Pten in Stromal Fibroblasts Suppresses Mammary Epithelial Tumors (original) (raw)
. Author manuscript; available in PMC: 2010 Apr 22.
Published in final edited form as: Nature. 2009 Oct 22;461(7267):1084–1091. doi: 10.1038/nature08486
SUMMARY
The tumor stroma is believed to contribute to some of the most malignant characteristics of epithelial tumors. However, signaling between stromal and tumor cells is complex and remains poorly understood. Here we show that the genetic inactivation of Pten in stromal fibroblasts of mouse mammary glands accelerated the initiation, progression and malignant transformation of mammary epithelial tumors. This was associated with the massive remodeling of the extra-cellular matrix (ECM), innate immune cell infiltration and increased angiogenesis. Loss of Pten in stromal fibroblasts led to increased expression, phosphorylation (T72) and recruitment of Ets2 to target promoters known to be involved in these processes. Remarkably, Ets2 inactivation in Pten stroma-deleted tumors ameliorated disruption of the tumor microenvironment and was sufficient to decrease tumor growth and progression. Global gene expression profiling of mammary stromal cells identified a Pten-specific signature that was highly represented in the tumor stroma of breast cancer patients. These findings identify the Pten-Ets2 axis as a critical stroma-specific signaling pathway that suppresses mammary epithelial tumors.
Coordinated signaling between different cell types of the ‘_normal stroma_’ is required during embryonic and adult development1. The stroma can be appropriately activated in response to extreme but normal physiological cues, such as wounding, inflammation or pregnancy2. The stroma can also be inappropriately activated in cancer3,4. In breast tumors, stromal fibroblasts are believed to adapt and continuously co-evolve along with tumor epithelial cells in order to foster transformation and tumor growth5. Fibroblasts are a principal constituent of the stroma responsible for the synthesis of growth and survival factors, angiogenic and immunological chemokines, and structural components of the ECM as well as enzymes that control its turnover6,7. Despite extensive evidence for a role of the tumor stroma in carcinogenesis, relatively little is known about the signaling pathways involved in the communication between the different cellular compartments of the microenvironment that contribute to the cancer phenotype.
Alterations in the phosphoinositide 3-kinase (PI3K) pathway are associated with the activation of tumor-associated stroma8, 9. One of the main regulators of PI3K signaling is the phosphatase and tensin homolog (PTEN), a tumor suppressor with lipid and protein phosphatase activity10, 11. PTEN inactivation disrupts multiple cellular processes associated with cell polarity, cell architecture, chromosomal integrity, cell cycle progression, cell growth and stem cell self–renewal12, 13. Germ-line inactivation of a single allele of PTEN in both humans and mice contributes to the genesis of a variety of tumor types of epithelial origin14. While tremendous progress in understanding PTEN function in tumor cells has been made since its discovery over a decade ago, relatively little is known about its potential role in the tumor stroma. Here, we show that Pten ablation in mammary stromal fibroblasts of mice results in massive remodeling of the ECM and tumor vasculature, recruitment of innate immune cells, and increased malignancy of mammary epithelial tumors. Gene expression profiling of _Pten-_deleted stromal fibroblasts identified an Ets2–specific transcription program associated with many of these aggressive tumor phenotypes. Remarkably, the concomitant inactivation of Ets2 in the mammary stroma reversed the increased malignancy caused by Pten deficiency. These findings expand _Pten_’s repertoire as a tumor suppressor by identifying the fibroblast as a key site from which it exerts its powerful tumor suppressive influence on the adjacent tumor epithelium.
RESULTS
Pten in stromal fibroblasts suppresses mammary tumors of epithelial origin
To rigorously evaluate the role of Pten in the tumor microenvironment of breast cancer we generated mice containing a mesenchymal-specific Fsp-cre transgene15 and conditional alleles of Pten (PtenloxP; Supplementary Fig. 1). Cell type-marker analysis using a β-galactosidase Rosa26LoxP reporter allele showed specific Fsp-cre expression in stromal fibroblasts surrounding the mammary epithelial ducts, with no expression in cytokeratin-positive epithelial cells, F4/80-positive macrophages and CD31-positive endothelial cells (Fig. 1a, Supplementary Fig. 2a, 2b). Western blot and PCR assays demonstrated efficient cre-mediated deletion of PtenloxP in stromal fibroblasts isolated from Fsp-cre;PtenloxP/loxP mammary glands (Fig. 1b, Supplementary Fig. 3a). Examination of mammary sections by immunohistochemistry (IHC) and immunofluorescence (IF) showed deletion of PtenloxP that was confined to stromal fibroblasts, with no collateral deletion in epithelial ducts or the adjacent myoepithelium (Fig. 1c, Supplementary Fig. 3b, 3c). Interestingly, this resulted in the expansion of the ECM, but did not lead to the transformation of the mammary epithelium (Fig 1c, 1e).
Figure 1. Stromal fibroblast-specific deletion of Pten.
(a) Whole mount, X-gal stained mammary glands from Fsp-cre;Rosa+/loxP and Rosa+/loxP (top inset) mice. Higher magnification of whole mount gland (bottom left) and a histological cross section (bottom right); scale bar represent 30 μm. lu, lumen; epi, epithelium; str, stroma.
(b) Representative Western blot analysis of mammary fibroblast lysates derived from 8 week-old PtenloxP/loxP mice with (+) without (−) Fsp-cre.
(c) Paraffin sections from 8 week-old female mammary glands stained with a Pten-specific antibody; lower panels represent higher magnification of boxed areas; scale bars for top panels represent 200 μm and for bottom panels 30 μm. lu, lumen; epi, epithelial compartment; str, stromal compartment; red dotted line indicates the border between the two compartments.
(d) Tumors collected at 26 weeks post-transplantation.
(e) Tumor development by 16 weeks in mammary glands with the indicated genotypes. Tumorigenicity was determined by palpation or histological presentation of adenoma/carcinoma at each implantation site and statistically analyzed using Fisher’s Exact test. (n), represents the total number of transplants.
(f) Total tumor burden at 26 weeks post-transplantation in mammary glands with the indicated genotypes. Differences were tested using the non-parametric Wilcoxon Rank Sum test.
(g) H&E stained sections of mammary glands harvested at time of transplantation (0 weeks) and indicated times post-transplantation; scale bars represent 100 μm.
We then examined the role of stromal Pten on mammary tumorigenesis using an established mouse model of breast cancer, MMTV-ErbB2/neu (ErbB2)16. To avoid possible confounding effects caused by Pten deletion in mesenchymal cells of other organs, mammary glands from Fsp-cre;PtenloxP/loxP, ErbB2;PtenloxP/loxP and ErbB2;Fsp-cre;PtenloxP/loxP donors were transplanted into syngeneic wild-type recipients17 and tumor development was monitored over the course of several months. By genetically marking the stroma with the Rosa26LoxP reporter allele, we demonstrated that both the epithelium and its associated stroma were effectively transplanted into host female mice (Supplementary Fig. 4). Loss of Pten in stromal fibroblasts dramatically increased the incidence of _ErbB2_-driven mammary tumors (Fig. 1d–f). By 16 weeks post-transplantation, these lesions progressed to adenoma, carcinoma in situ and invasive carcinoma (Fig. 1g) and by 26 weeks most females met the criteria for early removal due to excessive tumor burden (Fig. 1e). Histological examination showed that _ErbB2_-tumor cells in Pten stromal-deleted tumors retained their typical oncogene-specific morphology, with small nuclei, fine chromatin and abundant eosinophilic cytoplasm18. In contrast to non-deleted tumors 18,19, Pten stromal-deleted tumors had a significant amount of stroma surrounding and infiltrating the epithelial masses (Fig. 1g). PCR-based and immunohistochemical assays confirmed that tumors had intact PtenloxP alleles in the epithelial compartment (Supplementary Fig. 5a, 5b and data not shown). Moreover, we used the Rosa26loxP reporter allele to genetically mark early EMT events15 and found no evidence of EMT in tumors that either contained or lacked Pten in stromal fibroblasts (data not shown). Thus, the analysis of the ErbB2 breast cancer tumor model identified a potent tumor suppressor role for Pten in stromal fibroblasts of the mammary gland.
Pten in stromal fibroblasts controls ECM and innate immune functions
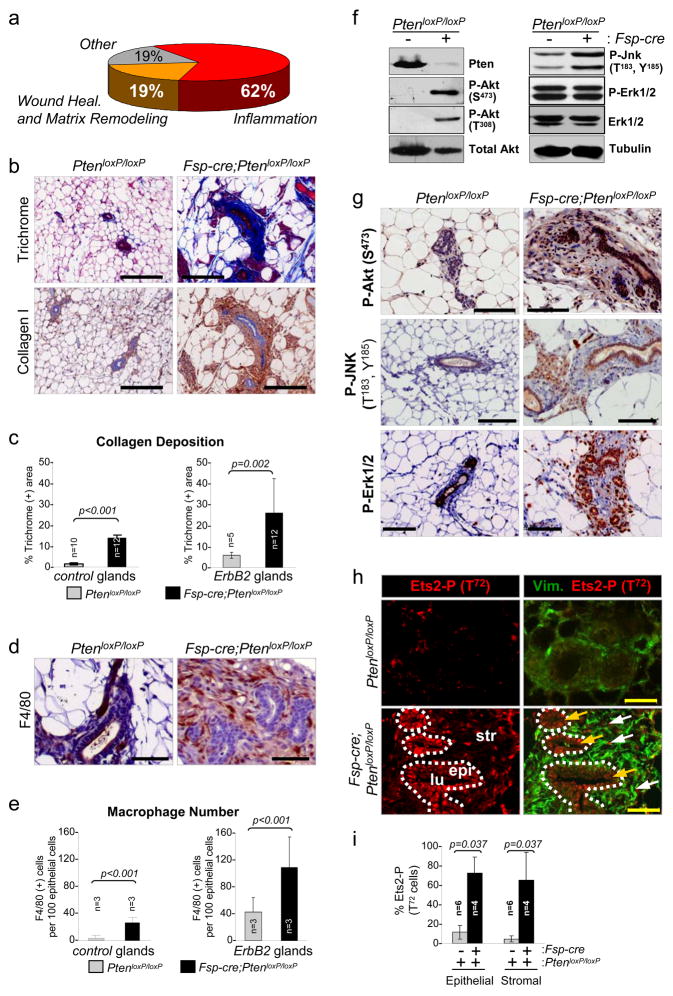
To investigate the tumor suppressive mechanism of Pten action in stromal fibroblasts, we profiled the transcriptome of mammary stromal fibroblasts isolated from PtenloxP/loxP and Fsp-cre;PtenloxP/loxP females. Details of sample collection, processing of Affymetrix oligo-arrays and expression data are available in the Methods and Supplementary Methods sections. Briefly, we implemented class comparison analyses of all probe sets on the Affymetrix mouse genome 430 2.0 array to identify genes differentially expressed between the two genetic groups. We also used an unbiased approach similar to Gene Set Enrichment Analysis20 to identify a priori defined groups of genes that were significantly differentially expressed. The analysis of over 14,000 mouse genes identified 129 upregulated and 21 downregulated unique genes in response to Pten deletion (Supplementary Fig. 6a, 6b; >4-fold at p <0.001; Supplementary Tables 1 and 2). Quantitative RT-PCR assays on a subset of genes confirmed >85% of these expression changes using independent fibroblast samples (Supplementary Fig. 6c, and Supplementary Table 3). Fibroblast samples used to probe the oligo-arrays lacked expression of macrophage-, endothelial- and epithelial-specific genes, confirming the purity of these fibroblast preparations (Supplementary Fig. 6d). Functional annotation21,22 (GO) of _Pten_-responsive targets revealed a remarkable bias toward genes encoding proteins involved in ECM remodeling, wound healing and chronic inflammation21,22 (Fig. 2a; Supplementary Tables 1 and 2). Given this unexpected convergence of function, we performed a more thorough cellular and molecular analysis of _Pten_-deleted stroma. Staining of consecutive mammary gland sections with H&E and Mason’s trichrome stains indicated enhanced deposition of collagen in _Pten_-deleted stroma, which was independent of _ErbB2_-oncogene expression (Fig. 2b, 2c, and Supplementary Fig. 7a). IHC and Western blot assays using collagen type-specific antibodies showed that the non-cellular material consisted mostly of type-I collagen and not the basement membrane type-IV collagen (Fig. 2b, 2c, and Supplementary Fig. 7b, 7c). There was significant infiltration of F4/80-positive macrophages into stromal _Pten_-deleted mammary glands (Fig. 2d, 2e), and this was also independent of ErbB2 expression (Supplementary Fig. 8a). The abundance of B- and T- cells did not change in response to stromal deletion of Pten (data not shown). From these experiments we conclude that ablation of Pten in stromal fibroblasts recapitulates two key events associated with tumor malignancy: increased ECM deposition and innate immune cell infiltration.
Figure 2. Characterization of ECM deposition and immune cell infiltration.
(a) Schematic representation of the biological processes affected by differentially expressed genes (<4 fold) in _Pten_-deleted stromal fibroblasts.
(b) Mammary gland paraffin sections stained with Masson’s Trichrome and Collagen I-specific antibodies, respectively; scale bars represent 200 μm.
(c) Trichrome stained sections were quantified for collagen deposition; mammary glands in the absence (left graph) or presence of ErBb2 (right graph) were analyzed, respectively. Values shown represent the mean with s.d.; Wilcoxon Rank Sum test was used for the comparison between groups.
(d) Mammary gland paraffin sections stained with the macrophage-specific marker F4/80; scale bars represent 50 μm.
(e) Quantification of F4/80 positive stained stromal cells in mammary glands in the absence (left graph) or presence of ErBb2 (right graph), respectively. Values shown represent the mean with s.d.; Wilcoxon Rank Sum test was used for the comparison between groups.
(f) Western blot analysis of whole-cell lysates derived from mammary stromal fibroblasts
(g) Mammary gland paraffin sections stained with the phospho-Akt473/308, phospho-JNK183/185 and phospho-Erk1/2 specific antibodies; scale bars represent 100 μm.
All analyses were performed using tissue or cells from 8 week-old females.
(h) Frozen mammary tissue sections stained with a phospho-Ets2T72-specific antibody. Note that loss of Pten in the mammary stroma increased Ets2 phosphorylation in both the stromal and epithelial compartments. Dotted-white line indicates the stromal-epithelial boundary. lu, lumen; epi, epithelium; str, stroma and scale bars represent 50 μm.
(i) Quantification of mammary epithelial and stromal cells that stained positive for nuclear phospho-Ets2T72. Values represent the mean with s.d.; Wilcoxon Rank Sum test was used for the statistical comparison between groups.
Loss of stromal Pten activates an Ets2-dependent transcriptional program
Along with the remarkable remodeling of the tumor microenvironment, loss of Pten in stromal fibroblasts resulted in activation of the Ras, JNK and Akt pathways. Western blot analysis using protein lysates derived from _Pten_-deleted stromal fibroblasts demonstrated an increase in the phospho-specific forms of Akt (T308 and S473) and JNK (T183 and Y185) (Fig. 2f; Supplementary Fig. 8b). Immunohistochemical assays confirmed the activation of Akt and JNK in stromal fibroblasts, and interestingly, also revealed a profound activation of these two pathways in ductal epithelial cells adjacent to the _Pten_-deleted stroma (Fig. 2g; Supplementary Fig. 8b). This analysis also showed increased levels of phospho-Erk1/2 in _Pten_-deleted stromal fibroblasts, however, this increase could not be detected in primary cultured fibroblasts (Fig. 2f, 2g), presumably due to the constitutive Pten-independent activation of Erk1/2 by serum-stimulation23.
Among the many expression changes observed in _Pten_-deleted stromal fibroblasts we noted that there was a significant increase of Ets2 mRNA levels (2.8 fold, p <0.001). This induction is notable because the Ets2 transcription factor is known to be transcriptionally induced by MAPK24,25 activation and its function to be post-translationally enhanced by the Akt- and JNK-mediated phosphorylation of its pointed domain at threonine 72 (Ets2T72)23,26. We confirmed the higher levels of Ets2 mRNA and protein in _Pten_-deleted fibroblasts (~3-fold, p<0.001; Supplementary Fig. 9a, 9b) and consistent with the activation of Akt and JNK in these mammary glands, there was a marked increase of P-Ets2T72 in stromal fibroblasts and adjacent epithelial ducts (Fig. 2h, 2i). Loss of Pten in stromal fibroblasts resulted in the induction of a number of genes involved in ECM remodeling and macrophage recruitment, two of which, Mmp9 and Ccl3, are known to be direct transcriptional targets of Ets227,28 (Supplementary Fig. 9c, 6c). The increase of Mmp9 expression appears to be of pathological relevance since in situ zymography29 showed robust Mmp9 activity in tumor samples (Supplementary Fig. 9d). Chromatin immunoprecipitation (ChIP) assays showed an increase in the loading of Ets2 onto the Mmp9 and Ccl3 promoters in _Pten-_deleted mammary fibroblasts (Supplementary Fig. 9e), suggesting a direct role for Ets2 in the transcriptional regulation of these two target genes in vivo. Together, these data illustrate the extensive molecular reprogramming that takes place in the tumor and its microenvironment in response to ablation of Pten in stromal fibroblasts.
Stromal Ets2 promotes mammary tumorigenesis
To determine whether Ets2 promotes a microenvironment conducive to tumor growth we analyzed the consequences of ablating a conditional allele of Ets2 (Ets2loxP)30 in mammary stromal fibroblasts of a well-characterized mouse model of breast cancer, MMTV-PyMT (PyMT)31. The PyMT oncogene initiates the rapid onset and progression of mammary tumors and thus represents an appropriate model for evaluating any potential delay that loss of Ets2 might have on tumorigenesis. The efficient Fsp-cre mediated ablation of Ets2 in stromal fibroblasts was facilitated by using mice carrying conventional and conditional knockout alleles of Ets2 (DNA-binding domain-Ets2db/LoxP)32 (Fig. 3a; Supplementary Fig. 10a, 10b). Ablation of Ets2 in these cells had no detectable physiological consequence on the development of mammary glands, either during puberty or pregnancy (MCO, unpublished observations). The evaluation of PyMT;Fsp-cre;Ets2db/loxP and control PyMT;Ets2db/loxP mice over a period of three months showed that ablation of Ets2 in mammary fibroblasts significantly reduced the tumor load (Fig. 3b) and slowed progression to adenoma and early carcinoma (Fig. 3c). Quantitative RT-PCR showed high levels of Mmp9 expression in tumor-associated fibroblasts containing Ets2 and low levels in _Ets2_-deleted fibroblasts (Fig. 3e). Because Mmp9 activity is known to mediate the release of matrix-bound VEGF-A to its active isoforms, including VEGF16433, we visualized the spatial distribution of VEGF164 and Mmp9 activity by IF. These assays showed that the accumulation of VEGF164, which was particularly acute within collagen1A-rich stromal locations overlapping Mmp9 activity, was significantly decreased in stromal-deleted Ets2 tumors (Fig. 3d, 3f). Given that VEGF164 is a specific ligand for VEGF Receptor 2 (VEGFR2; FLK-1; KDR), one of the most potent mediators of VEGF-induced endothelial signaling and angiogenesis34, we also evaluated VEGFR2 status by immuno-staining tumor sections with antibodies specific for CD31 and the phospho-activated form of the murine VEGF receptor (P-VEGFR2Y1173)35. This analysis revealed a four-fold decrease in the number of CD31/VEGFR2Y1173 double-positive cells in _Ets2-_deleted versus non-deleted tumor samples (Fig. 3f, 3g). Together, these data show that loss of Ets2 in stromal fibroblasts results in decreased Mmp9 activity in the tumor ECM and reduced VEGFR2Y1173-activation in the tumor vasculature.
Figure 3. Ets2 ablation in stroma fibroblasts restricts mammary tumorigenesis.
(a) Immunofluorescence staining of cultured mammary fibroblasts with Vimentin (green), p-Ets2(T72) (red) antibodies and counterstained with DAPI (blue).
(b) Total mammary tumor volume of PyMT;Ets2db//loxP (n = 20) and PyMT;Fsp-cre;Ets2db/loxP (n = 21) mice collected 30 days after tumor initiation. Values represent the mean with s.d. shown in parentheses.
(c) H&E staining of tumors harvested from PyMT;Ets2db//loxP or PyMT;Fsp-cre;Ets2db/loxP mice.
(d) Consecutive sections stained for (left to right): trichrome, Mmp9 gelatinase activity and VEGF164, and counterstained with DAPI from PyMT;Ets2db/loxP and PyMT;Fsp-cre;Ets2db/loxP mammary tumors.
(e) Quantification of Mmp9 mRNA expression by qRT-PCR.
(f) Quantification of EGF164 IF staining in tumor stroma (top graph) and tumor endothelial cells co-expressing CD31 and phospho-VEGFR2 (bottom graph).
(g) Tumor vascular endothelial cells visualized by IF double staining with CD31 (green) and p-VEGFR2(Tyr1173) (red), and counterstained with DAPI (blue) in mammary tumors collected one week post tumor initiation in PyMT;Ets2db/loxP and PyMT;Fsp-cre;Ets2db/loxP mice.
All analyses were performed using tissue or cells from 9–10 week-old females and all scale bars represent 50μm. Bar values in Fig. 3e and 3f represent the mean and error bars represent s.d. Student t-test test was used for all the statistical comparisons between groups.
Loss of Ets2 diminishes tumor formation in Pten stromal-deleted mammary glands
We then entertained the hypothesis that Ets2 may be contributing to the remodeling of the tumor microenvironment caused by stromal deletion of Pten. To directly test this possibility, we compared tumor incidence in PtenloxP/loxP, Fsp-cre;PtenloxP/loxP and Fsp-cre;PtenloxP/loxP;Ets2db/loxP mammary glands that were orthotopically injected with an established _ErbB2_-initiated mammary tumor cell line (NT 2.5)36. This orthotopic model recapitulated the consequences of deleting Pten in the mammary stroma that were observed in the genetically engineered _ErbB2_-mouse model described earlier in this study. Indeed, tumor incidence and tumor load in injected Fsp-cre;PtenloxP/loxP females was markedly higher than in control PtenloxP/loxP females (Fig. 4a, 4b). Importantly, mammary glands doubly deleted for stromal Pten and Ets2 had fewer and smaller tumors than glands deleted for Pten only. These mammary tumors had decreased number of macrophages and recruitment of new vasculature (Fig. 4c–f). Loss of Pten and Ets2, however, failed to fully reduce the tumor load and collagen deposition to control levels (Fig. 4b, Supplementary Fig. 11a, 11b), suggesting that additional effectors must contribute towards Pten’s tumor suppressor functions. From these data, we conclude that Ets2 is a major component of the Pten tumor suppressive axis that acts in the stromal fibroblast compartment of mammary glands.
Figure 4. Loss of Ets2 in stromal fibroblasts diminishes tumor growth in stromal _Pten_-deleted mammary glands.
(a) H&E sections of mammary glands after orthotopic injection of the _ErbB2_-expressing tumor cell line NT 2.5. T, tumor; str, stroma and scale bars represent 500 μm.
(b) Volumes of tumors collected 21 days after injection. PtenloxP/loxP;Ets2db/loxP (n=10) and PtenloxP/loxP (n=10) control groups were combined ((*), n=20) after it was determined that there was no statistical difference in tumor incidence/load between these two control groups. Values represent the mean with s.d shown in parentheses.
(c) Sections from mammary glands with the indicated genotypes stained with the macrophage-specific marker F4/80. Scale bars represent 100 μm.
(d) Quantification of stromal cells positive for F4/80 in mammary glands. Values shown represent the mean with s.d.
(e) Frozen mammary gland sections stained with the endothelial-specific antibody, CD31. Scale bars represent 50 μm.
(f) Quantification of CD31 positive staining.
Bar values in Fig. 4d and 4f represent the mean and error bars represent s.d. For all the statistical analyses, an ANOVA model with Bonferroni adjustment was used. Pairwise comparisons shown have a significant difference between marked genetic groups.
Mouse fibroblast Pten-signature distinguishes normal from tumor stroma in breast cancer patients
To determine the relevance of these findings to human breast cancer, we compared the mouse stromal fibroblast Pten-expression signature to the expression signatures derived from laser-captured tumor stroma (49 samples) and adjacent normal stroma (52 samples) in breast cancer patients37. This analysis identified 137 human orthologs from the 150 differentially expressed mouse genes detected by the Affymetrix oligo-arrays shown in Supplementary Fig. 6a and 6b. Of these 137 orthologs, 129 genes were represented in the expression platform used (Agilent) for the analysis of human patient stroma samples37. Only 70 of these 129 genes had highly variable gene expression across all human stromal samples (a variance cutoff of >0.5). The heat map generated for the human stroma dataset showed that this 70 gene-subset derived form the mouse Pten-signature was sufficient to distinguish normal from tumor stroma in all patients (Fig. 5a; p = 8e−5 as determined by a permutation test). Principal Component Analysis (PCA) also discriminated normal from tumor stroma perfectly (p<1e−10; Supplementary Fig. 12). Interestingly, 12 of the 70 human orthologs identified by the Pten-signature (Fig. 5a: gene names highlighted in red; Supplementary Fig. 13) were previously shown to be differentially expressed in the tumor stroma of breast cancer patients and to be associated with recurrence37 (Fig. 5b; p = 2.5e−8, Fisher’s Exact Analysis). These analyses suggest that the fibroblast Pten-expression signature identified by our stroma mouse model represents a significant subset of the total gene signature expressed in the stroma of human breast cancer. We interpret these results to mean that a portion of the transcriptome regulated by Pten in mammary stromal fibroblasts is dysregulated in the tumor stroma of breast cancer patients.
Figure 5. Pten-signature is represented in breast cancer stroma.
(a) Heat map displaying the expression of the human orthologs of the 70-gene subset in normal- and tumor-stroma from human breast cancer patients. The 70-gene subset derived from the mouse Pten-signature includes 57 genes upregulated (denoted by red bar on the y-axis) and 13 genes downregulated (denoted by the green bar on the y-axis). Red and green regions inside the heat map indicate relative gene expression levels (red, increased and green, decreased). The p-value indicates the ability of the mouse 70-gene signature to partition normal and tumor stroma in breast cancer patients (see Statistical Methods). The coded patient IDs are listed at the bottom.
(b) Venn diagram depicting the overlap between the mouse Pten-deleted fibroblast and human stroma microarray data sets. The 12 genes highlighted in red (Fig. 5a, right margin) are common between the mouse Pten 70 gene-signature and the human 163-gene signature that has been shown to associate with recurrence38. This overlap is highly significant (p-value=2.5e−8; Fisher’s Exact Test).
(c) Representative Pten, P-Ets2T72 and P-AktS473 IHC staining in human breast carcinoma samples from the tissue microarray. Scale bar represents 50 μm.
(d) Pearson correlations between Pten, P-Ets2T72 and P-AktS473 expression based on Allred scores of a tissue microarray (Fig 5c) containing 99 patients with advanced breast carcinoma
We also evaluated the relevance of the Pten Ets2 relationship in human cancer by immunohistochemical staining of breast cancer tissue microarrays (TMAs) with antibodies specific for Pten, P-Ets2T72 and P-AktS473 (Fig. 5c). From the analysis of 99 patient samples with invasive carcinoma, Pten expression was scored as absent or low in approximately fifty percent of samples. Importantly, Pten staining in the TMAs was negatively correlated with P-AktS473 and nuclear P-Ets2T72, whereas P-AktS473 and nuclear P-Ets2T72 showed a positive association (Fig. 5d). These results suggest that activation of P-Ets2T72 in human breast cancer stroma is a pathologic event that is favored by a reduction in Pten expression.
DISCUSSION
Histopathology and molecular studies suggest that malignant tumors consist of a complex cellular system that is dependent on reciprocal signaling between tumor cells and the adjacent stroma. However, the signaling pathways that mediate the communication between the various cell types in the tumor remain virtually unknown. We recently developed a mesenchymal-specific cre mouse15 and used it here to examine the consequences of inactivating Pten in mammary stromal fibroblasts. Using this system we show, for the first time, that Pten in stromal fibroblasts has a critical role in the suppression of epithelial mammary tumors that is, in part, mediated through an Ets2-regulated transcriptional program.
The tumor suppressor functions of PTEN have been extensively studied in the tumor cell38–40. We show here that genetic ablation of Pten in mammary stromal fibroblasts of mice alters the expression profile of these cells to increase ECM, chemokine and cytokine production in the tumor microenvironment. As a result, Pten stromal-deleted tumors exhibit high levels of collagen, macrophage recruitment and vascular networks, which together favor the initiation and progression of mammary epithelial tumors. Remarkably, side-by-side evaluation of histopathology by independent pathologists could not distinguish tumors between Pten stromal-deleted mice and human breast cancer patients, highlighting the importance of modeling stromal cell compartments of the tumor microenvironment. The mechanism by which Pten in the stroma exerts its tumor suppressor role likely involves the control of multiple signaling pathways, including components of the Ras, Akt and JNK networks, which together culminate in the regulation of Ets2 transcriptional activity. The fact that loss of Ets2 in mammary stromal fibroblasts diminished the oncogenic consequences of deleting Pten in these cells underscores the importance of the stromal Pten-Ets2 axis in stromal fibroblasts during tumor suppression. These observations are consistent with previous work from Oshima and colleagues that showed a critical cell non-autonomous role for Ets2 in the growth of mammary tumors in mice41 and with the identification of Ets2 activation as a key event associated with breast cancer in human patients having poor prognosis42–44. The relevance of this mouse Pten-Ets2 tumor suppressor axis to breast cancer is underligned by the high correspondence between the mouse and human stromal expression signatures. The observation that the dire consequences of targeting this _Ets2_-driven stromal program are tumor-specific, sparing normal mammary development, emphasizes the potential utility of stromal-specific strategies for therapeutic intervention in human breast cancer.
In summary, this work identifies Pten-Ets2 as a key regulatory axis in stromal fibroblasts that suppresses mammary epithelial tumors by profoundly attenuating some of the most malignant characteristics of the tumor microenvironment. This novel function of Pten may be relevant in the suppression of epithelial tumors of other organs, but may also extend beyond cancer, to conditions where the microenvironment may impact disease manifestation, such as in autoimmune syndromes45, lung fibrosis46 and neurodegeneration47. Interestingly, the stromal Pten expression signature identified here includes genes that have been causally linked to ECM deposition and inflammation in rheumatoid arthritis, lung fibrosis and neurodegeneration (Supplementary Table 1 and Table 2). These data offer a molecular basis for how altered Pten signaling in the tumor stroma may elicit broad responses in a variety of cells in the tumor microenvironment that contribute to disease manifestation.
METHODS SUMMARY
Transgenic mice
Generation of Fsp-cre mice has been described15. PtenloxP mice were created following the strategy described in Supplementary Fig. 1. Ets2loxP mice were generated by standard techniques30. Animals were maintained and euthanized following institutional guidelines. Tenth generation congenic (N10) FVB/N animals were used for transplantation and orthotopic injection studies.
Tissue processing, histology
Tissues were either embedded in OCT or fixed (4% PFA or formalin) and embedded in paraffin. Frozen sections were used for X-gal staining as previously described15. IHC on tissue microarray sections from 99 breast carcinoma patients were scored using Allred score’s system48.
Isolation of primary mammary fibroblasts
Primary mammary fibroblasts were purified following the protocol published previously with minor modifications49. Mammary glands were dissected from 8 week-old female mice, minced and digested with collagenase (0.15% Collagenase I, 160 U/ml Hyaluonidase, 1 μg/ml hydrocortisone and 10 μg/ml insulin with penicillin and streptomycin) in a 5% CO2 incubator overnight at 37°C. Collagenase was neutralized with 10% FBS-DMEM medium. Digested tissue was resuspended in medium and subjected to gravity for 12–15 min. Pellets were washed three times to collect epithelial organoids and supernatants were subjected four more times to gravity sedimentation and then cultured.
RNA and microarray analysis
RNA was harvested with Trizol according to manufacturer’s instructions (Invitrogen). RNA quality and concentration were assessed with Bioanalyzer and Nanodrop RNA 6000 nano-assays. RNA samples were hybridized to Affymetrix GeneChip Mouse genome 430 2.0 platform at the Microarray Shared Resource Facility, Ohio State University Comprehensive Cancer Center. The microarray data was deposited with GEO and can be viewed by going to the following link: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=zhmbrcoqaacyite&acc=GSE16073.
Supplementary Material
1
Acknowledgments
The authors thank Maysoon Rawahneh and Julie Moffitt for excellent histotechnical assistance, Karl Kornacker, Sean Cory and Indrani Vasudeva Murthy for bioinformatics assistance, Parul Gulati for statistics assistance, the OSUCCC Microarray, Nucleic Acids, Trangenics and Flow Cytometry Shared Facilities for technical assistance. MMTV-ErbB2 mice were kindly provided Dr. Muller. This work was funded by the National Institutes of Health to G.L. (R01CA85619, R01HD47470, P01CA097189) and to M.C.O. (P01CA097189), by the Terry Fox New Frontiers Group Grant to M.P., and by the Natural Science and Engineering Research Council of Canada Discovery Grants Program grant to M.H. F.L. and F.P. were funded by Department of Defense Pre-doctoral Fellowships, and J-L.C. was funded by a Department of Defense Postdoctoral Fellowship. G.L. is the recipient of the Pew Charitable Trusts Scholar Award and the Leukemia and Lymphoma Society Scholar Award. M.P. holds the Diane and Sal Guerrera Chair in Cancer Genetics at McGill University.
References
- 1.Wiseman BS, Werb Z. Stromal effects on mammary gland development and breast cancer. Science. 2002;296:1046–9. doi: 10.1126/science.1067431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2006;22:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mueller MM, Fusenig NE. Friends or foes - bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–49. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 4.Schedin P. Pregnancy-associated breast cancer and metastasis. Nat Rev Cancer. 2006;6:281–91. doi: 10.1038/nrc1839. [DOI] [PubMed] [Google Scholar]
- 5.Littlepage LE, Egeblad M, Werb Z. Coevolution of cancer and stromal cellular responses. Cancer Cell. 2005;7:499–500. doi: 10.1016/j.ccr.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 6.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–7. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 8.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–92. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 9.Bergamaschi A, et al. Extracellular matrix signature identifies breast cancer subgroups with different clinical outcome. J Pathol. 2008;214:357–67. doi: 10.1002/path.2278. [DOI] [PubMed] [Google Scholar]
- 10.Myers MP, et al. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci U S A. 1998;95:13513–8. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stambolic V, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 12.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–14. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Knobbe CB, Lapin V, Suzuki A, Mak TW. The roles of PTEN in development, physiology and tumorigenesis in mouse models: a tissue-by-tissue survey. Oncogene. 2008;27:5398–415. doi: 10.1038/onc.2008.238. [DOI] [PubMed] [Google Scholar]
- 14.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–55. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 15.Trimboli AJ, et al. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer Res. 2008;68:937–45. doi: 10.1158/0008-5472.CAN-07-2148. [DOI] [PubMed] [Google Scholar]
- 16.Guy CT, et al. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci U S A. 1992;89:10578–82. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cases S, et al. Development of the mammary gland requires DGAT1 expression in stromal and epithelial tissues. Development. 2004;131:3047–55. doi: 10.1242/dev.01158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrechek ER, et al. Amplification of the neu/erbB-2 oncogene in a mouse model of mammary tumorigenesis. Proc Natl Acad Sci U S A. 2000;97:3444–9. doi: 10.1073/pnas.050408497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desai KV, et al. Initiating oncogenic event determines gene-expression patterns of human breast cancer models. Proc Natl Acad Sci U S A. 2002;99:6967–72. doi: 10.1073/pnas.102172399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 21.Dennis G, Jr, et al. Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 22.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Weng LP, Brown JL, Baker KM, Ostrowski MC, Eng C. PTEN blocks insulin-mediated ETS-2 phosphorylation through MAP kinase, independently of the phosphoinositide 3-kinase pathway. Hum Mol Genet. 2002;11:1687–96. doi: 10.1093/hmg/11.15.1687. [DOI] [PubMed] [Google Scholar]
- 24.Fowles LF, et al. Persistent activation of mitogen-activated protein kinases p42 and p44 and ets-2 phosphorylation in response to colony-stimulating factor 1/c-fms signaling. Mol Cell Biol. 1998;18:5148–56. doi: 10.1128/mcb.18.9.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarthy SA, et al. Rapid phosphorylation of Ets-2 accompanies mitogen-activated protein kinase activation and the induction of heparin-binding epidermal growth factor gene expression by oncogenic Raf-1. Mol Cell Biol. 1997;17:2401–12. doi: 10.1128/mcb.17.5.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith JL, et al. ets-2 is a target for an akt (Protein kinase B)/jun N-terminal kinase signaling pathway in macrophages of motheaten-viable mutant mice. Mol Cell Biol. 2000;20:8026–34. doi: 10.1128/mcb.20.21.8026-8034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watabe T, et al. The Ets-1 and Ets-2 transcription factors activate the promoters for invasion-associated urokinase and collagenase genes in response to epidermal growth factor. Int J Cancer. 1998;77:128–37. doi: 10.1002/(sici)1097-0215(19980703)77:1<128::aid-ijc20>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 28.Wei G, et al. Activated Ets2 is required for persistent inflammatory responses in the motheaten viable model. J Immunol. 2004;173:1374–9. doi: 10.4049/jimmunol.173.2.1374. [DOI] [PubMed] [Google Scholar]
- 29.Ludwig T. Local proteolytic activity in tumor cell invasion and metastasis. Bioessays. 2005;27:1181–91. doi: 10.1002/bies.20306. [DOI] [PubMed] [Google Scholar]
- 30.Wei G, et al. Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood. 2009 doi: 10.1182/blood-2009-03-211391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin EY, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–26. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto H, et al. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998;12:1315–26. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S, Jilani SM, Nikolova GV, Carpizo D, Iruela-Arispe ML. Processing of VEGF-A by matrix metalloproteinases regulates bioavailability and vascular patterning in tumors. J Cell Biol. 2005;169:681–91. doi: 10.1083/jcb.200409115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Millauer B, et al. High affinity VEGF binding and developmental expression suggest Flk-1 as a major regulator of vasculogenesis and angiogenesis. Cell. 1993;72:835–46. doi: 10.1016/0092-8674(93)90573-9. [DOI] [PubMed] [Google Scholar]
- 35.Sakurai Y, Ohgimoto K, Kataoka Y, Yoshida N, Shibuya M. Essential role of Flk-1 (VEGF receptor 2) tyrosine residue 1173 in vasculogenesis in mice. Proc Natl Acad Sci U S A. 2005;102:1076–81. doi: 10.1073/pnas.0404984102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dakappagari NK, et al. Conformational HER-2/neu B-cell epitope peptide vaccine designed to incorporate two native disulfide bonds enhances tumor cell binding and antitumor activities. J Biol Chem. 2005;280:54–63. doi: 10.1074/jbc.M411020200. [DOI] [PubMed] [Google Scholar]
- 37.Finak G, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–27. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 38.Berns K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 39.Saal LH, et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008;40:102–7. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saal LH, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104:7564–9. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tynan JA, Wen F, Muller WJ, Oshima RG. Ets2-dependent microenvironmental support of mouse mammary tumors. Oncogene. 2005;24:6870–6. doi: 10.1038/sj.onc.1208856. [DOI] [PubMed] [Google Scholar]
- 42.Buggy Y, et al. Ets2 transcription factor in normal and neoplastic human breast tissue. Eur J Cancer. 2006;42:485–91. doi: 10.1016/j.ejca.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 43.Park ES, et al. Heterologous tissue culture expression signature predicts human breast cancer prognosis. PLoS ONE. 2007;2:e145. doi: 10.1371/journal.pone.0000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svensson S, et al. ERK phosphorylation is linked to VEGFR2 expression and Ets-2 phosphorylation in breast cancer and is associated with tamoxifen treatment resistance and small tumours with good prognosis. Oncogene. 2005;24:4370–9. doi: 10.1038/sj.onc.1208626. [DOI] [PubMed] [Google Scholar]
- 45.Pap T, et al. Activation of synovial fibroblasts in rheumatoid arthritis: lack of Expression of the tumour suppressor PTEN at sites of invasive growth and destruction. Arthritis Res. 2000;2:59–64. doi: 10.1186/ar69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White ES, et al. Negative regulation of myofibroblast differentiation by PTEN (Phosphatase and Tensin Homolog Deleted on chromosome 10) Am J Respir Crit Care Med. 2006;173:112–21. doi: 10.1164/rccm.200507-1058OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gibson GE, Huang HM. Oxidative processes in the brain and non-neuronal tissues as biomarkers of Alzheimer’s disease. Front Biosci. 2002;7:d1007–15. doi: 10.2741/A827. [DOI] [PubMed] [Google Scholar]
- 48.Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–68. [PubMed] [Google Scholar]
- 49.Soule HD, McGrath CM. A simplified method for passage and long-term growth of human mammary epithelial cells. In Vitro Cell Dev Biol. 1986;22:6–12. doi: 10.1007/BF02623435. [DOI] [PubMed] [Google Scholar]
- 50.Trimboli AJ, et al. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer Res. 2008;68:937–45. doi: 10.1158/0008-5472.CAN-07-2148. [DOI] [PubMed] [Google Scholar]
- 51.Wei G, et al. Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood. 2009 doi: 10.1182/blood-2009-03-211391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto H, et al. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998;12:1315–26. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 54.Cases S, et al. Development of the mammary gland requires DGAT1 expression in stromal and epithelial tissues. Development. 2004;131:3047–55. doi: 10.1242/dev.01158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dakappagari NK, et al. Conformational HER-2/neu B-cell epitope peptide vaccine designed to incorporate two native disulfide bonds enhances tumor cell binding and antitumor activities. J Biol Chem. 2005;280:54–63. doi: 10.1074/jbc.M411020200. [DOI] [PubMed] [Google Scholar]
- 56.Mook OR, Van Overbeek C, Ackema EG, Van Maldegem F, Frederiks WM. In situ localization of gelatinolytic activity in the extracellular matrix of metastases of colon cancer in rat liver using quenched fluorogenic DQ-gelatin. J Histochem Cytochem. 2003;51:821–9. doi: 10.1177/002215540305100613. [DOI] [PubMed] [Google Scholar]
- 57.Auer H, et al. Gene-resolution analysis of DNA copy number variation using oligonucleotide expression microarrays. BMC Genomics. 2007;8:111. doi: 10.1186/1471-2164-8-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu R, et al. Eos, MITF, and PU.1 recruit corepressors to osteoclast-specific genes in committed myeloid progenitors. Mol Cell Biol. 2007;27:4018–27. doi: 10.1128/MCB.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–68. [PubMed] [Google Scholar]
- 60.Westall PH, Young SS. Resampling-Based Multiple Testing: Examples and Methods for p-Value Adjustment. John Wiley & Sons, Inc; New York, NY: 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1