Tonic B cell antigen receptor signals supply an NF-κB substrate for prosurvival BLyS signaling (original) (raw)
. Author manuscript; available in PMC: 2009 Sep 15.
Published in final edited form as: Nat Immunol. 2008 Nov 2;9(12):1379–1387. doi: 10.1038/ni.1666
Abstract
The survival of transitional and mature B cells requires both the B cell antigen receptor (BCR) and BLyS receptor 3 (BR3), which suggests that these receptors send signals that are nonredundant or that engage in crosstalk with each other. Here we show that BCR signaling induced production of the nonclassical transcription factor NF-κB pathway substrate p100, which is required for transmission of BR3 signals and thus B cell survival. The capacity for sustained p100 production emerged during transitional B cell differentiation, the stage at which BCR signals begin to mediate survival rather than negative selection. Our findings identify a molecular mechanism for the reliance of primary B cells on continuous BR3 and BCR signaling, as well as for the gradual resistance to negative selection that is acquired during B cell maturation.
Primary B cells rely on signals from both the B cell antigen receptor (BCR) and B lymphocyte stimulator (BLyS1; also called BAFF2; A000383) receptor 3 (BR3; also called BAFFr; A000374) for survival. Most peripheral B cells die after BCR ablation regardless of BR3 sufficiency, which indicates a need for continuous ‘tonic’ signals through the BCR3. Conversely, the lack of either BLyS or BR3, both of which are members of the tumor necrosis factor (TNF) family, results in B cell deficiency despite normal BCR function4–6. The requirement for both BCR and BR3 becomes apparent during transitional B cell differentiation and affects survival at the transitional 2 (T2) and T3 differentiation stages, such that the BCR signaling thresholds for negative and positive selection are modulated by BLyS availability7,8. The molecular mechanism that underlies this codependence on BCR and BR3 is poorly understood.
NF-κB transcription factors are involved in both BCR and BR3 signaling9,10. The classical NF-κB pathway is rapidly activated by signals from several B cell surface molecules, including the BCR11–13. Mice with defects in either BCR signal propagation or the classical NF-κB pathway fail to produce most peripheral B cell subsets14,15. The nonclassical NF-κB pathway follows more protracted kinetics and is activated by a limited set of B cell surface molecules, including lymphotoxin receptors, BR3 and CD40 (refs. 11, 16). Mice that lack components of the nonclassical NF-κB pathway develop phenotypes similar to those of BLyS- or BR3-deficient mice, including a paucity of follicular B cells, no marginal zone B cells, altered germinal center kinetics and compromised T cell–dependent antibody formation10,17,18. These phenotypic similarities are not unexpected, as many of the prosurvival consequences of BR3 signaling, including the induction of members of the antiapoptosis protein Bcl-2 family, expression of Pim2 kinase and cytoplasmic retention of protein kinase C-δ, rely on the nonclassical NF-κB pathway19–21.
Despite a basic understanding of the NF-κB elements ‘downstream’ of BR3 and BCR, why continuous signaling through both receptors is necessary for B cell survival remains unclear. Findings suggest that the classical and nonclassical NF-κB pathways can be coupled through crosstalk involving various NF-κB binding partners22,23. For example, activation of the classical NF-κB pathway induces production of the nonclassical NF-κB substrate p100 (A002936) but does not mediate processing of p100 to the active p52 form10,24. Additionally, the loss of both classical and nonclassical pathways affects follicular B cell numbers more severely than loss of either pathway alone, which further suggests a commensal relationship14,21,25. Such reports led us to question whether such crosstalk might explain why B cell survival depends on both BR3 and BCR signaling.
Our results show that BCR signaling generated p100 for BR3-mediated processing and B cell survival. Thus, in the absence of concomitant BCR signals, BR3 signaling quickly depleted p100 stores and failed to induce long-term survival of splenic B cells. This crosstalk ability emerged during late transitional B cell development. Thus, unlike either late transitional (T2 and T3) or mature B cells, the least mature transitional (T1) B cells could not sustain p100 through BCR signaling. However, greater quantities of membrane cholesterol, a characteristic of mature follicular B cells, bestowed on T1 B cells the capacity for BCR-induced p100 expression. These observations explain why both BCR and BR3 signals are necessary for B cell survival beyond the T1 stage and establish a model in which NF-κB crosstalk integrates primary B cell selection and homeostasis.
RESULTS
BR3 promotes nonclassical NF-κB signaling
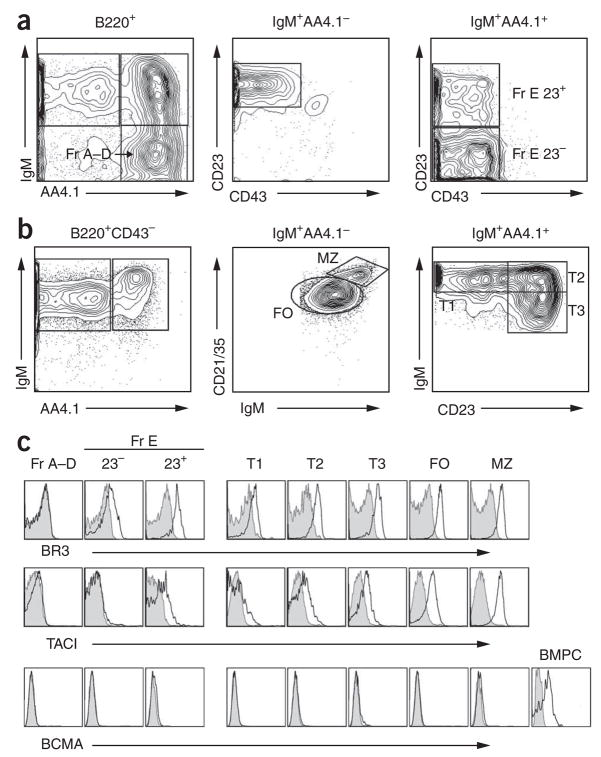
As all three BLyS family receptors (BR3, TACI (A002248) and BCMA (A000374)) can bind BLyS26, and as conflicting data exist about their expression in splenic B cell subsets, we assessed the expression patterns of BLyS receptors before assessing ‘downstream’ BLyS signaling pathways. Both BR3 and TACI were expressed on CD23+ immature B cells in the bone marrow and on all primary splenic B cell subsets beyond the T1 stage (Fig. 1). Real-time PCR analysis of sorted B cell subsets yielded data consistent with those findings (Supplementary Fig. 1a–c online). In contrast, BCMA was not expressed on any of the developing bone marrow or naive splenic B subsets. The failure to rigorously exclude plasma cells during previous analyses of T1 or bulk splenic B cells may explain contradictory results indicating BCMA mRNA expression in these populations27,28. In contrast to the generally similar expression of TACI mRNA and protein, we noted less BR3 mRNA than might have been predicted on the basis of the intensity of BR3 surface expression. Whether this reflected different sensitivities of surface-staining reagents or instead suggests post-transcriptional control of BR3 expression remains to be determined. Nonetheless, these results designate BR3 and TACI as the main BLyS receptors on transitional, follicular and marginal zone B cells.
Figure 1.
BLyS expression on splenic and bone marrow B cells. (a,b) Staining used to identify developmental B cell subsets in bone marrow (a) and splenic (b) populations. (c) Surface expression of BR3, TACI and BCMA on bone marrow and splenic B cell subsets (open histograms). Shaded histograms, isotype-matched control antibody. BMPC, bone marrow plasma cells (positive control for BCMA surface expression). Fr A–D, Hardy fractions A–D; Fr E, Hardy fraction E; 23−, CD23−; 23+, CD23+, FO, follicular; MZ, marginal zone. Data are representative of at least three independent experiments.
Because primary B cells express BR3 and TACI, as well as several additional receptors that can activate one or both NF-κB pathway(s), we first established the NF-κB signaling potential of BR3 and TACI in isolation. We cloned full-length cDNA encoding BR3 or TACI into the MigR1 expression plasmid and stably introduced the resultant constructs into NIH3T3 fibroblast cell lines (Supplementary Fig. 2a, b online). Only the BR3-expressing cell lines processed p100 when stimulated with BLyS (Supplementary Fig. 2c). Stimulation with APRIL (A000305), another TNF family member, failed to elicit p100 processing in BR3-expressing cell lines, consistent with the inability of APRIL to bind to BR3 (ref. 26). Although we occasionally found small increases in p52 expression in TACI-expressing cells after APRIL stimulation, no p100 degradation occurred. In contrast, BLyS and APRIL induced robust activation of the classical NF-κB pathway only in the TACI-expressing cell lines (Supplementary Fig. 2d). Thus, when isolated from other potential mediators of NF-κB activation, BR3 engagement ‘preferentially’ activated the nonclassical NF-κB pathway, whereas TACI ligation ‘favored’ the classical NF-κB pathway.
Quiescent B cells generate but do not process p100
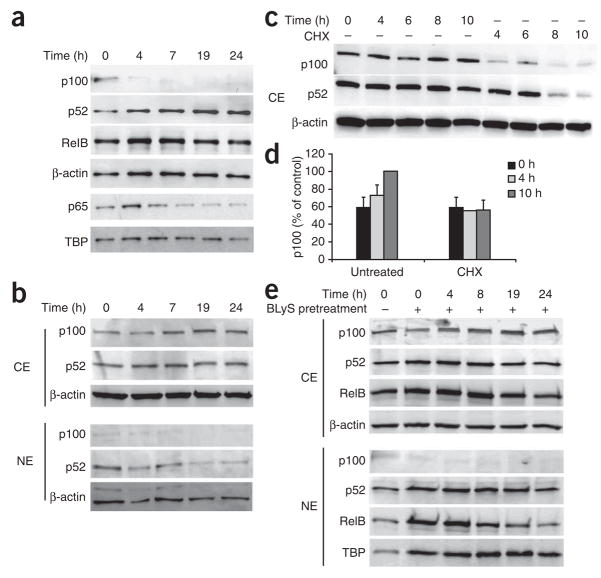
The results reported above suggested that in primary B cells, BLyS might be able to activate both classical and nonclassical NF-κB pathways through TACI and BR3, respectively. To test that idea, we treated CD43− splenic B cells with BLyS and monitored the nuclear localization of NF-κB family members. Over a time course of 24 h, we noted gradual accumulation of nuclear p52, a hallmark of the nonclassical NF-κB pathway (Fig. 2a). In contrast, nuclear localization of p65 (also called RelA), an indicator of activation of the classical pathway, was rapid and transient. We inferred that stimulation through TACI resulted in transient nuclear localization of p65, whereas stimulation through BR3 led to persistent p52 generation.
Figure 2.
Continuous BLyS signaling is required for p100 processing and sustained nuclear accumulation of p52. (a) Immunoblot analysis of nuclear extracts of CD43− splenic B cells cultured for 0–24 h (above lanes) with BLyS (100 ng/ml). Results are representative of five independent experiments. (b,c) Immunoblot analysis of cytoplasmic extracts (CE) and nuclear extracts (NE) of CD43− splenic B cells incubated for various times (above lanes) in medium alone (b) or in the presence of cycloheximide (CHX; 10 μg/ml; c). Results are representative of three experiments. (d) Expression of p100 in cytoplasmic extracts of B cells left untreated or treated with cycloheximide, presented as a proportion of the p100 expression in untreated cells at 10 h (control). Data are from three independent experiments (mean and s.d.). (e) Immunoblot analysis of cytoplasmic and nuclear extracts of CD43− splenic B cells left untreated (−) or pretreated for 24 h with BLyS (100 ng/ml; +), then washed extensively to remove exogenous BLyS and incubated for various times (above lanes) in medium alone. TATA-binding protein (TBP) and β-actin serve as loading controls. Data are representative of three independent experiments.
The activation of NF-κB-inducing kinase leads to phosphorylation and activation of inhibitor of NF-κB kinase-α (IKKα), which subsequently phosphorylates two key serine residues on p100 (ref. 11). The resulting ubiquitination and proteasomal degradation of p100 leaves a smaller p52 subunit and exposes a nuclear-localization sequence, which allows heterodimers of p52 and RelB to enter the nucleus11. The continued increase in p52 nuclear localization in BLyS-treated cells indicated either that starting p100 quantities were large enough to accommodate the rate at which processing occurred over the course of the assay or that continued p100 synthesis drove the increase in p52 nuclear localization. To distinguish between those possibilities, we measured p100 in nuclear extracts of BLyS-treated B cells. We found that p100 decreased rapidly after BLyS treatment and was barely detectable by 4–6 h of treatment (Fig. 2a). Thus, the continued increase in p52 accumulation during the first 24 h of BLyS treatment probably resulted from new p100 synthesis; presumably, the newly synthesized protein was continually phosphorylated and processed in the presence of BLyS, which thus precluded its accumulation. We therefore assessed whether p100 protein accumulated over time in quiescent B cells incubated in medium alone. We found increased p100 expression in the cytoplasm of these cells, particularly at later time points (Fig. 2b).
To determine whether p100 was newly synthesized in resting B cells, we blocked protein synthesis with cycloheximide. This treatment decreased the time-dependent accumulation of cytoplasmic p100 (Fig. 2c,d); the initial decrease in p100 signal intensity that occurred between 0 h and 4 h in cycloheximide-treated cells may have resulted from residual BLyS signaling in the purified cells. Of note, we could not extend this analysis to later time points because of cycloheximide toxicity (Supplementary Fig. 3a online). We inferred that the p100 accumulation in B cells cultured ex vivo was the result of new protein synthesis rather than selection for viable cells with high expression of p100. Our interpretation was further confirmed by higher p100 expression in cultured B cells transgenic for the antiapoptotic factor Bcl-xL, which are resistant to apoptosis29. Notably, newly synthesized p100 was restricted to the cytoplasm, and nuclear p52 quantities decreased gradually over time. The rate of p100 accumulation was not affected by pretreatment of B cells with BLyS (Fig. 2e and Supplementary Fig. 3b). After BLyS removal, cytoplasmic p100 steadily increased to the amount before BLyS treatment, which suggested involvement of ‘tonic’ BCR signaling in the generation of p100. When we removed BLyS from cultures after 24 h, nuclear p52 and RelB decreased over time, which indicated that activation of the nonclassical pathway is not self-sustaining and requires constant BR3 signaling to maintain nuclear localization of these heterodimers. We therefore concluded that B cells require constant production of p100 that can serve as a substrate for BLyS-mediated signaling through the nonclassical NF-κB pathway.
BCR signals generate p100 for BLyS-mediated processing
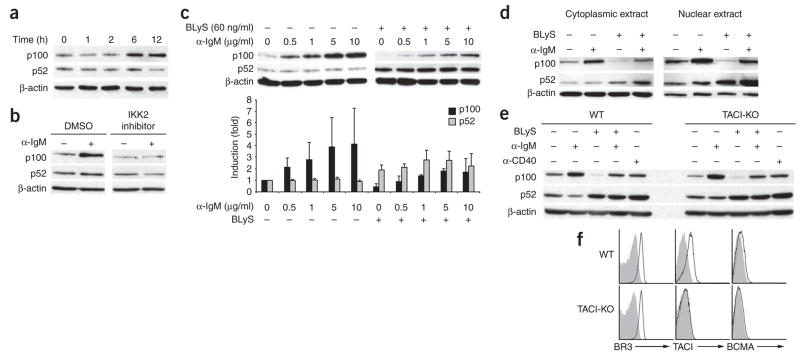
To understand the molecular basis of the p100 synthesis, we considered the following possibilities: new p100 protein may result from constitutive gene transcription and translation; alternatively, the gene encoding p100 may be the ‘downstream’ target of active signaling. Several studies have shown that B cells lacking the classical NF-κB pathway component p50 have less p100 and respond poorly to BLyS-dependent survival signals24,30. Because the gene encoding p100 has been proposed as a target of the classical NF-κB pathway, likely candidates for the induction of p100 transcription were either TACI or BCR signals31. To distinguish between those possibilities, we compared p100 expression in extracts of B cells stimulated through the BCR. We found that p100 increased after crosslinking of immunoglobulin M (IgM), typically within 6 h of stimulation (Fig. 3a). This increase was blocked by treatment with an inhibitor of the classical NF-κB pathway (Fig. 3b), which indicated that BCR-driven p100 generation relies on classical NF-κB elements30. Additional experiments with B cells from p50-deficient mice yielded similar results (data not shown). The magnitude of the increase in p100 expression was proportional to the strength of the BCR signal (Fig. 3c). Notably, in cells simultaneously treated with BLyS, we noted a range in which p100 was processed while p100 pools were maintained; these cells had more nuclear p52 localization than did those treated with BLyS alone (Fig. 3d). These data suggest that p100 expression can be sustained when BCR and BR3 signals balance p100 production and consumption.
Figure 3.
NF-κB crosstalk ‘downstream’ of BCR and BR3 signaling in follicular B cells. (a) Immunoblot analysis of lysates of CD23+ follicular cells cultured for various times (above lanes) with antibody to IgM F(ab′)2 (10 μg/ml). (b) Immunoblot analysis of lysates of CD23+ follicular B cells cultured for 18 h in medium alone (−) or with anti-IgM (α-IgM; 10 μg/ml; +) in the presence of an inhibitor of the classical pathway component IKK2 or an equivalent volume of DMSO. (c) Immunoblot analysis (top) of lysates of CD23+ follicular B cells cultured for 24 h with various concentrations (above lanes) of anti-IgM alone (left) or with BLyS (60 ng/ml; right). Below, band intensities normalized to β-actin and presented as expression relative to that of cells cultured in medium alone. (d) Immunoblot analysis of lysates of CD23+ B cells stimulated for 24 h with BLyS (60 ng/ml) and/or anti-IgM (10 μg/ml). (e) Immunoblot analysis of lysates of wild-type (WT) or TACI-deficient (TACI-KO) B cells cultured for 24 h with various stimuli (above lanes). β-actin serves as a loading control in all blots. (f) Flow cytometry of the surface expression of BLyS receptors on wild-type and TACI-deficient B cells (open histograms). Shaded histograms, isotype-matched control antibody. Data are representative of at least three independent experiments (a–e; mean and s.d., c) or ten experiments (f).
We obtained identical results with B cells isolated from TACI-deficient mice (Fig. 3e,f), which thus excluded TACI as a principal participant in p100 generation or processing. Furthermore, when we stimulated wild-type B cells with APRIL, we failed to find an increase in p100 expression (data not shown), even though our cell line data indicated that BLyS and APRIL can engender signals through TACI (Supplementary Fig. 2). Studies suggest that trimeric BLyS signals poorly through TACI32, and it is therefore possible that the signaling in our cell lines reflected an enhanced ability of BLyS to interact with TACI in the absence of competing BR3. These findings indicated that whereas TACI signaling contributes minimally, BCR signaling is a potent source of sustained p100 generation.
BCR-generated p100 facilitates BLyS-mediated survival
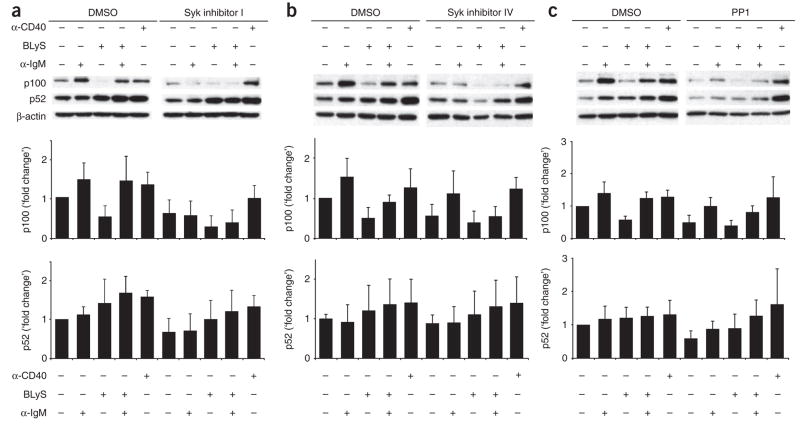
Having obtained evidence challenging the idea that TACI is involved, we further investigated BCR signaling as a mechanism for p100 synthesis. As the BLyS treatment reported above (Fig. 2) was done in the absence of IgM crosslinking, p100 generation may have occurred through ‘tonic’ BCR signaling, which is initiated in the apparent absence of a BCR-crosslinking ligand33. Thus, we assessed both constitutive and ligation-induced BCR signals as sources of sustained p100 generation. Activation of Src family tyrosine kinases and the Syk tyrosine kinase is an early event in BCR signal propagation. We therefore used two Syk inhibitors (Syk inhibitor I (ref. 34) and Syk inhibitor IV (ref. 35)), as well as the Src-family kinase inhibitor PP1 (ref. 36), to determine whether new p100 synthesis depended on BCR-initiated signals. All inhibitors decreased tyrosine phosphorylation and prevented blast formation after BCR crosslinking (Supplementary Fig. 4a,b online). We treated B cells with BLyS and/or antibody to IgM (anti-IgM) in the presence or absence of each inhibitor and measured p100 and p52 by immunoblot analysis (Fig. 4). Consistent with the idea that BCR signaling is necessary for p100 maintenance, all inhibitors decreased p100 in both unstimulated cells and cells treated with anti-IgM. As neither Syk nor Src family kinases are ‘downstream’ of BR3, BLyS-mediated p100 processing was unaffected by the inhibitors. In contrast, all inhibitors prevented concomitant BCR signaling from sustaining the amount of p100 in BLyS-treated cells. To rule out the possibility of global effects on p100 generation, we used CD40 ligation, which activates both classical and nonclassical NF-κB pathways independently of Syk. As expected, treatment with anti-CD40 induced p100 maintenance and processing regardless of the presence of inhibitor. The amount of p52 in whole-cell extracts was also decreased by inhibitor treatment, except in cells treated with anti-CD40. However, the decrease in p52 followed more protracted kinetics, especially in cells stimulated with anti-IgM, which probably reflected the p52 generated before the depletion of p100 stores. Indeed, the decrease in p52 was greater at 36 h (data not shown), but differences in the division of control and inhibitor-treated cells precluded rigorous comparisons at later time points.
Figure 4.
BCR signaling blockade decreases tonic and anti-IgM–stimulated p100 generation. Immunoblot analysis (top) of nuclear extracts of C57BL/6 CD23+ follicular B cells stimulated for 24 h with anti-IgM (10 μg/ml), BLyS (100 ng/ml) and/or anti-CD40 (10 μg/ml), along with a chemical inhibitor of Syk (I or IV) or Src (PP1) or an equivalent volume of DMSO. β-actin serves as a loading control. Below blots, band intensity relative to that of cells incubated in medium alone. Data are representative of at least three independent experiments (mean and s.d.).
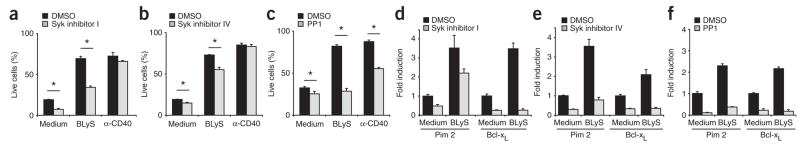
The results reported above are consistent with the idea that BR3 signaling relies on BCR-mediated maintenance of p100 substrate pools. If this is the case, then the enhanced survival and upregulation of antiapoptotic genes associated with BLyS treatment should be blocked by BCR signaling inhibitors despite the inability of these inhibitors to inhibit BR3 signaling itself. To test that hypothesis, we measured long-term (72-hour) cell survival and transcription of prosurvival genes (Fig. 5). Culture of B cells together with either of the two Syk inhibitors resulted in less BLyS-mediated survival but not less CD40-mediated survival (Fig. 5a,b). In agreement with those results, both Syk inhibitors also prevented the upregulation of Pim-2 and Bcl-xL mRNA normally associated with BR3 signaling (Fig. 5d,e). Global inhibition of Src kinase impaired BLyS-induced expression of Pim-2 and Bcl-xL but resulted in more death of cells stimulated with BLyS or anti-CD40 (Fig. 5c,f), which reflected the greater toxicity of this reagent.
Figure 5.
BCR signaling blockade decreases BLyS-mediated signaling and long-term survival. (a–c) Viability of CD23+ B cells cultured for 72 h with medium alone, BLyS (100 ng/ml) or anti-CD40 (10 μg/ml) in the presence of DMSO or a chemical inhibitor of Syk (I or IV) or Src (PP1), assessed by TO-PRO-3 exclusion. *, P, 0.05. (d–f) Real time PCR analysis of the expression of Pim2 mRNA and Bcl-xL mRNA in CD23+ B cells cultured for 24 h with medium alone, BLyS (100 ng/ml) or anti-CD40 (10 μg/ml) in the presence of DMSO or inhibitors of Syk or Src (as in a–c). Results are normalized to GAPDH expression and are presented relative to expression in cells incubated in medium alone (arbitrary units). Data are representative of at least two independent experiments (mean and s.d. of triplicate wells).
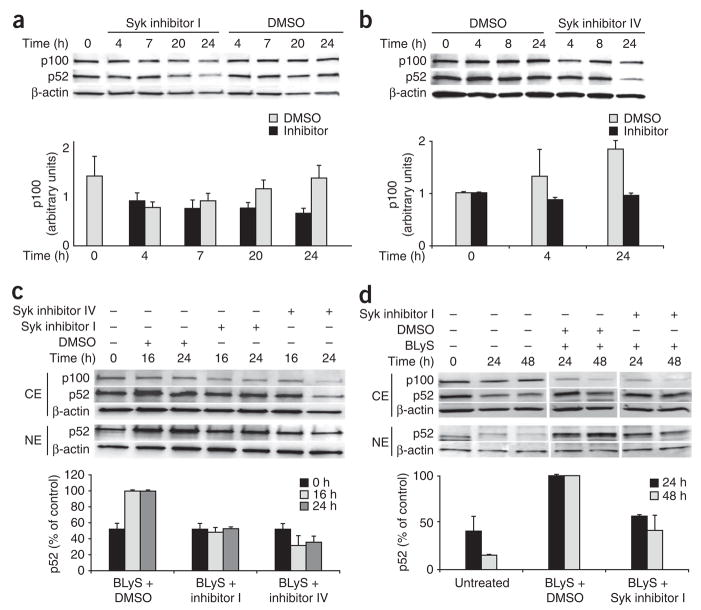
Having established that the inhibitors blocked ligation-induced BCR signals, we used them to determine whether tonic BCR signals contributed to p100 upregulation in quiescent B cells. We treated splenic B cells with the Syk inhibitors for various times and examined the effect on cytoplasmic p100 by immunoblot analysis. Although p100 increased in untreated cells or cells treated vehicle control (after an initial decrease at 4 h), we found no increase in cells treated with the inhibitors (Fig. 6a,b). These observations are all consistent with the interpretation that tonic BCR signals contribute to new synthesis of p100. We reasoned that if Syk-dependent p100 synthesis creates the substrate for effective BLyS signaling, then p52 would fail to accumulate after extended BLyS treatment of cells in which tonic BCR signals have been interrupted. To test our idea, we cultured B cells with BLyS in the presence or absence of the Syk inhibitors and measured nuclear p52. As expected, blockade of tonic BCR signaling by either inhibitor precluded nuclear accumulation of p52 in BLyS-treated B cells (Fig. 6c). A similar decrease in nuclear accumulation of p52 occurred in B cells from Bcl-xL-transgenic mice (Fig. 6d), which ruled out differential cell death as a cause of diminished p52 accumulation. We thus infer that sustained BR3 signaling in resting B cells relies on p100 provided by tonic BCR signals.
Figure 6.
Effect of blocking tonic BCR signals on nonclassical NF-κB components. (a,b) Immunoblot analysis (top) of cytoplasmic extracts of CD43− splenic B cells cultured with 10 μM Syk inhibitor I (a), 50 nM Syk inhibitor IV (b) or an equivalent volume of DMSO. β-actin serves as a loading control. Below blots, cytoplasmic p100, presented relative to β-actin. (c,d) Immunoblot analysis (top) of cytoplasmic and nuclear extracts of wild-type (c) or Bcl-xL-transgenic (d) splenic B cells incubated for various times (above lanes) with or without BLyS (100 ng/ml) in the presence or absence of Syk inhibitors I or IV or DMSO. Below blots, nuclear p52, presented as a proportion of the amount in cells treated with BLyS alone. β-actin serves as a loading control in all blots. Data are representative of two or more independent experiments (blots, a–d), five independent experiments (graph, a; mean and s.d.) or two independent experiments (graphs, b–d; mean and range).
BCR-mediated p100 maintenance arises with maturation
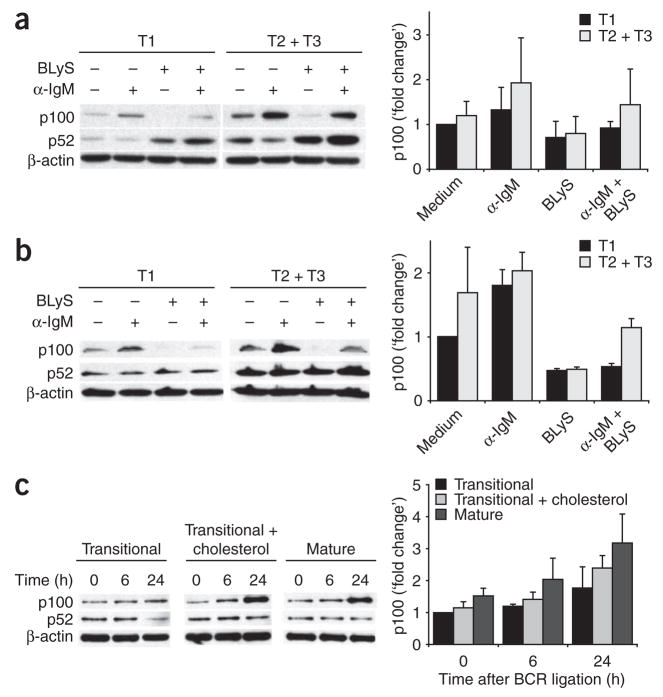
In parallel experiments, we consistently noticed little difference in the p100 quantities in resting Bcl-xL-transgenic B cells cultured with inhibitors (Supplementary Fig. 5a,b online). We reasoned that this might reflect a lack of selection for functional BR3 signaling, as enforced Bcl-xL expression ‘rescues’ BR3 mutations and restores transitional survival and maturation to an above-normal degree37. If this were the case, then wild-type transitional differentiation would be accompanied by a greater capacity to sustain p100 through BCR signaling. We therefore sought to determine whether the relationship between BCR-mediated p100 generation and BR3 signaling is established as cells pass through successive transitional differentiation subsets. We found that T1 B cells failed to induce p100 to the same extent as follicular B cells did after BCR crosslinking (Fig. 7a). In contrast, sorted T2 and T3 cells upregulated p100 to an extent similar to that of follicular B cells and could sustain p100 or p52 pools with concomitant BR3 signaling (Fig. 7a). As T1 cells are susceptible to greater cell death, we repeated the experiment with sorted T1 versus T2 and T3 B cells from Bcl-xL-transgenic mice. The results indicated that whereas BCR stimulation could lead to some p100 synthesis in T1 cells (Fig. 7b), it was insufficient to sustain p100 during concomitant BLyS signaling.
Figure 7.
T1 B cells have a diminished capacity to upregulate p100 after BCR stimulation in vitro. (a) Immunoblot analysis of whole-cell lysates of T1 B cells (B220+ AA4.1+ CD23−) or T2 and T3 B cells (B220+AA4.1+CD23+) sorted by flow cytometry from C57BL/6 splenocytes in low-pressure conditions (15 psi) and treated for 24 h with anti-IgM or BLyS. (b) Immunoblot analysis of splenic transitional B cells from Bcl-xL-transgenic mice irradiated (5 Gy) 13.5 d before cell collection to enrich for transitional B cells; cells were sorted, treated and analyzed as described in a. Right (a,b), p100 band intensity relative to that of untreated T1 cells. (c) Immunoblot analysis of CD43-depleted transitional B cells obtained from C57BL/6 spleens 13.5 d after 5 Gy of whole-body irradiation, followed by the addition of cholesterol by means of treatment with 5 mM methyl-β-cyclodextran, then stimulation of untreated transitional cells, follicular cells or transitional cells with added cholesterol with anti-IgM for 0–24 h (above lanes and below bars) and analysis as described in a. Right, p100 band intensity relative to that of untreated transitional cells at time 0. Data are representative of two or more independent experiments (blots, a–c), two independent experiments (graphs, a,b; mean and range) or three independent experiments (graph, c; mean and s.d.).
The observations reported above suggested that transitional differentiation includes a shift in the efficiency with which BCR signals generate p100, thus enabling greater capacity for BLyS-mediated survival. Because transitional B cells adopt the BCR signaling characteristics of follicular B cells when cholesterol is added to their membranes38, we sought to determine whether the addition of exogenous cholesterol would impart competence to generate p100. Whereas p100 upregulation in response to BCR signaling was poor among untreated transitional cells, cholesterol-enriched transitional cells had BCR-induced p100 production similar to that of follicular B cells (Fig. 7c). However, the addition of cholesterol did not induce phenotypic maturation of T1 cells, at least during their time in culture (Supplementary Fig. 6 online). Thus, the adoption of ‘upstream’ BCR signaling characteristics associated with maturation engenders the capacity for efficient downstream p100 generation.
DISCUSSION
Here we have shown that crosstalk between the classical and non-classical NF-κB pathways mediates the survival of follicular B cells. Tonic BCR signals generated p100, which was processed by BR3-induced signals. In addition, we have shown that the ability to sustain p100 through BCR signaling emerged during late transitional differentiation. Our findings provide mechanistic explanations for the involvement of tonic BCR signaling in mediating the survival and selection of primary B cells, as well as for the cross-modulation between BR3 and BCR signals20,21,24.
Most follicular B cells express both TACI and BR3, and these receptors ‘preferentially’ activate the classical and nonclassical NF-κB pathways, respectively. Our results have indicated that BCR-mediated activation of the classical pathway is critical in the generation and maintenance of substrate for the nonclassical pathway. We confirmed that TACI ligation effected degradation of the inhibitory cytoplasmic NF-κB chaperone IκBα in vitro, but we repeatedly failed to find p100 generation in primary B cells stimulated with APRIL (data not shown). This raises the question of why TACI, which can also bind BLyS and signal through the classical NF-κB pathway, cannot compensate for BCR signal blockage or ablation1,5,18. TACI signals may not engage the appropriate ‘downstream’ systems necessary for p100 generation in the context of primary B cells, despite activation of the classical NF-κB pathway. Many TNF receptor–activated factor combinations are ‘downstream’ of each BLyS receptor, and these may result in the generation of different NF-κB dimer combinations by each BLyS receptor and by the BCR39. Alternatively, studies suggest that oligomeric ligands can promote survival through TACI signaling32. These ligand forms may be present in low abundance or may be sequestered in vivo; the resulting sporadic TACI engagement may fail to maintain p100 quantities sufficient for BR3 signal propagation.
Our observations suggest a model in which continuous BCR signaling is needed to generate a substrate required for the transmission of BR3 signals; this capacity for BCR-induced p100 production is acquired during late transitional B cell differentiation. This model provides a molecular explanation for why neither receptor alone is sufficient for mature B cell survival and suggests that a developmentally regulated event facilitating this crosstalk promotes resistance to BCR-induced negative selection. Alternatively, the developmentally associated propensity for BCR-mediated p100 generation might reflect lower proportions of anergic B cells in successive transitional subsets. Indeed, anergic B cells have defective BCR signaling40, and two thirds of transitional B cells usually die, which reflects failed BCR and BR3 signaling41. Our finding that follicular B cells from Bcl-xL-transgenic mice had ‘muted’ tonic BCR-induced p100 generation is consistent with that idea, as these cells have bypassed the usual transitional selection for BR3-mediated survival37. These possibilities are not mutually exclusive, and both are consistent with the idea that only cells whose BCR signals maintain enough p100 to sustain BR3 signaling persist.
Whereas naive B cells are probably maintained by the crosstalk described here, a growing literature suggests that additional, bidirectional crosstalk points probably exist between these pathways. For example, BLyS-BR3 interactions can initiate classical NF-κB activation but with unusually slow kinetics21. Furthermore, sustained superphysiological amounts of active IKKβ can restore the B cell phenotype of mice lacking the key critical NF-κB pathway initiator NEMO (also called IKKγ) or BR3 (ref. 21). Such observations indicate that the stoichiometry of interacting components may be critical and that alterations in their ratios may shift or reverse particular signaling outcomes. For example, BR3 signaling through classical NF-κB is present in B cells mutant for the tyrosine kinase Btk30. Whether this reflects an intersection of the BR3 and BCR pathways that is exposed in the context of unusually weak BCR signaling or is instead a product of the crosstalk described here will require further study. Similarly, excess p100 can inhibit both classical and nonclassical pathways, which suggests that when the BLyS-mediated processing capacity is exceeded, the accumulating p100 may exert negative effects23. Consistent with that possibility, transitional B cell maturation is blocked in mice transgenic for a p100 construct incapable of undergoing processing to p52 (ref. 42), and we found more nuclear p100 after strong BCR crosslinking. Such a relationship, in which survival is fostered only when p100 generation and processing are balanced, might explain why strong BCR signals yield death at the transitional stages but can be reversed by signals that augment p100 processing, such as excess BLyS8,43 or CD40 ligation44.
METHODS
Plasmid construction
The plasmid pRK5B-hBR3 was from V. Dixit. The ‘hBR3’ region was excised from the parent vector with the restriction enzymes _Eco_RI and _Not_I (Invitrogen). The resulting fragment was then ligated into the plasmid MIGRI-GFP with complementary sites and was sequenced to show 100% base pair homology with the coding portion of the GenBank nucleotide sequence for human BR3 (TNFRSF13C mRNA; NM_052945). The BR3-pCR4-TOPO plasmid was cut with _Eco_RI to release the BR3 amplicon, which was cloned into the _Eco_RI site of plasmid MigRI to form plasmid BR3-MigRI. The MigR1 plasmid allows bicistronic expression of both green fluorescent protein and the protein of interest, which therefore allows the discernment of successfully transduced cells by flow cytometry. Plasmid TACI-MigRI was constructed in a similar way, with the primers MC7F (5′-AGTAATGAGTGGTGTAGGCC-3′) and MC7R (5′-CGTGTCTTCATCTGTCTCTTC-3′). The 900–base pair amplicon was cloned into vector pCR4-TOPO (Invitrogen) was sequenced to demonstrate 100% base pair homology with the coding portion of the GenBank nucleotide sequence for mouse TACI (Tnfrsf13b mRNA; NM_021349.1) and was cloned into the _Eco_RI site of plasmid MigRI to form plasmid TACI-MigRI.
Preparation of retroviral supernatant
Human embryonic kidney 293T cells were transfected with the BR3-MigRI or TACI-MigRI plasmid, together with helper plasmid45. Transfection ‘cocktails’ consisted of 4–6 μg helper plasmid, 8–12 μg MigRI plasmid and 36 μl FuGENE 6 transfection reagent (Roche) in a final total volume of 0.5 ml OptiMEM I medium (Gibco). Retroviral supernatants were collected at 24 h and 48 h after transfection and were stored at −80 °C.
Retroviral infection of NIH3T3 cells
NIH3T3 cells were infected with 0.5–1.0 ml retroviral supernatant as described45 in the presence of polybrene (4 μg/ml). After 36–48 h, cells were collected and their expression of green fluorescent protein was analyzed by flow cytometry. Additionally, surface protein expression was measured by flow cytometry with phycoerythrin-conjugated anti–human BR3 (8A7; eBioscience) and phycoerythrin-conjugated anti-TACI (FAB1041P; R&D Systems). In addition, cell lines were lysed and resolved by SDS-PAGE and expression was assessed by immunoblot analysis with polyclonal anti–human BR3 (14-6144-81; eBioscience) and anti-TACI (AF1041; R&D Systems).
Nuclear and cytoplasmic fractionation
Cells were collected by centrifugation and washed with cold PBS. Pellets (20 × 106 cells) were suspended in 200 μl buffer A (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5% (vol/vol) Nonidet P-40, 0.5 mM dithiothreitol and 0.5 mM phenylmethyl sulfonyl fluoride) and were incubated for 20 min on ice. After centrifugation at 6,000_g_ for 10 min at 4 °C, supernatants were collected as the cytoplasmic extract. Pellets were washed twice with buffer A and were resuspended in 20 μl buffer C (20 mM HEPES, pH 7.9, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% (vol/vol) glycerol, 0.5 mM dithiothreitol and 0.5 mM phenylmethyl sulfonyl fluoride) through the use of a pellet-homogenizing pestle. After incubation for 30 min on ice, lysates were centrifuged at 13,000 r.p.m. for 10 min at 4 °C and supernatants were collected as the nuclear extract. Then, 20 μl buffer D (20 mM HEPES, pH 7.9, 0.2 mM EDTA, 20% (vol/vol) glycerol, 0.5 mM dithiothreitol and 0.5 mM phenylmethyl sulfonyl fluoride) was added to the extract before quantification and use.
Cell culture and B cell enrichment
NIH3T3 cells were cultured in RPMI medium containing 10% (vol/vol) Fetalplex serum (Gemini Bio-Products), 1% (vol/vol) L-glutamine (Gibco) and 1% (vol/vol) penicilin-streptomycin solution (Gibco). CD23+ follicular splenic B cells were prepared by magnetic-activated cell sorting and cultured as described27,46. In some experiments, B cells were cultured with APRIL (315-13; Peprotech), antibody to IgM F(ab′)2 (115-006-020; Jackson ImmunoResearch), recombinant human BLyS (Human Genome Sciences) or anti-CD40 (3/23; BD Biosciences). BCR signaling was inhibited by the addition of 10 μM Syk inhibitor I (3-(1-methyl-1H-indol-3-yl-methlyene)-2-oxo-2,3-dihydro-1H-indole-5-sulfonamide; 574711; Calbiochem), 50 nM Syk inhibitor IV (2-(7-(3,4-dimethoxyphenyl)-imidazo(1,2-c)pyrimidin-5-ylamino)-nicotinamide, 2 HCl; BAY 61-3606; 574714; Calbiochem) or 10 μM PP1 (EI-275; Biomol International). As a control for the inhibitors, the equivalent volume of dimethyl sulfoxide (DMSO) was used. Classical NF-κB activation was inhibited by treatment with 20 μM InSolution IKK-2 Inhibitor (SC-514; Calbiochem) or an equivalent volume of DMSO. T cell depletion was achieved by magnetic-activated cell sorting as follows: splenocytes were incubated with biotin-conjugated anti-CD3ε (145-2c11; BD Biosciences) or, in some experiments, biotin-conjugated anti-CD43 (5–7; BD Biosciences), plus streptavadin-conjugated microbeads (Miltenyi Biotech), followed by passage through an LD negative-selection column (Miltenyi Biotech).
Cholesterol augmentation
C57BL/6 mice were irradiated with 5 Gy and were allowed to rest for 13.5 d to allow autoreconstitution to begin. Cholesterol was augmented in splenocyte samples depleted of CD43+ cells as described38,47. Cells were then treated with anti-IgM and p100 induction was analyzed by immunoblot.
SDS-PAGE and immunoblot analysis
Cells were collected and washed with cold PBS (Gibco), then lysed in TNT lysis buffer solution consisting of 150 mM NaCl, 50 mM Tris-HCl, pH 7.5 (Invitrogen), and 1% (vol/vol) Triton X-100 containing a Complete Protease Inhibitor Mini tablet (Roche). Proteins were quantified by BCA assay (Pierce Protein Assay Kit). Lysates were boiled for 3–5 min in loading buffer containing 50 mM dithiothreitol and 1% (wt/vol) SDS (Bio Rad). Samples were separated by 10% PAGE with a Tris-glycine system. Gels were blotted onto a Immobilon-P polyvinylidene difluoride or nitrocellulose membrane (Millipore or Protran, respectively) and were stained with rabbit anti-p100 (4882; Cell Signaling Technology), anti-RelB (C-19; sc-226; Santa Cruz Biotechnology), anti-p65 (3034; Cell Signaling Technology), anti-IκBα (C-21; sc-371; Santa Cruz Biotechnology) or anti-TBP (1TBP18; Abcam). Monoclonal antibody to β-actin (AC-15; Sigma) and monoclonal antibody to phosphorylated tyrosine (4G10; Upstate) were also used. Quantity One software (BioRad) or AlphaEaseFC Imaging software (Alpha Innotech) was used for gel densitometry.
Real-time PCR
Bone marrow or splenic cells were sorted for various populations either by enrichment with magnetic-activated cell sorting or with a FACSAria flow cytometer (Beckton Dickinson). RNA was extracted with the RNAeasy kit (Qiagen) and was reverse-transcribed by SuperScript II Reverse Transcriptase (Invitrogen). Bone marrow was sorted for Hardy fractions A–D (IgM−AA4.1+B220+), fraction E CD23+ (IgM+AA4.1+B220+CD23+) and fraction E CD23− (IgM+AA4.1+B220+CD23−). Splenocytes were isolated and sorted as T1 (B220+CD43−IgMhiAA4.1+CD23−), T2 (B220+CD43−IgMhiAA4.1+CD23+), T3 (B220+CD43−IgMloAA4.1+CD23+), follicular (B220+CD43−IgM+AA4.1− CD23+) and marginal zone (B220+CD43−IgMhiAA4.1−CD23−CD21/35+). Bone marrow plasma cells (Gr1−CD3−IgD−B220−CD19−, surface immunoglobulin–negative, CD138+) were sorted and further verified by detection of intracellular immunoglobulin staining and examination of cellular morphology. Real-time PCR Master Mix and the proprietary primer and probe sets for mouse GAPDH (glyceraldehyde phosphate dehydrogenase), Bcl-xL, Pim2, BR3, TACI and BCMA were from Applied Biosystems. The ABI 7300 system (Applied Biosystems) was used for real-time PCR, and the cycling threshold method (2−(ΔΔCt)) was used for relative quantification by comparison of the gene of interest to GAPDH (ABI PRISM 7700 user bulletin; Applied Biosystems).
BLyS receptor expression and cell viability
Bone marrow and splenic B cells were defined as described48,49. Recombinant human BLyS was provided by Human Genome Sciences. Phycoerythrin-conjugated anti-TACI (166010) and fluorescein isothiocyanate–conjugated anti-BCMA (161616) were from R&D Systems. Unlabeled anti-BR3 (7H22-E16; ALX-804-824-c100; Axxora) was visualized with fluorescein isothiocyanate–conjugated or phycoerythrin-conjugated mouse anti–rat IgG1 (KLH-G1-2-2; 0116-02; Southern Biotechnology). Cells were stained for 20 min with primary antibodies in flow cytometry buffer (0.5% (wt/vol) BSA and 0.5 M EDTA in PBS), followed by 10 min of incubation with secondary reagent. Samples were analyzed with a FACSCalibur (Becton Dickinson) and FlowJo software (TreeStar). Cell viability was measured by exclusion of the dye TO-PRO-3 as described50.
Mice
C57BL/6 or BALB/cJ mice were from The Jackson Laboratory or the animal facility of the National Institute on Aging. Mice with B cell–specific expression of the Bcl-xL transgene were provided by C. Thompson29. All procedures were in accordance with the Animal Welfare Act.
Statistics
Student’s _t_-test, two-tailed with equal variance was used for statistical analysis.
Supplementary Material
supp figs 1-7
Acknowledgments
We thank M. May, L. Solt and the University of Pennsylvania Flow Cytometry Core for assistance and advice; C.A. Hunter (University of Pennsylvania School of Veterinary Medicine) for NF-κB p50–deficient mice; C. Thompson (University of Pennsylvania) for mice with B cell–specific expression of the Bcl-xL transgene; V. Dixit (Genentech) for pRK5B-hBR3; and J.M. Stadanlick for critical reading of this manuscript. Supported by the US Public Health Service (R01AI073939 and R01AI0545488 to M.P.C.; T32HL07971 to J.E.S.; T32AI055428 to F.G.K.; and R01AI032592 to J.G.M.) and the Intramural Research Program of the National Institute on Aging.
Footnotes
Accession codes. UCSD-Nature Signaling Gateway (http://www.signaling-gateway.org): A000383, A000374, A002936, A002248, A000374 and A000305.
Note: Supplementary information is available on the Nature Immunology website.
AUTHOR CONTRIBUTIONS
J.E.S., M.K., F.G.K., J.L.S., J.P.M., W.J.Q., R.J.B., L.S.T. and K.A.J. did research, analyzed data and generated key reagents; J.E.S., M.K., J.G.M., R.S. and M.P.C. designed research and analyzed data; and J.E.S., R.S., M.K. and M.P.C. wrote the paper.
References
- 1.Moore PA, et al. BLyS: member of the tumor necrosis factor family and B lymphocyte stimulator. Science. 1999;285:260–263. doi: 10.1126/science.285.5425.260. [DOI] [PubMed] [Google Scholar]
- 2.Schneider P, et al. BAFF, a novel ligand of the tumor necrosis factor family, stimulates B cell growth. J Exp Med. 1999;189:1747–1756. doi: 10.1084/jem.189.11.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam KP, Kuhn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 4.Batten M, et al. BAFF mediates survival of peripheral immature B lymphocytes. J Exp Med. 2000;192:1453–1466. doi: 10.1084/jem.192.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan M, et al. Identification of a novel receptor for B lymphocyte stimulator that is mutated in a mouse strain with severe B cell deficiency. Curr Biol. 2001;11:1547–1552. doi: 10.1016/s0960-9822(01)00481-x. [DOI] [PubMed] [Google Scholar]
- 6.Thompson JS, et al. BAFF binds to the tumor necrosis factor receptor-like molecule B cell maturation antigen and is important for maintaining the peripheral B cell population. J Exp Med. 2000;192:129–135. doi: 10.1084/jem.192.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lesley R, et al. Reduced competitiveness of autoantigen-engaged B cells due to increased dependence on BAFF. Immunity. 2004;20:441–453. doi: 10.1016/s1074-7613(04)00079-2. [DOI] [PubMed] [Google Scholar]
- 8.Thien M, et al. Excess BAFF rescues self-reactive B cells from peripheral deletion and allows them to enter forbidden follicular and marginal zone niches. Immunity. 2004;20:785–798. doi: 10.1016/j.immuni.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Weil R, Israel A. T-cell-receptor- and B-cell-receptor-mediated activation of NF-κB in lymphocytes. Curr Opin Immunol. 2004;16:374–381. doi: 10.1016/j.coi.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-κB2 in maturing B cells. Nat Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- 11.Beinke S, Ley SC. Functions of NF-κB1 and NF-κB2 in immune cell biology. Biochem J. 2004;382:393–409. doi: 10.1042/BJ20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan M, et al. Identification of a receptor for BLyS demonstrates a crucial role in humoral immunity. Nat Immunol. 2000;1:37–41. doi: 10.1038/76889. [DOI] [PubMed] [Google Scholar]
- 13.Francis DA, Sen R, Rice N, Rothstein TL. Receptor-specific induction of NF-κB components in primary B cells. Int Immunol. 1998;10:285–293. doi: 10.1093/intimm/10.3.285. [DOI] [PubMed] [Google Scholar]
- 14.Cariappa A, Liou HC, Horwitz BH, Pillai S. Nuclear factor κB is required for the development of marginal zone B lymphocytes. J Exp Med. 2000;192:1175–1182. doi: 10.1084/jem.192.8.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasparakis M, Schmidt-Supprian M, Rajewsky K. IκB kinase signaling is essential for maintenance of mature B cells. J Exp Med. 2002;196:743–752. doi: 10.1084/jem.20020907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonizzi G, Karin M. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Yamada T, et al. Abnormal immune function of hemopoietic cells from alymphoplasia (aly) mice, a natural strain with mutant NF-κB-inducing kinase. J Immunol. 2000;165:804–812. doi: 10.4049/jimmunol.165.2.804. [DOI] [PubMed] [Google Scholar]
- 18.Sasaki Y, Casola S, Kutok JL, Rajewsky K, Schmidt-Supprian M. TNF family member B cell-activating factor (BAFF) receptor-dependent and -independent roles for BAFF in B cell physiology. J Immunol. 2004;173:2245–2252. doi: 10.4049/jimmunol.173.4.2245. [DOI] [PubMed] [Google Scholar]
- 19.Mecklenbrauker I, Kalled SL, Leitges M, Mackay F, Tarakhovsky A. Regulation of B-cell survival by BAFF-dependent PKCδ-mediated nuclear signalling. Nature. 2004;431:456–461. doi: 10.1038/nature02955. [DOI] [PubMed] [Google Scholar]
- 20.Enzler T, et al. Alternative and classical NF-κB signaling retain autoreactive B cells in the splenic marginal zone and result in lupus-like disease. Immunity. 2006;25:403–415. doi: 10.1016/j.immuni.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 21.Sasaki Y, et al. Canonical NF-κB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation. Immunity. 2006;24:729–739. doi: 10.1016/j.immuni.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Derudder E, et al. RelB/p50 dimers are differentially regulated by tumor necrosis factor-α and lymphotoxin-β receptor activation: critical roles for p100. J Biol Chem. 2003;278:23278–23284. doi: 10.1074/jbc.M300106200. [DOI] [PubMed] [Google Scholar]
- 23.Basak S, et al. A fourth IκB protein within the NF-κB signaling module. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatada EN, et al. NF-κB1 p50 is required for BLyS attenuation of apoptosis but dispensable for processing of NF-κB2 p100 to p52 in quiescent mature B cells. J Immunol. 2003;171:761–768. doi: 10.4049/jimmunol.171.2.761. [DOI] [PubMed] [Google Scholar]
- 25.Weih F, et al. p50-NF-κB complexes partially compensate for the absence of RelB: severely increased pathology in p50−/− relB−/− double-knockout mice. J Exp Med. 1997;185:1359–1370. doi: 10.1084/jem.185.7.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Day ES, et al. Selectivity of BAFF/BLyS and APRIL for binding to the TNF family receptors BAFFR/BR3 and BCMA. Biochemistry. 2005;44:1919–1931. doi: 10.1021/bi048227k. [DOI] [PubMed] [Google Scholar]
- 27.Hsu BL, Harless SM, Lindsley RC, Hilbert DM, Cancro MP. Cutting edge: BLyS enables survival of transitional and mature B cells through distinct mediators. J Immunol. 2002;168:5993–5996. doi: 10.4049/jimmunol.168.12.5993. [DOI] [PubMed] [Google Scholar]
- 28.Craxton A, Draves KE, Gruppi A, Clark EA. BAFF regulates B cell survival by downregulating the BH3-only family member Bim via the ERK pathway. J Exp Med. 2005;202:1363–1374. doi: 10.1084/jem.20051283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grillot DA, et al. Bcl-x exhibits regulated expression during B cell development and activation and modulates lymphocyte survival in transgenic mice. J Exp Med. 1996;183:381–391. doi: 10.1084/jem.183.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shinners NP, et al. Bruton’s tyrosine kinase mediates NF-κ B activation and B cell survival by B cell-activating factor receptor of the TNF-R family. J Immunol. 2007;179:3872–3880. doi: 10.4049/jimmunol.179.6.3872. [DOI] [PubMed] [Google Scholar]
- 31.Liptay S, Schmid RM, Nabel EG, Nabel GJ. Transcriptional regulation of NF-κB2: evidence for κB-mediated positive and negative autoregulation. Mol Cell Biol. 1994;14:7695–7703. doi: 10.1128/mcb.14.12.7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bossen C, et al. TACI, unlike BAFF-R, is solely activated by oligomeric BAFF and APRIL to support survival of activated B cells and plasmablasts. Blood. 2008;111:1004–1012. doi: 10.1182/blood-2007-09-110874. [DOI] [PubMed] [Google Scholar]
- 33.Monroe JG. Ligand-independent tonic signaling in B-cell receptor function. Curr Opin Immunol. 2004;16:288–295. doi: 10.1016/j.coi.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 34.Lai JY, et al. Potent small molecule inhibitors of spleen tyrosine kinase (Syk) Bioorg Med Chem Lett. 2003;13:3111–3114. doi: 10.1016/s0960-894x(03)00658-9. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto N, et al. The orally available spleen tyrosine kinase inhibitor 2-[7-(3,4-dimethoxyphenyl)-imidazo[1,2-c]pyrimidin-5-ylamino]nicotinamide dihydrochloride (BAY 61–3606) blocks antigen-induced airway inflammation in rodents. J Pharmacol Exp Ther. 2003;306:1174–1181. doi: 10.1124/jpet.103.052316. [DOI] [PubMed] [Google Scholar]
- 36.Hanke JH, et al. Discovery of a novel, potent, and Src family-selective tyrosine kinase inhibitor. Study of Lck- and FynT-dependent T cell activation. J Biol Chem. 1996;271:695–701. doi: 10.1074/jbc.271.2.695. [DOI] [PubMed] [Google Scholar]
- 37.Amanna IJ, Dingwall JP, Hayes CE. Enforced bcl-xL gene expression restored splenic B lymphocyte development in BAFF-R mutant mice. J Immunol. 2003;170:4593–4600. doi: 10.4049/jimmunol.170.9.4593. [DOI] [PubMed] [Google Scholar]
- 38.Karnell FG, Brezski RJ, King LB, Silverman MA, Monroe JG. Membrane cholesterol content accounts for developmental differences in surface B cell receptor compartmentalization and signaling. J Biol Chem. 2005;280:25621–25628. doi: 10.1074/jbc.M503162200. [DOI] [PubMed] [Google Scholar]
- 39.Bishop GA. The multifaceted roles of TRAFs in the regulation of B-cell function. Nat Rev Immunol. 2004;4:775–786. doi: 10.1038/nri1462. [DOI] [PubMed] [Google Scholar]
- 40.Merrell KT, et al. Identification of anergic B cells within a wild-type repertoire. Immunity. 2006;25:953–962. doi: 10.1016/j.immuni.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 41.Allman DM, Ferguson SE, Lentz VM, Cancro MP. Peripheral B cell maturation. II Heat-stable antigenhi splenic B cells are an immature developmental intermediate in the production of long-lived marrow-derived B cells. J Immunol. 1993;151:4431–4444. [PubMed] [Google Scholar]
- 42.Tucker E, et al. A novel mutation in the Nfkb2 gene generates an NF-κB2 “Super Repressor”. J Immunol. 2007;179:7514–7522. doi: 10.4049/jimmunol.179.11.7514. [DOI] [PubMed] [Google Scholar]
- 43.Hondowicz BD, et al. The role of BLyS/BLyS receptors in anti-chromatin B cell regulation. Int Immunol. 2007;19:465–475. doi: 10.1093/intimm/dxm011. [DOI] [PubMed] [Google Scholar]
- 44.Chung JB, Sater RA, Fields ML, Erikson J, Monroe JG. CD23 defines two distinct subsets of immature B cells which differ in their responses to T cell help signals. Int Immunol. 2002;14:157–166. doi: 10.1093/intimm/14.2.157. [DOI] [PubMed] [Google Scholar]
- 45.Pear W. Current Protocols in Molecular Biology. Wiley and Sons; San Francisco: 2001. Ch. 9 Unit 9 11. [Google Scholar]
- 46.Smith SH, Cancro MP. Cutting edge: B cell receptor signals regulate BLyS receptor levels in mature B cells and their immediate progenitors. J Immunol. 2003;170:5820–5823. doi: 10.4049/jimmunol.170.12.5820. [DOI] [PubMed] [Google Scholar]
- 47.Brezski RJ, Monroe JG. B cell antigen receptor-induced Rac1 activation and Rac1-dependent spreading are impaired in transitional immature B cells due to levels of membrane cholesterol. J Immunol. 2007;179:4464–4472. doi: 10.4049/jimmunol.179.7.4464. [DOI] [PubMed] [Google Scholar]
- 48.Allman D, et al. Resolution of three nonproliferative immature splenic B cell subsets reveals multiple selection points during peripheral B cell maturation. J Immunol. 2001;167:6834–6840. doi: 10.4049/jimmunol.167.12.6834. [DOI] [PubMed] [Google Scholar]
- 49.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Treml LS, et al. TLR stimulation modifies BLyS receptor expression in follicular and marginal zone B cells. J Immunol. 2007;178:7531–7539. doi: 10.4049/jimmunol.178.12.7531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supp figs 1-7