Minireview: Epigenetic Changes in Ovarian Cancer (original) (raw)
Abstract
Epigenetic aberrations, including DNA methylation, histone modifications, and micro-RNA dysregulation, are now well established in the development and progression of ovarian cancer, and their gradual accumulation is associated with advancing disease stage and grade. Epigenetic aberrations are relatively stable, associated with distinct disease subtypes, and present in circulating serum, representing promising diagnostic, prognostic, and pharmacodynamic biomarkers. In contrast to DNA mutations and deletions, aberrant gene-repressive epigenetic modifications are potentially reversible by epigenetic therapies, including inhibitors of DNA methylation or histone-modifying enzymes. Although epigenetic monotherapies have not shown activity against solid tumors, including ovarian cancer, preclinical studies suggest they will be effective when used in combination with one another or with conventional chemotherapeutics, and combinatorial epigenetic therapy regiments are being examined in cancer clinical trials. A greater understanding of the role of epigenetics in ovarian neoplasia will provide for improved interventions against this devastating malignancy.
A greater understanding of the role of epigenetics in ovarian cancer allows for improved interventions against this devastating malignancy.
Ovarian cancer is the most lethal gynecological cancer, causing an estimated 15,520 U.S. deaths in 2008 (1). Due to few early symptoms, most (>70%) patients are diagnosed with advanced-stage disease and 5-yr survival rates are less than 20%, with only modestly improved survival over the past 40 yr (1). Although most advanced-stage patients respond to standard chemotherapies, relapse occurs in over 70% of patients, resulting in chemoresistant, fatal disease (2).
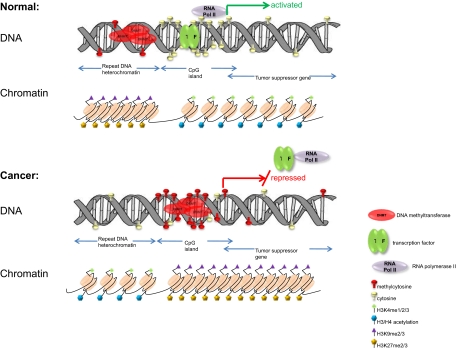
Altered epigenetic states are intimately associated with ovarian tumorigenesis. Epigenetics is defined as a heritable change in gene expression without alteration of the DNA sequence itself and includes DNA methylation, histone modification, nucleosome repositioning, and posttranscriptional gene regulation by micro-RNAs (miRNAs) (3,4). The most studied epigenetic alteration is DNA methylation, the addition of a methyl moiety to the cytosine-5 position within the context of a CpG dinucleotide, mediated by DNA methyltransferases (DNMTs) (3). Although most CpG sites in the human genome are methylated, CpG-dense regions known as CpG islands (often gene-associated) are typically unmethylated in normal tissue (Fig. 1, top). Additionally, DNA-associated histone proteins are subject to extensive modifications that mediate the assembly of transcriptionally permissive or repressive (i.e. open or closed) chromatin (Fig. 1). It is now recognized that DNA methylation and histone modifications are intimately linked (3). The overall epigenetic state (e.g. DNA methylation, histone modification, and miRNA expression) corresponding to a specific cell phenotype is now referred to as the epigenome (5). Although repressive epigenetic modifications (including DNA methylation) regulate genes in normal tissues (e.g. imprinted genes and female X-chromosome inactivation), these are significantly altered in cancer (3,6). Specifically, in cancer cells, global DNA hypomethylation and localized hypermethylation of promoter-associated CpG islands occur (Fig. 1, bottom), with the latter serving as a surrogate for point mutations or deletions to cause transcriptional silencing of tumor suppressor genes (3).
Figure 1.
Altered epigenetic states are intimately associated with ovarian tumorigenesis.
The most recently discovered epigenetic phenomenon is posttranscriptional gene down-regulation by small (21–23 nucleotides in length), non-protein-coding RNA molecules known as miRNAs (4,7). About 1000 miRNA genes have been computationally predicted in the human genome, with each miRNA targeting multiple protein-coding transcripts (7). Although miRNAs are vital to normal cell physiology, their misexpression has been linked to cancer development, and miRNA profiles have now been used to classify human cancers (8). The influence of miRNAs on the epigenetic machinery and the reciprocal epigenetic regulation of miRNA expression (9) strongly suggests that miRNA deregulation during tumorigenesis has important implications for global regulation of epigenetics (10).
Epigenetic Aberrations in Ovarian Cancer (Table 1)
Table 1.
Altered epigenetic regulation in ovarian cancer
| Epigenetic regulator | Modification | Type | Targets | Refs. |
|---|---|---|---|---|
| DNA methylation | Hypermethylation | Tumor suppressor genes | BRCA1 | 42 |
| p16 | 14 | |||
| hMLH1 | 15 | |||
| RASSF1A | 16 | |||
| OPCML | 17 | |||
| ARLTS1 | 87 | |||
| MYO18B | 86 | |||
| SPARC | 28 | |||
| CTGF | 30 | |||
| ANGPTL2 | 29 | |||
| Imprinted genes | ARH1 | 18 | ||
| PEG3 | 18 | |||
| Cell adhesion genes | ICAM-1 | 22 | ||
| CDH1 | 23 | |||
| Proapoptotic genes | LOT1 | 19 | ||
| DAPK | 15 | |||
| TMS1/ASC | 21 | |||
| PAR-4 | 20 | |||
| Arylsulfatase gene | Hsulf-1 | 24,25 | ||
| DNA damage repair gene | PALB2 | 26 | ||
| Class III β-tubulin | TUBB3 | 27 | ||
| Lipid phosphatase | PTEN | 85 | ||
| Development genes | HOXA10, HOXA11 | 31 | ||
| Ribosomal genes | 18S, 28S rDNAs | 92 | ||
| Hypomethylation | Tumor promoter | SNCG | 33 | |
| MCJ | 32 | |||
| IGF2 | 35 | |||
| BORIS | 34 | |||
| Claudin-4 | 36 | |||
| Histone modifications | Lysine acetylation (gene-activating) | Acetylated histones H3 and H4 | GATA4, GATA6 | 46 |
| p21/WAF1 | 48 | |||
| Cyclin B1 | 47 | |||
| Lysine methylation (gene-activating) | Trimethylated H3K4 | GATA4, GATA6 | 46 | |
| Lysine methylation (gene-repressing) | Dimethylated H3K9 | GATA4, GATA6 | 46 | |
| Adam19 | 49 | |||
| Trimethylated H3K27 | GATA4, GATA6 | 46 | ||
| Adam19 | 49 | |||
| RASSF1 | 50 | |||
| miRNAs | Up-regulation | miR-200a | BAP1, SIP1, ZEB1/2 | 8,54 |
| miR-299-5p | DLK1 | 8 | ||
| miR-135b | MSX2 | 8 | ||
| miR-141 | BAP1 | 8 | ||
| miR-200c | BAP1, ZEB1/2, SIP1 | 8,54 | ||
| miR-200b | BAP1, ZEB1/2, SIP1 | 8,54 | ||
| miR-214 | PTEN | 8,52 | ||
| miR-302d | VEGFA | 88 | ||
| miR-373 | VEGFA | 88 | ||
| Down-regulation | miR-199a | c-SRK, MMP13, and FGF2 | 8 | |
| miR-140 | c-SRK, MMP13, and FGF2, VEGFA | 8,88 | ||
| miR-145 | c-SRK, MMP13, and FGF2,PARP8, IRS1 | 8,89,90 | ||
| let-7i | Unknown | 53 | ||
| miR-15/16 | BCL2 | 91 |
Similar to all malignancies, aberrant DNA methylation, including global hypomethylation of heterochromatin and local CpG island methylation, occurs in ovarian cancer (11). Specific examples of hypomethylation include chromosome 1 satellite 2 and LINE-1 repetitive elements (11,12). A number of genes, including the classical tumor suppressors BRCA1 (_breast cancer susceptibility gene_-1) (13), p16 (14), and MLH1 (15) as well as putative tumor suppressor (RASSF1A and OPCMLI) (16,17), imprinted (ARH1 and PEG3I) (18) proapoptotic (LOT1, DAPK, TMS1/ASC, and _PAR_-4) (19,20,21) and cell adhesion (_ICAM_-1 and CDH1) (22,23) are hypermethylated and down-regulated in ovarian cancer. A newly identified gene, _HSulf_-1, encoding an arylsulfatase that acts on cell surface heparin sulfate proteoglycans and inhibits growth factor signaling and angiogenesis (24) was found methylated in over 50% of ovarian tumors and cell lines (25). PALB2 (partner and localizer of BRCA2) was reported to be hypermethylated similarly to BRCA1 (26) in inherited and sporadic breast and ovarian cancer, and hypermethylation of the class III β-tubulin TUBB3 gene may contribute to taxane resistance (27). Recently discovered, candidate tumor suppressor genes hypermethylated in ovarian cancer include SPARC (secreted protein acidic and rich in cysteine) (28), ANGPTL2 (_angiopoietin_-like protein 2) (29), and CTGF (connective tissue growth factor) (30), whereas methylation of the embryonic developmentally regulated genes HOXA10 and HOXA11 was also found to be highly discriminative between normal and malignant ovarian tissues (31).
In addition to repetitive elements and DNA satellites, a number of protein-coding genes are overexpressed in ovarian cancer, in association with promoter hypomethylation. These include MCJ (_methylation_-controlled DNAJ gene), associated with chemoresistance (32); SNCG (_synuclein_-γ), encoding an activator of the MAPK and Elk-1 signal cascades (33); and BORIS (brother of the regulator of imprinted sites), a cancer testis antigen-family candidate oncogene (34). Other ovarian cancer-hypomethylated genes include IGF2, an imprinted gene implicated in numerous malignancies (35), and _claudin_-4, whose overexpression leads to disrupted tight junctions between epithelial ovarian cancer cells (36).
Although candidate gene studies (as described above) have successfully identified a number of important epigenetically regulated genes in ovarian cancer, allowing greater insight into disease progression, we have also globally examined DNA hypermethylation (using methylation microarrays) to demonstrate that ovarian tumors contain a large number of hypermethylated loci (37). Such disease stage-specific methylated loci represent possible methylation signatures for classification and possible targets for therapy (38,39), and these and other (40) studies demonstrate that CpG island methylation is cumulative with ovarian cancer progression. Although the relationship between gene hypermethylation in ovarian cancer and altered DNMT RNA levels is not straightforward (41,42), functional validation of promoter DNA hypermethylation in ovarian carcinogenesis was further shown by Huang and co-workers (41) who, by down-regulating two DNA methyltransferase enzymes, demonstrated that extensive loss of CpG hypermethylation correlates significantly with ovarian cancer cell growth inhibition.
One of the most studied genes in ovarian cancer is BRCA1, due to its role in both inherited and sporadic forms of this disease (42). The clinical outcome for ovarian cancer patients having tumor-hypermethylated BRCA1 has recently been compared with patients with germline BRCA1 mutations or wild-type BRCA1 (43). BRCA1 hypermethylation occurs in 10–15% of sporadic disease cases, associates strongly with loss of BRCA1 RNA and protein (13), and significantly correlates with poor patient outcome (42). These studies suggest that BRCA1 hypermethylation, which has also been reported in ovarian cancer patient serum (16), may represent a minimally invasive approach for predicting patient response to standard therapies.
Although histone modifications (in addition to DNA methylation) regulate numerous normal ovarian functions, including estrogen synthesis, folliculogenesis, and luteal phase activity (44), ovarian cancer cells significantly alter their expression of chromatin-modifying proteins (45). Additionally, specific histone modifications (but not promoter DNA methylation) act to regulate the differentiation genes GATA4 and GATA6 (46) and the cell cycle regulatory proteins cyclinB1 (47) and _p21_WAF1/CIP1 (48). Similarly, we recently showed that in ovarian cancer cells refractory to TGF-β1, two repressive histone modifications (trimethyl-H3K27 and dimethyl-H3K9), in concert with histone deacetylase (HDAC) enzymes, down-regulate ADAM19 (a disintegrin and metalloprotease domain 19) in the absence of significant CpG island methylation (49). Those results demonstrate that aberrant TGF-β1 signaling can result in formation of a repressive chromatin environment, without DNA methylation, in ovarian cancer cells. Similarly, using a dominant-negative histone overexpression approach, our group showed that genome-wide loss of the repressive trimethyl-H3K27 mark associated with reduced global DNA methylation, allowing platinum resensitization of chemoresistant ovarian cancer cells, with one of the affected genes, RASSF1 shown to be a direct target of H3K27 methylation-mediated silencing (50). Because loss of H3K27 trimethylation has also been associated with poor prognosis in ovarian and other malignancies (51), and gene promoter DNA methylation can be maintained in the absence of this repressive mark (50), the above findings collectively demonstrate that complex epigenetic patterns, involving DNA methylation and histone modifications, contribute to ovarian cancer progression and drug resistance.
miRNAs represent the most recently discovered epigenetic phenomenon, and ovarian tumors were recently found to significantly up-regulate _miR_-199a, _miR_-200a, and _miR_-214 and down-regulate _miR_-100, and specifically, _miR_-214 was demonstrated to target the tumor suppressor PTEN and associate with platinum resistance (8,52). The miRNA _let_-7i was recently found to be a tumor suppressor significantly down-regulated in platinum-resistant ovarian tumors, and _let_-7i gain-of-function restored drug sensitivity of chemoresistant ovarian cancer cells, thus representing a candidate biomarker and therapeutic target (53). _miR_-429, _miR_-200a, and _miR_-200b were found to be clustered on a single primary transcript regulated by the epithelial-to-mesenchymal transition (EMT, a metastatic phenotype) repressor ZEB1/SIP1, with _miR_-200a and _miR_-200b negatively regulating ZEB1/SIP1 and creating a double-negative feedback cycle (54). In another study, 27 miRNAs significantly associated with chemotherapy response (55), showing that (similar to DNA methylation) miRNAs represent possible prognostic and diagnostic biomarkers for ovarian cancer. With respect to miRNA gene regulation, a group of six miRNAs clustered on chromosome 19, and seven clustered on chromosome 14, were up-regulated by the DNMT inhibitor decitabine (described below), demonstrating that miRNAs can be regulated by DNA methylation (56). Moreover, an overall, collective tumor-suppressive effect of miRNAs was suggested by down-regulation of Drosha and Dicer, two enzymes involved in miRNA processing, being significantly associated with advanced ovarian cancer stage and poor prognosis (57).
Epigenetic Therapies and Biomarkers for Ovarian Cancer
As mentioned above, ovarian cancer tumor progression is well characterized by a number of combinatorial epigenetic aberrations distinct to this malignancy, including (but not limited to) DNA methylation of RASSF1A, DAPK, _H_-_Sulf_-1, BRCA1, and HOXA10. Consequently, these methylated DNA sequences represent potential biomarkers for diagnosis, staging, prognosis (i.e. prognostic biomarkers), and monitoring of response to therapy (predictive biomarkers) (15). DNA methylation biomarkers hold a number of advantages over other biomarker types, such as proteins, gene expression, and DNA mutations, including their stability, ability to be amplified (thus greatly enhancing detection sensitivity), relatively low cost of assessment, and restriction to limited regions of DNA (CpG islands) (58). In the future, it is highly likely that DNA methylation analyses of resected ovarian tumors will be used to individually tailor treatment, similar to recently discovered predictive markers in stage I non-small-cell lung cancer (59). Although methylation assessment of single genes lacks sufficient specificity for ovarian cancer diagnostics, it is believed that panels of multiple methylation biomarkers may achieve the accuracy required for widespread population screening (58,60). Toward that objective, a panel of 112 methylated DNA markers was found to associate with ovarian cancer progression-free survival (39). Similarly, methylation of MCJ and hMLH1 correlates with chemotherapy response (38), and methylation of a four-gene panel was found to significantly predict overall survival and relapse (61). In ovarian cancer particularly, methylation biomarkers could likely augment the specificity of CA-125, similar to ongoing prostate cancer studies examining various prostate-specific antigen/biomarkers (62).
In addition to tissue analysis, methylated DNA has been detected in the serum and peritoneal fluid of ovarian cancer patients (15,16,38,61). Methylated DNA found in cancer patient serum correlated reasonably well with methylation levels in tumor tissue (16,61,63), and it is also believed that the source of serum DNA is necrotic tumor cells. Consequently, detection of methylated DNA biomarkers in body fluids has significant potential as a minimally invasive tool to regularly assess ovarian cancer patient response.
A major impediment to improving survival of ovarian cancer patients is the development of chemoresistance, and specifically, platinum resistance is strongly associated with methylation-induced silencing of various drug response genes and pathways. Whereas genetic mutations, deletions, or allelic losses are fixed and irreversible, epigenetic abnormalities can potentially be corrected (3,6). In this regard, several drugs that inhibit DNMT activity are now in clinical use. These drugs act by covalently and irreversibly binding to the DNMT enzyme active site, resulting in genomic hypomethylation (64). Although monotherapy of these agents is effective against hematological malignancies, their activity against solid tumors has been disappointing, suggesting greater promise in drug combinations (60,64). In preclinical studies, various DNMT inhibitors were found to elicit DNA hypomethylation and reverse chemoresistance of platinum-resistant ovarian cancer cells (65,66,67) and mouse xenografts (68), laying the foundation for the clinical evaluation of DNMT inhibitors for chemotherapy resensitization in ovarian cancer patients (69). To that end, we are currently conducting a phase I/II trial (NCT00477386, Study ID 0704-07, www.clinicaltrials.gov) of one DNMT inhibitor, decitabine (Dacogen; Eisai, Inc., Tokyo, Japan) paired with carboplatin, hypothesizing that low-dose decitabine derepresses silenced tumor suppressors to resensitize platinum-resistant ovarian tumors to carboplatin. The phase I component of this study is now complete, demonstrating safety of the combined regimen and biological activity in vivo, as assessed by decreased methylation of genome-wide repetitive elements and specific genes (70).
Because histone deacetylation is another transcriptional silencing mechanism in ovarian cancer, HDAC inhibitors (HDACIs) can relieve epigenetic gene repression and exert anticancer effects by inhibiting the deacetylation of nonhistone proteins (71). Similar to DNMT inhibitors, however, poor single-agent activity of HDACIs against solid tumors has been demonstrated. In a single-agent ovarian cancer trial of the HDACI vorinostat (Zolinza; Merck & Co., Inc.), only one of 27 patients experienced a partial response (72), suggesting that HDACIs may be more effective when used in combination with other agents (60,71). In one preclinical ovarian cancer study, the third-generation HDACI belinostat (CuraGen Corp., Branford, CT) resensitized platinum-resistant xenografts in mice (73). Likewise, our group also demonstrated that another rationally designed HDACI, AR-42 (Arno Therapeutics, Parsippany, NJ; www.arnothera.com), could resensitize platinum-resistant ovarian cancer cells and xenografts, even more potently than vorinostat (74); AR-42 is scheduled to begin clinical trials this year. Because additive or synergistic effects of HDAC and DNMT inhibitor combinations on silenced gene reexpression have been demonstrated (75), combining these two classes of epigenetic drugs with conventional therapies may be the most effective approach to use in the clinic (3,60,64). In support of this possibility, one preclinical study showed that a combination of decitabine with belinostat elicited greater platinum resensitization of resistant ovarian cancer xenografts than decitabine alone (76).
Future Directions
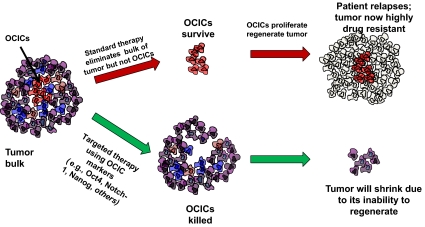
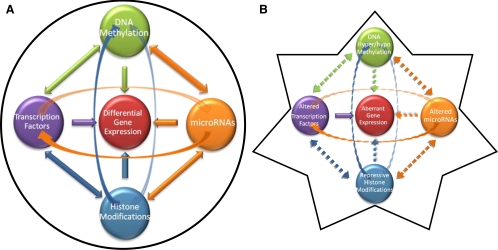
Recent findings suggest a role for tumor progenitors, known as cancer-initiating or cancer stem cells (CSCs), in the propagation of a drug-resistant phenotype in numerous malignancies (77), including ovarian cancer (78,79,80,81) (Fig. 2). The cancer stem cell hypothesis posits that conventional chemotherapies, targeted to highly mitotic cells, fail to destroy quiescent or slowly dividing CSCs, which then regrow the tumor (77). Because epigenetic therapies are well-established differentiating agents (60,64), they may also target poorly differentiated CSCs (3) (Fig. 2). Moreover, the realization of the Human Epigenome Project, an exhaustive annotation of all deoxcytosine and histone modifications throughout the human genome (82), could allow for the establishment of epigenetic ovarian cancer diagnostic, prognostic, and pharmacodynamic biomarkers (15,83,84) (Fig. 3). In summary, a greater understanding of the role of epigenetics in ovarian cancer will allow for improved interventions against this devastating malignancy.
Figure 2.
Targeting ovarian cancer-initiating/stem cells (OCIC).
Figure 3.
A, Normal cell epigenome; B, ovarian cancer cell epigenome.
Footnotes
This work was supported by National Institutes of Health, National Cancer Institute Grants CA085289 (to K.P.N.), CA113001 (to T.T.-M.H), CA133877 (to D.E.M), Ovar’coming Together (Indianapolis, IN; to C.B.), the Walther Cancer Institute (Indianapolis, IN; to K.P.N.), and Phi Beta Psi Sorority (Brownsburg, IN; to K.P.N.).
Disclosure Summary: C.B., F.F., D.E.M., T.H.-M.H., and K.P.N. have nothing to disclose.
First Published Online July 2, 2009
Abbreviations: CSC, Cancer stem cell; DNMT, DNA methyltransferase; HDAC, histone deacetylase; HDACI, HDAC inhibitor; miRNA, micro-RNA.
References
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ 2008 Cancer statistics, 2008. CA Cancer J Clin 58:71–96 [DOI] [PubMed] [Google Scholar]
- Liu CM 2005 Cancer of the ovary. N Engl J Med 352:1268–1269; author reply 1268–1269 [PubMed] [Google Scholar]
- Jones PA, Baylin SB 2007 The epigenomics of cancer. Cell 128:683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J, Percharde M, Coley HM, Webb A, Crook T 2009 The context and potential of epigenetics in oncology. Br J Cancer 100:571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES 2007 The mammalian epigenome. Cell 128:669–681 [DOI] [PubMed] [Google Scholar]
- Esteller M 2008 Epigenetics in cancer. N Engl J Med 358:1148–1159 [DOI] [PubMed] [Google Scholar]
- Schickel R, Boyerinas B, Park SM, Peter ME 2008 MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 27:5959–5974 [DOI] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Ménard S, Croce CM 2007 MicroRNA signatures in human ovarian cancer. Cancer Res 67:8699–8707 [DOI] [PubMed] [Google Scholar]
- Deng S, Calin GA, Croce CM, Coukos G, Zhang L 2008 Mechanisms of microRNA deregulation in human cancer. Cell Cycle 7:2643–2646 [DOI] [PubMed] [Google Scholar]
- Ventura A, Jacks T 2009 MicroRNAs and cancer: short RNAs go a long way. Cell 136:586–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M, Jiang G, Woods C, Müller HM, Fiegl H, Goebel G, Marth C, Muller-Holzner E, Zeimet AG, Laird PW, Ehrlich M 2004 DNA hypomethylation and ovarian cancer biology. Cancer Res 64:4472–4480 [DOI] [PubMed] [Google Scholar]
- Pattamadilok J, Huapai N, Rattanatanyong P, Vasurattana A, Triratanachat S, Tresukosol D, Mutirangura A 2008 LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. Int J Gynecol Cancer 18:711–717 [DOI] [PubMed] [Google Scholar]
- Press JZ, De Luca A, Boyd N, Young S, Troussard A, Ridge Y, Kaurah P, Kalloger SE, Blood KA, Smith M, Spellman PT, Wang Y, Miller DM, Horsman D, Faham M, Gilks CB, Gray J, Huntsman DG 2008 Ovarian carcinomas with genetic and epigenetic BRCA1 loss have distinct molecular abnormalities. BMC Cancer 8:17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milde-Langosch K, Ocon E, Becker G, Löning T 1998 p16/MTS1 inactivation in ovarian carcinomas: high frequency of reduced protein expression associated with hyper-methylation or mutation in endometrioid and mucinous tumors. Int J Cancer 79:61–65 [DOI] [PubMed] [Google Scholar]
- Balch C, Huang TH, Brown R, Nephew KP 2004 The epigenetics of ovarian cancer drug resistance and resensitization. Am J Obstet Gynecol 191:1552–1572 [DOI] [PubMed] [Google Scholar]
- Ibanez de Caceres I, Battagli C, Esteller M, Herman JG, Dulaimi E, Edelson MI, Bergman C, Ehya H, Eisenberg BL, Cairns P 2004 Tumor cell-specific BRCA1 and RASSF1A hypermethylation in serum, plasma, and peritoneal fluid from ovarian cancer patients. Cancer Res 64:6476–6481 [DOI] [PubMed] [Google Scholar]
- Sellar GC, Watt KP, Rabiasz GJ, Stronach EA, Li L, Miller EP, Massie CE, Miller J, Contreras-Moreira B, Scott D, Brown I, Williams AR, Bates PA, Smyth JF, Gabra H 2003 OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nat Genet 34:337–343 [DOI] [PubMed] [Google Scholar]
- Feng W, Marquez RT, Lu Z, Liu J, Lu KH, Issa JP, Fishman DM, Yu Y, Bast Jr RC 2008 Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer 112:1489–1502 [DOI] [PubMed] [Google Scholar]
- Cvetkovic D, Pisarcik D, Lee C, Hamilton TC, Abdollahi A 2004 Altered expression and loss of heterozygosity of the LOT1 gene in ovarian cancer. Gynecol Oncol 95:449–455 [DOI] [PubMed] [Google Scholar]
- Pruitt K, Ulkü AS, Frantz K, Rojas RJ, Muniz-Medina VM, Rangnekar VM, Der CJ, Shields JM 2005 Ras-mediated loss of the pro-apoptotic response protein Par-4 is mediated by DNA hypermethylation through Raf-independent and Raf-dependent signaling cascades in epithelial cells. J Biol Chem 280:23363–23370 [DOI] [PubMed] [Google Scholar]
- Terasawa K, Sagae S, Toyota M, Tsukada K, Ogi K, Satoh A, Mita H, Imai K, Tokino T, Kudo R 2004 Epigenetic inactivation of TMS1/ASC in ovarian cancer. Clin Cancer Res 10:2000–2006 [DOI] [PubMed] [Google Scholar]
- Arnold JM, Cummings M, Purdie D, Chenevix-Trench G 2001 Reduced expression of intercellular adhesion molecule-1 in ovarian adenocarcinomas. Br J Cancer 85:1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuecheng Y, Hongmei L, Xiaoyan X 2006 Clinical evaluation of E-cadherin expression and its regulation mechanism in epithelial ovarian cancer. Clin Exp Metastasis 23:65–74 [DOI] [PubMed] [Google Scholar]
- Backen AC, Cole CL, Lau SC, Clamp AR, McVey R, Gallagher JT, Jayson GC 2007 Heparan sulphate synthetic and editing enzymes in ovarian cancer. Br J Cancer 96:1544–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub J, Chien J, Pan Y, Qian X, Narita K, Aletti G, Scheerer M, Roberts LR, Molina J, Shridhar V 2007 Epigenetic silencing of HSulf-1 in ovarian cancer: implications in chemoresistance. Oncogene 26:4969–4978 [DOI] [PubMed] [Google Scholar]
- Potapova A, Hoffman AM, Godwin AK, Al-Saleem T, Cairns P 2008 Promoter hypermethylation of the PALB2 susceptibility gene in inherited and sporadic breast and ovarian cancer. Cancer Res 68:998–1002 [DOI] [PubMed] [Google Scholar]
- Izutsu N, Maesawa C, Shibazaki M, Oikawa H, Shoji T, Sugiyama T, Masuda T 2008 Epigenetic modification is involved in aberrant expression of class III β-tubulin, TUBB3, in ovarian cancer cells. Int J Oncol 32:1227–1235 [DOI] [PubMed] [Google Scholar]
- Socha MJ, Said N, Dai Y, Kwong J, Ramalingam P, Trieu V, Desai N, Mok SC, Motamed K 2009 Aberrant promoter methylation of sparc in ovarian cancer. Neoplasia 11:126–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi R, Tsuda H, Kozaki K, Kanai Y, Kasamatsu T, Sengoku K, Hirohashi S, Inazawa J, Imoto I 2008 Frequent inactivation of a putative tumor suppressor, angiopoietin-like protein 2, in ovarian cancer. Cancer Res 68:5067–5075 [DOI] [PubMed] [Google Scholar]
- Kikuchi R, Tsuda H, Kanai Y, Kasamatsu T, Sengoku K, Hirohashi S, Inazawa J, Imoto I 2007 Promoter hypermethylation contributes to frequent inactivation of a putative conditional tumor suppressor gene connective tissue growth factor in ovarian cancer. Cancer Res 67:7095–7105 [DOI] [PubMed] [Google Scholar]
- Fiegl H, Windbichler G, Mueller-Holzner E, Goebel G, Lechner M, Jacobs IJ, Widschwendter M 2008 HOXA11 DNA methylation: a novel prognostic biomarker in ovarian cancer. Int J Cancer 123:725–729 [DOI] [PubMed] [Google Scholar]
- Strathdee G, Vass JK, Oien KA, Siddiqui N, Curto-Garcia J, Brown R 2005 Demethylation of the MCJ gene in stage III/IV epithelial ovarian cancer and response to chemotherapy. Gynecol Oncol 97:898–903 [DOI] [PubMed] [Google Scholar]
- Czekierdowski A, Czekierdowska S, Wielgos M, Smolen A, Kaminski P, Kotarski J 2006 The role of CpG islands hypomethylation and abnormal expression of neuronal protein synuclein-gamma (SNCG) in ovarian cancer. Neuro Endocrinol Lett 27:381–386 [PubMed] [Google Scholar]
- Woloszynska-Read A, James SR, Link PA, Yu J, Odunsi K, Karpf AR 2007 DNA methylation-dependent regulation of BORIS/CTCFL expression in ovarian cancer. Cancer Immun 7:21 [PMC free article] [PubMed] [Google Scholar]
- Murphy SK, Huang Z, Wen Y, Spillman MA, Whitaker RS, Simel LR, Nichols TD, Marks JR, Berchuck A 2006 Frequent IGF2/H19 domain epigenetic alterations and elevated IGF2 expression in epithelial ovarian cancer. Mol Cancer Res 4:283–292 [DOI] [PubMed] [Google Scholar]
- Litkouhi B, Kwong J, Lo CM, Smedley 3rd JG, McClane BA, Aponte M, Gao Z, Sarno JL, Hinners J, Welch WR, Berkowitz RS, Mok SC, Garner EI 2007 Claudin-4 overexpression in epithelial ovarian cancer is associated with hypomethylation and is a potential target for modulation of tight junction barrier function using a C-terminal fragment of Clostridium perfringens enterotoxin. Neoplasia 9:304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SH, Chen CM, Strathdee G, Harnsomburana J, Shyu CR, Rahmatpanah F, Shi H, Ng SW, Yan PS, Nephew KP, Brown R, Huang TH 2002 Methylation microarray analysis of late-stage ovarian carcinomas distinguishes progression-free survival in patients and identifies candidate epigenetic markers. Clin Cancer Res 8:2246–2252 [PubMed] [Google Scholar]
- Barton CA, Hacker NF, Clark SJ, O'Brien PM 2008 DNA methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecol Oncol 109:129–139 [DOI] [PubMed] [Google Scholar]
- Wei SH, Balch C, Paik HH, Kim YS, Baldwin RL, Liyanarachchi S, Li L, Wang Z, Wan JC, Davuluri RV, Karlan BY, Gifford G, Brown R, Kim S, Huang TH, Nephew KP 2006 Prognostic DNA methylation biomarkers in ovarian cancer. Clin Cancer Res 12:2788–2794 [DOI] [PubMed] [Google Scholar]
- Watts GS, Futscher BW, Holtan N, Degeest K, Domann FE, Rose SL 2008 DNA methylation changes in ovarian cancer are cumulative with disease progression and identify tumor stage. BMC Med Genomics 1:47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu YW, Rahmatpanah F, Shi H, Wei SH, Liu JC, Yan PS, Huang TH 2003 Double RNA interference of DNMT3b and DNMT1 enhances DNA demethylation and gene reactivation. Cancer Res 63:6110–6115 [PubMed] [Google Scholar]
- Baldwin RL, Nemeth E, Tran H, Shvartsman H, Cass I, Narod S, Karlan BY 2000 BRCA1 promoter region hypermethylation in ovarian carcinoma: a population-based study. Cancer Res 60:5329–5333 [PubMed] [Google Scholar]
- Hilton JL, Geisler JP, Rathe JA, Hattermann-Zogg MA, DeYoung B, Buller RE 2002 Inactivation of BRCA1 and BRCA2 in ovarian cancer. J Natl Cancer Inst 94:1396–1406 [DOI] [PubMed] [Google Scholar]
- LaVoie HA 2005 Epigenetic control of ovarian function: the emerging role of histone modifications. Mol Cell Endocrinol 243:12–18 [DOI] [PubMed] [Google Scholar]
- Ozdađ H, Teschendorff AE, Ahmed AA, Hyland SJ, Blenkiron C, Bobrow L, Veerakumarasivam A, Burtt G, Subkhankulova T, Arends MJ, Collins VP, Bowtell D, Kouzarides T, Brenton JD, Caldas C 2006 Differential expression of selected histone modifier genes in human solid cancers. BMC Genomics 7:90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caslini C, Capo-chichi CD, Roland IH, Nicolas E, Yeung AT, Xu XX 2006 Histone modifications silence the GATA transcription factor genes in ovarian cancer. Oncogene 25:5446–5461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valls E, Sánchez-Molina S, Martínez-Balbás MA 2005 Role of histone modifications in marking and activating genes through mitosis. J Biol Chem 280:42592–42600 [DOI] [PubMed] [Google Scholar]
- Richon VM, Sandhoff TW, Rifkind RA, Marks PA 2000 Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci USA 97:10014–10019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MW, Huang YW, Hartman-Frey C, Kuo CT, Deatherage D, Qin H, Cheng AS, Yan PS, Davuluri RV, Huang TH, Nephew KP, Lin HJ 2008 Aberrant transforming growth factor β1 signaling and SMAD4 nuclear translocation confer epigenetic repression of ADAM19 in ovarian cancer. Neoplasia 10:908–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbosh PH, Montgomery JS, Starkey JA, Novotny M, Zuhowski EG, Egorin MJ, Moseman AP, Golas A, Brannon KM, Balch C, Huang TH, Nephew KP 2006 Dominant-negative histone H3 lysine 27 mutant derepresses silenced tumor suppressor genes and reverses the drug-resistant phenotype in cancer cells. Cancer Res 66:5582–5591 [DOI] [PubMed] [Google Scholar]
- Wei Y, Xia W, Zhang Z, Liu J, Wang H, Adsay NV, Albarracin C, Yu D, Abbruzzese JL, Mills GB, Bast Jr RC, Hortobagyi GN, Hung MC 2008 Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Mol Carcinog 47:701–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, Cheng JQ 2008 MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res 68:425–433 [DOI] [PubMed] [Google Scholar]
- Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K, Liang S, Leminen A, Deng S, Smith L, Johnstone CN, Chen XM, Liu CG, Huang Q, Katsaros D, Calin GA, Weber BL, Bützow R, Croce CM, Coukos G, Zhang L 2008 MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res 68:10307–10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ 2008 A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res 68:7846–7854 [DOI] [PubMed] [Google Scholar]
- Boren T, Xiong Y, Hakam A, Wenham R, Apte S, Chan G, Kamath SG, Chen DT, Dressman H, Lancaster JM 2009 MicroRNAs and their target messenger RNAs associated with ovarian cancer response to chemotherapy. Gynecol Oncol 113:249–255 [DOI] [PubMed] [Google Scholar]
- Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, Johnstone CN, Megraw MS, Adams S, Lassus H, Huang J, Kaur S, Liang S, Sethupathy P, Leminen A, Simossis VA, Sandaltzopoulos R, Naomoto Y, Katsaros D, Gimotty PA, DeMichele A, Huang Q, Bützow R, Rustgi AK, Weber BL, Birrer MJ, Hatzigeorgiou AG, Croce CM, Coukos G 2008 Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci USA 105:7004–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, Deavers MT, Mourad-Zeidan A, Wang H, Mueller P, Lenburg ME, Gray JW, Mok S, Birrer MJ, Lopez-Berestein G, Coleman RL, Bar-Eli M, Sood AK 2008 Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med 359:2641–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW 2003 The power and the promise of DNA methylation markers. Nat Rev Cancer 3:253–266 [DOI] [PubMed] [Google Scholar]
- Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S, Glöckner S, Piantadosi S, Gabrielson E, Pridham G, Pelosky K, Belinsky SA, Yang SC, Baylin SB, Herman JG 2008 DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med 358:1118–1128 [DOI] [PubMed] [Google Scholar]
- Balch C, Montgomery JS, Paik HI, Kim S, Kim S, Huang TH, Nephew KP 2005 New anti-cancer strategies: epigenetic therapies and biomarkers. Front Biosci 10:1897–1931 [DOI] [PubMed] [Google Scholar]
- Su HY, Lai HC, Lin YW, Chou YC, Liu CY, Yu MH 2009 An epigenetic marker panel for screening and prognostic prediction of ovarian cancer. Int J Cancer 124:387–393 [DOI] [PubMed] [Google Scholar]
- Parekh DJ, Ankerst DP, Troyer D, Srivastava S, Thompson IM 2007 Biomarkers for prostate cancer detection. J Urol 178:2252–2259 [DOI] [PubMed] [Google Scholar]
- Mori T, O'Day SJ, Umetani N, Martinez SR, Kitago M, Koyanagi K, Kuo C, Takeshima TL, Milford R, Wang HJ, Vu VD, Nguyen SL, Hoon DS 2005 Predictive utility of circulating methylated DNA in serum of melanoma patients receiving biochemotherapy. J Clin Oncol 23:9351–9358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F, Brown R 2005 DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst 97:1498–1506 [DOI] [PubMed] [Google Scholar]
- Li Y, Hu W, Shen DY, Kavanagh JJ, Fu S 2009 Azacitidine enhances sensitivity of platinum-resistant ovarian cancer cells to carboplatin through induction of apoptosis. Am J Obstet Gynecol 200:177.e1–e9 [DOI] [PubMed] [Google Scholar]
- Lenzi R, Frost P, Abbruzzese JL 1994 Modulation of cisplatin resistance by 2′-deoxy-5-azacytidine in human ovarian tumor cell lines. Anticancer Res 14:247–251 [PubMed] [Google Scholar]
- Balch C, Yan P, Craft T, Young S, Skalnik DG, Huang TH, Nephew KP 2005 Antimitogenic and chemosensitizing effects of the methylation inhibitor zebularine in ovarian cancer. Mol Cancer Ther 4:1505–1514 [DOI] [PubMed] [Google Scholar]
- Plumb JA, Strathdee G, Sludden J, Kaye SB, Brown R 2000 Reversal of drug resistance in human tumor xenografts by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res 60:6039–6044 [PubMed] [Google Scholar]
- Appleton K, Mackay HJ, Judson I, Plumb JA, McCormick C, Strathdee G, Lee C, Barrett S, Reade S, Jadayel D, Tang A, Bellenger K, Mackay L, Setanoians A, Schätzlein A, Twelves C, Kaye SB, Brown R 2007 Phase I and pharmacodynamic trial of the DNA methyltransferase inhibitor decitabine and carboplatin in solid tumors. J Clin Oncol 25:4603–4609 [DOI] [PubMed] [Google Scholar]
- Nephew KP, Matei DE, Balch C, Fang F, Schilder J 2009 DNA methylation inhibitors for chemotherapy resensitization of solid tumors. Proc 100th Annual Meeting of the American Association for Cancer Research, Denver, CO, 2009 (Abstract 25) [Google Scholar]
- Minucci S, Pelicci PG 2006 Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 6:38–51 [DOI] [PubMed] [Google Scholar]
- Modesitt SC, Sill M, Hoffman JS, Bender DP 2008 A phase II study of vorinostat in the treatment of persistent or recurrent epithelial ovarian or primary peritoneal carcinoma: a Gynecologic Oncology Group study. Gynecol Oncol 109:182–186 [DOI] [PubMed] [Google Scholar]
- Qian X, LaRochelle WJ, Ara G, Wu F, Petersen KD, Thougaard A, Sehested M, Lichenstein HS, Jeffers M 2006 Activity of PXD101, a histone deacetylase inhibitor, in preclinical ovarian cancer studies. Mol Cancer Ther 5:2086–2095 [DOI] [PubMed] [Google Scholar]
- Yang YT, Balch C, Kulp S, Mand MR, Nephew KP, Chen CS 2009 A rationally designed histone deacetylase inhibitor with distinct antitumor activity against ovarian cancer. Neoplasia 11:552–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpf AR, Jones DA 2002 Reactivating the expression of methylation silenced genes in human cancer. Oncogene 21:5496–5503 [DOI] [PubMed] [Google Scholar]
- Steele N, Finn P, Brown R, Plumb JA 2009 Combined inhibition of DNA methylation and histone acetylation enhances gene re-expression and drug sensitivity in vivo. Br J Cancer 100:758–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan CT, Guzman ML, Noble M 2006 Cancer stem cells. N Engl J Med 355:1253–1261 [DOI] [PubMed] [Google Scholar]
- Baba T, Convery PA, Matsumura N, Whitaker RS, Kondoh E, Perry T, Huang Z, Bentley RC, Mori S, Fujii S, Marks JR, Berchuck A, Murphy SK 2009 Epigenetic regulation of CD133 and tumorigenicity of CD133+ ovarian cancer cells. Oncogene 28:209–218 [DOI] [PubMed] [Google Scholar]
- Bapat SA, Mali AM, Koppikar CB, Kurrey NK 2005 Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res 65:3025–3029 [DOI] [PubMed] [Google Scholar]
- Kusumbe AP, Mali AM, Bapat SA 2009 CD133-expressing stem cells associated with ovarian metastases establish an endothelial hierarchy and contribute to tumor vasculature. Stem Cells 27:498–508 [DOI] [PubMed] [Google Scholar]
- Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, Yan PS, Huang TH, Nephew KP 2008 Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res 68:4311–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Association for Cancer Research Human Epigenome Task Force; European Union, Network of Excellence, Scientific Advisory Board 2008 Moving AHEAD with an international human epigenome project. Nature 454:711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero-Navarro S, Esteller M 2008 Epigenetic biomarkers for human cancer: the time is now. Crit Rev Oncol Hematol 68:1–11 [DOI] [PubMed] [Google Scholar]
- Paige AJ, Brown R 2008 Pharmaco(epi)genomics in ovarian cancer. Pharmacogenomics 9:1825–1834 [DOI] [PubMed] [Google Scholar]
- Schöndorf T, Ebert MP, Hoffmann J, Becker M, Moser N, Pur S, Göhring UJ, Weisshaar MP 2004 Hypermethylation of the PTEN gene in ovarian cancer cell lines. Cancer Lett 207:215–220 [DOI] [PubMed] [Google Scholar]
- Yanaihara N, Nishioka M, Kohno T, Otsuka A, Okamoto A, Ochiai K, Tanaka T, Yokota J 2004 Reduced expression of MYO18B, a candidate tumor-suppressor gene on chromosome arm 22q, in ovarian cancer. Int J Cancer 112:150–154 [DOI] [PubMed] [Google Scholar]
- Petrocca F, Iliopoulos D, Qin HR, Nicoloso MS, Yendamuri S, Wojcik SE, Shimizu M, Di Leva G, Vecchione A, Trapasso F, Godwin AK, Negrini M, Calin GA, Croce CM 2006 Alterations of the tumor suppressor gene ARLTS1 in ovarian cancer. Cancer Res 66:10287–10291 [DOI] [PubMed] [Google Scholar]
- Ye W, Lv Q, Wong CK, Hu S, Fu C, Hua Z, Cai G, Li G, Yang BB, Zhang Y 2008 The effect of central loops in miRNA:MRE duplexes on the efficiency of miRNA-mediated gene regulation. PLoS ONE 3:e1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A 2004 A combined computational-experimental approach predicts human microRNA targets. Genes Dev 18:1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B, Sepp-Lorenzino L, Prisco M, Linsley P, deAngelis T, Baserga R 2007 Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J Biol Chem 282:32582–32590 [DOI] [PubMed] [Google Scholar]
- Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D 2008 miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer 123:372–379 [DOI] [PubMed] [Google Scholar]
- Chan MW, Wei SH, Wen P, Wang Z, Matei DE, Liu JC, Liyanarachchi S, Brown R, Nephew KP, Yan PS, Huang TH 2005 Hypermethylation of 18S and 28S ribosomal DNAs predicts progression free survival in patients with ovarian cancer. Clin Cancer Res 11:7376–7383 [DOI] [PubMed] [Google Scholar]