Viral Capsid DNA Aptamer Conjugates as Multivalent Cell Targeting Vehicles (original) (raw)
. Author manuscript; available in PMC: 2010 Aug 12.
Published in final edited form as: J Am Chem Soc. 2009 Aug 12;131(31):11174–11178. doi: 10.1021/ja903857f
Abstract
Nucleic acid aptamers offer significant potential as convenient and evolvable targeting groups for drug delivery. To attach them to the surface of a genome-free viral capsid carrier, an efficient oxidative coupling strategy has been developed. The method involves the periodate-mediated reaction of phenylene diamine substituted oligonucleotides with aniline groups installed on the outer surface of the capsid shells. Up to 60 DNA strands can be attached to each viral capsid with no apparent loss of base-pairing capabilities or protein stability. The ability of the capsids to bind specific cellular targets was demonstrated through the attachment of a 41-nucleotide sequence that targets a tyrosine kinase receptor on Jurkat T cells. After the installation of a fluorescent dye on the capsid interior, capsids bearing the cell-targeting sequence showed significant levels of binding to the cells relative to control samples. Colocalization experiments using confocal microscopy indicated that the capsids were endocytosed and trafficked to lysosomes for degradation. These observations suggest that aptamer-labeled capsids could be used for the targeted drug delivery of acid-labile prodrugs that would be preferentially released upon lysosomal acidification.
Introduction
Multivalent and targeted delivery vehicles offer great promise for drug administration and diagnostic imaging. A number of core scaffolds, including polymers,1,2,3 dendrimers,4,5 inorganic nanoparticles,6,7,8 and liposomes,9,10 have been used with considerable success in these applications. In terms of biomolecule-based vectors, engineered heat shock cages11 and viral capsids12 have also been developed to house drug molecules on their interior. For each of these carrier types, a critically important consideration is the installation of receptor-binding groups that enable the selective association of the carriers with targeted tissue types. The most common molecular strategies for this purpose have involved folic acid,13,14 cobalamin,15,16 carbohydrates,17 peptides and antibodies,18,19 and nucleic acid aptamers.20 The rich chemical diversity of these molecules, added to the desire to attach multiple copies of each to scaffolds of varying composition, calls for chemical reactions that are exquisitely functional-group tolerant and proceed under physiological conditions.
An increasing number of chemoselective coupling reactions have been advanced for the labeling of full-sized biomolecules.21,22,23,24,25 All of the reported methods have their particular strengths and ideal usages, and with the addition of each technique, new possibilities have arisen for the generation of complex structures comprising multiple biomolecular components. To add to this list, we have reported a highly efficient oxidative coupling reaction that occurs between anilines and phenylene diamines in the presence of aqueous sodium periodate.26 This reaction has shown exceptional chemoselectivity to date, and proceeds rapidly at micromolar concentrations and at neutral pH. In this report, we apply this method to attach 20–60 copies of DNA aptamers to the surface of genome-free viral capsids, as outlined in Figure 1a. The resulting multivalent assemblies bind to tyrosine kinase receptors on the surface of Jurkat cells and are readily endocytosed. Finally, we show that this chemistry can be combined with other bioconjugation methods that could install functional drug molecules within the carriers. The ability of the oxidative coupling strategy to prepare these heterobiomolecular structures bodes well for its use in the preparation of many different types of delivery vehicles.
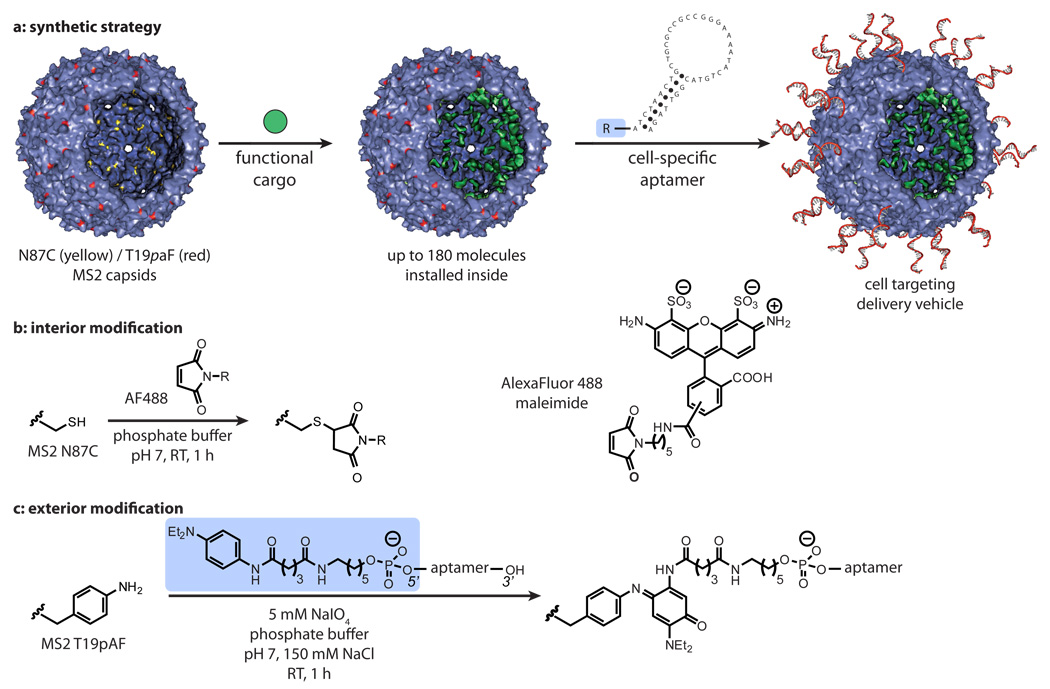
Figure 1. Dual-surface modification of capsids for targeted delivery.
The overall synthetic strategy is shown in (a). (b) For interior surface modification, an N87C mutation on the MS2 coat protein allows for site specific alkylation. Up to 180 cargo molecules can be installed in these locations. (c) For exterior surface modification, the aptamer is first modified with a phenylene diamine group. A T19_p_aF mutation on the capsid allows for the attachment of the modified DNA to the exterior surface of MS2 by a NaIO4 mediated oxidative coupling reaction.
Results and Discussion
Bacteriophage MS2 provides a readily available scaffold for the construction of targeted delivery agents. The protein coat of this virus consists of 180 sequence-identical monomers that are arranged in a hollow spherical structure.27,28 The coat protein monomer can be expressed and self-assembled readily in E. coli, yielding robust, non-toxic and biodegradable structures that are genome-free.29 As MS2 capsids possess thirty-two pores that allow access to the inside of the capsid, selective modification can be achieved on both the interior and the exterior surfaces using orthogonal bioconjugation reactions.30,31 In previous reports, we have shown that tyrosine-based chemistry can be used to install F-18 PET tracers32 and Gd-based MRI contrast enhancement agents33,34 to allow their use in imaging applications.
To endow the capsids with specific targeting capabilities, we have developed an efficient synthetic method to attach nucleic acid aptamers to their surfaces.35 Using the SELEX (Systematic Evolution of Ligands by Exponential Enrichment) process, aptamer sequences can be evolved to bind virtually any target,36,37 and once the composition has been identified, they can be obtained readily using automated solid phase synthesis techniques. Their synthesis is also amenable to the introduction of modified backbones that can improve stability38,39 or impart novel functionality.40 In addition, aptamers can often match or even surpass the specificity and affinity of antibodies, with the added convenience of smaller size. These qualities make them attractive tools for the development of targeted therapeutics and imaging platforms with widely varied targeting capabilities. This potential has been realized in a number of recent reports.41,42,43,44,45,46,47,48,49,50
In order to install oligonucleotide aptamers on the surface of the MS2 capsids, we chose a previously reported NaIO4− mediated oxidative coupling strategy.26,51 This method entails the chemoselective coupling of an N,_N_-diethyl-_N_’-acylphenylene diamine moiety (Figure 1c) to an aniline in the presence of NaIO4. In previous studies, sodium periodate has been used to oxidize the 1,2-diol at the 3’-end of RNA to introduce aldehyde functionalities, but was not observed to degrade the RNA strand.52 Although this observation suggests that RNA-based aptamers could be used with our method, we chose to proceed with DNA due to its enhanced stability towards hydrolysis during the isolation and handling steps.
The aniline coupling partners can be introduced on the exterior surface of MS2 capsids either through direct chemical modification26 or through the introduction of an unnatural amino acid, _p_-aminophenylalanine (_p_aF), into position 19 of the MS2 coat protein using the amber stop codon suppression system.29,53 Using the latter route, we have reported previously that the oxidative coupling strategy can introduce over 100 copies of peptide chains bearing phenylene diamine groups. For this report, we again used the suppression method to produce capsids bearing the T19_p_aF mutation (MS2-_p_aF19), thus providing 180 aniline groups for coupling to the exterior surface. In this case, however, we also introduced an N87C mutation to provide 180 sulfhydryl groups on the interior surface for cargo installation (Figure 1b). The resulting MS2-_p_aF19-N87C double mutant was obtained with a yield of 10 mg/L of culture.
To develop the reaction conditions, a 20-base DNA sequence, strand A (Table 1), was first selected. Starting from an amine-terminated version of this sequence, the N,_N_-diethyl-_N_-acylphenylene diamine moiety was introduced through acylation with an NHS-ester containing precursor. Next, reaction conditions were screened by varying the concentrations of reacting DNA, sodium periodate, and reaction time. Upon analysis by SDS-PAGE, the DNA conjugates exhibited a significant gel shift relative to the unmodified monomers (Figure 2a), allowing facile quantification of the reaction conversion using densitometry after Coomassie staining. Optimal DNA coupling was achieved using 10–20 equivalents (200–400 µM) of the DNA conjugate relative to MS2-_p_aF19 coat protein (20 µM, based on monomer) and 250 equivalents of periodate (5 mM) at room temperature for 1 h. An additional increase in the coupling efficiency was observed when the reaction was run at higher concentrations of sodium chloride, which is likely due to the ability of the higher ionic strength to shield the buildup of negative charge density on the capsid surface as more DNA is attached (see Supplementary Figure S1). For strand A, SDS-PAGE and Coomassie staining, followed by optical densitometry, indicated that 32% of the capsid monomers had been modified with a single strand of DNA. As there are 180 monomers in each capsid, this corresponds to 55 DNA strands on the surface of each. Longer sequences showed slightly lower conversion, most likely due to increased steric effects as well as electrostatic repulsion.
Table 1.
DNA sequences used for attachment to MS2 capsids.a
| Strand | Sequence |
|---|---|
| A | 5’-TCATACGACTCACTCTAGGGA-3’ |
| B | 5’-ATCTAACTGCTGCGCCGCCGGGAAAATACTGTACGGTTAGA-3’ |
| C | 5’-CCCTAGAGTGAGTCGTATGACCCTAGAGTGAGTCGTATGAA-3’ |
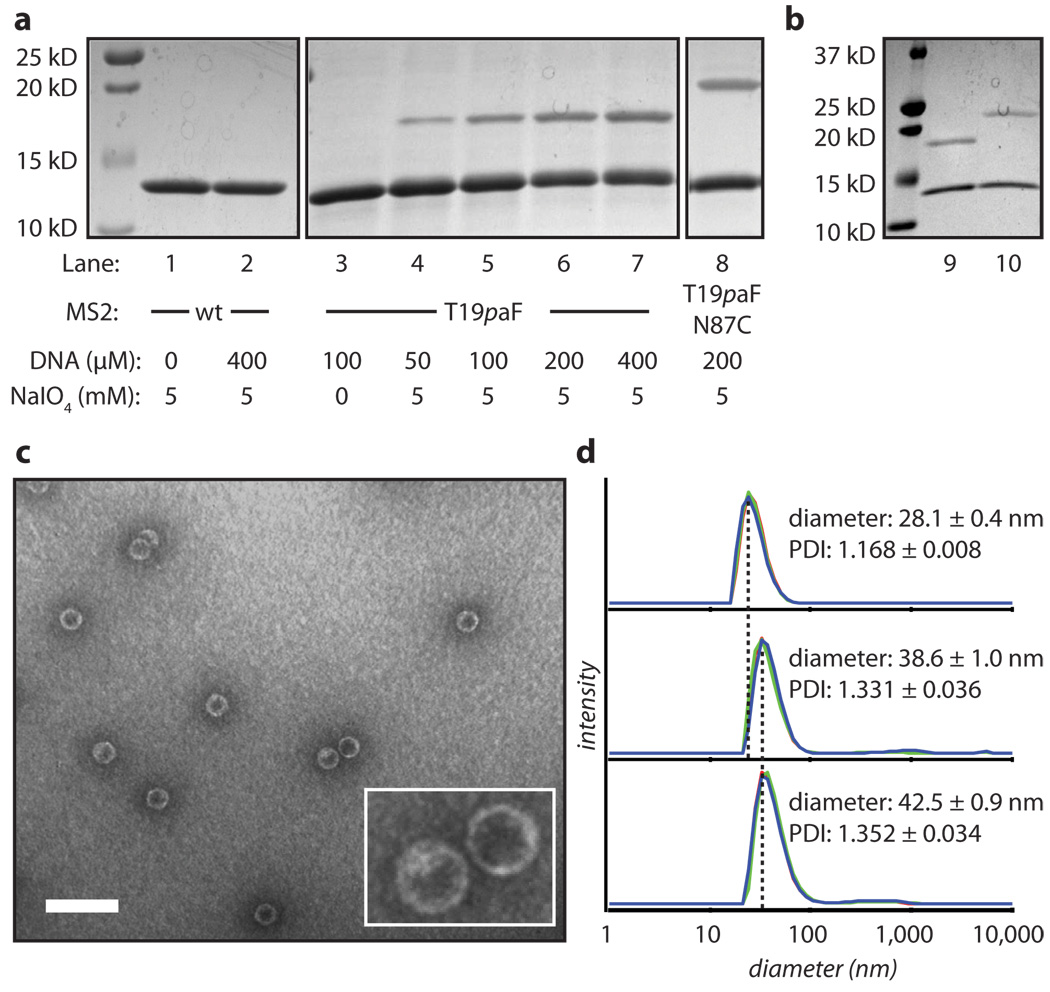
Figure 2. Analysis of DNA attachment to the exterior surface of MS2.
(a) MS2-DNA conjugates were analyzed by SDS-PAGE followed by Coomassie staining. Lanes 1–7 were reacted with strand A. Lane 8 shows the reaction with strand B. (b) A gel-shift assay confirmed DNA competency for base-pairing after conjugation to MS2. Lane 10 includes the complementary sequence to strand A with an additional 20 adenine bases for an increased eletrophoretic shift, while lane 9 has no additional DNA added. Transmission electron micrograph images (c) and dynamic light scattering analysis (d) showed intact capsids after DNA conjugation. In addition, DLS showed a significant increase in the diameter upon conjugation of strand A, as well as an additional increase in diameter upon addition of the 20-base complementary sequence to strand A (without the additional 20-base overhang). The scale bar in the TEM image represents 100 nm.
Following the reaction, the modified capsids could be separated from excess DNA using either size exclusion chromatography or centrifugal concentrators with 100 kDa molecular weight cutoffs. As a technical note, care must be taken to remove all glycerol, ethylene glycol or other vicinal diols from the samples to prevent them from reacting with the periodate.
As shown in Figure 2c,d, the resulting capsids remained intact by transmission electron microscopy (TEM) and dynamic light scattering (DLS). DLS showed an increase of 10.5 ± 0.7 nm in the hydrodynamic diameter upon conjugation of strand A to the capsids. When the complementary strand was introduced, the diameter increased by an additional 3.9 ± 1.0 nm, suggesting that the DNA was still capable of Watson-Crick-Franklin base pairing when conjugated to the exterior of the capsid. The ability to base pair also provides good evidence that the DNA strands are stable throughout the oxidative coupling conditions. Using denatured capsid monomers, base-pairing of the conjugated DNA was further confirmed by SDS-PAGE using a gel-shift assay (Figure 2b).
For functionalization of the interior surface of MS2, standard cysteine bioconjugation was chosen. MS2 contains two native cysteines that have proven to be inaccessible under normal maleimide bioconjugation conditions. Therefore, a double mutant (MS2-_p_aF19-N87C) was expressed to introduce a cysteine on the interior surface. The reactivity of the cysteine mutant is shown in Supplementary Figure S2, where MS2 becomes fluorescently labeled in the presence of Alexa Fluor 488 maleimide (AF488). For this sample, MALDI-TOF MS shows near-quantitative conversion to the singly-modified product. MS2-_p_aF19 capsids lacking the cysteine mutation showed no dye incorporation under identical conditions.
To finish synthesizing the delivery vehicle shown in Figure 1a, a 41-nucleotide DNA aptamer that targets a specific cell surface marker on Jurkat cells (strand B, Table 1) was chosen as the exterior targeting group. Strand B, previously reported as sgc8c, was isolated using a cell-SELEX process, and its binding partner was determined to be protein tyrosine kinase 7 (PTK7).54,55,56 PTK7 is a transmembrane protein that is present on the surface of Jurkat T leukemia cells, as well as many other leukemia cell lines, and has been proposed as a potential biomarker for T cell acute lymphoblastic leukemia.56 Using the oxidative coupling strategy, 20–40 copies of diethyl phenylenediamine-labeled strand B were attached to each capsid, as determined by SDS-PAGE and densitometry analysis (Figure 2a, lane 8). To detect the capsids in cell-binding assays, the interior was modified with AF488 chromophores as described above prior to DNA attachment.
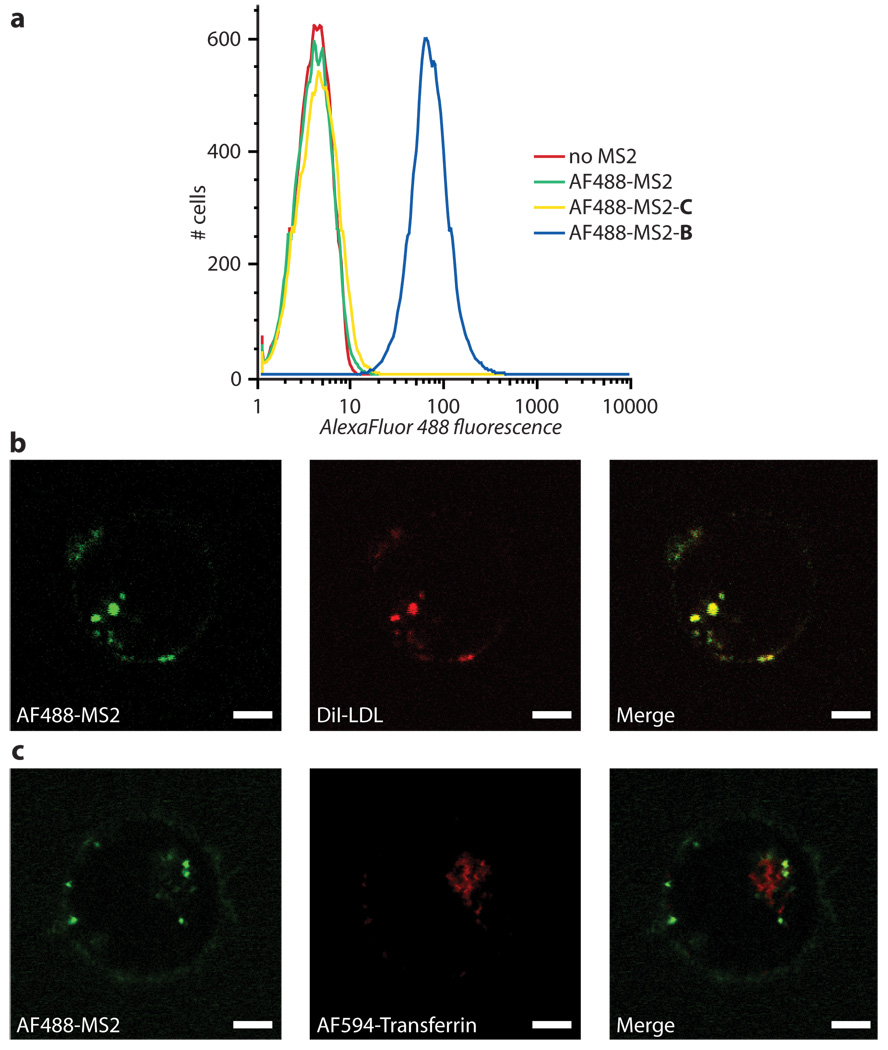
We tested the targeting specificity of these capsids by incubating them with Jurkat cells at 37 °C for 30–60 min in culture media. Subsequent analysis using flow cytometry revealed that samples of cells exposed to MS2 capsids bearing strand B (11 nM in capsids) showed a significant increase in mean fluorescence intensity compared to background cellular autofluorescence, Figure 3a. For negative controls, we synthesized AF488-modified capsids with no exterior modification (AF488-MS2), as well as ones modified with a 41-nt strand of a randomized sequence (C, Table 1). Both control capsids did not give rise to an increase in mean fluorescence intensity, confirming the role of the specific aptamer sequence in cell-binding.
Figure 3. Cellular targeting and uptake with aptamer-labeled capsids.
(a) Cell targeting was confirmed using flow cytometry. Only MS2 capsids modified with strand B bound to Jurkat cells (blue trace). Capsids that were unmodified on the exterior and capsids modified with strand C (green and yellow traces, respectively) showed only background autofluorescence (red trace). Live confocal images showed a colocalization of B-labeled capsids with LDL-labeled endosomes (b), but not with transferrin-labeled endosomes (c). Scale bars represent 3 µm.
Having validated the targeting of B-labeled capsids in flow cytometry experiments, we investigated the cellular internalization of the modified capsids with confocal microscopy. After incubation for 30–60 min at 37 °C with Jurkat cells, the presence of capsids labeled with strand B could be detected as brightly fluorescent dots within the cells, Figure 3b. Co-staining experiments with fluorescent endocytic markers indicated that the B-labeled capsids co-localized with low-density lipoprotein (LDL) particles, but not transferrin. While both transferrin and LDL are known endocytic markers, they traffic through different pathways once inside the cell. Transferrin has been shown to indicate endosomes that are directed back to the surface through the recycling pathway, while vesicles associated with LDL eventually traffic to lysosomes.57 In combination with the targeting specificity of the α-PTK7 aptamer, the lysosomal fate of B-labeled capsids is encouraging for the targeted drug delivery of acid-labile prodrugs that would be preferentially released in these compartments.
Conclusion
For any delivery vehicle composition, the attachment of unprotected biomolecular targeting agents will likely be of key importance to achieve tissue specificity. This report demonstrates the utility of a chemoselective oxidative coupling reaction for this purpose. In principle, the MS2-based vehicle described herein can now be targeted to any receptor for which a binding aptamer has been determined. For the purposes of diagnostic imaging it may not be necessary for the capsids to be internalized; however the observed uptake is envisioned to be highly beneficial for drug delivery applications. In current experiments, we are adding anticancer drugs to these carriers, as well as radionuclides and contrast agents that can be used to determine the location of cellular markers in vivo.
Supplementary Material
1_si_001
Acknowledgements
The development of the coupling reaction described in this work was supported by the NIH (R01 GM072700). Our studies of modified viral capsids as targeted imaging agents are supported by the DOD Breast Cancer Research Program (BC061995). Bryan C. Dickinson is gratefully acknowledged for lending his cell imaging expertise, and we thank Carolyn Bertozzi and her research group for cell culture assistance. The Schultz laboratory is gratefully acknowledged for providing the plasmids required to introduce _p_aF using the amber codon suppression technique.
References
- 1.Liu S, Maheshwari R, Kiick KL. Macromolecules. 2009;42:3–13. doi: 10.1021/ma801782q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R. Proceedings of the National Academy of Sciences. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ. Proceedings of the National Academy of Sciences. 2008;105:17356–17361. doi: 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee CC, MacKay JA, Frechet JMJ, Szoka FC. Nat. Biotech. 2005;23:1517–1526. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- 5.Almutairi A, Rossin R, Shokeen M, Hagooly A, Ananth A, Capoccia B, Guillaudeu S, Abendschein D, Anderson CJ, Welch MJ, Fréchet JMJ. Proceedings of the National Academy of Sciences. 2009;106:685–690. doi: 10.1073/pnas.0811757106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liong M, Lu J, Kovochich M, Xia T, Ruehm SG, Nel AE, Tamanoi F, Zink JI. ACS Nano. 2008;2:889–896. doi: 10.1021/nn800072t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medarova Z, Pham W, Farrar C, Petkova V, Moore A. Nat. Med. 2007;13:372–377. doi: 10.1038/nm1486. [DOI] [PubMed] [Google Scholar]
- 8.Javier DJ, Nitin N, Levy M, Ellington A, Richards-Kortum R. Bioconjugate Chemistry. 2008;19:1309–1312. doi: 10.1021/bc8001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee R, Low P. J. Biol. Chem. 1994;269:3198–3204. [PubMed] [Google Scholar]
- 10.Medinai OP, Zhu Y, Kairemo K. Current Pharmaceutical Design. 2004;10:2981–2989. doi: 10.2174/1381612043383467. [DOI] [PubMed] [Google Scholar]
- 11.Flenniken ML, Willits DA, Harmsen AL, Liepold LO, Harmsen AG, Young MJ, Douglas T. Chemistry & Biology. 2006;13:161–170. doi: 10.1016/j.chembiol.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Flenniken M, Uchida M, Liepold L, Kang S, Young M, Douglas T. Viruses and Nanotechnology. 2009;327:71–93. doi: 10.1007/978-3-540-69379-6_4. [DOI] [PubMed] [Google Scholar]
- 13.Hilgenbrink AR, Low PS. Journal of Pharmaceutical Sciences. 2005;94:2135–2146. doi: 10.1002/jps.20457. [DOI] [PubMed] [Google Scholar]
- 14.Sudimack J, Lee RJ. Advanced Drug Delivery Reviews. 2000;41:147–162. doi: 10.1016/s0169-409x(99)00062-9. [DOI] [PubMed] [Google Scholar]
- 15.Gupta Y, Kohli D, Jain S. Critical Reviews In Therapeutic Drug Carrier Systems. 2008;25:347–379. doi: 10.1615/critrevtherdrugcarriersyst.v25.i4.20. [DOI] [PubMed] [Google Scholar]
- 16.Petrus A, Fairchild T, Doyle R. Angewandte Chemie International Edition. 2009;48:1022–1028. doi: 10.1002/anie.200800865. [DOI] [PubMed] [Google Scholar]
- 17.Darbre T, Reymond J. Curr. Top. Med. Chem. 2008;8:1286–1293. doi: 10.2174/156802608785849058. [DOI] [PubMed] [Google Scholar]
- 18.Brown KC. Current Opinion in Chemical Biology. 2000;4:16–21. doi: 10.1016/s1367-5931(99)00045-9. [DOI] [PubMed] [Google Scholar]
- 19.Schrama D, Reisfeld RA, Becker JC. Nat. Rev. Drug Discov. 2006;5:147–159. doi: 10.1038/nrd1957. [DOI] [PubMed] [Google Scholar]
- 20.Lee JF, Stovall GM, Ellington AD. Current Opinion in Chemical Biology. 2006;10:282–289. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Kolb HC, Finn MG, Sharpless KB. Angewandte Chemie International Edition. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 22.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Proc. Natl. Acad. Sci. U S A. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jencks WP. Journal of the American Chemical Society. 1959;81:475–481. [Google Scholar]
- 24.Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. Journal of the American Chemical Society. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 25.Saxon E, Bertozzi CR. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 26.Hooker JM, Esser-Kahn AP, Francis MB. Journal of the American Chemical Society. 2006;128:15558–15559. doi: 10.1021/ja064088d. [DOI] [PubMed] [Google Scholar]
- 27.Valegard K, Liljas L, Fridborg K, Unge T. Nature. 1990;345:36–41. doi: 10.1038/345036a0. [DOI] [PubMed] [Google Scholar]
- 28.Mastico RA, Talbot SJ, Stockley PG. J. Gen. Virol. 1993;74:541–548. doi: 10.1099/0022-1317-74-4-541. [DOI] [PubMed] [Google Scholar]
- 29.Carrico ZM, Romanini DW, Mehl RA, Francis MB. Chem. Commun. 2008:1205–1207. doi: 10.1039/b717826c. [DOI] [PubMed] [Google Scholar]
- 30.Hooker JM, Kovacs EW, Francis MB. Journal of the American Chemical Society. 2004;126:3718–3719. doi: 10.1021/ja031790q. [DOI] [PubMed] [Google Scholar]
- 31.Kovacs EW, Hooker JM, Romanini DW, Holder PG, Berry KE, Francis MB. Bioconjugate Chemistry. 2007;18:1140–1147. doi: 10.1021/bc070006e. [DOI] [PubMed] [Google Scholar]
- 32.Hooker JM, O’Neil JP, Romanini DW, Taylor SE, Francis M. Molecular Imaging and Biology. 2008;10:182–191. doi: 10.1007/s11307-008-0136-5. [DOI] [PubMed] [Google Scholar]
- 33.Hooker JM, Datta A, Botta M, Raymond KN, Francis MB. Nano Letters. 2007;7:2207–2210. doi: 10.1021/nl070512c. [DOI] [PubMed] [Google Scholar]
- 34.Datta A, Hooker JM, Botta M, Francis MB, Aime S, Raymond KN. Journal of the American Chemical Society. 2008;130:2546–2552. doi: 10.1021/ja0765363. [DOI] [PubMed] [Google Scholar]
- 35.For an alternative approach for the attachment of DNA strands to the cowpea mosaic virus, see: Strable E, Johnson JE, Finn MG. Nano Letters. 2004;4:1385–1389..
- 36.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 37.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 38.Keefe AD, Cload ST. Current Opinion in Chemical Biology. 2008;12:448–456. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 39.Eulberg D, Klussmann S. ChemBioChem. 2003;4:979–983. doi: 10.1002/cbic.200300663. [DOI] [PubMed] [Google Scholar]
- 40.Li M, Lin N, Huang Z, Du L, Altier C, Fang H, Wang B. Journal of the American Chemical Society. 2008;130:12636–12638. doi: 10.1021/ja801510d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farokhzad OC, Jon S, Khademhosseini A, Tran TT, LaVan DA, Langer R. Cancer Res. 2004;64:7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 42.Nimjee SM, Rusconi CP, Sullenger BA. Annu. Rev. Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 43.McNamara JO, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Nat. Biotech. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 44.Bagalkot V, Farokhzad OC, Langer R, Jon S. Angewandte Chemie International Edition. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 45.Bagalkot V, Zhang L, Levy-Nissenbaum E, Jon S, Kantoff PW, Langer R, Farokhzad OC. Nano Letters. 2007;7:3065–3070. doi: 10.1021/nl071546n. [DOI] [PubMed] [Google Scholar]
- 46.Chu TC, Marks JW, Lavery LA, Faulkner S, Rosenblum MG, Ellington AD, Levy M. Cancer Res. 2006;66:5989–5992. doi: 10.1158/0008-5472.CAN-05-4583. [DOI] [PubMed] [Google Scholar]
- 47.Chu TC, Twu KY, Ellington AD, Levy M. Nucl. Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hicke BJ, Stephens AW, Gould T, Chang Y, Lynott CK, Heil J, Borkowski S, Hilger C, Cook G, Warren S, Schmidt PG. J. Nucl. Med. 2006;47:668–678. [PubMed] [Google Scholar]
- 49.Bunka DHJ, Stockley PG. Nat. Rev. Micro. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 50.Huang Y, Shangguan D, Liu H, Phillips JA, Zhang X, Chen Y, Tan W. ChemBioChem. 2009;10:862–868. doi: 10.1002/cbic.200800805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.For an additional example of aromatic amino acid activation using periodate, see: Liu B, Burdine L, Kodadek T. Journal of the American Chemical Society. 2006;128:15228–15235. doi: 10.1021/ja065794h..
- 52.Wu TP, Ruan KC, Liu WY. Nucleic Acids Res. 1996;24:3472–3473. doi: 10.1093/nar/24.17.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mehl RA, Anderson JC, Santoro SW, Wang L, Martin AB, King DS, Horn DM, Schultz PG. Journal of the American Chemical Society. 2003;125:935–939. doi: 10.1021/ja0284153. [DOI] [PubMed] [Google Scholar]
- 54.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Proceedings of the National Academy of Sciences. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shangguan D, Tang Z, Mallikaratchy P, Xiao Z, Tan W. ChemBioChem. 2007;8:603–606. doi: 10.1002/cbic.200600532. [DOI] [PubMed] [Google Scholar]
- 56.Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan W. Journal of Proteome Research. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maxfield FR, McGraw TE. Nat. Rev. Mol. Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1_si_001