNA, it would still be possible to detect the mRNA. In contrast, the reliance of LNA-ELF-FISH on the hybridization of a single LNA probe means that nonspecific binding would result in a false-positive detection and inefficient hybridization would result in an underestimate of target mRNA. Therefore, it becomes extremely important to optimize the experimental conditions by comparing the average copy number per cell to quantitative RT-PCR measurements. It should be noted that MP-FISH also offers the advantage of multiplexing. The ability to select optically distinct fluorophores can allow multiple mRNA targets to be visualized simultaneously. Conversely, the current availability of only a single ELF substrate limits this approach to imaging only a single RNA per cell sample.
Although MP-FISH and LNA-ELF-FISH both offer their unique advantages in regards to imaging mRNA and miRNA, perhaps these approaches would be the most useful when used in unison. We envision that the ability to visualize the interplay between miRNA expression and the expression of target mRNAs with high spatial resolution could provide important insight into miRNA function and control. As a result, this may help us understand how miRNAs influence central signaling pathways and cell cycle control.
FUNDING
The National Institutes of Health (NCI) R21-CA125088 and R21-CA116102; the National Science Foundation BES-0616031; and the American Cancer Society RSG-07-005-01. Funding for open access charge: The National Institutes of Health (NCI) R21-CA125088 and R21-CA116102; the National Science Foundation BES-0616031; and the American Cancer Society RSG-07-005-01.
Conflict of interest statement. None declared.
REFERENCES
- 1.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 4.Couzin J. Cancer biology. A new cancer player takes the stage. Science. 2005;310:766–767. doi: 10.1126/science.310.5749.766. [DOI] [PubMed] [Google Scholar]
- 5.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N. Engl. J. Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 6.Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl Acad. Sci. USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dostie J, Mourelatos Z, Yang M, Sharma A, Dreyfuss G. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA. 2003;9:180–186. doi: 10.1261/rna.2141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 13.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neely LA, Patel S, Garver J, Gallo M, Hackett M, McLaughlin S, Nadel M, Harris J, Gullans S, Rooke J. A single-molecule method for the quantitation of microRNA gene expression. Nat. Methods. 2006;3:41–46. doi: 10.1038/nmeth825. [DOI] [PubMed] [Google Scholar]
- 17.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 18.Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2005;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Politz JC, Zhang F, Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc. Natl Acad. Sci. USA. 2006;103:18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silahtaroglu AN, Nolting D, Dyrskjot L, Berezikov E, Moller M, Tommerup N, Kauppinen S. Detection of microRNAs in frozen tissue sections by fluorescence in situ hybridization using locked nucleic acid probes and tyramide signal amplification. Nat. Protoc. 2007;2:2520–2528. doi: 10.1038/nprot.2007.313. [DOI] [PubMed] [Google Scholar]
- 21.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 22.Chou LS, Meadows C, Wittwer CT, Lyon E. Unlabeled oligonucleotide probes modified with locked nucleic acids for improved mismatch discrimination in genotyping by melting analysis. Biotechniques. 2005;39:644. doi: 10.2144/000112050. 646, 648 passim. [DOI] [PubMed] [Google Scholar]
- 23.Johnson MP, Haupt LM, Griffiths LR. Locked nucleic acid (LNA) single nucleotide polymorphism (SNP) genotype analysis and validation using real-time PCR. Nucleic Acids Res. 2004;32:e55. doi: 10.1093/nar/gnh046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You Y, Moreira BG, Behlke MA, Owczarzy R. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res. 2006;34:e60. doi: 10.1093/nar/gkl175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 27.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paragas VB, Zhang YZ, Haugland RP, Singer VL. The ELF-97 alkaline phosphatase substrate provides a bright, photostable, fluorescent signal amplification method for FISH. J. Histochem. Cytochem. 1997;45:345–357. doi: 10.1177/002215549704500302. [DOI] [PubMed] [Google Scholar]
- 30.Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, et al. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat. Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]