Adenosine deamination in human transcripts generates novel microRNA binding sites (original) (raw)
Abstract
Animals regulate gene expression at multiple levels, contributing to the complexity of the proteome. Among these regulatory events are post-transcriptional gene silencing, mediated by small non-coding RNAs (e.g. microRNAs), and adenosine-to-inosine (A-to-I) editing, generated by adenosine deaminases that act on double-stranded RNA (ADAR). Recent data suggest that these regulatory processes are connected at a fundamental level. A-to-I editing can affect Drosha processing or directly alter the microRNA (miRNA) sequences responsible for mRNA targeting. Here, we analyzed the previously reported adenosine deaminations occurring in human cDNAs, and asked if there was a relationship between A-to-I editing events in the mRNA 3′ untranslated regions (UTRs) and mRNA:miRNA binding. We find significant correlations between A-to-I editing and changes in miRNA complementarities. In all, over 3000 of the 12 723 distinct adenosine deaminations assessed were found to form 7-mer complementarities (known as seed matches) to a subset of human miRNAs. In 200 of the ESTs, we also noted editing within a specific 13 nucleotide motif. Strikingly, deamination of this motif simultaneously creates seed matches to three (otherwise unrelated) miRNAs. Our results suggest the creation of miRNA regulatory sites as a novel function for ADAR activity. Consequently, many miRNA target sites may only be identifiable through examining expressed sequences.
INTRODUCTION
Adenosine-to-inosine (A-to-I) RNA editing catalyzed by dsRNA-specific ADAR (adenosine deaminase acting on RNA) refers to the conversion of adenosine to inosine in double-stranded (ds) or stem-loop regions of precursor mRNAs (1). Experimental evidence demonstrates that, whether found in a codon, anticodon or mature miRNA, inosine, like guanine, preferentially base-pairs with cytosine (2). Several characterized examples of amino acid changes created by adenosine deamination show that ADARs can regulate gene expression by directing the synthesis of distinct proteins from a single open reading frame (1,3). Recent work by Li et al. (4) confirms that editing events occur at a much higher frequency within non-coding regions. Comparisons of human EST and genomic sequences have identified thousands of distinct ADAR deaminations occurring in many different genes (5). Possible functions for editing events include altered splicing, RNA localization, nuclear retention, mRNA stability and translational efficiency (reviewed in 6). Interestingly, most editing sites occur in Alu elements (5,7–9), the majority of which are in untranslated regions (UTRs) (5,9).
Experimental evidence suggests that miRNA-mediated post-transcriptional gene silencing and A-to-I editing are interrelated (10–13). MiRNA transcripts have been found to undergo ADAR deamination with editing affecting Drosha processing, Dicer processing or mRNA targeting (11,13,14). Work by Kawahara et al. (13) showed that ADAR deamination of the seed region of miR-376 alters the gene set regulated by the edited versus the unedited miRNA. In this work, we asked if A-to-I editing of the target mRNA, rather than the miRNA, could impact mRNA:miRNA binding by creating seed matches. We examined the previously reported 12 723 distinct ADAR editing sites (5), and find A-to-I editing creates perfect complementarities to human miRNA seeds.
RESULTS
Adenosine deamination creates miRNA complementarities
ADAR-mediated conversion of adenosine to inosine allows inosine:cytosine pairing because inosine is chemically similar and functionally equivalent to guanosine (Fig. 1A). A well-established participant in regulating RNA:RNA interactions through altering sequence complementarity, the preferential base pairing of inosine to cytosine was described several decades ago in codon:anticodon interactions (2). More recently, the direct ADAR deamination of a miRNA (miR-376) was found to alter miRNA target selection (13). Over 12 000 A-to-I editing sites have been identified in human mRNAs with nearly 90% of these occurring in UTRs (5,7,8). Because 3′-UTRs are widely accepted as the predominant site of miRNA:mRNA association, we asked whether deamination of A-to-I editing sites (5) significantly altered their complementarity to currently annotated human miRNAs.
Figure 1.
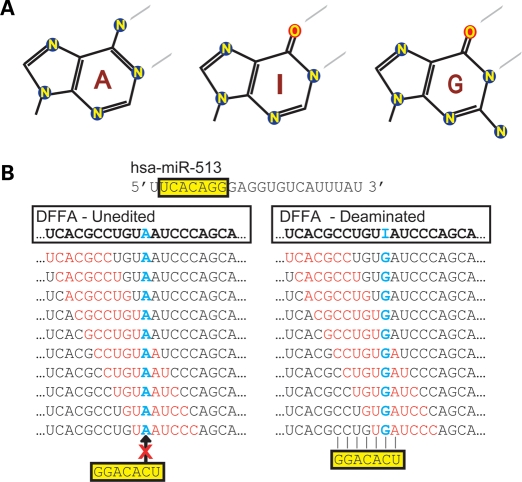
ADARs deaminate adenosine to inosine, potentially altering miRNA complementarities. (A) A cartoon depicting adenosine, deaminated adenosine (inosine) and guanine. (B) A representative deamination site occurring in the 3′-UTR of DNA fragmentation factor α (DFFA) is shown in both the unedited (left) and edited (right) state. For this transcript, the 7 nt sequence scan (red) of the 200 nts flanking the deamination site (blue) identified a miR-513 seed sequence (yellow). This was repeated for all miRNA seeds (miRBase 13.0) for all known edited transcripts.
Although miRNAs are generally ∼21–22 nt in length, their association with target mRNAs is typically mediated through a seven base pair (bp) interaction involving base pairs 2–8 (5′ to 3′) of the mature miRNA (15). This 7 nt sequence constitutes a miRNA ‘seed’ and its reverse complement in a target mRNA, a ‘seed match’ (16). Using a simple 7 bp seed scan of the 100 bp 5′ and 3′ of the 12 723 distinct deamination sites (5), we identified miRNA seed matches that were created or lost. All sites were screened once with a central adenosine (unedited and lost) and once with a central guanosine (edited and created) (Fig. 1B). Using this approach, we identified seed matches to 30 miRNA families that were significantly enriched (P ≤ 1.8 × 10−5) in sequences bearing a central G position (Table 1 and Supplementary Material, Table S1). Strikingly, over 3000 of the 12 723 sites form perfect miRNA seed complements if deaminated. We coined these miRNA associating if deaminated (MAID) sites, and find that most are localized to the noncoding regions (Supplementary Material, Table S2). While editing can also destroy sites (Supplementary Material, Table S1), we focus here on MAIDs and their ability to confer miRNA-mediated regulation.
Table 1.
miRNA seed matches markedly enriched by adenosine deamination
| miRNA | Seed | Totala/createdb/expectedc | _P_-value* |
|---|---|---|---|
| mir-513a-5pd | TCACAGG | 377/258/13.5 | 2.0E−226 |
| mir-769-3p/-450b-3pd | TGGGATC | 400/252/14.4 | 1.2E−212 |
| mir-140-3p | ACCACAG | 348/139/12.5 | 1.2E−92 |
| mir-129-5p | TTTTTGC | 322/105/11.6 | 4.0E−62 |
| mir-936 | CAGTAGA | 269/94/9.7 | 2.5E−58 |
| mir-412 | CTTCACC | 199/83/7.1 | 1.6E−57 |
| mir-326/-330 | CTCTGGG | 196/82/7.0 | 6.1E−57 |
| mir-646 | AGCAGCT | 311/91/11.2 | 2.7E−50 |
| mir-371-3p | AGTGCCG | 97/57/3.5 | 6.2E−48 |
| mir-763/-1207-3p | CAGCTGG | 413/99/14.8 | 3.9E−47 |
| mir-1229 | TCTCACC | 256/74/9.2 | 6.8E−41 |
| mir-630 | GTATTCT | 146/56/5.2 | 1.6E−37 |
| mir-325-5p | CTAGTAG | 161/57/5.8 | 2.3E−36 |
| mir-99ab/-100 | ACCCGTA | 46/27/1.7 | 1.4E−23 |
| mir-518a-dfe | AAAGCGC | 57/27/2.1 | 3.2E−21 |
| mir-643 | CTTGTAT | 66/28/2.4 | 1.0E−20 |
| mir-518ee | AAGCGCT | 61/27/2.2 | 1.7E−20 |
| mir-548c-3p | AAAAATC | 419/59/15.0 | 8.1E−18 |
| mir-578 | TTCTTGT | 140/32/5.0 | 7.8E−16 |
| mir-28-5p/-708 | AGGAGCT | 176/35/6.3 | 2.2E−15 |
| mir-149 | CTGGCTC | 160/30/5.7 | 8.8E−13 |
| mir-1306 | AGGAGCT | 176/28/6.3 | 2.0E−10 |
| mir-1289 | GGAGTCC | 92/20/3.3 | 4.3E−10 |
| mir-344-5p/-484 | CAGGCTC | 311/36/11.2 | 2.9E−9 |
| mir-324-3p | CTGCCCC | 134/22/4.8 | 9.3E−9 |
| mir-615-3p | CCGAGCC | 29/11/1.0 | 1.5E−8 |
| mir-1281 | CGCCTCC | 754/57/27.1 | 3.6E−7 |
| mir-1298 | TCATTCG | 5/5/0.2 | 1.3E−6 |
| mir-197 | TCACCAC | 440/38/15.8 | 1.5E−6 |
| mir-1247 | CCCGTCC | 15/7/0.5 | 1.6E−6 |
MiR-513 and miR-769-3p/-450b-3p specifically target deamination sites
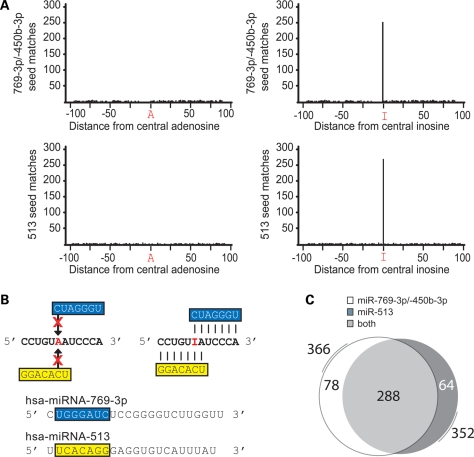
We first examined the greatest outliers, miR-513 and miR-769-3p/-450b-3p, in greater detail. In the 12 723 data set representing unedited sequences, the average number of seed matches to miR-769-3p/-450b-3p at any 7 nt position was 0.79 (max = 4). This strongly contrasts the 252 miR-769-3p/-450b-3p seed matches unique to the edited 3′-UTR data set (Table 1 and Fig. 2A). Similarly, the average number of seed matches to miR-513 at any position was 0.63 (max = 4) versus 257 when comparing the unedited to the edited 3′-UTR flanking sequence and edit site. Therefore, for these mRNAs, miR-513 and miR-769-3p/-450b-3p preferentially target deaminated sequences.
Figure 2.
A-to-I edits create miR-513 and miR-769-3p/-450b-3p complementarities. (A) 12 723 unique EST sequences (www.cgen.com), each consisting of a central A-to-I deamination and 100 nt flanks [i.e. n100 (A or I) n100], were screened for complementarity to human miRNAs. All human miRNA seed matches were identified within the individual 201 nt sequences originally identified as an A-to-I transition by Compugen (statistical significance is addressed in Table 1). The top two panels represent all miR-769-3p (and miR-450b-3p) seed matches occurring at each position in both the unedited (left) and edited (right) states. The lower panels represent all miR-513 seed matches occurring in unedited (left) and edited (right) states. (B) A cartoon of miR-513 and miR-769-3p complementarities to a miRNA Associating If Deaminated (MAID) site in both unedited (left) and edited (right) state is shown. The seed of miR-450b-3p (not shown) is identical to miR-769-3p. Perfect seed matches to miR-769-3p/-450b-3p (blue) and miR-513 (yellow) are significantly enriched in sequences containing characterized deaminations (red). Vertical lines indicate complementary base pairing. (C) Venn diagram depicting the overlap between miR-513 and miR-769-3p/-450b-3p target sites matching the full 13-bp MAID motif created by adjacent miR-513 and miR-769/-450-3p seeds and a G:U wobble. Nearly 100 additional sequences are identified by allowing for the G:U wobble immediately 3′ to the deamination.
Upon closer examination, we found that ∼190 of the matches to the miR-513 seed (3′-GGACACU-5′) and miR-769-3p/-450b-3p seed (3′-CUAGGGU-5′) were created by a single deamination within a common 12 nt motif (5′-CCUGUIAUCCCA-3′) (Fig. 2B). Finding an invariant guanine immediately 3′ of these 12 nt, and allowing for a single G:U wobble at an adenosine or guanine immediately 3′ to the deamination site, extended the miRNA-513/-769-3p/-450b-3p MAID to 5′-CCUGUIRUCCCAG-3′. Thus, the simple scanning approach used identified 288 distinct sites within this 13 nt motif, which when edited forms seed matches to miR-513 and miR-769-3p/-450b-3p (Fig. 2C). Thus, MAIDs containing miR-513 and miR-769-3p/-450b-3p seed matches are significantly enriched in a subset of the deamination sites originally identified by Levanon et al. (5) (Supplementary Material, Table S3). Of note, this result was repeated using a standalone TargetScan program (16) without considering conservation of seed matches as a ranking criterion.
MiR-513 and miR-769-3p repress deaminated sequences
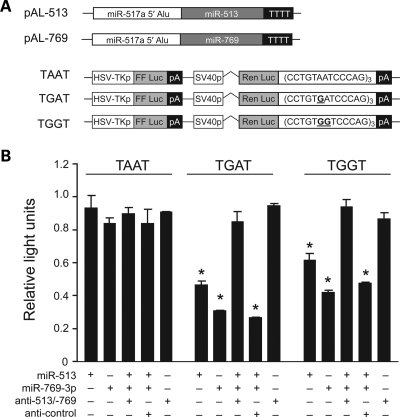
To test whether MAID sequences could serve as miR-513 and/or miR-769-3p/-450b-3p targets, we constructed a series of luciferase reporters possessing unedited sequences, or ‘edited’ 13 bp MAIDs specific to miR-513/miR-759-3p/-450-3p downstream of Renilla luciferase (Fig. 3A). MiR-513 expression vectors repressed TGAT (edited) target activity by ∼50% and TGGT target activity by ∼40% when co-transfected with the reporters (Fig. 3B). Replacing miR-513 with miR-769 in similar experiments resulted in ∼70% and ∼60% reductions in Renilla luciferase activity, respectively (Fig. 3B). Importantly, reporter activity from the ‘unedited’ reporter construct TAAT was not affected when transfected with either miRNA (similar to control, data not shown). We next repeated these experiments with pooled miRNA expression vectors and either miRNA-specific or control miRNA inhibitors (anti-miRs) (17). As shown in Figure 3B, activity was restored to near normal levels in the presence of specific miR-513 and miR-769-3p inhibitors. These data demonstrate that MAIDs can be specifically repressed by miR-513 and miR-769-3p.
Figure 3.
MiR-513 and miR-769-3p target MAID sites but not the corresponding unedited sequence. (A) A diagram shows hairpin expression vectors and MAID reporter constructs. pAL-513 and pAL-769-3p reporter vectors have miR-513 and miR-769-3p hairpins downstream of the miR-517 Pol-III promoter (28). TAAT, TGAT and TGGT reporters contain three tandem copies of the 13 bp MAID sequence in the 3′-UTR of Renilla luciferase for testing activity in the unedited (TAAT) or edited (TGAT and TGGT) states. Guanines mimicking A-to-I edits are bolded and underscored. (B) Renilla luciferase activity (normalized to firefly luciferase and presented as percent mock transfected control) following co-transfection of miR-513, miR-769-3p, pooled miR-513 and miR-769-3p inhibitors and/or control miRNA inhibitor with the indicated reporters into HEK 293 cells (n = 3) is illustrated. *P < 0.005.
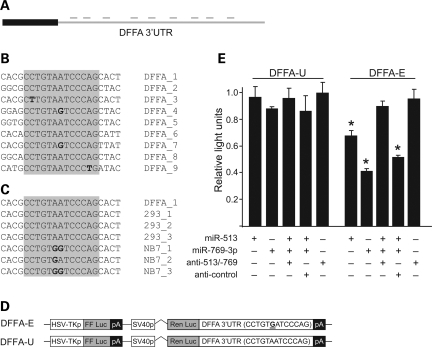
To test whether MAIDs can confer miRNA regulation to endogenous mRNAs, we examined the 3′-UTR of DFFA (DNA fragmentation factor alpha—also referred to as ICAD). DFFA was selected for three reasons: one, the presence of nine 5′-CCUGUIRUCCCAG-3′ motifs within the 3′-UTR (Fig. 4A and B) (18); two, the prevalence of ESTs within the NCBI data set showing ADAR activity in the 3′-UTR (19); and three, our own sequencing confirmation of DFFA deamination in cDNAs cloned from neuroblastoma (NB7) versus HEK 293 cells (Fig. 4C). Together, these characteristics present DFFA as a possible target for MAID regulation.
Figure 4.
Endogenous MAIDs are targets for miR-513 and miR-769-3p repression. (A) A cartoon depicts the DFFA 3′-UTR and the localization of nine distinct MAIDs (lines above the 3′-UTR). (B) Alignment of the nine DFFA 3′-UTR MAIDs commonly deaminated in ESTs is represented. MAID sequences are shaded. Four MAIDs contain G:U wobbles from consensus (bold). (C) Alignment of DFFA_1 sequences from three independent DFFA clones isolated from HEK 293 cells and NB7 cells is shown. DFFA_1 was deaminated in NB7s (bold) but not in HEK 293 cells. RT reactions were performed using a thermostable reverse transcriptase. (D) A diagram of DFFA 3′-UTR reporter constructs is shown. In DFFA-Edited (-E) and DFFA-Unedited (-U), the Renilla luciferase 3′-UTRs are the cloned DFFA 3′-UTRs from NB7 and HEK 293 cells, respectively. DFFA-E nucleotides differing from DFFA-U are bolded and underscored (compare NB7_2 and 293_1 detailed in panel (C). (E) Luciferase assays performed identically to those in Fig. 3B except for the reporter constructs illustrated (n = 3). *P < 0.005.
Two luciferase reporters were constructed to evaluate the ability of miR-513 and/or miR-769-3p to repress DFFA (Fig. 4D). These vectors encode Renilla luciferase harboring either the DFFA edited 3′-UTR cloned from NB7 cells (DFFA-E), or the unedited DFFA 3′-UTR cloned from HEK 293 cells (DFFA-U). DFFA-U and DFFA-E constructs differ by a single adenosine deamination (293_2 versus NB7_2; Fig. 4C). As shown in Figure 4E, miR-513 and miR-769-3p co-transfection repressed DFFA-E reporter activity by ∼30 and 60%, respectively. In contrast, DFFA-U reporter expression was not repressed when transfected with either miRNA (similar to control, data not shown; Fig. 4E). Experiments with pooled miRNA expression vectors and either miRNA-specific or control miRNA inhibitors (anti-miRs) confirmed miRNA targeting activity specific to the edited state. For DFFA-E, but not DFFA-U, Renilla activity was significantly increased (to near normal levels) in the presence of anti-miR-513 and anti-miR-769-3p (Fig. 4E). These experiments indicate that miR-513 and miR-769-3p can regulate the DFFA mRNA 3′-UTR in an adenosine deamination-dependent manner.
MiR-769-3p represses DFFA expression specifically in cells that deaminate the DFFA 3′-UTR
Having confirmed that miR-513 and miR-769-3p selectively repress deaminated DFFA 3′-UTRs within the context of our reporters assays, we next tested whether they could repress endogenous DFFA protein expression. Because miR-513 and miR-769-3p were either undetectable or expressed at very low levels in NB7 cells (data not shown and Fig. 5A), we used over-expression plasmids. MiR-769-3p overexpression in NB7 cells resulted in ∼60% reduction in DFFA, in contrast to miR-769-3p overexpression in HEK 293 cells (Fig. 5B and C). Co-transfection of anti-miR-769-3p, but not a control anti-miR, abrogated this repression (Fig. 5B). To assure these findings were the consequence of miRNA expression, we repeated these experiments using commercially synthesized miRNA precursor RNAs and found these similarly capable of silencing endogenous DFFA levels in NB7 cells (data not shown). The cell line-specific reduction of endogenous DFFA supports the hypothesis that miR-769-3p can regulate the deaminated DFFA 3′-UTRs.
Figure 5.
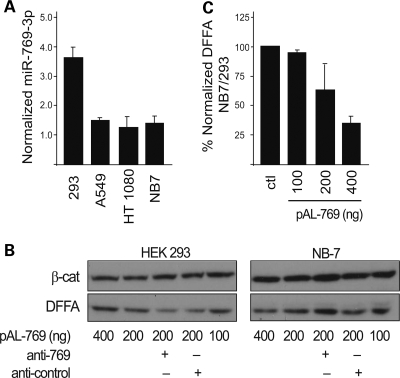
MiR-769-3p selectively represses DFFA protein. (A) Relative mature miR-769-3p levels in HEK 293, A549, HT1080 and NB7 cell lines are shown as determined by quantitative PCR. MiR-513 was not detected in these cell lines. (B) Western blot analysis of endogenous DFFA in HEK 293 and NB7 cell lysates following transfection of miR-769-3p as indicated. Representative blots for DFFA and β-catenin (loading control) are shown. (C) Relative DFFA levels calculated as band intensity ratios of DFFA to β-catenin and normalized to mock (left most bar in each graph). 400, 200 and 100 refer to ng of miR-769-3p expression vector.
DISCUSSION
In this work, we demonstrate that A-to-I editing can create miRNA target sites and that these sites are functional in vitro. On the surface, our findings appear to contradict a recent hypothesis report indicating no correlation between miRNA target sites and A-to-I editing (20); however, our data are largely in agreement, and the apparent conflict can be explained by the scope of our study. The prior work by Liang and Landweber (20) searched for a relationship between ADAR deaminations and 73 conserved miRNA families. In contrast, we evaluated all human miRNAs. By extending the study beyond conserved miRNAs, we identify 325 miRNA families with complementarities enriched at deamination sites. Notably, the majority of miRNAs identified is primate specific and would, therefore, not be identified in an analysis of evolutionarily conserved miRNAs. In more recent work, Li et al. (4) also found no enrichment of editing sites in miRNA target genes when sites within Alu elements were excluded.
While the simple scanning approach we used identified a number of 7 bp miRNA seed matches that were formed upon editing, our results are likely an underestimation. Our approach does not consider the formation (or loss) of a1 sites, which is defined by TargetScan5.0 as having a 6 bp perfect match (nts 2–7 of the seed) and an adenosine immediately 3′ of the seed match. Similarly, our scan would not identify sites with an edited adenosine in position 1 of an 8mer seed match. Even with these possible differences in absolute numbers of created MAIDs or miRNA-targeting sites lost upon editing, data from TargetScan5.0 resulted in the same overall trends for the transcripts profiled.
Alu sequences in transcripts are notable for harboring human miRNA seed complementarities (21,22) and we extend that complementarity to A-to-I edited sites. The MAIDs identified in our work overlap with adenosines that are edited more frequently than those at other positions, as quantified by Kim et al. (8) (Supplementary Material, Fig. S1). Interestingly, the sequences surrounding and containing these sites are highly conserved among Alu families (23). Together with our experimental and informatics analyses, these findings suggest that one outcome of primate mRNA 3′-UTR adenosine deamination is the modulation of miRNA target sites.
Editing occurring within 3′-UTR-resident Alu elements can result in nuclear retention of the transcripts (24); however, 3′-UTR-edited mRNAs can also be associated with polysomes (9), suggesting that not all edited transcripts are retained in the nucleus. Furthermore, a more recent bioinformatics study demonstrated that deletion of sequences between paired Alus can occur, potentially removing miRNA target sites (25). Relevant to our work, Osenberg et al. (25) did not detect cleavage of human DFFA 3′-UTRs, suggesting that our predicted sites remain intact in the final transcript. It will be interesting in further work to determine the sequence isoform of DFFA and other edited 3′-UTRs as to their cytoplasmic expression (or nuclear retention) between cells of different origin, cells in variable states or among different tissues.
Finally, although miRNA:target interactions have been catalogued using bioinformatic approaches, target validation still requires empirical validation. Factors such as G:U base-pairing, local secondary structure, target accessibility and position effects due to nucleotide composition clearly complicate accurate target prediction (26). Here, we demonstrate that some miRNA target sequences are not detectable by querying only the genomic sequence. For the miRNAs we identify as complementary to deaminated sequences, as well as miRNAs potentially targeting other post-transcriptional editing events, accurate target identification may only be possible through evaluating expressed sequences directly. More recent data showing editing events between tissues of an individual (4), and deep sequencing that reveal rare editing events in the developing brain (27), will be useful to analyze further the extent of editing that confers miRNA target site modulation.
MATERIALS AND METHODS
Informatics evaluation of ADAR deamination sites
The full, publically available Compugen deamination data set (composed of 12 723 characterized human deamination sites with 200 nt of flanking sequence (100 nt 5′ and 3′) were obtained from www.cgen.com/research/Publications. Perfect seed matches to all currently annotated human miRNAs were identified in each of the Compugen sequences using either an in-house Perl script which counted seed match occurrence and recorded the position of each within individual 201 nt Compugen sequences (as illustrated in Fig. 1), or an in-house Excel-based analysis, and using the standalone version of Targetscan (Targetscan 5.0). Each 7-mer sequence was queried for perfect identity to the reverse and complement of the 7nt seed sequence for human miRNAs (miRBASE 13.0). Statistical significance of individual miRNA seed occurrence was determined by Poisson distribution. Significance of A-to-I editing site and MAID occurrence in human cDNA databases was determined by chi-square distribution test.
Vector construction
Unless otherwise indicated, PCR amplifications were performed in 40 µl reactions at standard concentrations (1.5 mm MgCl2, 0.2 mm dNTP, 1×Biolase PCR buffer, 0.5 U Taq (Biolase USA, Inc., Randolph, MA, USA), 0.5 µM each primer) and using standard cycling parameters [94°C, 3 min (94°C, 30 s; 55°C, 30 s; 72°C, 60 s) ×30 cycles, 72°C, 3 min] then cloned into Topo TA PCR 2.1 (Invitrogen, Carlsbad, CA, USA). RT–PCRs were performed at 55°C using RetroScript III Reverse Transcriptase (Invitrogen) and oligo-dT 20mers. UTR amplifications were cloned into Topo TA PCR 2.1 and sequenced. Antisense reporters (TAAT, TAGT, TGAT, TGGT, Concensus-A and Concensus-G) were constructed by oligonucleotide primer extension (amplifications performed as above except cycle number was decreased to 25 and extensions to 10s) with primers containing 5′ Xho-I and 3′ Spe-I restriction enzyme sites. Following digestion, amplicons were ligated into the Renilla luciferase 3′-UTR of psiCheck2 (Promega, Madison, WI, USA) vector linearized with Xho-I and Spe-I then incubated with antarctic phosphatase (NEB, Ipswich, MA, USA). The presence of an independently transcribed firefly luciferase in these reporters allowed normalization for transfection efficiency.
Luciferase assays
HEK 293s were cultured in DMEM (10% FBS and 1% PS) in 12-well plates. At 90% confluency, cells were transfected following the Lipofectamine 2000 (Invitrogen) protocol. As indicated, luciferase assays (n = 3) were performed on HEK 293 lysates following cotransfections of psiCheck2 (Promega) luciferase reporters with Alu promoter expression vectors [pAL-513, pAL-769-3p or pAL-1-control (28)] and/or antagomirs (Ambion, Foster City, CA, USA) following manufacturer recommended guidelines. At 35 h, existing media was replaced with 1 ml fresh media. At 36 h, cells were scraped from well bottoms and transferred to 1.5 ml Eppendorf tubes. Eppendorfs were centrifuged at 2000 RCF for 3 min, followed by supernatant aspiration and cell resuspension in 300 µl of PBS. Cells were lysed by three freeze thaws and debris removed by centrifuging at 3000 RCF for 3 min. Fifty microliter of supernatant was transferred to a 96-well MicroLite plate (MTX Lab Systems, Vienna, VA, USA) then firefly and Renilla luciferase activities measured using the Dual-glo Luciferase® Reporter System (Promega) and a 96-well plate luminometer (Dynex, Worthing, West Sussex, UK). RLUs were calculated as the quotient of Renilla luciferase/firefly luciferase RLU and normalized to mock.
Western blotting
Cells were cultured and transfected as described for luciferase assays. At 36 h, existing media was replaced with 100 µl of lysis buffer containing protease inhibitors and incubated for 15 min at 4°C after which cells were scraped from well bottoms and transferred to 1.5 ml Eppendorf tubes. Proteins were electrophoresed through an 8% SDS–polyacrylamide gel (BioRad, Hercules, CA, USA) and transferred to Immobilon-P PVDF membranes (Millipore, Billerica, MA, USA). Membranes were blocked for 1 h in 2% (w/v) non-fat milk in phosphate-buffered saline containing 0.05% Tween 20, washed and incubated with primary antibody overnight at 4°C using the following dilution: DFFA—1:3000 (ab16258, Abcam, Cambridge, MA, USA) and β-catenin—1:6000 (ab2982, Abcam). Membranes were washed and incubated with goat anti-rabbit peroxidase-conjugated secondary antibody (111-035-144, Jackson ImmunoResearch, West Grove, PA, USA). Immunoreactive bands were visualized with ECL Plus (Amersham, Piscataway, NJ, USA) and quantified using a Fluorochem densitometer (Alpha Innotech Corp., San Leandro, CA, USA).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by the National Institutes of Health (B.L.D.: HD44093 and NS50210; D.B.: ES013679) and the Roy J Carver Trust (B.L.D.). G.M.B., R.M.S. and W.L. were supported by Training awards from the National Institutes of Health (T32 HL007638, T32 GM073610 and T32 GM98629, respectively). Funding to pay the Open Access publication charges for this article was provided by discretionary funds.
Supplementary Material
[Supplementary Data]
ACKNOWLEDGEMENTS
The authors would like to thank Ji Wan for his assistance with the statistical analysis.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Bass B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshida M., Kaziro Y., Ukita T. The modification of nucleosides and nucleotides. X. Evidence for the important role of inosine residue in codon recognition of yeast alanine tRNA. Biochim. Biophys. Acta. 1968;166:646–655. [PubMed] [Google Scholar]
- 3.Burns C.M., Chu H., Rueter S.M., Hutchinson L.K., Canton H., Sanders-Bush E., Emeson R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 4.Li J.B., Levanon E.Y., Yoon J.K., Aach J., Xie B., Leproust E., Zhang K., Gao Y., Church G.M. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 5.Levanon E.Y., Eisenberg E., Yelin R., Nemzer S., Hallegger M., Shemesh R., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D., et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 6.Chen L.L., Carmichael G.G. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7:3294–3301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- 7.Athanasiadis A., Rich A., Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim D.D., Kim T.T., Walsh T., Kobayashi Y., Matise T.C., Buyske S., Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hundley H.A., Krauchuk A.A., Bass B.L. C. elegans and H. sapiens mRNAs with edited 3′-UTRs are present on polysomes. RNA. 2008;14:2050–2060. doi: 10.1261/rna.1165008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luciano D.J., Mirsky H., Vendetti N.J., Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang W., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scadden A.D. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 13.Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawahara Y., Megraw M., Kreider E., Iizasa H., Valente L., Hatzigeorgiou A.G., Nishikura K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai E.C. Micro RNAs are complementary to 3′-UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genetics. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 16.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 17.Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 18.Hubbard T.J., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang H., Landweber L.F. Hypothesis: RNA editing of microRNA target sites in humans? RNA. 2007;13:463–467. doi: 10.1261/rna.296407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehnert S., Van Loo P., Thilakarathne P.J., Marynen P., Verbeke G., Schuit F.C. Evidence for co-evolution between human microRNAs and Alu-repeats. PLoS ONE. 2009;4:e4456. doi: 10.1371/journal.pone.0004456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smalheiser N.R., Torvik V.I. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Ray D.A., Batzer M.A. Tracking Alu evolution in New World primates. BMC Evol. Biol. 2005;5:51. doi: 10.1186/1471-2148-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L.L., DeCerbo J.N., Carmichael G.G. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osenberg S., Dominissini D., Rechavi G., Eisenberg E. Widespread cleavage of A-to-I hyperediting substrates. RNA. 2009;15:1632–1639. doi: 10.1261/rna.1581809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smalheiser N.R., Torvik V.I. Complications in mammalian microRNA target prediction. Methods Mol. Biol. 2006;342:115–127. doi: 10.1385/1-59745-123-1:115. [DOI] [PubMed] [Google Scholar]
- 27.Wahlstedt H., Daniel C., Enstero M., Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borchert G.M., Lanier W., Davidson B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]