4E-BP1 is a target of Smad4 essential for TGFβ-mediated inhibition of cell proliferation (original) (raw)
Abstract
Assembly of the multi-subunit eukaryotic translation initiation factor-4F (eIF4F) is critical for protein synthesis and cell growth and proliferation. eIF4F formation is regulated by the translation-inhibitory protein 4E-BP1. While proliferation factors and intracellular pathways that impinge upon 4E-BP1 phosphorylation have been extensively studied, how they control 4E-BP1 expression remains unknown. Here, we show that Smad4, a transcription factor normally required for TGFβ-mediated inhibition of normal cell proliferation, enhances 4E-BP1 gene-promoter activity through binding to a conserved element. 4E-BP1 expression is specifically modulated by treatment with TGFβ and by manipulations of the natural Smad4 regulators (co-Smads) in cells isolated from Smad4+/+ human tumours, whereas no response is observed in cells isolated from Smad4−/− human tumours or in cells where Smad4 has been knocked down by specific siRNAs. In addition, cells where 4E-BP1 has been knocked down (inducible shRNAs in human pancreatic cancer cells or siRNAs in non-malignant human keratinocytes) or has been knocked out (mouse embryonic fibroblasts isolated from 4E-BP1−/− mice) proliferate faster and are resistant to the antiproliferative effect of TGFβ. Thus, 4E-BP1 gene appears critical for TGFβ/Smad4-mediated inhibition of cell proliferation.
Keywords: 4E-BP1, pancreas, Smad4, TGFβ, translation initiation
Introduction
In eukaryotic cells, most mRNAs are translated through the cap-binding protein eukaryotic translation initiation factor-4E (eIF4E), which interacts with the ribosome-associated protein eIF4G, thus docking the ribosome at the mRNA 5′ end (Gingras et al, 1999; Gebauer and Hentze, 2004). eIF4E's function is controlled by the translation-inhibitory protein 4E-BP1 (Richter and Sonenberg, 2005), which competes with eIF4G for a common binding site on eIF4E. Upon various stimuli, 4E-BP1 is hyper-phosphorylated and frees eIF4E, which can in turn stimulate protein synthesis and consequent cell growth and proliferation (Bjornsti and Houghton, 2004; Sonenberg and Pause, 2006). While the signalling pathways (mainly the PI3K/AKT/mTOR cascade) that control 4E-BP1 phosphorylation have been extensively studied (Armengol et al, 2007), little is known about the regulation of its expression. Yet, PI3K also impinges upon 4E-BP1 gene transcription (Azar et al, 2008), and regulation of 4E-BP1 gene transcription appears critical for resistance to stress (Tettweiler et al, 2005; Yamaguchi et al, 2008). Consistently, in addition to inactivation by phosphorylation, alterations in 4E-BP1 levels have been described under different pathological conditions. For instance, eIF4E gain of function consequent to downregulation of the amount of 4E-BP1 has been shown to participate in the genesis and maintenance of the cancer phenotype (Avdulov et al, 2004). Similarly, increased levels of eIF4E stimulates protein synthesis, induces cell growth and proliferation (De Benedetti and Graff, 2004; Mamane et al, 2004), and promotes tumour formation in animal models (Ruggero et al, 2005; Wendel et al, 2007). To gain insight into the molecular mechanisms that control 4E-BP1 expression, we first performed in silico analysis (Genomatix) of its genomic sequence. Among the transcription factors identified (Supplementary Figure S1), the p53 and Smad4 tumour suppressors appeared as highly probable candidates. The use of cells with or without p53 expression indicated that differences in the activity of p53 have no effect on 4E-BP1 promoter activity (A Elia, MJ Clemens, R Azar and S Pyronnet, data not shown). Many arguments, however (see below), identified Smad4 as a stronger candidate.
Smad4 is a transcription factor also termed DPC4 (Deleted in Pancreatic Cancer locus 4) because it was first identified as a candidate tumour-suppressor gene frequently altered in human pancreatic adenocarcinomas (Hahn et al, 1996). Smad4 acts as a tumour suppressor likely through its function in TGFβ signalling (Siegel and Massagué, 2003; ten Dijke and Hill, 2004). For instance, upon pancreatic damage, production of TGFβ exerts a protective effect through a Smad4-dependent transcriptional programme, which blocks epithelial-cell proliferation and maintains a polarized phenotype. Following Smad4 loss, epithelial cells are no longer protected and are susceptible to transformation. Thus, given that (i) 4E-BP1 gene sequence (exon-1) contains a putative Smad4-binding element (Supplementary Figure S1), (ii) Smad4 is a pancreatic tumour suppressor and (iii) pancreas is the organ that expresses the most 4E-BP1 mRNA (Tsukiyama-Kohara et al, 1996), we hypothesized that 4E-BP1 could be a transcriptional target of Smad4 important for TGFβ's antiproliferative action, at least in pancreatic cells.
Results and discussion
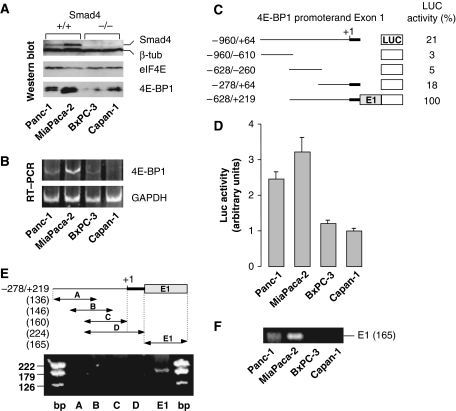
To test the possibility that 4E-BP1 acts downstream of TGFβ in a Smad4-dependent manner, we first analysed the expression levels of 4E-BP1 and Smad4 in four different cell lines (Panc-1, MiaPaca-2, BxPC-3 and Capan-1) that exhibit various Smad4 and/or TGFβ-receptor status (Grau et al, 1997; Giehl et al, 2000). Whereas the TGFβ-Smad4 signalling cascade is fully active in Panc-1 cells, it is altered in the three other cell lines as TGFβ-RII receptor (MiaPaca-2 cell line) or Smad4 (BxPC-3 and Capan-1 cell lines) is absent, as described in the Supplementary Table S1. The data we obtained showed that the amounts of 4E-BP1 protein and mRNA were elevated in Smad4+/+ cell lines (Panc-1 and MiaPaca-2), but low in Smad4−/− cell lines (BxPC-3 and Capan-1; Figure 1A and B). To explore the molecular mechanism that could link Smad4 to 4E-BP1 transcription, we have first delineated the 4E-BP1 genomic sequence that permits efficient transcription following transient transfection of Smad4+/+ pancreatic cells. A series of deletion mutants spanning from nucleotide −960 (relative to the putative +1 transcription start site) to the end of exon-1 (+219) has been generated and fused to a firefly LUC reporter open reading frame (ORF; Figure 1C). In Smad4+/+ Panc-1 cells, the higher transcriptional activity was obtained when a promoter fragment of 4E-BP1 gene was extended to the end of exon-1 (Figure 1C, −628/+219). Similar data were obtained with the other Smad4+/+ cell line MiaPaca-2 (Figure 1D). Furthermore, the activity of such extended promoter fragment reflected cell-line-dependent variability of endogenous 4E-BP1 and Smad4 levels (compare Figure 1D to A and B), whereas other fragments lacking exon-1 did not (not shown), suggesting that Smad4 responsiveness resided in 4E-BP1 exon-1. This hypothesis was further supported by chromosome immunoprecipitation assays (ChIP). We first showed that endogenous Smad4 binding to endogenous 4E-BP1 promoter was detected in Smad4+/+ MiaPaca-2 (Figure 1E) or Panc-1 (not shown) cells solely when primers encompassing exon-1 were used. Consistently, no PCR fragment was obtained following anti-Smad4–ChIP assay of DNA extracted from Smad4−/− cells, and the amount of Smad4 bound to 4E-BP1 promoter was in direct proportion to the endogenous level of Smad4 protein in the different cell lines (Figure 1F).
Figure 1.
4E-BP1 and Smad4 in pancreatic epithelial cell lines. (A) Immunoblotting of equal amounts of proteins from different cell lines using antibodies against Smad4 and β-tubulin (internal control) (top), eIF4E (middle) and 4E-BP1 (bottom). (B) Semi-quantitative RT–PCR of total RNAs from different cell lines using primers specific to 4E-BP1 and GAPDH (internal control). (C) Transcriptional activity of 4E-BP1 promoter deletion mutants inserted upstream of firefly LUC. LUC activity was assayed 36 h following Panc-1 cell transfection. Transfection efficiency was normalized to the concurrent transfection of a CMV–Renilla LUC reporter plasmid. LUC activities are representative of three independent experiments performed in triplicates, and are expressed as percents of the value obtained for the −628/+219 promoter fragment after normalization to the residual firefly LUC activity of pGL2-Basic. (D) Cell-line-dependent transcriptional activity of the 4E-BP1 −628/+219 promoter fragment. Firefly LUC activity normalized to the concurrent transfection of a CMV–Renilla LUC reporter plasmid are the means (±s.e.) of three separate experiments performed in triplicates. (E) ChIP assay of MiaPaca-2 cell extracts using a specific antibody against Smad4 and a set of primers that covers the −278/+219 4E-BP1 promoter fragment. (F) ChIP assay of DNA of different cell lines using a specific antibody against Smad4 and the E1 primers described in panel E.
Interestingly, sequence alignments of the 4E-BP1 nucleotide sequences from different species indicated that the putative Smad4-binding element (SBE: CAGCCAGA) in exon-1 is conserved among mammals (Supplementary Figure S1). To confirm that this putative sequence is a true Smad4-binding element, the human sequence was mutated (Figure 2A, hMut) and tested in different cell lines where Smad4 status was manipulated. The data first revealed that mutations in the putative SBE rendered 4E-BP1 promoter much less active in Smad4+/+ cells (Figure 2B, left, mock), but had no effect in Smad4−/− cells (Figure 2B, right, mock). Then, inactivation of Smad4 by transfection with a dominant-negative truncated form (DN) in Smad4+/+ cells clearly inhibited wild-type 4E-BP1 promoter activity but had no effect on the mutated promoter (Figure 2B, left). Conversely, re-expression of the transcription factor in Smad4−/− cells stimulated wild-type 4E-BP1 promoter activity, but had no effect on the mutated sequence (right). In addition, activity of the mutated 4E-BP1 promoter in Smad4+/+ cells became equivalent to that of the wild-type promoter in Smad4−/− cells (Figure 2C). Finally, the use of specific siRNAs showed that silencing of Smad4 substantially diminished endogenous 4E-BP1 expression (Figure 2D). These data indicated that binding of Smad4 to a responsive element located in 4E-BP1 exon-1 enhanced 4E-BP1 promoter activity.
Figure 2.
4E-BP1 exon-1 contains a functional Smad4-binding element (SBE). (A) Alignment of 4E-BP1 exon-1 sequences from rat, mouse and human, which contain a conserved computer-predicted SBE (box). Mutated nucleotides that leave unchanged the human amino-acid sequence (hMut) but destroy the putative SBE are underlined. (B) SBE destruction renders 4E-BP1 promoter insensitive to Smad4. Left: Firefly LUC activity normalized as in Figure 1D was assayed in Smad4+/+ cells (Panc-1 and MiaPaca-2) 36 h following co-transfection with either wild-type (hWT) or mutated (hMut) −628/+219 4E-BP1 promoter fragment and vector alone (mock) or a dominant negative form of Smad4 (DN). Right: Firefly LUC activity normalized as in Figure 1E was assayed in Smad4−/− cells (BxPC-3 and Capan-1) 36 h following co-transfection with either wild-type (hWT) or mutated (hMut) −628/+219 4E-BP1 promoter fragment and vector alone (mock) or Smad4. Values are the means (±s.e.) of three separate experiments performed in triplicates. (C) SBE destruction or absence of Smad4 has a similar effect on 4E-BP1 promoter activity. Firefly LUC activity normalized as in Figure 1E was assayed in Smad4+/+ (Panc-1 and MiaPaca-2) or Smad4-/- (BxPC-3 and Capan-1) cells 36 h following transfection with either wild-type (hWT) or mutated (hMut) −628/+219 4E-BP1 promoter fragment. Values are the means (±s.e.) of three separate experiments performed in triplicates. (D) Smad4 silencing alters 4E-BP1 expression. MiaPaca-2 cells were transfected with either a nonspecific siRNA (NS) or a siRNA targeting Smad4 mRNA for 48, 72 or 96 h, and equal amounts of proteins were examined by SDS–PAGE immunoblotting as shown.
Smad4 belongs to a large family of proteins (Xu, 2006), which also includes Smad1, 2, 3, 5, 6, 7 and 8 (also termed co-Smads). Smad1, 2, 3, 5 and 8 are phosphorylated by TGFβ-receptor kinases and are, therefore, referred to as R-Smads (receptor-activated Smads). Upon phosphorylation, R-Smads interact with Smad4, and the neo-formed heterodimeric complex translocates to the nucleus and targets genes that contain Smad4-binding elements. Smad1, 5 and 8 are activated by BMPs (bone morphogenetic proteins), whereas Smad2 and 3 (which were used in this study) act specifically downstream of TGFβ. Smad6 and 7 belong to the I-Smads (inhibitory Smads) subgroup. They bind to Smad4 and preclude its association with R-Smads, and hence inhibit TGFβ signalling.
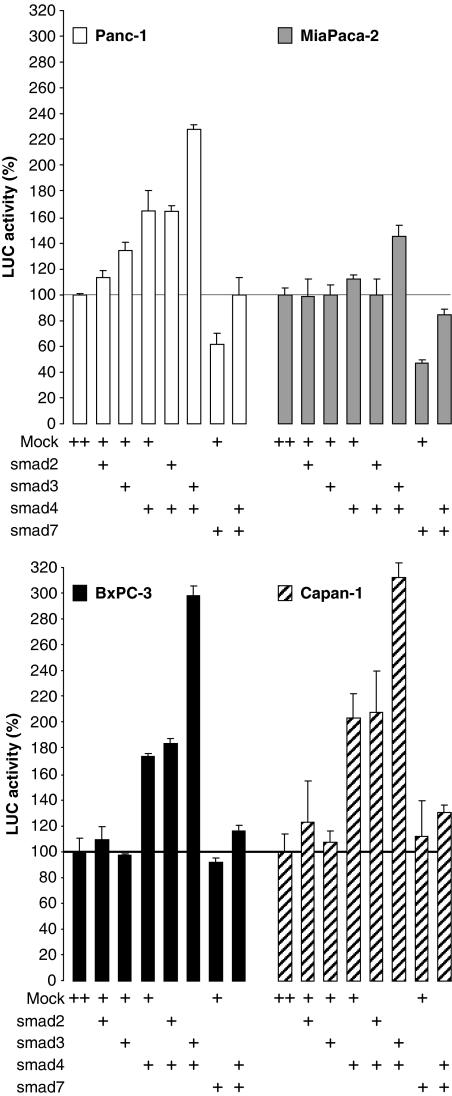
To explore the possibility that 4E-BP1 transcription could be specifically modulated by Smad4 downstream of TGFβ, we first focused on the Smad4 activators Smad2 and 3, and on the Smad4 inhibitor Smad7. Cell lines were transfected with different combinations of Smads and 4E-BP1 promoter activity was monitored (Figure 3). Expression of Smad2 or Smad3 alone had no (or poor) effect on 4E-BP1 promoter activity in either cell line. Expression of Smad4 alone slightly (160%) induced 4E-BP1 promoter activity in Panc-1 cells (which already contain Smad4), but was not significant in MiaPaca-2 cells (which contain the highest level of Smad4 and which carries a defective TGFβ-RII receptor). Induction of 4E-BP1 promoter activity following transfection with Smad4 was more pronounced in the Smad4−/− cell lines BxPC-3 (180%) and Capan-1 (200%). In addition, Smad4-dependent induction of 4E-BP1 promoter activity was not affected by coexpression of Smad2, but was substantially enhanced by coexpression of Smad3 in every cell line. Conversely, expression of Smad7 inhibited 4E-BP1 promoter activity in Smad4+/+ cells (Panc-1 and MiaPaca-2), had no effect in Smad4−/− cells (BxPC-3 and Capan-1) and precluded Smad4-dependent stimulation of 4E-BP1 promoter activity observed following transfection of Smad4 in every cell line. These data indicated that 4E-BP1 promoter activity was sensitive to the co-Smads known to act specifically downstream of TGFβ.
Figure 3.
Regulation of 4E-BP1 promoter activity by co-Smads. Firefly LUC activity normalized as in Figure 1E was assayed in different cell lines co-transfected with the −628/+219 4E-BP1 promoter fragment and various combinations of Smad2, 3, 4 or 7 as described. Values are the means (±s.e.) of three separate experiments performed in triplicates.
We have then tested more directly whether 4E-BP1 expression and/or activity can be regulated by TGFβ in a Smad4-dependent manner. As suspected, TGFβ had no effect on 4E-BP1 in the Smad4−/− cell line Capan-1 (Figure 4A). However, treatment of Smad4+/+ Panc-1 cells with TGFβ provoked accumulation of 4E-BP1 and a corresponding enhancement in 4E-BP1 activity as attested by increased binding of 4E-BP1 to eIF4E (Figure 4A). Strikingly, a reporter assay showed that TGFβ actually induced 4E-BP1 promoter activity in Panc-1 cells, whereas no induction was detected in the three other cell lines lacking either TGFβ-RII (MiaPaca-2) or Smad4 (BxPC-3 and Capan-1) (Figure 4B). Such stimulation in Panc-1 cells was specific to TGFβ as it was precluded by co-incubation with anti-TGFβ blocking antibodies (anti-TGFβ; Figure 4B). Similarly, the enhanced stimulation of 4E-BP1 promoter activity observed upon TGFβ treatment in Panc-1 cells transfected with Smad4 was precluded with co-treatment with anti-TGFβ blocking antibodies (Supplementary Figure S2). TGFβ-stimulated 4E-BP1 promoter activity was also precluded either by mutations that destroy the SBE (hMut) or by coexpression of the dominant negative truncated form (DN) of Smad4 (Figure 4C). Conversely, restoration of Smad4 in Smad4−/− BxPC-3 and Capan-1 cells rescued 4E-BP1 promoter responsiveness to TGFβ (Figure 4D). These data indicated that TGFβ activated 4E-BP1 promoter activity through Smad4.
Figure 4.
TGFβ controls 4E-BP1 activity through Smad4-dependent induction of 4E-BP1 promoter. (A) Regulation of 4E-BP1 expression and binding to eIF4E by TGFβ. Capan-1 or Panc-1 cells were treated with TGFβ and equal amounts of proteins were subjected to immunoprecipitation and/or examined by SDS–PAGE immunoblotting as shown. (B) Regulation of 4E-BP1 promoter activity by TGFβ in different cell lines. Firefly LUC activity normalized as described in Figure 1E was assayed in different cell lines following transfection with −628/+219 4E-BP1 promoter fragment for 36 h and treatment with TGFβ alone or together with blocking TGFβ antibodies (αTGFβ) for 24 h. Values are the means (±s.e.) of three separate experiments performed in triplicates. (C) Requirement of Smad4 in TGFβ-induced 4E-BP1 promoter activity. Firefly LUC activity normalized as described in Figure 1E was assayed in Panc-1 cells following co-transfection with wild-type (hWT) or mutated (hMUT) −628/+219 4E-BP1 promoter fragment and vector alone (mock) or a dominant-negative (DN) form of Smad4 for 36 h and treatment with TGFβ for 24 h. Values are the means (±s.e.) of three separate experiments performed in triplicates. (D) Overexpression of Smad4 enhances 4E-BP1 promoter induction by TGFβ. Firefly LUC activity normalized as described in Figure 1E was assayed in BxPC-3 or Capan-1 cells following co-transfection with the −628/+219 4E-BP1 promoter fragment and vector alone (mock) or a Smad4 for 36 h and treatment with TGFβ for 24 h. Values are the means (±s.e.) of three separate experiments performed in triplicates.
To test for the implication of 4E-BP1 in the inhibition of epithelial cell proliferation by TGFβ, we have created a pancreatic cell line where the level of endogenous 4E-BP1 protein can be manipulated on demand, by using a doxicyclin-inducible vector and RNA interference properties. First, an siRNA complementary to the 4E-BP1 3′UTR (siBP1) was validated for its ability to actually silence 4E-BP1 expression at least for 72 h following transient transfection of Panc-1 cells, as compared with a nonspecific siRNA directed against LUC (siNS; Figure 5A). Then, to avoid inconveniences due to transient transfection of siRNAs, we have generated a cell line (derived from Panc-1 cells) stably transfected with a doxycycline-inducible vector driving simultaneous expression of GFP (used as a marker protein) and either a nonspecific microRNA (directed against LacZ; miRNeg) or a microRNA directed against 4E-BP1 (derived from siBP1; miRBP1; Supplementary Figure S3). Panc-1 cells were chosen because they possess both Smad4 and functional TGFβ receptors (Supplementary Table S1), and because their proliferation has been shown to be inhibited by TGFβ in a Smad4-dependent manner (Grau et al, 1997). Dose–response (Figure 5B, right, top) and time-course (right, bottom) assays showed that efficient extinction of 4E-BP1 was achieved following treatment with 100 ng/ml doxycycline for 72 h. Extinction was specific as doxycycline treatment of miRNeg-stably transfected cells had no effect on 4E-BP1 amount (Figure 5B, left). A proliferation assay then revealed that 4E-BP1 protein silencing increased proliferation rate (Figure 5C), thus showing that 4E-BP1 attenuates pancreatic epithelial-cell proliferation. Finally, the importance of 4E-BP1 in the antiproliferative effect of TGFβ was evaluated. As expected, TGFβ was capable of inhibiting Panc-1-cell proliferation (Figure 5D, compare miRNeg with miRNeg+TGFβ). Interestingly, silencing 4E-BP1 protein in contrast accelerated proliferation (compare miRNeg with miRBP1) and prevented the inhibitory effect of TGFβ (compare miRNeg+TGFβ with miRBP1+TGFβ). Taken together, these data indicate that 4E-BP1 is a new target of Smad4 that is important for the antiproliferative effect of TGFβ in pancreatic cancer cells.
Figure 5.
Involvement of 4E-BP1 in TGFβ-mediated inhibition of pancreatic cancer-cell proliferation. (A) Validation of 4E-BP1 siRNAs. Panc-1 cells were transiently transfected with either a nonspecific (siNS) or a 4E-BP1 (siBP1) siRNA and equal amounts of protein was examined by SDS–PAGE immunoblotting as shown. (B) Validation of Panc-1 inducible cell lines. Panc-1 cells stably transfected with inducible miRNeg (left) or miRBP1 (right) were treated for 72 h with increasing concentrations of doxycycline (top), or for different times with 100 ng/ml of doxycyclin. Equal amounts of proteins were examined by SDS–PAGE immunoblotting as shown. (C) 4E-BP1 knockdown accelerates MiaPaca-2-cell proliferation. MiaPaca-2 cells stably transfected with inducible miRNeg or miRBP1 were incubated in the presence of 100 ng/ml doxycycline or vehicle alone and cell proliferation was monitored. Values are the means (±s.e.) of three separate experiments performed in triplicates. (D) 4E-BP1 is involved in TGFβ-mediated proliferation inhibition of Panc-1 cells. Panc-1 cells stably transfected with inducible miRNeg or miRBP1 were all incubated in the presence of 100 ng/ml doxycycline, and untreated or treated with TGFβ. Cell proliferation was monitored as described under Materials and methods. Values are the means (±s.e.) of three separate experiments performed in triplicates.
Finally the implication of 4E-BP1 in TGFβ-mediated inhibition of cell proliferation was tested outside the tumour situation. To this end, we used two distinct models of cultured cells, which were not isolated from tumours and whose proliferation has been shown to be inhibited by TGFβ: human immortalized keratinocytes (HaCaT cells; Kortlever et al, 2008) and mouse embryonic fibroblasts (MEFs; Datto et al, 1999). TGFβ-mediated inhibition of HaCat cells' proliferation was almost completely prevented upon silencing of 4E-BP1 by specific siRNAs (Figure 6A), and 4E-BP1-knockout (4E-BP1−/−) MEFs are partially resistant to the antiproliferative effect of TGFβ as compared with wild-type (4E-BP1+/+) MEFs (Figure 6B). This is consistent with our observations that 4E-BP1 knockdown accelerates progression through the G1 cell-cycle phase (data not shown), and that TGFβ, in contrast, slows cell-cycle progression through downregulation of the G1-phase cyclin-D1 (Ko et al, 1995; Halder et al, 2005; Kolb et al, 2007, and data not shown).
Figure 6.
Role of 4E-BP1 in TGFβ-mediated inhibition of keratinocyte- and fibroblast-cell proliferation. (A) 4E-BP1 is involved in TGFβ-mediated inhibition of HaCaT-cell proliferation. HaCaT cells transfected with nonspecific siRNAs (siNS) or with siRNAs specific to 4E-BP1 (siBP1) were untreated or treated with TGFβ and cell proliferation was monitored as described under Materials and methods. Values are the means (±s.e.) of three separate experiments performed in triplicates (top). TGFβ upregulates 4E-BP1 in HaCat cells. HaCaT cells transfected as above were untreated or treated with TGFβ for 48 h. Equal amounts of proteins were then subjected to SDS–PAGE immunoblotting as shown (bottom). (B) 4E-BP1 is involved in TGFβ-mediated inhibition of MEF proliferation. 4E-BP1+/+ and 4E-BP1−/− MEFs were untreated or treated with TGFβ and cell proliferation was monitored as described under Materials and methods. Values are the means (±s.e.) of three separate experiments performed in triplicates (top). TGFβ upregulates 4E-BP1 in MEFs. 4E-BP1+/+ and 4E-BP1−/− MEFs were untreated or treated with TGFβ for 48 h. Equal amounts of proteins were then subjected to SDS–PAGE immunoblotting as shown (bottom).
These data, taken together, establish a role of 4E-BP1 in mediating the antiproliferative effects of TGFβ. It is likely that through 4E-BP1 induction, the TGFβ/Smad4 signalling pathway controls translational expression of critical regulators of cell growth and proliferation (such as cyclin-D1). In this regard, the inducible 4E-BP1–shRNAs pancreatic cancer cell line we have created and the 4E-BP1-null MEF model provide useful systems to identify novel routes of TGFβ-mediated inhibition of cell proliferation.
Materials and methods
Cell lines, culture, treatment and proliferation assay
Pancreatic cancer cell lines (Panc-1, MiaPaca-2, BxPC-3 and Capan-1) were cultured as described (Puente et al, 2001). To obtain Panc-1 and MiaPaca-2 cells stably expressing inducible miRNeg or miRBP1, cells were first transfected with the pTet-on advanced vector (Clontech) using Exgen 500 (Euromedex) and cultured in the presence of genetecin (Invivogen). Resistant clones were isolated, transiently transfected with pTRE-Tight-LUC (Clontech) and assayed for LUC activity following treatment with doxycycline (Sigma). Clones that gave satisfactory induction of LUC upon doxycycline treatment were then amplified and co-transfected with linear hygromycin selection marker (Clontech) and with either pTRE-Tight-miRNeg or pTRE-Tight-miRBP1, and cultured in the presence of hygromycin. Resistant clones were then isolated and clones that gave satisfactory induction of GFP and concomitant knockdown of endogenous 4E-BP1 upon treatment with doxycycline were selected and amplified.
HaCaT and immortalized MEFs isolated from 4E-BP1/4E-BP2 double-knockout mice (4E-BP1−/− cells) or from their normal littermates (4E-BP1+/+; Le Bacquer et al, 2007) were grown in Dulbecco's Modified Eagle's Medium (Gibco BRL) with 10% heat-inactivated fetal bovine serum (Gibco BRL) and penicillin–streptomycin (Gibco BRL).
Proliferation assays with Panc-1 cells were performed in the presence of 100 ng/ml doxycycline as follows: cells were plated and allowed to grow in six-well dishes for 24 h, serum-starved for 24 h and incubated with 1% serum in the absence or presence of 10 ng/ml TGFβ (Euromedex). HaCat cells were plated, allowed to grow in six-well dishes for 24 h and transfected with siRNAs targeting 4E-BP1 (Applied Biosystems, forward 5′-CAAGAACGAACCCUUCCUU-3′ and reverse), using the siPort NeoFx transfection reagent (Applied Biosystems), according to the manufacturer's instructions. Twenty-four hours following transfection, cells were serum-starved for 24 h and incubated with 1% serum in the absence or presence of 10 ng/ml TGFβ (Euromedex). 4E-BP1+/+ and 4E-BP1−/− MEFs were plated and allowed to grow in six-well dishes for 24 h, serum-starved for 24 h and incubated with 0.5% serum in the absence or presence of 10 ng/ml TGFβ (Euromedex). At the end of each experiment, cells were trypsinized and counted with a Coulter counter (Coulter Electronics) at the indicated times.
Immunoprecipitation and western blot analysis
Cells were harvested, lysed, the protein concentration of extracts was measured using Protein Assay reagent (Bio-Rad) and equal amounts of proteins were either immunoprecipitated with mouse monoclonal antibody to eIF4E (Santa Cruz Biotechnology) as described previously (Cormier et al, 2001) or subjected directly to SDS–PAGE and electroblotted onto Immobilon-P membranes (Millipore) as described previously (Pyronnet et al, 2000, 2001). Membranes were incubated with mouse monoclonal antibodies to β-tubulin (Sigma) or Smad4 (Santa Cruz Biotechnology), or with rabbit polyclonal antibodies to 4E-BP1 (Cell Signaling), eIF4E (Cell Signaling) or GFP (Ab-cam). Bound antibodies were detected with peroxidase-coupled goat antibodies to mouse or rabbit IgG (Pierce) using chemiluminescence detection reagent (Pierce). Figures represent contiguous lanes.
Semi-quantitative RT–PCR and ChIP
For RT–PCR, RNA was extracted with RNeasy (Qiagen) and equal amounts of RNAs were subjected to RT–PCR using the Superscript One Step RT–PCR kit (Invitrogen) according to the manufacturers' instructions. Primers used were as follows: 4E-BP1—forward primer: 5′-ATGTCCGGGGGCAGCAGCTGCAGCCAG-3′, reverse primer: 5′-TTAAATGTCCATCTGAAACTGTGACTC-3′ and GAPDH—forward primer: 5′-GTCGGAGTCAACGGATTTGG-3′, reverse primer: 5′-AAAAGCAGCCCTGGTGACC-3′. For ChIP assay, genomic DNA was extracted, fragmented and subjected to immunoprecipitation using antibodies to Smad4, and immunoprecipitates were PCR-amplified using a ChIP kit (Upstate) according to the manufacturer′s instructions.
Constructs, mutagenesis, transfections and LUC activities
Fragments of the human 4E-BP1 promoter were PCR-amplified using primers extended by _Kpn_I (forward primers) or by _Xho_I or _Hin_dIII (reverse primers) restriction sites, digested by _Kpn_I/_Xho_I or _Kpn_I/_Hin_dIII restriction endonucleases and inserted into the _Kpn_I/_Xho_I or _Kpn_I/_Hin_dIII-linearized pGL2-Basic (Promega) luciferase (LUC) reporter vector (see Figure 2D for details). The 4E-BP1 exon-1 has been cloned in frame with the LUC ORF. Mutations were generated by PCR using a Quick Change mutagenesis kit (Stratagene) according to the manufacturer's instructions. Smad2, 3, 4 and 7 vectors were as described previously (Jonckheere et al, 2004). pRL-CMV (Promega) containing the Renilla LUC coding sequence driven by the CMV promoter was used as a transfection efficiency internal control. Following co-transfection of pRL-CMV with any pGL2 or Smad construct using Exgen 500 (Euromedex), cells were harvested in Passive Lysis Buffer (Promega) and extracts were assayed for firefly and Renilla LUC activities using the Dual Luciferase Reporter Assay System (Promega), and detected with a Luminoscan Ascent Microplate Luminometer (Thermo Labsystems), as described previously (Lahlou et al, 2005). Cells were transfected with siRNAs targeting Smad4 (Applied Biosystems, forward 5′-CAUCCUAGUAAAUGUGUUA-3′ and reverse) or 4E-BP1 (see above), using siPort NeoFx transfection reagent (Applied Biosystems), according to the manufacturer's instructions.
Supplementary Material
Supplementary Figure S1–S3 and Table S1
Legends to supplemental material
Review Process File
Acknowledgments
We thank Professor Mike Clemens, Professor David Lane and Professor Nahum Sonenberg for critical reading and helpful suggestions. This work was supported by grants from INSERM, La Ligue contre le cancer (comités de Haute-Pyrénées et de Lot-et-Garonne), Cancéropôle Grand Sud-Ouest, Agence Nationale de la Recherche (ANR; #RPV06174BSA) and Association pour la Recherche contre le Cancer (ARC; #3899 to C. Bousquet and #3633 to S Pyronnet). R Azar was a recipient of fellowships from ARC, Fondation pour la Recherche Médicale (FRM) and Agence Universitaire de la Francophonie (AUF). A Alard was a recipient of fellowships from the French Ministery of Research and ARC.
Footnotes
The authors declare that they have no conflict of interest.
References
- Armengol G, Rojo F, Castellví J, Iglesias C, Cuatrecasas M, Pons B, Baselga J, Ramón y Cajal S (2007) 4E-binding protein 1: a key molecular ‘funnel factor' in human cancer with clinical implications. Cancer Res 67: 7551–7555 [DOI] [PubMed] [Google Scholar]
- Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, Perlman DM, Manivel JC, Sonenberg N, Yee D, Bitterman PB, Polunovsky VA (2004) Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell 5: 553–563 [DOI] [PubMed] [Google Scholar]
- Azar R, Najib S, Lahlou H, Susini C, Pyronnet S (2008) Phosphatidylinositol 3-kinase-dependent transcriptional silencing of the translational repressor 4E-BP1. Cell Mol Life Sci 65: 3110–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsti MA, Houghton PJ (2004) Lost in translation: dysregulation of cap-dependent translation and cancer. Cancer Cell 5: 519–523 [DOI] [PubMed] [Google Scholar]
- Cormier P, Pyronnet S, Morales J, Mulner-Lorillon O, Sonenberg N, Bellé R (2001) eIF4E association with 4E-BP decreases rapidly following fertilization in sea urchin. Dev Biol 232: 275–283 [DOI] [PubMed] [Google Scholar]
- Datto MB, Frederick JP, Pan L, Borton AJ, Zhuang Y, Wang XF (1999) Targeted disruption of Smad3 reveals an essential role in transforming growth factor beta-mediated signal transduction. Mol Cell Biol 19: 2495–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedetti A, Graff JR (2004) eIF-4E expression and its role in malignancies and metastases. Oncogene 23: 3189–3199 [DOI] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW (2004) Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 5: 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl K, Seidel B, Gierschik P, Adler G, Menke A (2000) TGFbeta1 represses proliferation of pancreatic carcinoma cells which correlates with Smad4-independent inhibition of ERK activation. Oncogene 19: 4531–4541 [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963 [DOI] [PubMed] [Google Scholar]
- Grau AM, Zhang L, Wang W, Ruan S, Evans DB, Abbruzzese JL, Zhang W, Chiao PJ (1997) Induction of p21waf1 expression and growth inhibition by transforming growth factor beta involve the tumor suppressor gene DPC4 in human pancreatic adenocarcinoma cells. Cancer Res 57: 3929–3934 [PubMed] [Google Scholar]
- Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE (1996) DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 271: 350–353 [DOI] [PubMed] [Google Scholar]
- Halder SK, Beauchamp RD, Datta PK (2005) Smad7 induces tumorigenicity by blocking TGF-beta-induced growth inhibition and apoptosis. Exp Cell Res 307: 231–246 [DOI] [PubMed] [Google Scholar]
- Jonckheere N, Perrais M, Mariette C, Batra SK, Aubert JP, Pigny P, VanSeuningen I (2004) A role for human MUC4 mucin gene, the ErbB2 ligand, as a target of TGF-beta in pancreatic carcinogenesis. Oncogene 23: 5729–5738 [DOI] [PubMed] [Google Scholar]
- Kolb S, Fritsch R, Saur D, Reichert M, Schmid RM, Schneider G (2007) HMGA1 controls transcription of insulin receptor to regulate cyclin D1 translation in pancreatic cancer cells. Cancer Res 67: 4679–4686 [DOI] [PubMed] [Google Scholar]
- Kortlever RM, Nijwening JH, Bernards R (2008) Transforming growth factor-beta requires its target plasminogen activator inhibitor-1 for cytostatic activity. J Biol Chem 283: 24308–24313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko TC, Sheng HM, Reisman D, Thompson EA, Beauchamp RD (1995) Transforming growth factor-beta 1 inhibits cyclin D1 expression in intestinal epithelial cells. Oncogene 10: 177–184 [PubMed] [Google Scholar]
- Lahlou H, Fanjul M, Pradayrol L, Susini C, Pyronnet S (2005) Restoration of functional gap junctions through internal ribosome entry site-dependent synthesis of endogenous connexins in density-inhibited cancer cells. Mol Cell Biol 25: 4034–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bacquer O, Petroulakis E, Paglialunga S, Poulin F, Richard D, Cianflone K, Sonenberg N (2007) Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest 117: 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N (2004) eIF4E—from translation to transformation. Oncogene 23: 3172–3179 [DOI] [PubMed] [Google Scholar]
- Puente E, Saint-Laurent N, Torrisani J, Furet C, Schally AV, Vaysse N, Buscail L, Susini C (2001) Transcriptional activation of mouse sst2 somatostatin receptor promoter by transforming growth factor-beta. Involvement of Smad4. J Biol Chem 276: 13461–13468 [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Dostie J, Sonenberg N (2001) Suppression of cap-dependent translation in mitosis. Genes Dev 15: 2083–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Pradayrol L, Sonenberg N (2000) A cell cycle-dependent internal ribosome entry site. Mol Cell 5: 607–616 [DOI] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N (2005) Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433: 477–480 [DOI] [PubMed] [Google Scholar]
- Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP (2005) The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med 10: 484–486 [DOI] [PubMed] [Google Scholar]
- Siegel PM, Massagué J (2003) Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer 3: 807–821 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Pause A (2006) Signal transduction. Protein synthesis and oncogenesis meet again. Science 314: 428–429 [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Hill CS (2004) New insights into TGF-beta–Smad signalling. Trends Biochem Sci 29: 265–273 [DOI] [PubMed] [Google Scholar]
- Tettweiler G, Miron M, Jenkins M, Sonenberg N, Lasko PF (2005) Starvation and oxidative stress resistance in Drosophila are mediated through the eIF4E-binding protein, d4E-BP. Genes Dev 19: 1840–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Vidal SM, Gingras AC, Glover TW, Hanash SM, Heng H, Sonenberg N (1996) Tissue distribution, genomic structure, and chromosome mapping of mouse and human eukaryotic initiation factor 4E-binding proteins 1 and 2. Genomics 38: 353–363 [DOI] [PubMed] [Google Scholar]
- Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Teruya-Feldstein J, Pelletier J, Lowe SW (2007) Dissecting eIF4E action in tumorigenesis. Genes Dev 21: 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L (2006) Regulation of Smad activities. Biochim Biophys Acta 1759: 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Ishihara H, Yamada T, Tamura A, Usui M, Tominaga R, Munakata Y, Satake C, Katagiri H, Tashiro F, Aburatani H, Tsukiyama-Kohara K, Miyazaki J, Sonenberg N, Oka Y (2008) ATF4-mediated induction of 4E-BP1 contributes to pancreatic beta cell survival under endoplasmic reticulum stress. Cell Metab 7: 269–276 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1–S3 and Table S1
Legends to supplemental material
Review Process File