Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators (original) (raw)
. Author manuscript; available in PMC: 2010 Aug 14.
Published in final edited form as: J Mol Biol. 2009 Jun 3;391(2):341–358. doi: 10.1016/j.jmb.2009.05.078
Abstract
NusG is a conserved regulatory protein that interacts with elongating complexes (ECs) of RNA polymerase (RNAP), DNA, and RNA to modulate transcription in multiple and sometimes opposite ways. In E. coli, NusG suppresses pausing and increases elongation rate, enhances termination by E. coli ρ and phage HK022 Nun protein, and promotes antitermination by λN and in ribosomal RNA operons. We report NMR studies that suggest E. coli NusG consists of two largely independent N- and C- terminal structural domains, NTD and CTD. Based on tests of the functions of the NTD and CTD and variants of NusG in vivo and in vitro, we find that NTD alone is sufficient to suppress pausing and enhance transcript elongation in vitro. However, neither domain alone can enhance ρ-dependent termination or support antitermination, indicating that interactions of both domains with ECs are required for these processes. We propose that the two domains of NusG mediate distinct interactions with ECs: the NTD interacts with RNAP and the CTD interacts with ρ and other regulators, providing NusG with different combinations of interactions to effect different regulatory outcomes.
Introduction
NusG is an exceptionally conserved regulator of gene expression that exerts complex and sometimes opposite effects on transcript elongation by RNA polymerase (RNAP). However, little is known about how NusG interacts with elongation complexes (ECs) to generate these effects on transcription. Alone, E. coli NusG (_Ec_NusG) increases the elongation rate of E. coli RNAP both in vivo and in vitro 1; 2; 3, primarily by suppressing transcriptional pauses that involve backtracking by RNAP 3; 4. However in B. subtilis NusG actually enhances rather than suppresses transcriptional pausing 5. The role of NusG in termination is also complex. _Ec_NusG enhances ρ-dependent transcription termination, whereas, in combination with NusA, NusB, NusE, and either λN or other cellular factors, NusG can modify RNAP to create specialized “antitermination” complexes that resist pausing and termination. These complexes are required for bacteriophage λ growth or for efficient transcription of ribosomal RNA operons 6; 7. This ability of NusG to inhibit rather than stimulate termination cannot be explained as a difference between NusG acting alone on RNAP versus acting as part of a multi-protein assemblage. Indeed, not all NusG-containing multi-factor complexes are resistant to termination. During λ infection of an E. coli HK022 lysogen, NusG, complexed with the other Nus factors and the phage HK022 Nun protein, promotes transcription termination 8. NusG is essential in wild-type E. coli K12 because it enhances ρ-dependent termination upstream of the rac prophage gene kil; nusG can be deleted if kil also is deleted or inactivated 9; 10. The essentiality of NusG is species-specific: NusG is dispensable in S. aureus, and both NusG and ρ are dispensable in B. subtilis 11; 12; 13. This extensive and complex involvement of NusG in transcriptional processes with different transcriptional outcomes as well as the apparent species-specific differences in NusG activity have stymied understanding of how NusG regulates ECs. Additionally, the regions of NusG that participate in interactions with RNAP or transcription factors have not yet been determined.
X-ray or NMR structures were recently obtained for NusG from Thermus thermophilus (Tt) and Aquifex aeolicus (Aa) 14; 15; 16; 17, for the NusG paralog RfaH 18, and for the NGN (NusG N-terminal) domain of S. cerevisiae Spt5 19. All NusGs are composed of conserved N-terminal and C-terminal domains (NTD and CTD) connected by a flexible linker that contain, respectively, an NGN motif and a KOW motif with potential to interact with either protein or nucleic acid 20; 21. The NTD of _Tt_NusG contains a flexible loop that is replaced by a third protein domain in _Aa_NusG and is absent in Spt5. Based on analogy to _Ec_RfaH 18 and on bacterial two-hybrid interaction assays 22, the NusG NTD is proposed to bind RNA polymerase (RNAP) via interaction with the loop between two anti-parallel helices in the β’ clamp domain (the β’ clamp helices; β’CH). A hydrophobic pocket on the NusG NTD is the proposed site of β’CH interaction 18. However, neither this proposal nor any other function of NusG’s NTD have been tested experimentally.
The C-terminal domain (CTD) of NusG forms a Tudor domain in _Tt_NusG and _Aa_NusG, within which is embedded the KOW sequence motif. Based on similarities to other KOW or Tudor domain-containing proteins, the NusG CTD is proposed to interact with either protein or nucleic acid 15. Despite the large number of potential interaction partners to mediate the plethora of NusG activities, no interaction partners with the CTD have been identified. Interestingly, the crystal lattice of one _Aa_NusG structure generates a domain-swapped dimer formed by two NusG molecules 16. Based on this structure and strong evidence that NusG is a monomer 4; 16, Knowlton et al. proposed that the NusG CTD binds to the NTD to create a spring-loaded state that is released upon interaction with RNAP 16. Although such a two-state model appears to operate for _Ec_RfaH (which has a structurally non-homologous CTD), NMR analysis of the _Tt_NTD with and without CTD revealed no changes in NTD NMR signal and thus does not lend support to the existence of the intense CTD-NTD interaction predicted by the spring-loaded model 17.
Better understanding of the structure and function of _Ec_NusG is important to understand how transcription is regulated in other organisms as well. NusG is highly conserved, and genes encoding NusG or close homologs exist in all eubacterial genomes sequenced to date, although the majority of what is known about NusG function results from biochemical studies of _Ec_NusG. The S. cerevisea NusG homolog, Spt5, is essential for growth, and complexes of Spt5 and Spt4 stimulate elongation by both RNAPI and RNAPII 23; 24. In metazoans, Spt5 complexes with Spt4 to form the dimeric DRB-sensitivity-inducing factor (DSIF) 25. Like bacterial NusG, DSIF also has complex effects on transcription: it mediates negative-elongation-factor (NELF)-dependent blocks to transcript elongation in promoter-proximal regions but can also mediate pause suppression once NELF is removed by P-TEFb phosphorylation of DSIF, NELF, and the RNAPII C-terminal repeat domain 26.
Since the two domains of NusG may have distinct interaction partners (e.g., NTD with RNAP, CTD with ρ, Nus factors, or RNA), we hypothesized that expression of the NusG NTD or CTD separately or of full-length (FL) NusG containing substitutions that eliminated interactions of one domain without affecting the interactions of the other could inhibit wild-type function. In addition, we tested the phenotype of the various mutants in strains lacking NusG. To allow accurate structure-function analysis of these and other substitutions, we also determined NMR structures for the NTD and CTD of _Ec_NusG.
Results
The solution structure of E. coli NusG
The 1H, 15N HSQC spectrum of FL E. coli NusG showed the high dispersion of resonances expected for a well-folded protein. The large differences in signal intensities combined with the detection of increasing transversal relaxation rates as determined by spin-echo experiments in NusG above 400 µM concentration indicates a tendency of the protein to aggregate at higher concentrations. Whereas signals from the CTD (R123-A181) could be completely assigned using standard triple resonance NMR experiments, the NTD could not be assigned due to very low and often missing signal intensities. This is because the two domains exhibit different relaxational behavior, a consequence of different rotational tumbling of the two domains. This observation suggests that (1) _Ec_NusG consists of at least two domains that are connected via a flexible linker and (2) the NTD tends to aggregate at very high concentration.
Expression and purification of the NTD (M1-R123) yielded a protein that showed a well-dispersed 1H, 15N HSQC spectrum that again showed aggregation tendency at concentrations above 400 µM. In contrast to the FL, the NTD exhibited reduced transverse relaxation to an extent sufficient to allow assignment of all backbone resonances and more than 90 % of the sidechain resonances (Table 1). Chemical shifts close to random coil values and lack of medium-and long range NOEs for residues M1-K6, E47-G67, and V118-R123 indicate structural disorder for these regions (Figure 1).
Table 1.
Experimental restraints for structure calculation
| NusG NTD | NusG CTD | ||
|---|---|---|---|
| Distance restraints | total | 860 | 543 |
| intraresidual | 31 | 21 | |
| sequential | 189 | 142 | |
| medium range | 254 | 64 | |
| long range | 386 | 252 | |
| hydrogen bonds a | - | 32 | |
| Dihedral restraints | 8 | 17 | |
| Restraint violations | |||
| rms distance violation (Å) | 0.003 | 0.003 (± 0.002) | |
| max. distance violation (Å) | 0.11 | 0.16 | |
| rms dihedral violation (°) | 0 | 0.09 (± 0.01) | |
| max. dihedral violation (°) | 0 | 0.5 | |
| rmsd bond length (Å) | 0.0005 (± 0.00004) | 0.0004 (± 0.00005) | |
| rmsd bond angle (°) | 0.09 (± 0.003) | 0.08 (± 0.004) | |
| Atomic coordinate | |||
| precision | |||
| backbone atoms (Å) | 0.54 c | 0.42 b | |
| all heavy atoms (Å) | 0.97 c | 1.00 b | |
| Ramachandran plot | |||
| statistics d | |||
| most favored regions (%) | 89.3 | 83.6 | |
| additional allowed regions (%) | 10.0 | 16.3 | |
| generously allowed regions (%) | 0.5 | 0.1 | |
| disallowed regions (%) | 0.2 | 0.0 |
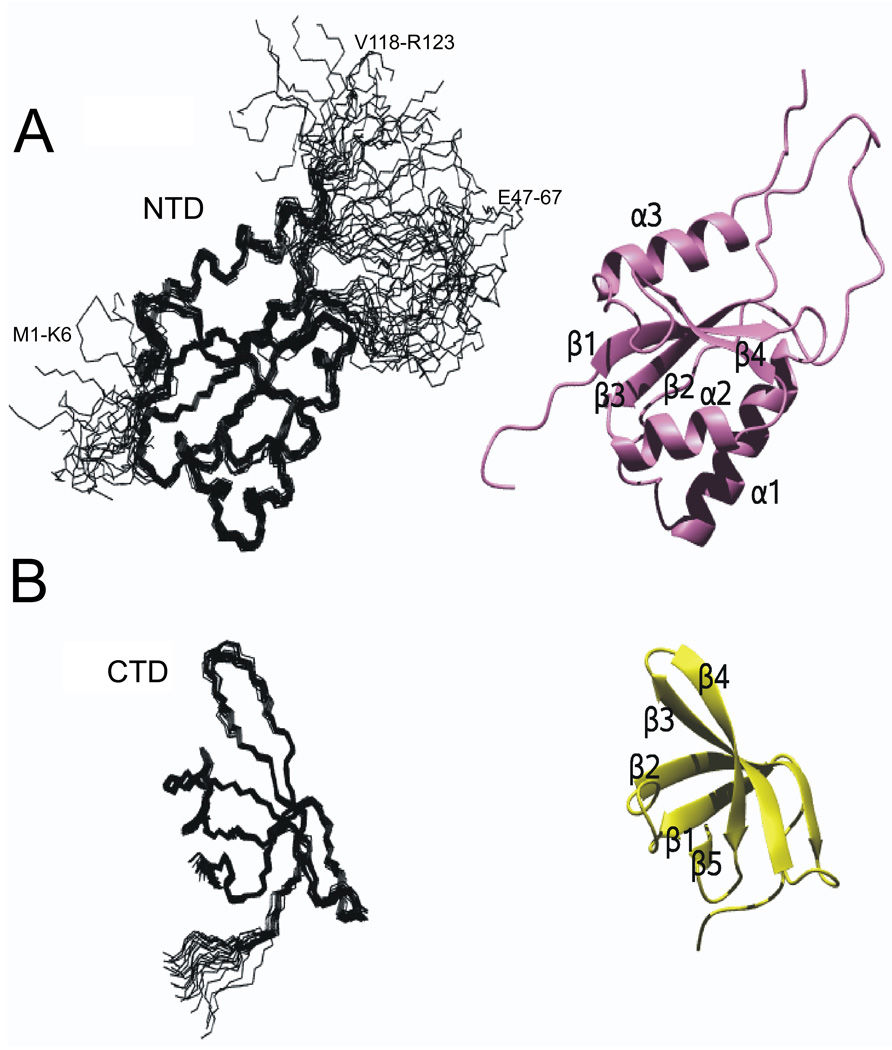
Figure 1. Solution structure of E. coli NusG NTD and CTD.
(A) Solution structure of _Ec_NusG NTD
(left) Structural ensemble of 20 accepted structures; regions of disorder are labeled by residue numbers (M1-K6, P45-F65, V1180-R123).
(right) Ribbon representation of representative structure. The structure was determined with experimental NMR restraints obtained from the NTD construct.
(B) Solution structure of NusG CTD.
(left) Structural ensemble of 20 accepted structures.
(right) Ribbon representation of representative structure. The structure was determined with NMR restraints obtained from experiments with FL NusG.
624 restraints for the structure calculation of the CTD could be derived from the NMR experiments with FL NusG (Table 1). The ensemble resulting from the final structure calculation shows only distance restraint violations less than 0.16 Å and only dihedral restraint violations less than 0.5°. Coordinate precision was 0.42 Å backbone rmsd, and all stereochemical properties were valid (Table 1, Methods). 868 restraints were obtained for the NTD from the NMR data. The final structure calculation yielded only distance restraint violations less than 0.1 Å and only dihedral restraint violations less than 0.44°. Coordinate precision was 0.62 Å backbone rmsd, and all stereochemical properties were valid.
Structure description
The secondary structure of the _Ec_NusG NTD consists of three α-helices (α1:E19-H33, α2: D77-S85, α3: D105-Q116) and four β-strands (β1:R8-A14, β2:E41-M43, β3:Y68-Q72, β4: V90-I93) (Figure 1 and Figure 2). The β-strands form an antiparallel β-sheet with strand order β2-β3-β1-β4, and helices α1 and α2 are arranged nearly antiparallel, packing against one side of the β-sheet, while α3 packs on the opposite side of the β-sheet. Numerous hydrophobic interactions are found between the helices and the β-sheet. The sequence region P45-F65 shows chemical shifts typical for unstructured residues and no medium or long-range NOE cross signals for these residues were observed, suggesting that this sequence region forms a highly flexible loop. Comparison of the solution structure of _Ec_NusG NTD with the corresponding domain of _Aa_NusG shows backbone coordinate rmsds of about 1.8 Å. This demonstrates that the solution conformation of this domain is very similar to that seen in crystal structures of _Aa_NusG and in the solution structure of the _Tt_NusG, as expected from the high sequence similarity. Also, the large flexible loop between strands β2 and β3 of _Tt_NusG is also found in EcNusG.
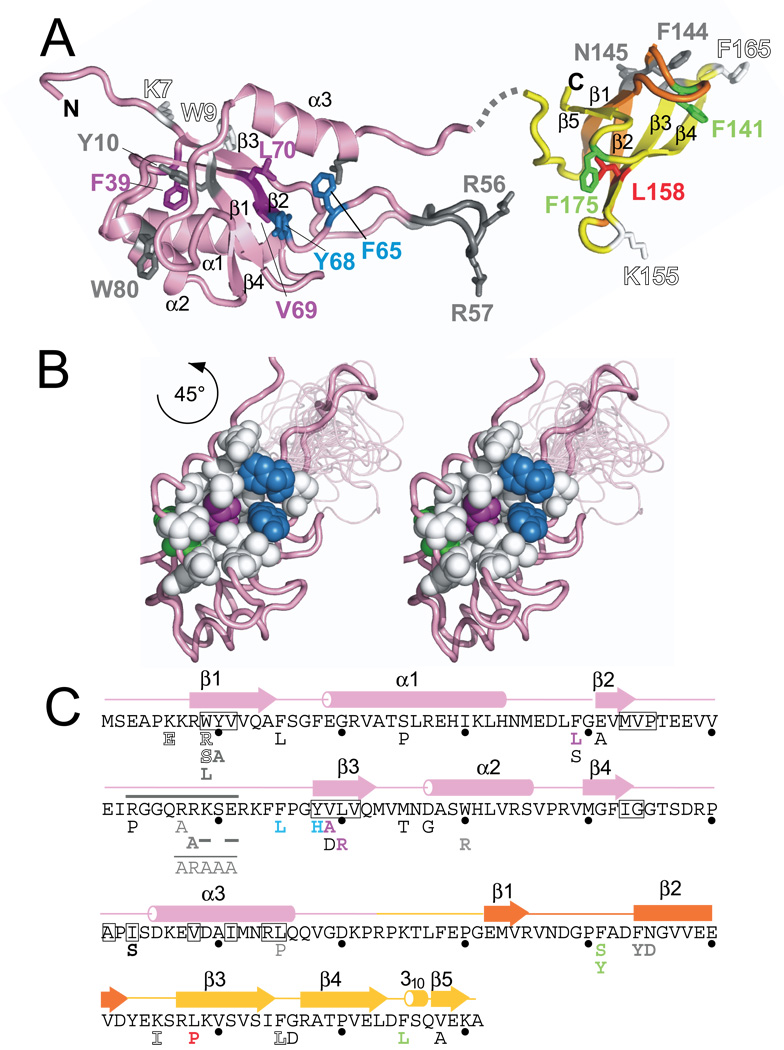
Figure 2. Structure of E. coli NusG.
(A) Structures of E. coli NusG. NusG has two main structural domains separated by a linker. The NTD (pink) consists entirely of the NusG N-terminal (NGN) motif, identified only in NusG and its homologs. The CTD (yellow and orange) contains the KOW sequence motif (orange), which has been found in NusG and homologs as well as in ribosomal proteins L24, L26, L27. Mutations characterized in this paper and in previous work (Table 3) are mapped onto the structure and color-coded as follows: green residues fail to complement Δ_nusG in vivo_ but function in vitro, magenta residues are defective for both elongation and termination, blue residues work only at high concentration, the red residue is defective for termination but not elongation, white (outlined) residues exhibited overexpression toxicity but allowed complementation of Δ_nusG in vivo_ and functioned in in vitro assays, and gray residues were previously characterized 8 16 50. Gray dashed line indicates the linker between the two domains and is not intended to suggest its actual position.
(B) (left and center) Stereo view showing mutations on one face of the NTD that are part of a hydrophobic pocket, coloring as in (A);
(C) Amino acid sequence of _Ec_NusG shown with corresponding structure (above) and substitutions (below). Substitutions are colored as in (A).
The secondary structure of the _Ec_NusG CTD consists of five β-strands (β1: E132-V136, β2: F144-D152, β3: R157-I164, β4: R167-L173, β5: V178-K180) that form an antiparallel barrel-type β-sheet with strand order β5-β1-β2-β3-β4 (Figure 1 and Figure 2). Comparison of the solution structure of the _Ec_NusG CTD with the corresponding domain of _Aa_NusG (pdb:1m1h) and _Tt_NusG (pdb:1nz9) resulted in backbone rmsds in the range 1.6–2.1 Å. This is within the limit found for different molecules in the asymmetric units of the crystal structures, confirming a high degree of structural similarity.
Over-expression of NusG NTD inhibits cell growth
The multiple roles of NusG in transcription regulation and the two-domain structure suggest each domain could have a distinct function. To test this, we separately investigated the properties of the individual domains, both in vivo and in vitro. If the individual domains make distinct interactions with components of the EC or other regulators to influence elongation, over-expression of the individual domains in a wild-type strain may be dominant-lethal if that domain out-competes the wild-type protein for binding to an interaction partner.
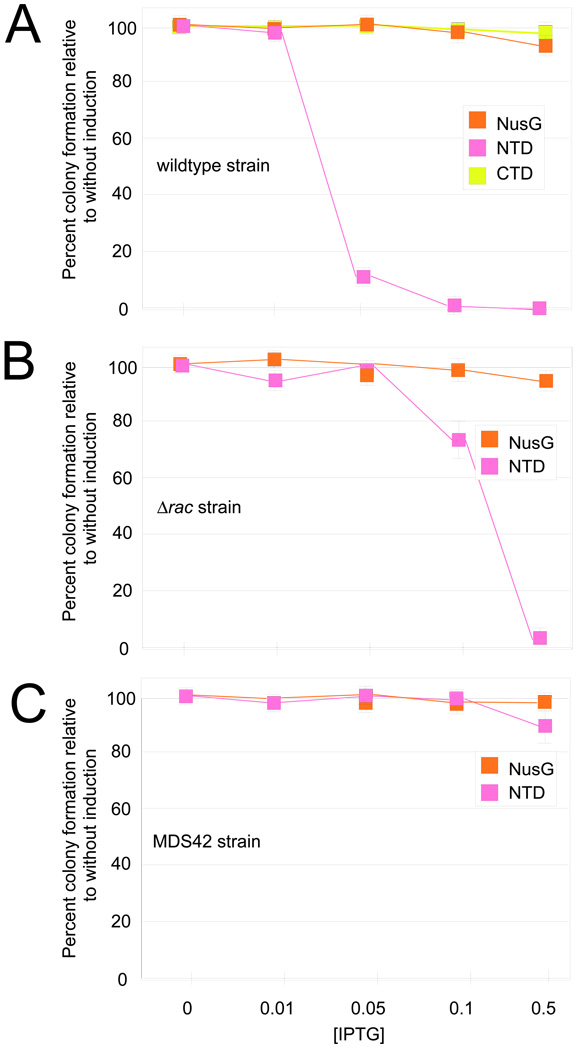
To determine the toxicity of the individual domains, we used plasmids encoding FL NusG or individual domains of NusG under control of the trc promoter which is induced by IPTG (FL NusG, pRM431; NTD, pRM442; and CTD, pRM443; Table 2). We then measured the number of colonies able to grow on plates with increasing amounts of IPTG (Figure 3A). Over-expression of either FL NusG or CTD did not reduce colony numbers, and thus was not toxic to the bacteria. However, NTD over-expression resulted in a clear defect in cell growth; colony numbers decreased as IPTG concentrations were raised. The number of colonies at 0.5 mM IPTG compared to the number without IPTG was < 1 × 10−6 (plating efficiency). This is consistent with the idea that the NTD maintains partial function, i.e. binds to RNAP, but is defective in another essential NusG activity. One possible explanation for the toxicity of the NTD is that it out-competes FL NusG in binding ECs but blocks ρ-dependent termination because it cannot interact with ρ.
Table 2.
Strains, plasmids, primers, and transcription templates
| Name | Description | Source or note |
|---|---|---|
| XL1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F proAB lacIq lacZ_Δ_M15 Tn10 (TetR )] | Stratagene |
| BLR(DE3) | F- _ompT hsdS_B(rB− mB− ) gal dcm (DE3) Δ(srl-recA)306::Tn10 (TetR) | Novagen |
| RL301 | CY15001; W3110 trpR tnaA2 | 45 |
| MG49 | nusG∷kan | 9 |
| MG1655 | Wild-type E. coli | 46 |
| 10422 | MG1655 pRM442 | This work |
| 10423 | MG1655 pRM443 | This work |
| N8499 | N99 c1857 λpRM-cI-rex-_t_imm-N::lacZ | This work |
| 10437 | N8499 ptrc99c | This work |
| 10438 | N8499 pRM442 | This work |
| 10439 | N8499 pRM443 | This work |
| 10440 | N8499 pRM431 | This work |
| N9441 | N99-λpR-cro-nutR-t_R1-cII::l_acZ cI857 | This work |
| 10461 | N9441 ptrc99c | This work |
| 10462 | N9441 pRM442 | This work |
| 10463 | N9441 pRM443 | This work |
| 10464 | N9441 pRM431 | This work |
| RSW472 | MG1655 Δr_ac_::CamR | 10 |
| MDS42 | Reduced genome E. coli | 47 |
| Plasmids | ||
| ptrc99c | Vector plasmid | 48 |
| pRM431 | p_trc_-His6-nusG M13 ori , NusG expression plasmid | This work |
| pRM442 | p_trc_-His6-NTD (NusG aa 1–118), NTD expression plasmid | This work |
| pRM443 | p_trc_-CTD (NusG aa 126–181), CTD expression plasmid | This work |
| pRM567 | CTD (aa 126–181)::MBP, CTD::MBP expression plasmid | This work |
| pCB111 | T7 ρ expression plasmid | 40 |
| pIA146 | pT7A1-668nt transcript template plasmid | 44 |
| pIA251 | ops pause template plasmid | 34 |
| pIA267 | λ _t_R1 terminator template plasmid | 34 |
| Primers | ||
| 021 | 5´ -TTTTCCAGCTGGCGAATGTG −3´ | rpoB ds |
| 645 | 5´ -CAGTTCCCTACTCTCGCATG-3´ | T7 A1 us |
| 947 | 5´ -GGAGAGACAACTTAAAGAG-3´ | T7 A1 ds |
| 3071 | 5´- CGTTAAATCTATCACCGCAAG GG-3´ | λ Pr us |
| In vitro templates | ||
| ops pause | PCR of pIA251 with 947 and 645 | 34 |
| Elongation assay | PCR of pIA146 with 947 and 021 | 44 |
| λ _t_R1 terminator | PCR of pIA267 with 645 and 3071 | 34 |
Figure 3. Plating efficiency of cells expressing FL NusG, NTD, or CTD.
(A) Wild-type MG1655 was tested for growth with plasmids encoding FL NusG, NTD, or CTD. Cells were plated onto media containing different amount of IPTG to induce protein expression. The number of colonies was determined and expressed as a fraction of the number of colonies formed without IPTG (without protein induction). The vertical bars represent the propagated errors from these calculations.
(B) Plating efficiency of the Δ_rac_ strain (RSW472, Table 1) with plasmids encoding FL NusG or NTD, as described above.
(C) Plating efficiency of the MDS42 strain with plasmids encoding FL NusG or NTD, as described above.
NusG is essential in wild-type E. coli K12 because it enhances ρ-dependent termination upstream of the rac prophage, preventing transcription of the downstream kil gene 10. We reasoned that if NTD toxicity was due to competition for FL NusG and interference with termination, particularly of this toxic gene, toxicity should be decreased in a strain in which the rac prophage was deleted (Δ_rac_ strain, Figure 3B; Table 2, RSW472). We performed the plating assay in this strain background and saw a significant decrease in toxicity of NTD expression, though it was still toxic to cells at higher levels (at 0.5 mM IPTG, the plating efficiency was 3 × 10−2). We next assayed NTD expression toxicity in a reduced genome E. coli strain that is deleted for the rac prophage as well as many other mobile DNA elements and cryptic virulence genes (Figure 3C, Table 2, MDS42). We found that toxicity was dramatically relieved in this strain, even at the highest concentration of IPTG (plating efficiency of 9.1 × 10−1). The difference in toxicity between the Δ_rac_ and MDS42 strains suggests that there is another gene like kil that is contained within the regions deleted in MDS42 and whose transcription is normally blocked by NusG-promoted ρ-dependent termination.
NTD can compete with FL NusG for ρ-termination in vivo
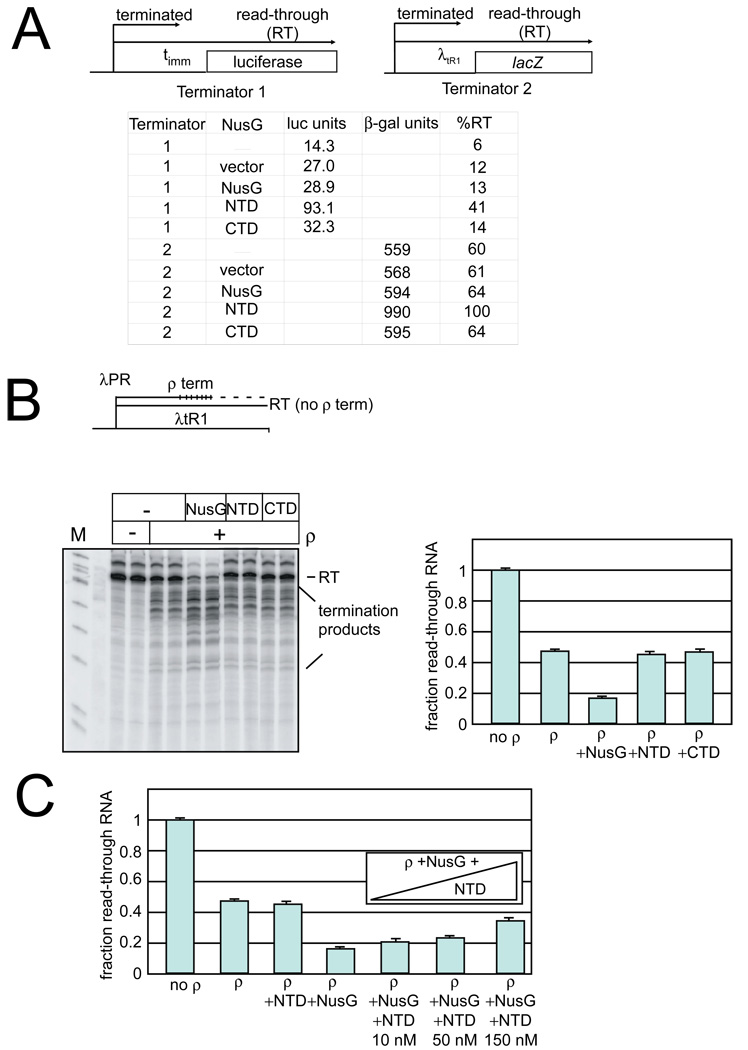
To test the idea that NusG NTD can interfere with ρ-dependent termination, we asked how NTD over-expression affected termination at two known ρ-dependent λ terminators, λ_t_imm and λ_t_R1. We did this in a strain containing chromosomally encoded, wild-type NusG that acts to promote termination. By expressing the NTD from a plasmid in this strain, we could ask if in vivo NusG-aided termination was suppressed by NTD expression. Read-through of _t_imm was monitored in a λPRm – _t_imm - lacZ fusion using a luciferase-linked β-galactosidase assay (Terminator 1 in Figure 4A; Table 3, N8499). Strains carrying either the vector plasmid, FL, or CTD gave similar amounts of terminator read-through (∼27, 29, and 32 million luciferase units respectively), about 2-fold higher than the level seen without any plasmid (∼14 million units, Figure 4A). Thus neither CTD nor FL NusG affected termination at _t_imm. In contrast, NTD overproduction yielded ∼93 million luciferase units, 3 times more than the vector alone, representing 41% read-through (Figure 4A, Methods). We conclude that NTD over-expression partially suppresses termination at _t_imm because NTD competes with FL NusG in EC binding, but is defective for ρ interaction.
Figure 4. In vivo and in vitro ρ-termination.
(A) In vivo ρ-termination
(Top) In vivo ρ-termination was measured on two terminators, _t_imm (terminator 1) and λtR1 (terminator 2) with downstream reporters.
(Bottom) Reporter expression for strains containing plasmids expressing NusG, NTD, and CTD, vector alone, or no plasmid. The %RT is determined by comparison of the levels of expression compared to the levels when ρ is inhibited by bicyclomycin. Luciferase units (RLU) are given as × 106 values. β-galactosidase values are Miller units 43.
(B) In vitro ρ-termination
(Top) Schematic of termination template.
(Bottom) In vitro transcription reactions were performed in the presence and absence of ρ, FL NusG, NTD, or CTD as indicated. The read-through RNA product is indicated at the top of the gel (labeled RT). Addition of ρ stimulates termination, as seen by a decrease in the amount of the RT product and an increase in the amounts of shorter products (labeled termination products). NusG additionally increases ρ -termination at earlier template positions. (Right). Quantitation of ρ–termination assays. The fraction read-through RNA is determined as the amount of the read-through RNA as a fraction of the total RNA (read-through RNA plus the terminated RNAs). Bars represent averages from three experiments; error is the standard deviation of these averages.
(C) Termination assays were performed as in (B), but increasing amounts of NTD (10, 50, and 150 nM) were added together with FL NusG (at 10 nM). NTD competes for RNAP binding with FL NusG as evidenced by the decreased NusG stimulation of termination with increasing amounts of competing NTD. Bars represent averages from three experiments; error is the standard deviation of these values.
Table 3.
Properties of NusG variants
| Plasmid Number or reference | Substitution | Comp. Δ_nusG_ | Inc. elongation rate | Inc. ρ term. | Δ_rac_ Δ_nusG_ viability | λ phage growth | HK022 exclusion of λ |
|---|---|---|---|---|---|---|---|
| pRM431 | wild-type | + | + | + | + | + | + |
| pRM442 | NTD | − | + | − | − | ||
| pRM443 | CTD | − | − | − | − | ||
| 2785 | K7E | + | + | + | + | + | + |
| 2786 | W9R | + | + | + | + | + | − |
| 2787 | W9S | + | + | + | |||
| 49 | W9L | ||||||
| 49 | Y10A | ||||||
| 2788 | F15L | + | |||||
| 2789 | S25P | + | + | + | + | ||
| 2790 | F39L | − | − | − | + | + | + |
| 2791 | F39S | − | − | ||||
| 2792 | V42A | + | |||||
| 2793 | R53P | + | |||||
| 49 | Δ(I52-E61) | − | |||||
| 49 | R57-E61 >ARAAA | + | |||||
| 49 | R57A | + | |||||
| 49 | R58A Δ(59 –61) | − | |||||
| 2794 | F65L | + | +, at [high]a | + | + | + | + |
| 2795 | Y68H | + | +, at [high] a | + | |||
| 2796 | V69A | + | − | − | + | + | + |
| 2797 | V69D | − | |||||
| 2798 | L70R | − | − | − | − | ||
| 2799 | M75T | + | |||||
| 2800 | D77G | + | |||||
| 8 | W80R | + | − | ||||
| 2801 | I103S | + | + | + | − | ||
| 8 | L115P | + | |||||
| 2802 | F141S | − | + | ||||
| 2803 | F141Y | − | + | + | + | + | + |
| 8 | F144Y | + | − | ||||
| 8 | N145D | − | |||||
| 2804 | K155I | + | + | + | + | + | + |
| 2805 | L158P | − | + | − | + | + | + |
| 50 | S163F | + | |||||
| 2806 | F165L | + | + | + | + | + | − |
| 16 | F165V | + | |||||
| 16 | F165Y | + | |||||
| 16 | F165D | − | |||||
| 16 | F165T | − | |||||
| 16 | F165A | cs | |||||
| 2807 | G166D | + | − | ||||
| 2808 | F175L | − | + | + | |||
| 2809 | V178A | + | + | + | − |
Termination at λ_t_R1, which is known to be stimulated by NusG 27, was determined by assaying expression of lacZ downstream of a λpR-cro-λ_t_R1 terminator cassette (Terminator 2 in Figure 4A). Although λ_t_R1 in isolation is ∼90% efficient, translation of cro in the cassette enhances read-through to ∼60% (N9441 in Table 2, Figure 4A). Importantly, this was done in the presence of a wild-type chromosomally encoded nusG and rho so the levels of lacZ expression are determined by chromosomal NusG-enhanced ρ-dependent termination. Strains carrying no plasmid, empty plasmid, plasmid expressing FL NusG, or plasmid expressing CTD yielded equivalent levels of β-galactosidase activity (Figure 4A), so having additional FL NusG or CTD does not interfere with the NusG-enhanced termination that occurs on this cassette. In contrast, the strain that over-expressed NTD (N10462) displayed nearly twice the amount of β-galactosidase activity, which represents complete read-through of the λ_t_r1 terminator. Thus, NTD inhibits termination both at _t_R1 and _t_imm and, presumably, generally suppresses ρ termination in vivo. This is consistent with a model in which the NTD competes with FL NusG for binding to RNAP, and suggests that physical linkage of the CTD to the NTD is required for NusG to enhance ρ-EC interaction and termination.
Individual NusG domains have no direct effect on ρ-termination in vitro
To exclude a direct effect of the NTD on ρ-termination, we assayed the ability of the purified individual domains to modulate ρ termination in vitro on a λ_t_R1-containing template (Figure 4B). In the absence of NusG, ρ terminated ∼50% of the transcripts before the end of the template (Figure 4B). Addition of FL NusG to the reaction mixture significantly enhanced termination efficiency. Comparison of the two transcription reactions reveals that NusG both decreased the amount of read-through product and shifted termination to more promoter-proximal sites. Enhancement of early termination has been reported previously for NusG action at _t_R1 4; 27; 28. In contrast to the strong enhancement of termination by FL NusG, neither NTD nor CTD modulated ρ-activity (Fig. 4B). We next asked if the two domains could stimulate ρ-activity without being tethered. Simultaneous addition of purified NTD and CTD had no effect on ρ termination, suggesting the two domains must be tethered to stimulate ρ-activity (data not shown). That NTD did not reduce termination efficiency indicates that this domain does not directly inhibit ρ. Instead, our in vitro data is consistent with the model we proposed to explain the effects of the NTD in vivo: NTD not tethered to CTD inhibits ρ termination by competing with FL NusG for interaction with EC.
NTD competes with FL NusG for EC interaction at a ρ-dependent terminator
To test further the model that NTD competes for EC interaction and to confirm our in vivo findings, we next tested for direct competition between NTD and FL NusG in vitro (Figure 4C). NTD added in equimolar amounts to FL NusG (10 nM each) only slightly inhibited NusG activation of ρ. NTD inhibition increased in a concentration-dependent manner. Interestingly, even at the highest concentration of protein tested, 150 nM, NTD was unable to inhibit NusG stimulation of termination completely. This result can be explained if NTD binding to the EC is slightly (less than 10 times) weaker than FL NusG binding. Possibly, interaction of NusG with the EC involves additional contacts that increase the affinity of FL NusG relative to NTD, or if ρ stabilizes FL NusG but not NTD binding through EC contacts.
NusG NTD but not CTD enhances elongation rate and suppresses pausing
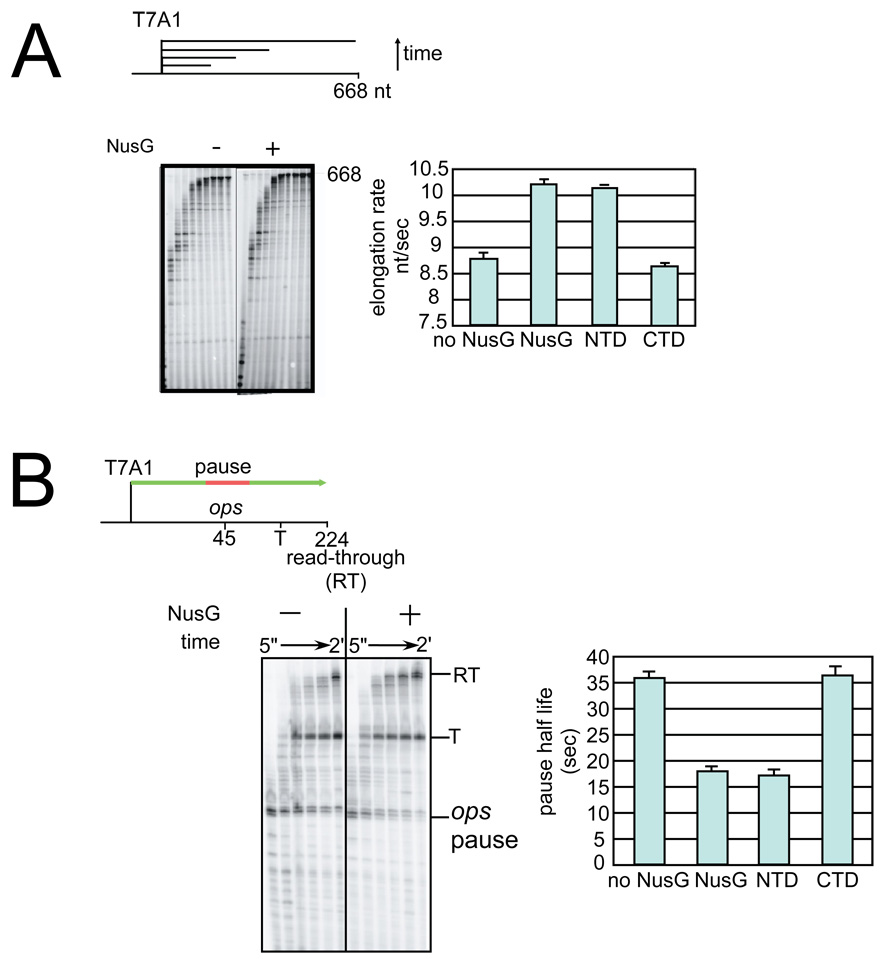
The finding that NTD can compete with FL NusG for EC interaction raises the possibility that it could affect transcript elongation. To test this, we investigated the individual domains or FL NusG using a transcription template on which the strong T7 A1 promoter initiates a 668 nt transcript (Figure 5A). By measuring the rate of appearance of the 668-nt RNA, we determined the elongation rate and found that FL NusG and NTD both enhanced this rate by ∼20%. In contrast, CTD had no significant effect on elongation rate. We conclude that NTD contains the elongation enhancement activity of NusG, and can, by itself, bind ECs and stimulate elongation.
Figure 5. The effect of FL NusG, NTD, and CTD on elongation enhancement.
(A) Elongation rate assay
(top) Schematic of transcription template. Transcription was performed using a template containing the strong T7A1 promoter followed by a U-less cassette allowing formation of halted complexes in the presence of ATP, CTP, and 32P-GTP.
(left) After halted complex formation, all four NTPs were added to resume transcription and samples were removed at 5”, 10”, 15”, 20”, 30”, 45”, 1’, 1.5’, 2’, and 4’. Stimulation of transcription elongation by NusG is seen by the faster accumulation of the 668nt product (labeled 668).
(right) Quantification of elongation rate in nucleotides synthesized per second for reactions performed in the absence of added factor or with FL NusG, NTD, or CTD. Values plotted are the results from at least three independent experiments; error bars indicate the standard deviation of these averaged values.
(B) Pause assay
(top) Schematic of _ops_-containing transcription template.
(left)Transcription assays were initiated by forming halted complexes as described above. All four NTPs were then added to resume transcription from the halted complexes through the ops pause sequence; samples were removed at 5”, 15”, 30”, 45”, 1’, and 2’. The effect of NusG is seen by the faster decrease in RNAs paused at the ops pause (labeled ops pause) as ECs escape the pause and the faster accumulation of the longer RNA products (labeled T for products terminating at the downstream terminator or RT for the read-through products).
(right) Quantification of pause half-life determined from reactions performed in the absence of added factor, FL NusG, NTD, or CTD. Pause half-life is the amount of time it takes half of the complexes to escape the pause. Values plotted are the results from at least three independent experiments; error bars indicate the standard deviation of these averaged values. Pause efficiencies did not vary significantly with experimental conditions (80–90%).
We next asked if NTD, like NusG, could suppress the pause at ops, a well-characterized pause site that induces RNAP backtracking (Figure 5B; 3). NusG suppression is evident in synchronized transcription experiments by the faster appearance of RNA at the downstream terminator (T) and read-through (RT) positions and by the accelerated disappearance of RNA at the pause position (labeled ops pause in Figure 5B). Similar to its effect on the overall elongation rate (Figure 5A), NTD suppressed pausing as efficiently as FL NusG, whereas CTD had no effect. Thus the interaction of RNAP with NTD is necessary and sufficient for elongation rate enhancement and pause suppression.
Neither NTD nor CTD can substitute for FL NusG in a wild-type background
NusG is required in wild-type E. coli K12 cells because it promotes ρ–dependent termination upstream of the toxic rac prophage kil gene10. Since neither NTD nor CTD were able to stimulate ρ-dependent termination, we predicted that neither NTD nor CTD could replace FL NusG in rac+ cells. To test this prediction, we asked if a plasmid expressing the individual domains of NusG could compensate for a chromosomal disruption of nusG by a _kan_R insertion. Accordingly, we grew P1 phage on a strain carrying this disruption and a suppressing NusG+ plasmid 9; 29. The P1 lysate was used to transduce strains containing plasmids expressing FL NusG, NTD (at non-toxic levels), or CTD to kanamycin-resistance. As expected, cells expressing FL NusG readily accepted the nusG::kanR allele, whereas cells expressing either NTD or CTD alone could not be transduced to kanamycin-resistance (Table 3). We conclude that the individual domains cannot replace FL NusG in wild-type cells.
Screen for dominant lethal nusG variants
To define more precisely NusG interactions with EC, we looked for single amino-acid NusG substitutions that were dominant-lethal when over-expressed, similar to NTD toxicity. Based on the effects of the individual domains, we expected to identify variant proteins defective for some, but not all NusG functions. We predicted we could find NTD mutants that still allowed CTD interactions and CTD mutants that would still allow NTD interactions. A variant NusG protein that maintained one interaction could compete with wild-type NusG for this interaction, but be non-functional because of the lack of the ability to make the other interactions. We created a bank of pTrc99-based plasmids carrying nusG amplified by PCR using conditions designed to promote misincorporation 30 (Methods). We then transformed this bank of plasmids into a nusG+ strain and screened transformants for toxicity when plasmid-encoded NusG expression was induced with IPTG. Plasmids that expressed toxic NusG variants (dominant-lethals) were isolated and sequenced. Because the PCR conditions were designed to introduce errors, many of the isolated plasmids had multiple substitutions. These were discarded, and we focused on characterizing 25 single, unique substitutions (Table 3). These substitutions were distributed throughout the sequence of the protein, in both domains, and did not cluster in any obvious pattern (Figure 2D).
A subset of the dominant-lethal mutants were unable to support growth
We next asked if these dominant-lethal NusG mutants could serve as the sole source of NusG. Although these variants are toxic at high levels, we considered the possibility that some of these variants might be defective at high level due to increased interactions (gain-of-function) and might be able to provide some NusG function if expressed at lower levels. Accordingly, we expressed the mutant protein at low levels in wild-type cells and transduced the cells with P1 grown on the nusG::kanR strain (Table 2). Although several mutants failed to replace NusG+, other substitutions, surprisingly, were able to complement the nusG disruption (Table 3, complementation of Δ_nusG_).
Characterization of the NusG variants in vitro
A subset of these NusG mutants was selected for further characterization. These included variants with substitutions in the NTD or CTD domains, and mutants that could or could not replace chromosomal nusG. Substitutions K7E, W9R, W9S, F39L, F65L, Y68H, V69A, L70R, F141S, F141Y, K155I, L158P, F165L, and F175L were thus selected for further biochemical characterization. The variants of interest were expressed at low levels and purified from the soluble fraction of cells to avoid potential complications introduced by protein refolding (Methods).
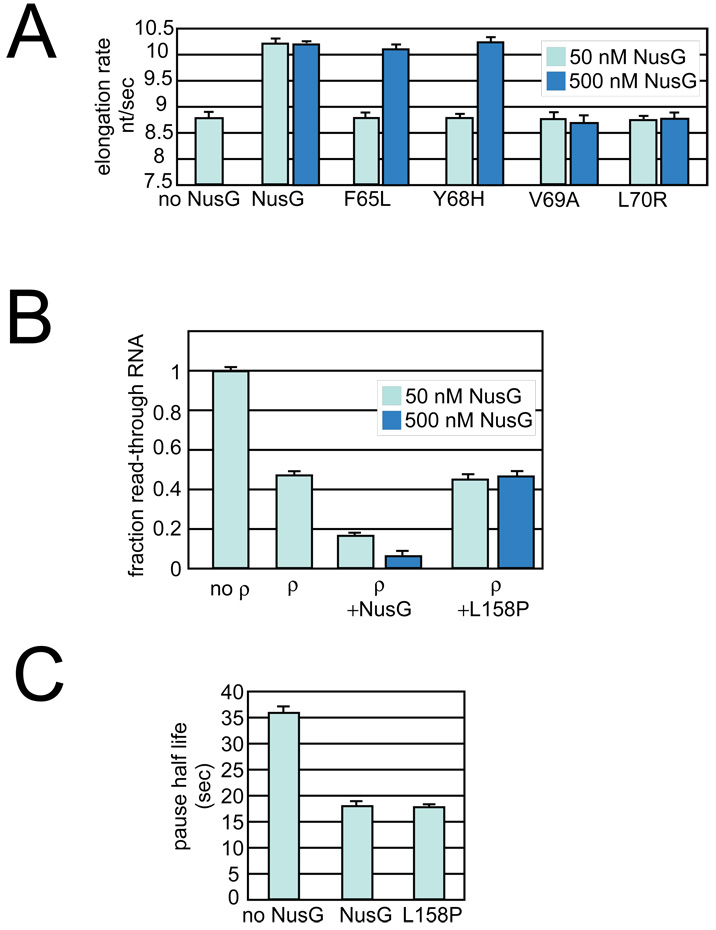
We first asked if the variants enhanced the elongation rate (Elongation rate enhancement, Table 3). Many NusG variants were able to stimulate elongation as well as wild-type NusG, whereas other mutants did not significantly enhance elongation rates. To determine if these latter mutants were defective because of decreased affinity to EC, we repeated the assay at higher concentrations of F39L, F65L, Y68H, V69A, and L70R. Elongation stimulation by two of the mutants, F65L and Y68H, was restored at high protein levels (Table 3; Figure 6, Supplemental Figure 3A). This suggests that these two mutants are defective in binding EC. Although it is possible that only a fraction of the mutant NusG is properly folded, we were able to purify all mutant NusG proteins from the soluble fraction. F39L, V69A, and L70R are located within the hydrophobic core of the NTD and are likely to interfere with proper domain folding. These mutants did not enhance elongation even at high concentrations. Interestingly, the mutants defective in binding (F65L and Y68H, colored blue in Fig. 2A and Fig. 2B) cluster on one side of the NTD, and together with residues F92, I93, A101, I103, P102, A110, and I111 form a hydrophobic, concave surface that is likely the site of interaction of NTD with RNAP (Fig. 2C).
Figure 6. The effect of NusG mutants on elongation and termination.
(A) Mutants near the hydrophobic binding site were tested at higher concentrations for elongation rate enhancement (experiment as shown in Figure 5AFigure 5A). Light blue, 50 nM NusG; Dark blue, 500 nM NusG.
(B) The L158P mutation was tested at higher concentrations for enhancement of ρ–termination (experiment as shown in Figure 4BFigure 4B). Light blue, 50 nM NusG; Dark blue, 500 nM NusG.
(C) The L158P mutation is able to enhance elongation at a rate comparable to that of wildtype NusG.
We next determined if the purified NusG variant proteins stimulated ρ-termination. Again, some of the mutants enhanced ρ-dependent termination as efficiently as wild-type NusG, whereas other mutants were defective (Table 3). In general, the effects of the substitutions in elongation enhancement and ρ-termination assays correlated positively. NusG variants that were unable to enhance elongation rate, even at high concentration, were also unable to enhance termination, e.g. F39L, V69A, and L70R, whereas all of the mutants, except for L158P, that enhanced elongation were also able to stimulate termination (Figure 6 and Table 3). L158P was able to suppress pausing, but was completely defective for stimulation of termination (Figure 6B and Figure 6C, Supplemental Figure 3B). In the NusG structure, L158 forms part of the hydrophobic core of CTD and the L158P substitution is likely to disrupt the folding of this domain (Figure 2). Like the NTD alone, the L158P mutant suppresses pausing and enhances elongation but is defective for ρ-dependent termination, even at high concentration, suggesting that the L158P substitution significantly disrupts CTD structure and function (Supplemental Figure 3B). The results with the L158P mutation further support a model in which NTD interaction with EC is sufficient for elongation rate enhancement, but interaction of CTD with ρ (and possibly additional components of the EC) is required for NusG to stimulate ρ-dependent termination.
Some NusG mutants did not allow function in multi-protein complexes in vivo
We also assayed the variant NusG proteins in vivo in processes requiring NusG’s participation in multi-protein complexes. NusG in combination with NusA, NusB, and NusE is required for two phage transcription reactions: antitermination with λN protein and termination with HK022 phage Nun protein 6; 8; 9; 31. Accordingly, we introduced plasmids expressing the variant NusG proteins into the Δ_rac_ Δ_nusG_ strain. This allowed us to test their function in the absence of wild- type NusG. For reasons that are not yet understood, this strain can only maintain ColE1 plasmids that express NusG with some activity; the vector plasmid cannot be maintained. Three of the variants, F39S, L70R, and G166D could not be introduced into Δ_rac_ Δ_nusG_; presumably they lack this essential function (Table 3, viability in Δ_rac_ Δ_nusG_).
Productive infection of E.coli by λ is dependent on NusG function 6. Terminators preceding essential λ genes are suppressed when NusG, along with the other Nus factors and λ N protein, assemble into an antitermination complex that modifies RNAP. Modified RNAP transcribes through the terminators and into the downstream genes. Thus, the Δ_rac_ Δ_nusG_ strain fails to support λ growth in the absence of a plasmid-expressed NusG that can assemble into a functional antitermination complex. Strikingly, all variants that we introduced into the Δ_rac_ Δ_nusG_ strain could propagate λ, suggesting that the NusG proteins formed functional antitermination complexes. Thus, to date, no NusG point mutants have been isolated that are defective in λN antitermination; this may be because a direct selection for such mutants has not yet been performed.
We also investigated a second phage process that similarly requires NusG as a component of a large protein assemblage: HK022 Nun termination. HK022 Nun terminates transcription on the λ chromosome and blocks phage growth. To assay HK022 Nun activity, we introduced a HK022 prophage into the Δ_rac_ Δ_nusG_ strains containing the NusG plasmids and infected with λ. As expected, λ did not grow on the strain expressing wild-type NusG. Of the variants tested, four failed to exclude λ, W9R, I103S, F165L, V178A, indicating that they are defective in the NusG function required for Nun termination (Table 3, HK022 exclusion of λ (Nun function)). Three other NusG substitutions had been previously identified as being defective for Nun function: W80R, F144Y, and N145D 8. Interestingly, three of the seven known Nun-defective-substitutions cluster on one face of the CTD (Figure 1A, F144, N145, and F165) suggesting that part of the CTD may be an important interaction surface for Nun. Another is in a different region of the CTD, and the other three substitutions lie in the NTD and do not appear to define a particular surface.
Discussion
NusG binds RNAP, accelerates RNA transcription elongation, suppresses pausing, and stimulates ρ-termination. We report that these distinct activities and protein contacts are contained in two non-interacting domains connected by a flexible linker. This picture is supported by NMR structures of the individual domains of E. coli NusG, the functional properties of these domains assayed in isolation, and the properties of NusG variants both in vivo and in vitro. We arrive at five main conclusions about NusG structure/function: (1) NTD and CTD mediate distinct interactions required for NusG activity; (2) a hydrophobic pocket on the NTD is required for RNAP interaction; (3) E. coli NusG does not necessarily operate via a spring-loaded mechanism; (4) the requirements for NusG function as a component of multiprotein complexes required for λ N antitermination and Nun termination are different; and (5) differences between the NTD of E. coli NusG and its homologs affect the properties of the proteins.
NTD and CTD mediate distinct interactions required for NusG function
We find that NTD alone stimulates the rate of transcript elongation and suppresses pausing in vitro as efficiently as FL NusG. NTD is toxic to cells and suppresses ρ-termination in vivo. Additionally, NTD competes with FL NusG for binding to RNAP and blocks enhancement of ρ-termination by FL NusG in vitro. We propose that bound NTD, although it can suppress pausing and accelerate elongation, is unable to interact with ρ without covalently-attached CTD. We conclude that enhancement of ρ-dependent termination requires both NusG domains, and that elongation enhancement and stimulation of termination are separable functions of NusG.
Consistent with the analysis of the individual domains, a CTD mutation (L158P) eliminated NusG stimulation of ρ-dependent termination without abrogating elongation rate enhancement. Thus at least two interactions are required for full NusG activity: an NTD-EC interaction and a second interaction, mediated by CTD. Since NusG was shown previously to form a stable and specific complex with ρ, we think it likely that the CTD helps recruit ρ to the EC 4; 32. NusG may also interact with RNA, perhaps mediated by the KOW sequence motif in the CTD 4; 15; 16. The interactions of NTD with the EC and of CTD with ρ and perhaps RNA would facilitate the interaction of ρ and RNAP and thus stimulate EC dissociation during termination. A requirement for tethering to the EC normally provided by the NTD may explain why we don’t see a deleterious effect on growth when the CTD alone is expressed.
A hydrophobic concavity on NTD allows binding to RNAP
Examination of the _Ec_NusG structure model reveals a substantial hydrophobic surface on the NTD (Figure 2B), and many of the residues involved were altered in our genetic screen for dominant-negative NusG mutants. NusG mutants F65L and Y68H were defective for RNAP binding, and could only function in the elongation assay when provided in excess (500 nM NusG for 25 nM transcription complexes; Figure 6A). This apparent decrease in the ability of these mutants to bind EC strongly supports direct involvement of the hydrophobic pocket of NusG in the interaction surface of NusG with RNAP. This concavity also is complementary to the convex tip of RNAP’s β’ clamp helices (RNAP β’ residues 260–309 in E. coli) shown to interact with NusG in a bacterial two-hybrid assay 22. It is notable that despite their toxicity and in vitro defects, these amino-acid substitutions are mostly conservative. More disruptive changes in this region, V69A and L70R, yielded completely non-functional proteins in vitro, although they were selected as dominant-negative variants. The in vitro activity of the NTD taken together with the mutant analysis suggests that this domain interacts with RNAP via its hydrophobic concavity.
No evidence for NusG operation via a spring-loaded mechanism was found
Based on the structure of a NusG dimer, a model for NusG function has been proposed in which an NTD-CTD interaction occurs within a single molecule of NusG 16. In this model, the interaction keeps NusG in an inactive form; structural rearrangement breaks this interaction and allows NusG to bind RNAP. This model is consistent with what is proposed to regulate the activity of the NusG paralog RfaH (see below)18. The spring-loaded NusG model predicts that the NTD should bind RNAP more tightly than FL NusG. We found, however, that similar concentrations of NTD or FL NusG have similar effects on elongation rate and pause suppression (Fig. 5). This implies that the E. coli CTD does not significantly inhibit NTD-EC association. Instead, experiments show that NTD must be added at higher concentrations than FL NusG to block enhancement of ρ-termination (Figure 4C). Thus, CTD seems to promote NTD-EC interaction rather than to inhibit it. Additionally, the severely reduced NMR signal intensity of the NTD compared to the CTD in FL NusG shows that the two domains exhibit different rotational behavior, suggesting uncorrelated movements of these domains, inconsistent with the proposed spring-loaded state. This behavior is similar to that reported for the α-subunit of E. coli RNAP, a protein whose NTD and CTD are connected by a flexible linker and are known not to interact 33. Finally, to make the contacts necessary for the spring-loaded state, residues in the NTD:CTD interface should exhibit changes in chemical shifts or relaxation rates when compared to the isolated domains. Recent NMR analysis performed on _Tt_NusG did not reveal such effects 17. Taken together, these data argue against the spring-loaded mechanism of NusG function.
NusG participation in different multi-protein complexes has different features
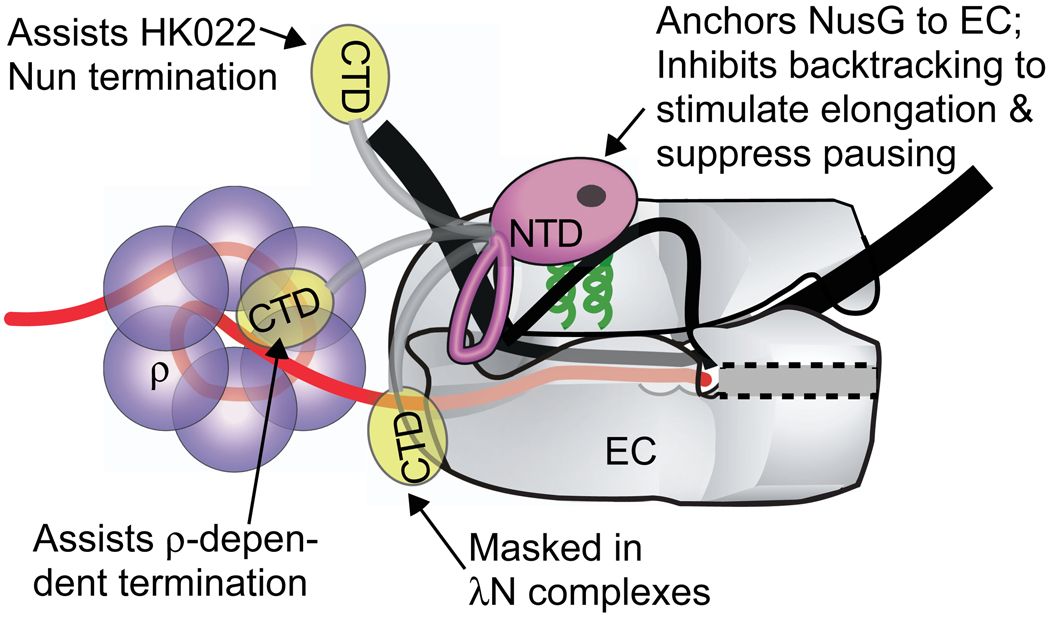
NusG is required for at least three processes that require the formation and function of large multi-protein complexes: phage λ antitermination, phage HK022 Nun termination, and ribosomal RNA antitermination. Here we investigated two of these processes, λN antitermination and HK022 Nun termination, in which NusG contributes to dramatically different regulatory outcomes. Interestingly, although NusG is required for λ antitermination 6, none of the NusG point mutants tested in this study or reported previously were defective for this process. This is consistent with the view that NusG functions largely as a scaffolding protein in λN complexes, and that a wide range of substitutions in NusG may be tolerated while still permitting assembly with the other λ antitermination complex components. This is in sharp contrast to our results with HK022 Nun termination complexes. We isolated four NusG point mutations that eliminated Nun termination, adding to the group of three previously described NusG substitutions with the same property. This difference in NusG effects on the λ N and HK022 Nun complexes is remarkable, given that both share many common protein components (NusA, NusB, NusE, NusG). One possibility is that this difference reflects direct NusG-Nun interaction in the HK022 Nun complex. If so, the multiple mutants located in the NusG CTD that affect Nun termination suggest the CTD may be important for this contact, though the non-overlapping effects of CTD mutants on Nun- and ρ-termination suggest ρ and Nun use different interaction surfaces on the CTD. Additionally, there are two NTD mutants that affect Nun function. This raises the possibility that Nun additionally makes contacts to the NTD as well as the CTD or that some aspect of Nun function may depend on a NusG-RNAP interaction that is mediated by the NTD. It is important to note that expression of the CTD alone was not toxic, unlike NTD expression. Although we know that the CTD is expressed (as evidenced by protein gel) and at least some of that expressed protein is soluble, we cannot exclude the possibility that our in vivo expression conditions did not result in enough functional protein to titrate the CTD’s interaction partners. It is also possible that effective binding of the CTD to such factors may require the tethering to the EC that is normally provided by the linkage to the NTD that binds RNAP. An attractive model for the different effects of NusG in different complexes with different regulatory outcomes is that NusG anchors its NTD to RNAP but allows independent and alternative interactions of its CTD (Fig. 7). Masking of the CTD in complexes like the λ N antermination complex could explain their resistance to ρ action. Elucidation of the structures of ECs bound by NusG, particularly of termination and antitermination complexes, should refine this picture.
Figure 7. Model of NusG interaction.
Model of NusG interactions with EC. NusG NTD (pink) is shown interacting with the β’ clamp helices (green) near the upstream edge of the transcription bubble. Alternative interactions of NusG CTD (yellow), for example with ρ, generate NusG’s regulatory complexity. The location of the flexible loop in the NTD and the precise orientation of the domains remain to be determined.
NTD specialization of NusG, RfaH, and Spt5
The structure of the NusG paralog RfaH reveals a NTD that is strikingly similar to that of NusG, including an extensive hydrophobic patch 18. However, RfaH CTD differs dramatically from that of NusG. Rather than forming a β-strand domain as in NusG, RfaH CTD folds into an α-helical coiled-coil 18. In the RfaH structure, the NTD is bound by the CTD, effectively concealing the hydrophobic, putative RNAP-interaction surface. Removal of RfaH CTD enhances NTD binding to ECs, as predicted from a spring-loaded model (see above) 18. Importantly, the hydrophobic patch of RfaH NTD is significantly larger than that of NusG (Figure S1), which likely explains why RfaH CTD shields this patch from interaction with RNAP, or from other, non-productive interactions. Consistent with this model, free RfaH NTD aggregates, but binds tightly to ECs once interaction is established 18.
The structural similarity of RfaH and NusG NTDs and the fact that both NTDs contain a hydrophobic patch that likely mediates interaction with RNAP implies that they affect ECs in a similar fashion. Indeed, both NusG NTD and RfaH NTD enhance the rate of transcription elongation (Figure 5)18. However, both the FL proteins and the NTDs of NusG and RfaH are functionally distinct. FL RfaH or RfaH NTD enhances pausing at ops and suppresses the his pause 18; 34. In contrast, FL NusG or NusG NTD suppresses pausing at ops and has no effect at the his pause 3, Figure 5B). Thus, despite the structural similarities of the two NTDs and the likelihood that they both interact with the β’ clamp helices of RNAP, the interactions of NusG and RfaH with ECs induce different transcriptional outcomes. These differences might reflect the way the two proteins bind RNAP or how they alter the structure of an EC.
A recent finding shows that NusG from different organisms can have distinct effects on transcription. Thus, _Bs_NusG, unlike EcNusG, increases pausing at his5. This provides further evidence of the regulatory plasticity of NusG. We do not yet know if _Bs_NusG NTD alone induces pausing or if _Bs_NTD is structurally similar to _Ec_NusG and RfaH NTDs. Finally, the importance of NusG as a transcription regulator is suggested by the retention of a eukaryotic homolog, Spt5. Spt5 binds Spt4 to form the heterodimeric transcription factor DSIF that enhances RNAPII processivity 23. The recently solved crystal structure of the complex of yeast Spt5 NGN domain with its interaction partner Spt4 shows an NGN domain structurally similar to NusG NTD, as expected from the high sequence similarity 19; 21. Interestingly, the surface of Spt5 NGN domain corresponding to the proposed RNAP interaction site of NusG and RfaH is hydrophobic and remains accessible in the Spt5-Spt4 complex (Fig. S1). Further studies are needed to determine if this Spt5 surface similarly mediates interaction with the RNAP or if other Spt5 contacts are required 35.
Methods
Strains and plasmids
Plasmids and strains used in this study are listed in Table 2. NusG mutants are listed in Table 3.
NMR spectroscopy and structure determination
NMR samples contained 0.4–0.5 mM protein in 50 mM sodium chloride, 10 mm sodium phosphate, pH 6.4. 10 % D2O was added as lock reference. Experiments were recorded at a sample temperature of 298 K.NMR experiments for solution structure determination of E. coli NusG domains were recorded on Bruker Avance spectrometers operating at 1H frequencies of 600–900 MHz partially equipped with cryogenically cooled probes. Standard through bond double and triple resonance experiments were performed for resonance assignments 36. Distance restraints for structure calculation were derived from 15N-NOESY-HSQC and 13C-NOESY-HSQC spectra with mixing times of 100–120 ms. NOESY cross peaks were classified according to their relative intensities and converted to distance restraints with upper limits of 3.0 Å (strong), 4.0 Å (medium), 5.0 Å (weak), and 6.0 Å (very weak). For ambiguous distance restraints the r−6 summation over all assigned possibilities defined the upper distance limit. Experimental NOESY spectra were validated semi-quantitatively against back-calculated spectra to confirm the assignment and to avoid bias of upper distance restraints by spin diffusion. Hydrogen bonds were included for backbone amide protons in regular secondary structure if the amide proton did not show a water exchange cross peak in the 15N -edited NOESY spectrum. The structure calculations were performed with the program XPLOR-NIH 1.2.1 37; 38 using a three-step simulated annealing protocol with floating assignment of prochiral groups including a conformational database potential. The structures were analyzed with the programs XPLOR-NIH 1.2.1, PROCHECK, and Molprobity. The Ramachandran values calculated by PROCHECK are presented in Table 1. The Ramachandran values calculated by Molprobity for the NTD are 95.4 % in the favored regions, 99.2 % in allowed regions, and 0.8% outliers; the Molprobity values for the CTD are 96.7 % in the favored regions, 99.6 % in allowed regions, and 0.4 % outliers.
Isolation of nusG mutants
Mutagenesis was performed using the template plasmid pRM431 encoding His6-NusG expressed from the IPTG-inducible trc promoter (Table 2). Error-prone PCR was done using flanking primers to amplify the nusG coding sequence in 50 independent PCR reactions. NTP concentrations, shown to reduce bias of substitutions and resulting in a more uniform mutagenesis, were used 30. Independent reactions were pooled, extracted with phenol/chloroform, and spermine-precipitated 39. Purified DNA was digested with Nco I and Hind III and ligated into similarly cut pRM431, yielding a pool of plasmids variant only in nusG. The mixture of plasmids was transformed into XL1Blue with selection on LB-Amp plates (100 µg ampicillin/ml). Individual transformants were patched onto LB-Amp plates, grown at 37°C and then replica-plated onto the same medium and medium containing 1 mM IPTG to induce NusG protein expression. Plates were examined after growth at 37°C to identify potential growth defects. Plasmids were isolated from slow- growing transformants and sequenced.
Proteins
Plasmids encoding nusG were grown in BLR (Table 2) in LB with 100 µg/mL ampicillin at 30° or 37° C to an OD600 of 0.35–0.5, induced for protein expression by addition of IPTG to a final concentration of 10 mM, grown for an additional 2.5–3 hours, harvested by centrifugation, and stored at −80° C until purification. NusG protein was then purified from the frozen cell pellets as described 4 except that a Ni2+ affinity chromatography step was done after the lysing and Polymin P precipitation steps, followed by Q ion-exchange chromatography 4. All NusG proteins used in this study were purified from the soluble cell fraction. For biochemical studies, NTD and CTD of NusG were purified from pRM442 and pRM567 respectively. CTD::MBP was purified by binding to amalylose resin, washing, and eluting the protein as described in the product description for pMALC2X (New England Biolabs). ρ protein was expressed from pCB111 as described in 40 and purified as described 41. RNAP holoenzyme was purified from MRE600 E. coli cells as described previously 42.
Assay for NusG variant activity
To determine if NusG variants could serve as the sole source of NusG in cells, P1 transduction was done using a lysate grown on a strain containing a chromosomal disruption of nusG by a kanamycin resistance cassette (MG49, Table 2). RL301 cells containing the NusG plasmid to be tested were grown in LB containing 5 mM CaCl2 and 100 mg/ml ampicillin, mixed with P1 phage, and then plated on LB plates containing 100 mg/ml ampicillin, 20 mg/ml kanamycin, 1.25 mM sodium pyrophosphate, and IPTG at 10, 25, 50, 100, 250, and 500 µM. This range was chosen as an induction level low enough to avoid toxicity, yet sufficient to provide protein. W3110 containing a plasmid expressing wild-type NusG could be successfully transduced at all concentrations of IPTG tested, from 10 µM to 1mM.
β-Galactosidase Assay (_t_imm function)
N8499 cultures (Table 2), bearing the λpRM-cI857-t_imm-N::lacZ fusion_, and bearing no plasmid, vector or plasmids expressing FL NusG, NTD, or CTD (strains N8499, 10437, 10438, 10439, and 10440), were grown at 32 °C with shaking for 3 hours at which time plasmid-containing strains were induced with 1mM IPTG. All cultures were grown at 32 °C for 1 hour post induction to final O.D. 600= 0.3–0.7. β-Galactosidase activity was then measured in a luminometer using the β-glo assay system (Promega, Madison, WI) according to manufacturer’s instructions. Activity was expressed as relative luminescence units (RLU) divided by O.D. 600. The %RT is determined by comparison of the levels of expression compared to the levels seen when ρ is inhibited by bicyclomycin.
β-Galactosidase Assay (_t_R1 function)
N9441 cultures (Table 2), containing the fusion λcI857-pR-cro-nutR-tR1-cII::lacZ and bearing no plasmid or the following plasmids: vector, NusG, NTD, or CTD (strains N9441, 10461, 10462, 10463, and 10464) were grown at 32 °C with shaking for 1 hour, at which time 10461, 10462, 10463, and 10464 were induced with 1mM IPTG. All 5 cultures were shifted to 42 °C and incubated for 1 hour. The shift from 32 °C to 42 °C inactivates the λcI857 repressor and initiates transcription at pR promoter. Samples were assayed for β-galactosidase activity as described 43. The percent read-through (%RT) is determined dividing β-galactoside activity by the activity when ρ is inhibited by bicyclomycin.
Plating Efficiency Assay
Single colonies from fresh transformants of MG1655, Δ_rac_, or MDS42 strains (Table 2) were inoculated by toothpick into LB media supplemented with 100 µg/ml ampicillin media and then serially diluted. Three ul of each dilution was then transferred to LB + ampicillin plates containing 0 mM, 0.01 mM, 0.05 mM, 0.1 mM, or 0.5 mM IPTG. This was performed for 3- 6 independent isolates. Plates were incubated for ∼15 hours at 37 °C. The plating efficiency for each strain/plasmid was determined by the number of colonies on IPTG-containing plates divided by the colony number on a plate without IPTG; these values are presented in Figure 3. The vertical bars are the propagated error for each calculated value.
In vitro transcription assays
Templates for all transcription assays were generated by PCR amplification (Table 2) and purified by phenol extraction and spermine precipitation 39. Halted A29 ECs (for the elongation rate assay and the ops pause assay) were formed at 37 °C by mixing 40 nM RNAP, 25 nM template, 150 µM ApU, and 2.5–10 µM ATP, GTP, CTP, and either 32P-CTP or 32P-GTP in transcription buffer. NusG, when present, was added to ECs at a final concentration of 100 nM unless otherwise indicated, and incubated at 37 °C for three minutes to allow binding. All 4 NTPs and 100 µg/mL Rif were then added to allow elongation to resume. The elongation rate assay was performed as described previously with NTPs at 150 µM 44. The ops pause assay was performed and quantitated as described previously with ATP, CTP, and UTP at 150 µM and GTP at 10 µM 34 44. The pause-half lives are presented in the figure; pause efficiencies were 80–90%. Termination assays were performed by forming 25 nM halted complexes at A26 at 37 °C with 150 µM ApU, 2.5 µM ATP and UTP, 1 µM GTP, and [α32P]GTP (3000 Ci/mmol in 40 mM Tris-HCl, 50 mM KCl, 5 mM MgCl2, 0.1 mM dithiothreitol, 3% glycerol, pH 7.9). 5 nM ρ and FL NusG (or NTD and CTD) at 10 nM were added followed by 3 min incubation at 37 °C. Transcription was restarted by adjustment of GTP, CTP, ATP and UTP to 40 µM, and rifampicin at 100 µg/ml, incubated at 37 °C for 15 min, and stopped by mixing with equal volume STOP buffer (8 M urea, 98 mM Tris-borate at pH 8.3, 10 mM Na2 EDTA, 0.2% bromophenol blue, 0.2% xylene cyanol), separated by denaturing PAGE (8–10%; 0.5x TBE), and analyzed using a PhosphorImager. To determine if NTD could compete with FL NusG for binding ECs, FL (10 nM) and NTD (10, 50, and 150 nM) were added simultaneously.
Lambda and HK022 phage plating
Plasmids encoding the NusG variants were transformed into the Δ_rac_ Δ_nusG_ strain and assayed as the sole source of NusG for the ability of these strains to plate λ. Phage growth requires NusG modulation of antitermination factor N function. The activity of termination factor Nun was tested for the NusG mutants in the Δ_rac_ Δ_nusG_ strain by making a HK022 lysogen and spotting λ. Functional Nun terminates transcription on the λ chromosome and thus blocks λ growth (Nun+).
Supplementary Material
01. Supplemental Figure 1. Comparison of structures of E. coli NusG, RfaH, and Spt5.
(Top) Solution structure of _Ec_NusG NTD (this work). The hydrophobic residues defining the pocket are colored green; a ribbon structure representation with key residues labeled is presented on the left, a space-fill representation is on the right.
(Middle) RfaH NTD 18. Colors and labeling as above.
(Bottom) Spt5 NGN. 19; 21 . Colors and labeling as above. The region of Spt4 that interacts with Spt5 in the co-crystal is shown in cyan. This does not overlap with the hydrophobic patch on Spt5.
02. Supplemental Figure 2. In vitro transcription assays with NTD and CTD.
(A) Elongation rate assay with no factor, NTD, or CTD. The experimental design is shown above the gel. The quantitation of this and two other experiments is presented in Figure 5A. NTD allows RNAP to reach the end of the template faster than reactions that contain either no additional factor or the CTD alone.
(B) Pausing at ops with no factor, NTD, or CTD. The experimental design is shown above the gel. The quantitation of this and two other experiments is presented in Figure 5B. NTD can suppress pausing and allows RNAP to reach the end of the template faster than reactions that contain either no additional factor or the CTD alone.
03. Supplemental Figure 3. In vitro transcription assays with selected NusG point mutants.
(A) Elongation rate assay. The experimental scheme is shown above the gel. Transcription rate was not stimulated by addition of either the F65L or Y68H mutants at 50 nM, but at 500 nM these two mutants could stimulate elongation similarly to wild-type NusG as evidenced by the faster accumulation of the full-length product (668 nt). The data from this experiment and two others is quantitated and presented in Figure 6A.
(B) L158P does not function for termination but can still suppress pausing
(left) Termination assay with either wild-type or L158P NusG at either 50 nM or 500 nM. Neither condition allowed enhancement of termination. The data from this experiment and others is quantitated and presented in Figure 6B.
(right) Pausing assay with the L158P variant. Despite being defective for termination, the L158P mutant can suppress pausing at the ops pause site. The data from this experiment and two others is quantitated and presented in Figure 6C.
Acknowledgements
RAM thanks Shannon Mooney for technical assistance with the NusG mutant screen. MG thanks April Rose for valuable assistance with phage plating and toxicity assays and Robert Washburn for helpful discussions. This work was supported by NIH grant GM38660 to RL, a grant from Deutsche Forschungsgemeinschaft (DFG RO 617/16–1) to PR, and NIH grant GM037219 to MG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Codes
NMR resonance assignments were deposited in the BMRB under the accession codes 15642 (NTD) and 15490 (CTD). Coordinates and restraints for structure calculation were deposited in the PDB under accession 2K06 (E. coli NusG NTD) and 2JVV (E. coli NusG CTD).
References
- 1.Burova E, Gottesman ME. NusG overexpression inhibits Rho-dependent termination in Escherichia coli. Mol Microbiol. 1995;17:633–641. doi: 10.1111/j.1365-2958.1995.mmi_17040633.x. [DOI] [PubMed] [Google Scholar]
- 2.Burns CM, Richardson JP. NusG is required to overcome a kinetic limitation to Rho function at an intragenic terminator. Proc Natl Acad Sci U S A. 1995;92:4738–4742. doi: 10.1073/pnas.92.11.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci U S A. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasman Z, von Hippel PH. Regulation of rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry. 2000;39:5573–5585. doi: 10.1021/bi992658z. [DOI] [PubMed] [Google Scholar]
- 5.Yakhnin AV, Yakhnin H, Babitzke P. Function of the Bacillus subtilis transcription elongation factor NusG in hairpin-dependent RNA polymerase pausing in the trp leader. Proc Natl Acad Sci U S A. 2008;105:16131–16136. doi: 10.1073/pnas.0808842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y, Filter JJ, Court DL, Gottesman ME, Friedman DI. Requirement for NusG for transcription antitermination in vivo by the lambda N protein. J Bacteriol. 2002;184:3416–3418. doi: 10.1128/JB.184.12.3416-3418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres M, Balada JM, Zellars M, Squires C, Squires CL. In vivo effect of NusB and NusG on rRNA transcription antitermination. J Bacteriol. 2004;186:1304–1310. doi: 10.1128/JB.186.5.1304-1310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burova E, Hung SC, Chen J, Court DL, Zhou JG, Mogilnitskiy G, Gottesman ME. Escherichia coli nusG mutations that block transcription termination by coliphage HK022 Nun protein. Mol Microbiol. 1999;31:1783–1793. doi: 10.1046/j.1365-2958.1999.01315.x. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan SL, Gottesman ME. Requirement for E coli NusG protein in factor-dependent transcription termination. Cell. 1992;68:989–994. doi: 10.1016/0092-8674(92)90041-a. [DOI] [PubMed] [Google Scholar]
- 10.Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E coli. Science. 2008;320:935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia M, Lunsford RD, McDevitt D, Iordanescu S. Rapid method for the identification of essential genes in Staphylococcus aureus. Plasmid. 1999;42:144–149. doi: 10.1006/plas.1999.1422. [DOI] [PubMed] [Google Scholar]
- 12.Quirk PG, Dunkley EA, Jr, Lee P, Krulwich TA. Identification of a putative Bacillus subtilis rho gene. J Bacteriol. 1993;175:647–654. doi: 10.1128/jb.175.3.647-654.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingham CJ, Dennis J, Furneaux PA. Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis. Mol Microbiol. 1999;31:651–663. doi: 10.1046/j.1365-2958.1999.01205.x. [DOI] [PubMed] [Google Scholar]
- 14.Andrykovitch M, Guo W, Routzahn KM, Gu Y, Anderson DE, Reshetnikova LS, Knowlton JR, Waugh DS, Ji X. Crystallization and preliminary X-ray diffraction studies of NusG, a protein shared by the transcription and translation machines. Acta Crystallogr D Biol Crystallogr. 2002;58:2157–2158. doi: 10.1107/s0907444902015810. [DOI] [PubMed] [Google Scholar]
- 15.Steiner T, Kaiser JT, Marinkovic S, Huber R, Wahl MC. Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. Embo J. 21:4641–4653. doi: 10.1093/emboj/cdf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knowlton JR, Bubunenko M, Andrykovitch M, Guo W, Routzahn KM, Waugh DS, Court DL, Ji X. A spring-loaded state of NusG in its functional cycle is suggested by X-ray crystallography and supported by site-directed mutants. Biochemistry. 2003;42:2275–2281. doi: 10.1021/bi0272508. [DOI] [PubMed] [Google Scholar]
- 17.Reay P, Yamasaki K, Terada T, Kuramitsu S, Shirouzu M, Yokoyama S. Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus. Proteins. 2004;56:40–51. doi: 10.1002/prot.20054. [DOI] [PubMed] [Google Scholar]
- 18.Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovitch I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo M, Xu F, Yamada J, Egelhofer T, Gao Y, Hartzog GA, Teng M, Niu L. Core structure of the yeast spt4-spt5 complex: a conserved module for regulation of transcription elongation. Structure. 2008;16:1649–1658. doi: 10.1016/j.str.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kyrpides NC, Woese CR, Ouzounis CA. KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem Sci. 1996;21:425–426. doi: 10.1016/s0968-0004(96)30036-4. [DOI] [PubMed] [Google Scholar]
- 21.Ponting CP. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 2002;30:3643–3652. doi: 10.1093/nar/gkf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nickels BE. Genetic assays to define and characterize protein-protein interactions involved in gene regulation. Methods. 2009;47:53–62. doi: 10.1016/j.ymeth.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider DA, French SL, Osheim YN, Bailey AO, Vu L, Dodd J, Yates JR, Beyer AL, Nomura M. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc Natl Acad Sci U S A. 2006;103:12707–12712. doi: 10.1073/pnas.0605686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu W, Wada T, Okabe S, Taneda T, Yamaguchi Y, Handa H. DSIF contributes to transcriptional activation by DNA-binding activators by preventing pausing during transcription elongation. Nucleic Acids Res. 2007;35:4064–4075. doi: 10.1093/nar/gkm430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nehrke KW, Zalatan F, Platt T. NusG alters rho-dependent termination of transcription in vitro independent of kinetic coupling. Gene Expr. 1993;3:119–133. [PMC free article] [PubMed] [Google Scholar]
- 28.Burns CM, Nowatzke WL, Richardson JP. Activation of Rho-dependent transcription termination by NusG Dependence on terminator location and acceleration of RNA release. J Biol Chem. 1999;274:5245–5251. doi: 10.1074/jbc.274.8.5245. [DOI] [PubMed] [Google Scholar]
- 29.Burova E, Hung SC, Sagitov V, Stitt BL, Gottesman ME. Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J Bacteriol. 1995;177:1388–1392. doi: 10.1128/jb.177.5.1388-1392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cadwell RC, Joyce GF. Mutagenic PCR. PCR Methods Appl. 1994;3:S136–S140. doi: 10.1101/gr.3.6.s136. [DOI] [PubMed] [Google Scholar]
- 31.Li J, Horwitz R, McCracken S, Greenblatt J. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage lambda. J Biol Chem. 1992;267:6012–6019. [PubMed] [Google Scholar]
- 32.Li J, Mason SW, Greenblatt J. Elongation factor NusG interacts with termination factor rho to regulate termination and antitermination of transcription. Genes Dev. 1993;7:161–172. doi: 10.1101/gad.7.1.161. [DOI] [PubMed] [Google Scholar]
- 33.Jeon YH, Yamazaki T, Otomo T, Ishihama A, Kyogoku Y. Flexible linker in the RNA polymerase alpha subunit facilitates the independent motion of the C-terminal activator contact domain. J Mol Biol. 1997;267:953–962. doi: 10.1006/jmbi.1997.0902. [DOI] [PubMed] [Google Scholar]
- 34.Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 35.Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. NMR Spectrosc. 1999;34:93–158. [Google Scholar]
- 37.Tjandra N, Garrett DS, Gronenborn AM, Bax A, Clore GM. Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy. Nat Struct Biol. 1997;4:443–449. doi: 10.1038/nsb0697-443. [DOI] [PubMed] [Google Scholar]
- 38.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. "The Xplor-NIH NMR Molecular Structure Determination Package,". J.Magn. Res. 2003;160:66–74. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 39.Hoopes BC, McClure WR. Studies on the selectivity of DNA precipitation by spermine. Nucleic Acids Res. 1981;9:5493–5504. doi: 10.1093/nar/9.20.5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burgess BR, Richardson JP. RNA passes through the hole of the protein hexamer in the complex with the Escherichia coli Rho factor. J Biol Chem. 2001;276:4182–4189. doi: 10.1074/jbc.M007066200. [DOI] [PubMed] [Google Scholar]
- 41.Nowatzke W, Richardson L, Richardson JP. Purification of transcription termination factor Rho from Escherichia coli and Micrococcus luteus. Methods Enzymol. 1996;274:353–363. doi: 10.1016/s0076-6879(96)74030-2. [DOI] [PubMed] [Google Scholar]
- 42.Hager DA, Jin DJ, Burgess RR. Use of Mono Q high-resolution ion-exchange chromatography to obtain highly pure and active Escherichia coli RNA polymerase. Biochemistry. 1990;29:7890–7894. doi: 10.1021/bi00486a016. [DOI] [PubMed] [Google Scholar]
- 43.Miller J. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 44.Ederth J, Mooney RA, Isaksson LA, Landick R. Functional interplay between the jaw domain of bacterial RNA polymerase and allele-specific residues in the product RNA-binding pocket. J Mol Biol. 2006;356:1163–1179. doi: 10.1016/j.jmb.2005.11.080. [DOI] [PubMed] [Google Scholar]
- 45.Yanofsky C, Horn V. Rifampin resistance mutations that alter the efficiency of transcription termination at the tryptophan operon attenuator. J Bacteriol. 1981;145:1334–1341. doi: 10.1128/jb.145.3.1334-1341.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 47.Posfai G, Plunkett G, 3rd, Feher T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, Burland V, Harcum SW, Blattner FR. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 48.Amann E, Ochs B, Abel KJ. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 49.Richardson LV, Richardson JP. Identification of a structural element that is essential for two functions of transcription factor NusG. Biochim Biophys Acta. 2005;1729:135–140. doi: 10.1016/j.bbaexp.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan SL, Ward DF, Gottesman ME. Effect of Escherichia coli nusG function on lambda N-mediated transcription antitermination. J Bacteriol. 1992;174:1339–1344. doi: 10.1128/jb.174.4.1339-1344.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01. Supplemental Figure 1. Comparison of structures of E. coli NusG, RfaH, and Spt5.
(Top) Solution structure of _Ec_NusG NTD (this work). The hydrophobic residues defining the pocket are colored green; a ribbon structure representation with key residues labeled is presented on the left, a space-fill representation is on the right.
(Middle) RfaH NTD 18. Colors and labeling as above.
(Bottom) Spt5 NGN. 19; 21 . Colors and labeling as above. The region of Spt4 that interacts with Spt5 in the co-crystal is shown in cyan. This does not overlap with the hydrophobic patch on Spt5.
02. Supplemental Figure 2. In vitro transcription assays with NTD and CTD.
(A) Elongation rate assay with no factor, NTD, or CTD. The experimental design is shown above the gel. The quantitation of this and two other experiments is presented in Figure 5A. NTD allows RNAP to reach the end of the template faster than reactions that contain either no additional factor or the CTD alone.
(B) Pausing at ops with no factor, NTD, or CTD. The experimental design is shown above the gel. The quantitation of this and two other experiments is presented in Figure 5B. NTD can suppress pausing and allows RNAP to reach the end of the template faster than reactions that contain either no additional factor or the CTD alone.
03. Supplemental Figure 3. In vitro transcription assays with selected NusG point mutants.
(A) Elongation rate assay. The experimental scheme is shown above the gel. Transcription rate was not stimulated by addition of either the F65L or Y68H mutants at 50 nM, but at 500 nM these two mutants could stimulate elongation similarly to wild-type NusG as evidenced by the faster accumulation of the full-length product (668 nt). The data from this experiment and two others is quantitated and presented in Figure 6A.
(B) L158P does not function for termination but can still suppress pausing
(left) Termination assay with either wild-type or L158P NusG at either 50 nM or 500 nM. Neither condition allowed enhancement of termination. The data from this experiment and others is quantitated and presented in Figure 6B.
(right) Pausing assay with the L158P variant. Despite being defective for termination, the L158P mutant can suppress pausing at the ops pause site. The data from this experiment and two others is quantitated and presented in Figure 6C.