Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo (original) (raw)
Abstract
Nuclear factor interleukin-3 (Nfil3; also known as E4-binding protein 4) is a basic region leucine zipper transcription factor that has antiapoptotic activity in vitro under conditions of growth factor withdrawal. To study the role of Nfil3 in vivo, we generated gene-targeted _Nfil3_-deficient (Nfil3−/−) mice. Nfil3−/− mice were born at normal Mendelian frequency and were grossly normal and fertile. Although numbers of T cells, B cells, and natural killer (NK) T cells were normal in Nfil3−/− mice, a specific disruption in NK cell development resulted in severely reduced numbers of mature NK cells in the periphery. This defect was NK cell intrinsic in nature, leading to a failure to reject MHC class I–deficient cells in vivo and reductions in both interferon γ production and cytolytic activity in vitro. Our results confirm the specific and essential requirement of Nfil3 for the development of cells of the NK lineage.
E4-binding protein 4 (E4bp4) was initially isolated by its ability to recognize the proximal activating transcription factor binding site of the adenovirus E4 promoter (Cowell et al., 1992). Subsequently, E4bp4 was independently identified as nuclear factor IL-3 (Nfil3), a protein expressed in T cells that binds to the 5′ flanking region of the human IL-3 promoter (Zhang et al., 1995).
Nfil3 shares sequence identity in its basic DNA-binding domain with members of the proline- and acidic amino acid–rich (PAR) subfamily of mammalian bZIP (basic region leucine zipper) transcription factors, a subfamily that includes HLF (hepatic leukemia factor; Ishida et al., 2000), DBP (albumin gene promoter D-box binding protein; Mueller et al., 1990), and TEF (thyrotroph embryonic factor; Drolet et al., 1991). Structurally, the PAR bZIP factors are closely related to CES-2, a neuron-specific cell death specification protein in the nematode Caenorhabditis elegans (Metzstein et al., 1996). This similarity implies that mammalian PAR proteins may be involved in cell fate commitment. Indeed, we have previously demonstrated that both E2A-HLF (Inaba et al., 1992) and Nfil3 play critical roles in the regulation of apoptosis in mammalian pro–B lymphocytes (Ikushima et al., 1997; Kuribara et al., 1999). In the murine pro–B cell lines Baf-3 and FL5.12, Nfil3 is a delayed-early IL-3–responsive gene whose expression depends on de novo protein synthesis. Moreover, in these IL-3–dependent pro–B cells, enforced expression of human Nfil3 complementary DNA (cDNA) promotes cell survival, indicating that Nfil3 induction is a mechanism by which IL-3 suppresses apoptosis (Ikushima et al., 1997).
Since the publication of these findings, Nfil3 has been implicated in a diverse range of processes, including the antiinflammatory response (Wallace et al., 1997), intracellular signal transduction (Kuribara et al., 1999), and the mammalian circadian oscillatory mechanism (Mitsui et al., 2001; Ohno et al., 2007). The plethora of regulatory pathways that impinge on Nfil3, including control by Ras (via IL-3) in murine B cells (Kuribara et al., 1999), thyroid hormone during Xenopus laevis tail resorption (Brown et al., 1996; Furlow and Brown, 1999), glucocorticoids in murine fibroblasts (Wallace et al., 1997), and calcium in rat smooth muscle cells (Nishimura and Tanaka, 2001), reflect the many diverse functions that have been attributed to this transcription factor. While this manuscript was under review for publication, E4BP4 was reported as being essential for mature NK (mNK) cell development (Gascoyne et al., 2009).
In this study, we show that Nfil3 is highly expressed in cells of the NK lineage, starting at the immature NK (iNK) cell stage. We can confirm that the absence of Nfil3 in Nfil3−/− mice severely reduces the number of mNK cells present in the periphery and that this disturbance in NK cell maturation is NK cell intrinsic.
Defects in NK cell development have previously been reported in several gene knockout mice, including those lacking genes encoding cytokines or their receptors (for review see Boos et al., 2008), downstream targets such as Jak3 (Park et al., 1995), or transcription factors such as Ets1 (Barton et al., 1998), Gata3 (Samson et al., 2003), or Id2 (Boos et al., 2007). However, all these mutants also exhibit defects in other hematopoietic cell lineages such as T and NK T cells. Although Kim et al. (2000) have described a transgenic mouse model with a selective NK cell deficiency, _Nfil3_−/− mice, as described by Gascoyne et al. (2009) and ourselves, are currently the only gene-targeted animals reported to exhibit an NK cell–specific developmental defect.
RESULTS AND DISCUSSION
Gene targeting and characterization of Nfil3 expression
To determine the physiological role of Nfil3, we created _Nfil3_-deficient (_Nfil3_−/−) mice using conventional gene-targeting strategies in embryonic stem (ES) cells, replacing the single Nfil3 coding exon (exon 2) with the neoR gene cassette (Fig. S1 A). _Nfil3_−/− mice were viable and fertile and did not show any obvious abnormalities after backcrossing for six generations to C57BL/6. We also generated an Nfil3 reporter mouse in which to assess Nfil3 expression in normal mouse tissues. We created this animal by inserting an IRES β-geo cassette (Murakami et al., 1997) into exon 2 of the Nfil3 gene (Fig. S1 B). This approach allowed detection of Nfil3 in tissues of Nfil3+/−-IRES β-geo mice by staining with X-Gal and assaying for β-galactosidase activity. Deletion of Nfil3 in _Nfil3_−/− mice was confirmed using both Southern (Fig. S1, C and D) and Northern (Fig. S1 E) blotting.
To establish the pattern of expression of endogenous Nfil3 in normal mice, we performed quantitative real-time PCR on mouse tissues. Nfil3 was almost ubiquitously expressed and was present at relatively high levels in lung, liver, and BM (Fig. S2 A, top). In contrast, Nfil3 was low in unfractionated spleen. Analysis of messenger RNA (mRNA) levels by RT-PCR in sorted cell populations revealed low levels of Nfil3 in T and B cells but high expression of Nfil3 in elicited peritoneal macrophages and BM-derived DCs. Up-regulation of Nfil3 by IL-3 in BM-derived mast cells was used as a positive control (Fig. S2 A, top). Analysis of Nfil3 expression levels in NK, NK T, and CD3+ T cells purified from BM revealed higher levels of Nfil3 in mNK cells (CD3−CD122+NK1.1+) than in CD3+ T cells (Fig. S2 A, bottom).
Previous studies using in situ hybridization in mouse brain have demonstrated significant Nfil3 mRNA expression in the suprachiasmatic nucleus, hippocampus, gyrus dentatus, and piriform cortex (Mitsui et al., 2001). In our hands, X-Gal staining of brain tissues from Nfil3+/−-IRES β-geo mice revealed a similar pattern of Nfil3 expression. Purkinje cells in the cerebellum showed prominent X-Gal staining of their complete cell bodies and dendrites (Fig.S2, B and C), as did the hippocampus, dentate, and cingular gyri (Fig. S2 D) and the olfactory bulbs and olfactory tract (not depicted). Other tissues that stained strongly positive with X-Gal included skeletal and smooth muscle, as evident in the longitudinal and circular muscle layers in the bowel wall (Fig. S2 E), the peripheral lobular cells of the liver (Fig. S2 F), and the medulla (but not the cortex) of the kidney (Fig. S2 G). Tissues weakly positive for X-Gal staining and, thus, Nfil3 expression included the testis (sperm), salivary glands, and the islets (but not the exocrine portion) of the pancreas (unpublished data).
mNK cells are specifically and markedly decreased in _Nfil3_−/− mice
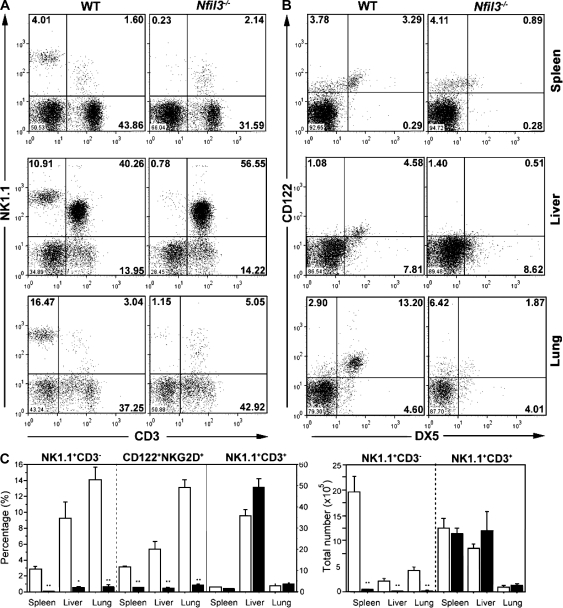
We next examined the consequences of an absence of Nfil3 on the immune system. Consistent with our finding of very low Nfil3 expression in total spleen and isolated pure T and B cells, gross hematological analysis showed no significant differences between WT and _Nfil3_−/− mice in circulating blood cell counts, total T and B cell numbers, and T and B cell subsets in lymphoid organs (Table I). T and B cell activation and proliferation in vitro were also normal in the absence of Nfil3 (Fig. S3). Strikingly, however, both the percentages and absolute numbers of mNK cells (defined as NK1.1+CD3− or CD122+DX5+) in spleen, lung, and liver were markedly decreased in _Nfil3_−/− mice (Fig. 1 and Table S1). Numbers of NK T cells (NK1.1+CD3+) in the same tissues were unaffected.
Table I.
Comparison of blood cell parameters in WT and _Nfil3_−/− mice
| Population | Marker and parameter | WT | _Nfil3_−/− |
|---|---|---|---|
| PBMC | WBC, ×103/µl | 5.97 ± 2.55 | 7.48 ± 4.53 |
| RBC, ×106/µl | 9.06 ± 0.65 | 8.86 ± 1.83 | |
| HGB, g/dl | 14.83 ± 0.80 | 14.49 ± 1.15 | |
| HCT, % | 52.26 ± 2.97 | 50.36 ± 7.85 | |
| MCV, fL | 57.54 ± 2.07 | 56.68 ± 2.94 | |
| MCH, pg | 16.34 ± 0.28 | 16.50 ± 1.19 | |
| MCHC, g/dl | 28.41 ± 0.68 | 29.27 ± 3.73 | |
| PLT, ×103/µl | 1311 ± 150 | 1279 ± 164 | |
| BM lymphocytes, ×107 | 3.64 ± 1.09 | 3.49 ± 0.39 | |
| B220+CD43+, % | 14.28 ± 1.24 | 13.13 ± 0.76 | |
| B220+CD25+, % | 17.29 ± 2.76 | 19.0 ± 1.31 | |
| B220+IgM+, % | 16.55 ± 3.94 | 16.41 ± 3.41 | |
| Liver lymphocytes, ×107 | 0.24 ± 0.05 | 0.23 ± 0.10 | |
| Lung lymphocytes, ×107 | 0.29 ± 0.05 | 0.34 ± 0.10 | |
| Splenocytes, ×107 | 6.60 ± 0.56 | 7.14 ± 1.245 | |
| B220+CD3−, % | 48.80 ± 1.22 | 46.51 ± 5.05 | |
| B220+IgM+, % | 34.64 ± 9.34 | 38.54 ± 4.69 | |
| B220−CD3+, % | 33.87 ± 1.86 | 34.40 ± 3.72 | |
| CD8+CD4−, % | 41.92 ± 1.75 | 42.33 ± 4.45 | |
| CD8−CD4+, % | 46.57 ± 2.22 | 45.86 ± 1.11 | |
| TCR-αβ+, % | 95.33 ± 0.61 | 95.83 ± 0.44 | |
| TCR-γδ+, % | 1.43 ± 0.56 | 1.37 ± 1.23 | |
| Thymocytes, ×107 | 10.68 ± 2.23 | 10.24 ± 1.21 | |
| CD4+CD8−, % | 9.1 ± 0.20 | 9.54 ± 1.45 | |
| CD4−CD8+, % | 3.87 ± 0.61 | 2.94 ± 0.27 | |
| CD4+CD8+, % | 84.02 ± 1.29 | 84.48 ± 1.69 | |
| CD4−CD8−, % | 3.03 ± 0.58 | 3.01 ± 0.64 | |
| CD25−CD44+, % | 15.38 ± 4.37 | 19.05 ± 2.01 | |
| CD25+CD44+, % | 4.15 ± 1.86 | 5.17 ± 1.56 | |
| CD25+CD44−, % | 64.13 ± 4.63 | 62.42 ± 2.02 | |
| CD25−CD44−, % | 14.32 ± 2.73 | 13.31 ± 0.49 |
Figure 1.
_Nfil3_−/− mice exhibit defective NK cell populations. (A and B) Representative flow cytometric histograms of NK (A, NK1.1+CD3−; B, CD122+DX5+) and NK T (NK1.1+CD3+) cells present in the indicated organs from WT and _Nfil3_−/− mice. Percentages of cells in each quadrant are representative of three mice analyzed per genotype. (C) Quantification of the percentages (left) and total numbers (right) of lymphocyte-gated NK and NK T cells in the indicated organs from WT and _Nfil3_−/− mice. Data shown are the mean ± SEM of three animals analyzed per genotype. NK cell percentages and numbers were significantly reduced in Nfil3−/− mice. *, P < 0.05; **, P < 0.01. Gating was identical to the FACS profiles shown in A and B.
The NK cell abnormality in _Nfil3_−/− mice is the result of an intrinsic defect in NK cell progenitors
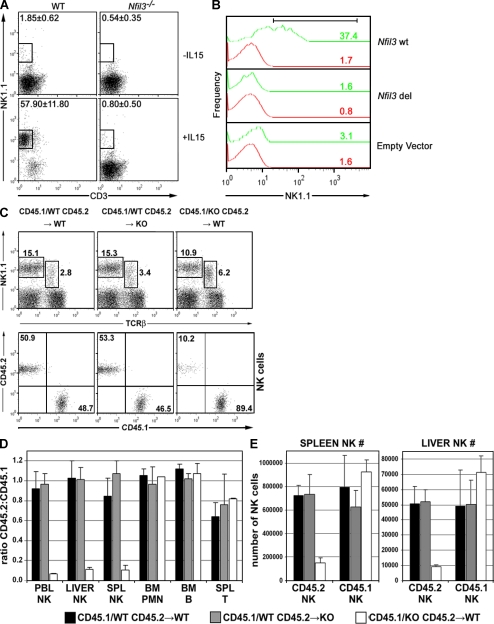
Murine multipotent progenitors (Kondo et al., 1997) give rise to mNK cells in vitro after appropriate cytokine stimulation (Shimozato et al., 2002). We applied an in vitro NK cell generation assay to a lineage-negative (Lin−) population of BM cells. Lin− BM cells prepared from WT and _Nfil3_−/− mice were stimulated in vitro for 2 wk with a combination of murine steel factor (mSlf), murine FMS-like tyrosine kinase 3 ligand (mFlt3L), and mIL-7, with or without mIL-15. In the presence of mIL-15, more than half (57.9 ± 11.8; mean ± SD) of WT Lin− BM cells became NK1.1+CD3− cells (Fig. 2 A), compared with <1% (0.8 ± 0.5%; P = 0.001) of _Nfil3_−/− Lin− BM cells. To determine whether this defect was specifically related to the loss of Nfil3, we retrovirally reintroduced the WT Nfil3 gene into _Nfil3_−/− fetal liver cells (embryonic day [E] 13.5) and stimulated the transduced cells in culture for 2 wk with mIL-15. Over 35% of the transduced cells gave rise to NK1.1-expressing cells (Fig. 2 B, Nfil3 wt). This result contrasted dramatically with the lack of NK1.1+ cells generated from _Nfil3_−/− fetal liver cells transduced with a retrovirus containing an Nfil3 gene lacking its DNA binding domain (Fig. 3 B, Nfil3 del) or with cells transduced with empty vector. Thus, hematopoietic progenitors cannot carry out normal generation of NK cells in the absence of Nfil3.
Figure 2.
The defect in NK cell development in Nfil3−/− mice is NK cell intrinsic. (A) Impaired IL-15–induced in vitro generation of NK cells. Lin− BM cells from WT or _Nfil3_−/− mice were cultured in medium containing mSlf, mFlt3L, and mIL-7, with or without mIL-15. NK cell expansion was assessed by flow cytometry. Results shown are representative of two independent experiments. (B) Rescue by WT Nfil3. Using GFP as a marker for transfected gene expression, NK1.1 expression was determined on gated CD3−GFP+ (green) or CD3−GFP− (red) Nfil3−/− fetal liver cells (105) that had been retrovirally transduced with either the WT Nfil3 gene (wt) or a gene encoding the WT Nfil3 gene lacking its DNA binding domain (deleted [del]). Transduced cells were cultured with mIL-15. Numbers indicate the percentage of NK1.1+ cells generated. (C–E) The developmental defect in Nfil3−/− mice is NK cell intrinsic. (C) Donor-derived liver NK cell populations after BMT showing overall percentages of NK cells (top) and the percentage of CD45.1 and CD45.2 NK cells (bottom). Data shown are representative of three mice per group. (D) The ratio of CD45.2/CD45.1 cells in the various indicated lymphoid cell populations after BMT. (E) The absolute numbers of CD45.2 and CD45.1 NK cells present in spleen and liver, as calculated from the total cellularity of the lymphoid organs and the percentages of CD45.2 or CD45.1 NK1.1+CD3− cells as determined by flow cytometry. Results shown are the mean ± SD for three mice per group.
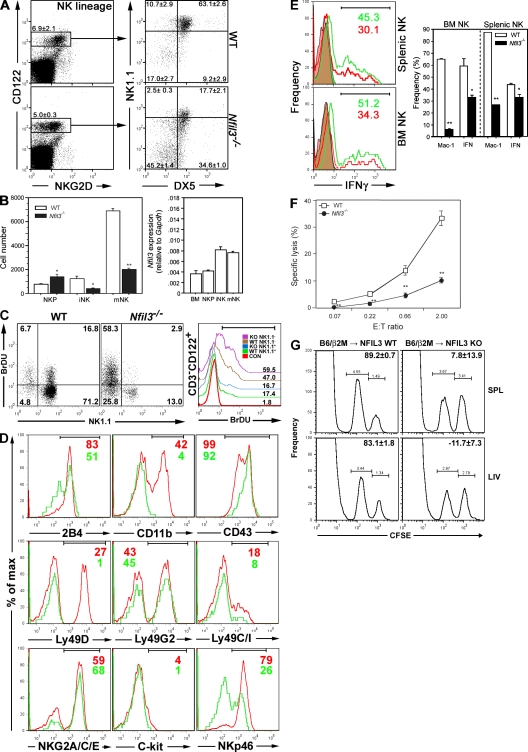
Figure 3.
Impaired development, maturation, and function of _Nfil3_−/− NK cells. (A) The percentages of cells at each NK developmental stage in WT and Nfil3−/− BM were determined by flow cytometry. NK lineage cells were gated on BM precursors expressing equivalent levels of CD122. Figures in quadrants indicate mean percentage ± SD for three mice per group, and dot plots are representative of multiple experiments. (B, Left) Absolute numbers of cells at each NK developmental stage in BM (mean ± SD, n = 3 mice/group). Absolute numbers of NK populations were significantly reduced in Nfil3−/− BM. *, P < 0.05; **, P < 0.01. (B, Right) Real-time RT-PCR analysis of Nfil3 mRNA expression in sorted WT BM cells at each NK developmental stage. Nfil3 expression was normalized to Gapdh. Data shown are the mean ± SD for triplicate determinations. (C) Proliferation of BM NK subsets. The left shows CD3−CD122+ gated BM cells (105), analyzed for NK1.1+ (mNK) or NK1.1− (iNK) expression and BrdU incorporation. Numbers in bold indicate the number of cells in each quadrant. The right shows iNK (NK1.1+) or mNK (NK1.1−) cells (5 × 103) and histograms for anti-BrdU staining overlaid. The control histogram was gated on CD3−CD122+NK1.1+ cells from an untreated WT mouse. Numbers indicate the percentage of BrdU+ cells as specified by the gate. Results are representative of three separate experiments. (D) Developmental markers on BM CD3−CD122+ NK cells. BM enriched for NK cells was analyzed by flow cytometry. Histogram overlays shown are gated on CD3−CD122+NKG2D+ BM cells to include both iNK and mNK populations. Red, WT; green, Nfil3−/−. Numbers indicate percentages of cells within the indicated gates (top). Fluorescence intensity values are cited in the text. Data shown are representative of three independent experiments. (E) Impaired NK cell maturation and IFN-γ production. Left, flow cytometric determination of intracellular IFN-γ production in splenocytes or NK1.1+CD3− BM cells stimulated ex vivo with mIL-2 plus mIL-12. WT, green; Nfil3−/−, red. Filled histograms indicate baseline IFN-γ production. Right, quantification of the frequencies of CD11b-expressing cells (Mac-1) and cells showing intracellular IFN-γ production (IFN). Data shown are the mean ± SD for three mice per group. (F) Impaired in vitro cytolytic activity. Yac-1 cell lysis by poly I:C–activated NK1.1+CD3− splenocytes from WT and Nfil3−/− mice was compared. The E/T ratio (2 × 104 Yac-1 cells/well) was analyzed after 4 h of co-culture. For E and F, data are the mean ± SEM of triplicate experiments. *, P < 0.05; **, P < 0.01. (G) Impaired rejection in vivo. WT and Nfil3−/− mice received CFSE-labeled β2-microglobulin–deficient and WT splenocytes, and preferential killing of the MHC class I–deficient targets in spleen (SPL) and liver (LIV) was examined by flow cytometry. NK cells from Nfil3−/− mice failed to reject the donated cells. Data shown are representative of two independent experiments.
To assess whether the defect in NK cell development in vivo and expansion in vitro was a phenomenon intrinsic to NK cell progenitors or was caused by a defect in the microenvironment of _Nfil3_−/− BM, we performed a mixed chimeric BM transfer (BMT). We reconstituted lethally irradiated WT or _Nfil3_−/− mice with an equal mix of WT B6.SJL-Ptprc Pep3/BoyJ (CD45.1) BM and either WT BM (CD45.2) or _Nfil3_−/− BM (CD45.2, backcrossed six times to C57BL/6). By 8 wk after BMT, the mix of WT CD45.1 plus WT CD45.2 donor cells had repopulated (in equal proportions) the NK cell compartments of the spleen, BM, peripheral blood (not depicted), and liver (Fig. 2 C) in WT recipients. The same pattern of reconstitution was observed in _Nfil3_−/− recipients, indicating that a BM microenvironment deficient in Nfil3 is fully able to support NK cell development. However, when WT CD45.1 BM was cotransferred with _Nfil3_−/− CD45.2 BM, the _Nfil3_−/− cells were unable to contribute to the repopulation of the BM of WT recipients, and the WT BM was the source of the vast majority of NK cells present (Fig. 2, C–E). This defect in BM reconstitution was NK cell specific because _Nfil3_−/− BM was able to support the normal repopulation of NK T cells (Fig. 2 C) as well as splenic T, BM B, and BM polymorphonuclear neutrophil cells (Fig. 2 D). These findings demonstrate that the NK cell abnormality observed in _Nfil3_−/− mice is caused by an intrinsic defect in the Nfil3-deficient NK cell progenitors themselves and is not the result of an Nfil3-deficient tissue microenvironment.
NK cell development in _Nfil3_−/− mice is disrupted primarily at the iNK stage
To dissect in detail the effect of Nfil3 deficiency on NK cell development, we evaluated CD122, NKG2D, NK1.1, and DX5 expression on NK lineage cells enriched from total BM cells of WT and _Nfil3_−/− mice. According to the current literature (Kim et al., 2002; Vosshenrich et al., 2005; Di Santo, 2006; Huntington et al., 2007; for review see Boos et al., 2008), the development of NK lineage cells (CD122+NKG2D+ BM cells) progresses through three major stages defined by NK1.1 and DX5 expression: NK progenitor (NKP; NK1.1−DX5−), iNK (NK1.1+DX5−), and mNK (NK1.1+DX5+) cells. We performed flow cytometric analyses in which CD3−CD122+ cells were gated. Because of the slightly reduced levels of NKG2D present on Nfil3−/− CD122+ cells, equivalent populations of CD122+NKG2D+/− cells were gated and expression levels of NK1.1 and DX5 were then used to determine the developmental maturity of NK lineage cells in WT and Nfil3−/− BM populations (Fig. 3 A). In the case of _Nfil3_−/− CD122+ BM cells, the total number of NKP cells was increased (_Nfil3_−/−, 1,400.81 ± 383.92; WT, 772.44 ± 62.25; P = 0.049), whereas significant reductions had occurred in the iNK population (_Nfil3_−/−, 405.31 ± 77.32; WT, 1,227.56 ± 342.55; P = 0.015) and the mNK population (_Nfil3_−/−, 1,995.69 ± 163.94; WT, 6,908.33 ± 373.60; P < 0.001; Fig. 3 B, left). These data indicate that NK cell development in _Nfil3_−/− mice is impaired before the iNK stage.
Real-time RT-PCR was used to determine the relative levels of Nfil3 mRNA in fractionated WT NK cells sorted by flow cytometry. In WT BM, Nfil3 expression was higher in iNK and mNK cells than in NKP cells or in unfractionated BM (Fig. 3 B, right). The apparent block in NK cell development at the iNK stage therefore corresponds to the relative increase in Nfil3 mRNA expressed at this stage compared with the NKP stage.
Analysis of proliferation of BM NK cells in vivo was assessed via BrdU incorporation. BrDU incorporation was comparable between WT and Nfil3−/− iNK and mNK cells (Fig. 3 C), indicating that proliferation of NK precursors and residual mNK cells present in _Nfil3_−/− mice is intact. Flow cytometric analysis of a panel of NK cell developmental markers revealed that Nfil3−/− CD3−CD122+ BM cells exhibited lower levels of the activatory receptors 2B4, Ly49D, and Nkp46 and the integrin CD11b than did WT controls (Fig. 3 D). The mean fluorescence intensities of cells staining positively for each marker were the following (WT vs. Nfil3−/−): 2B4, 948 versus 694; CD11b, 3,850 versus 3,509; CD43, 3,148 versus 4,408; Ly49D, 7,971 versus 2,750; Ly49G2, 5,606 versus 6,932; NKG2A/C/E, 3,945 versus 3,607; c-kit, 764 versus 1,224; and NKp46, 2,407 versus 1,430. These data confirm the immature nature of the NK lineage cells present in _Nfil3_−/− mice.
NK cell effector functions are disturbed in _Nfil3_−/− mice
To determine if the immaturity of _Nfil3_−/− NK cells affected their functionality, we examined IFN-γ production by residual _Nfil3_−/− BM and splenic NK cells. When these NK cells were stimulated ex vivo with IL-2 plus IL-12, the percentage of _Nfil3_−/− NK cells able to produce IFN-γ was significantly decreased compared with the WT in both the BM (_Nfil3_−/−, 33.22 ± 2.95; WT, 59.09 ± 11.44; P = 0.019;) and splenic (_Nfil3_−/−, 33.27 ± 4.14%; WT, 43.44 ± 2.90%; P = 0.025; Fig. 3 E) compartments. To assess the cytolytic capacity of _Nfil3_−/− residual splenic NK cells, we performed a standard in vitro cytolytic assay against Yac-1 target cells. As shown in Fig. 3 F, the cytolytic activity of _Nfil3_−/− residual splenic NK cells was significantly lower than that of equivalent numbers of WT NK cells. In addition, although WT mice were able to reject >80% of i.v.-transferred CFSE-labeled β2-microglobulin–deficient spleen cells by 24 h after transfer, _Nfil3_−/− mice were unable to do so (Fig. 3 G). These results demonstrate that Nfil3 is required for both the phenotypic and functional maturation of NK cells in both the BM and spleen. In particular, the residual NK cells present in _Nfil3_−/− mice were unable to eliminate cells that lacked expression of functional MHC class I.
Like the work of Gascoyne et al. (2009), our study demonstrates that mNK cells are markedly reduced in number, maturity, and functional capability in the absence of Nfil3 but that other hematopoietic cell lineages are not adversely affected. We also show that retroviral expression of Nfil3 in murine Nfil3−/− fetal liver cells is able to restore the development of NK1.1-expressing cells and that this activity is dependent on the intact DNA binding domain of this transcription factor. To our knowledge, aged _Nfil3_−/− mice do not manifest increased development of spontaneous cancers or immune system disorders (unpublished data). Thus, the absence of NK cells does not appear to increase susceptibility to these conditions in mice housed under SPF conditions. Although outside of the scope of this brief initial paper, further experiments on _Nfil3_−/− mice, such as viral challenge assays, in vivo tumor transplantation, and/or crosses with tumor initiation models, should prove helpful in elucidating the nonredundant roles of NK cells in vivo. The results of such studies may be highly significant, especially given the importance of NKG2D in immune surveillance (Guerra et al., 2008).
In conclusion, our study shows that Nfil3 is highly expressed in cells of the NK cell lineage and promotes the development and functional maturation of NK cells. Further studies designed to identify the target genes regulated by Nfil3 in NK cells, either in an Id2-dependent or -independent fashion (Gascoyne et al., 2009), will help to elucidate the molecular pathways underlying the development and survival of NK cells.
MATERIALS AND METHODS
Gene targeting.
A cDNA fragment of murine Nfil3 (GenBank Accession no. U83148) was used to screen a bacterial artificial chromosome library made from the 129/SvJ mouse strain (Genome Systems). The knockout targeting vector was constructed by replacing a 2.2-kb fragment containing Nfil3 exon 2 with a neomycin resistance cassette. The IRES β-geo reporter vector was constructed by replacing a 1.7-kb fragment containing Nfil3 exon 2 with an IRES β-geo promoterless reporter-resistance selection gene cassette. To create Nfil3−/− mice, 20 µg of linearized knockout targeting vector was electroporated into 107 RW4 ES cells (129/SvJ; Genome Systems) and transfectants were selected 24 h later in 260 µg/ml G418 (Sigma-Aldrich) plus 0.2 µM FIAU (Bristol-Myers Squibb). To create Nfil3+/−-IRES β-geo mice, 20 µg of linearized reporter vector was electroporated into 107 RW4 ES cells and transfectants were selected 24 h later in 200 µg/ml G418. In both cases, transfectants that had undergone homologous recombination were identified by Southern blotting. Heterozygous ES clones with a normal karyotype were injected into C57BL/6 blastocysts, which were subsequently implanted into the uteri of pseudopregnant F1 B/CBA foster mothers and allowed to develop to term. Male chimeras (selected by agouti coat color) that carried the mutant allele were mated to C57BL/6 females to obtain germline transmission. Disruption of the Nfil3 locus in F1 progeny was confirmed by Southern blotting of tail DNA, and heterozygous F1 males and females were interbred to generate F2 offspring that were backcrossed to C57/Bl6 mice for six generations before use in experiments. To generate Nfil3−/− ES cells, one Nfil3+/− ES clone bearing the β-geo construct was cultured in increased G418 (750 µg/ml) for 22 d.
Animals.
All mice used in experiments were 6–8 wk old. WT, Nfil3−/−, and Nfil3+/− IRES β-geo mice were described in the previous section. B6.129P2-β2mtm/Unc/J β2 microglobulin-deficient mice and B6.SJL-Ptprc Pep3/BoyJ CD45.1 mice were obtained from The Jackson Laboratory. All animal experiments were approved by the University Health Network Animal Care Committee and performed in accordance with the institutional guidelines of the Canadian Council for Animal Care.
X-Gal staining of Nfil3 expression in WT and Nfil3+/−-IRES β-geo mice.
X-Gal staining of frozen sections was performed as follows. In brief, animals were perfused transcardially at 4°C under deep anesthesia with a 2% paraformaldehyde solution in PIPES buffer (34.74 g/liter PIPES disodium salt, 0.41 g/liter MgCl2.6H2O, and 0.76 g/liter EGTA, pH 6.9; all chemicals were obtained from Sigma-Aldrich unless otherwise stated), post-fixed for 1 h, dissected, and rinsed overnight in 30% sucrose in PBS. Frozen sections were prepared, stained overnight with an X-Gal–containing buffer (1 mg/ml X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside]; GIBCO BRL) predissolved in DMF (N,N-dimethylformamide; 0.65 g/400 ml potassium ferricyanide, 0.84 g/400 ml potassium ferrocyanide.3H2O, 0.16 g/400 ml MgCl2.6H2O, 80 µl/400 ml Nonidet P-40, and 0.04 g/400 ml sodium deoxycholate), and counterstained with eosin-y for 10 s.
Detection of Nfil3 gene expression in NKPs and mouse tissues.
NKPs (at least 1.5 × 104 cells per developmental stage) were sorted from BM cells of 60 WT mice. cDNA for real-time RT-PCR was prepared using the Advantage RT-for-PCR kit (Takara Bio Inc.), and total mRNA was isolated using the RNeasy mini kit (QIAGEN). Real-time RT-PCR assays to detect Nfil3 and Gapdh were performed in triplicate using the 7900HT Sequence Detection System and Taqman assays (Applied Biosystems). RNA from tissues was prepared using the RNeasy mini kit and cDNA was prepared using the iScript cDNA kit (Bio-Rad Laboratories). Real-time RT-PCR was performed as described in this section but using SYBR green reagent (Applied Biosystems).
Primers for real-time PCR.
The primers Mm00600292-s1 for Nfil3 and Mm99999915-g1 for Gapdh were premade by Applied Biosystems. The parameter threshold cycle (_C_t) is defined as the fractional cycle number at which the fluorescence generated by cleavage of the probe exceeds a fixed threshold above baseline.
Blood and lymphoid cell preparation.
Peripheral blood cells were obtained from the femoral arteries of 6-wk-old WT and _Nfil3_−/− mice. Blood was analyzed using an automated complete blood cell counter (Coulter counter for mice; model ATVIA #120; Bayer). Single cell suspensions of total splenocytes and thymocytes were obtained by passing organs through 70-µm cell strainers (BD), followed by treatment with red blood cell (RBC) lysis buffer (Sigma-Aldrich). Lymphocytes from BM were isolated from femurs and tibias and also treated with RBC lysis buffer. Mononuclear lymphocytes from homogenized liver and lung were recovered from the interface of a 70%/40% Percoll centrifugation gradient (Amersham Biosciences). Live lymphocyte counts were performed using an automated cell viability analyzer (Vi-cell XR 2.03; Beckman Coulter).
Antibodies.
Cells from murine lymphoid tissues were analyzed using mAbs recognizing the following: NK1.1 (clone PK136), CD3 (17A2), CD49b (DX5), CD122 (TM-β1), NKG2D (CX5), CD11b (M1/70), CD117 (2B8), 2B4 (2B4), CD43 (S7), Ly49D (4E5), Ly49G2 (Cwy-3), Ly49C (5E6), NKG2A/C/E (20d5), NKp46 (29A1.4), CD45.1 (A20), CD45.2 (104), or IFN-γ (XMG1.2). All antibodies, except for anti-NKG2D (FITC conjugated [BioLegend] and APC conjugated [eBioscience]) and CD3 eFluor780 and NKp46 eFluor710 (eBioscience), were purchased from BD. Antibodies were incubated with cells for 15 min at 4°C after blocking with purified anti-CD16/CD32 mAb (2.4G).
Flow cytometric analysis.
Single cell suspensions from BM, spleen, liver, and lung were incubated with mouse mAbs recognizing specific markers. Cells were analyzed using FACSCalibur or Canto flow cytometers (BD) and FlowJo software (Tree Star, Inc.). NK and NK T populations were quantified based on lymphocyte gating of total lymphoid organ populations.
IL-15–induced in vitro generation of NK cells.
Lin− BM cells (105) were cultured for 2 wk with 50 ng/ml mSlf, 50 ng/ml mFlt3L, and 5 ng/ml mIL-7, with or without 20 ng/ml mIL-15 (all cytokines were obtained from R&D Systems).
Retroviral Nfil3 expression in fetal liver cells.
Fetal liver cells from E13.5 Nfil3−/− embryos were cultured in 20% FCS, 100 mg/ml mSlf, 250 mg/ml mFlt3L, and 50 ng/ml mTPO as previously described (Pellegrini et al., 2005). The WT Nfil3 gene and Nfil3 lacking its DNA binding domain (synthesized using information provided by T. Inaba, Hiroshima University, Hiroshima, Japan) were subcloned in MSCV-ires-GFP and transfected into Phoenix packaging cells (G. Nolan, Stanford University, Stanford, CA). The retrovirus-containing supernatant was precoated on RetroNection-coated six-well plates (Cambrex), and the cytokine-stimulated fetal liver cells were cultured in these plates in the presence of 50 ng/ml mSlf, 50 ng/ml mFlt3L, 5 ng/ml mIL-7, and 20 ng/ml mIL-15.
Lin− BM cell preparation.
Lin− BM cells were prepared using the IMag NK cell enrichment kit (BD). Lin−CD34+ BM cells did not express CD3, CD19, Gr-1, Mac-1, or Ter119.
Adoptive cell transfer.
WT and _Nfil3_−/− recipients were irradiated with 6Gy on days 0 and 1. On day 2, the mice were transplanted i.v. with 5 × 106 B6.SJL-Ptprc Pep3/BoyJ (CD45.1) BM cells mixed equally with 5 × 106 C57BL/6 WT (CD45.2) or Nfil3−/− (CD45.2) BM cells. A 1:1 ratio of CD45.1/CD45.2 cells was confirmed by flow cytometry before injection. At 8 wk after transfer, blood, spleen, BM, and liver were harvested and single cell suspensions prepared. Liver mononuclear cells were isolated using Percoll density gradients. After erythrocyte lysis, lymphoid cell populations were counted and NK, polymorphonuclear neutrophil, B, and T cell numbers were assessed by flow cytometry using mAbs against NK1.1, TCR-β, B220, CD11b, Gr-1, CD45.1, or CD45.2.
In vivo proliferation of BM NK cells.
WT (n = 2) and Nfil3−/− (n = 4) mice were fed 0.8 mg/ml BrdU orally in drinking water for 3 d. Pooled BM was depleted of TER119+, CD19+, CD3+, CD4+, CD8+, and Gr-1+ cells using the iMag magnetic bead system (BD), and the remaining cells were stained with mAbs against CD3, CD122, or NK1.1. Proliferation of CD3−CD122+ cells was determined using the APC BrdU flow cytometry kit (BD) according to the manufacturer's instructions.
IFN-γ production in NK cells.
106 enriched NK cells from BM and spleen were incubated with 50 ng/ml mIL-2 plus 50 ng/ml mIL-12 (both from R&D Systems) for 12 h, treated with 1 µl/ml GolgiPlug (BD), and cultured for an additional 4 h with IL-2 and IL-12. IFN-γ production was assessed by intracellular flow cytometry (Prussin and Metcalfe, 1995).
In vitro NK cytotoxicity assay.
Mice were injected i.p. with 150 µg poly I:C 16 h before sacrifice, and NK cytotoxicity was analyzed in FACS-sorted NK1.1+CD3− splenocytes using a flow cytometric method as previously described (Souza et al., 2001).
In vivo NK cytotoxicity assay.
WT or Nfil3−/− recipients were injected i.p. with 200 µg poly I:C. After 24 h, 30 × 106 spleen cells, consisting of an equal mix of differentially CFSE-labeled C57BL/6 (0.5 µM) and B6.129P2-β2mtm/Unc/J (5 µM) spleen cells, were transferred i.v. into the recipients. After 24 h, spleen and liver mononuclear cells were isolated and examined by flow cytometry. The percent killing of differentially labeled donor cells was calculated as follows: % killing = [1−(%β2m−/− CFSEhi × %C57BL/6 CFSElo)/(%C57BL/6 CFSElo × %β2m−/− CFSEhi)] × 100.
Statistical analysis.
An unpaired Student's t test was used to determine the significance of differences between the means of parameters measured in WT and _Nfil3_−/− mice.
T cell activation assay.
Splenic T cells purified by negative selection (Imag streptavidin particles; BD) were left untreated or stimulated for 24 h in plates coated with anti-CD3_ϵ_ (145-2C11; BD) plus anti-CD28 (37.51; BD) or in plates containing 20 ng/ml PMA plus 100 ng/ml ionomycin (both from Sigma-Aldrich). Recovered cells were immunostained with antibodies against CD25, CD44, and CD69 and activation was assessed by flow cytometry.
T and B cell proliferation assay.
Purified splenic T cells were stimulated for 48 h in plates coated with 1 µg/ml of anti-CD3_ϵ_ and/or anti-CD28 antibodies, with or without 40 µg/ml of soluble IL-2 (R&D Systems), or with 10 ng/ml PMA plus 50 ng/ml ionomycin. Proliferation was determined by 3H-thymidine incorporation. Purified splenic B cells were stimulated for 48 h with 5 µg/ml of soluble anti-IgM (BD) and/or 5 µg/ml of anti-CD40 antibody (BD), or with 20 µg/ml LPS (Sigma-Aldrich). Proliferation was determined by 3H-thymidine incorporation.
Online supplemental material.
Fig. S1 shows generation of Nfil3−/− and Nfil3+/−-IRES β-geo mice. Fig. S2 shows endogenous Nfil3 expression patterns. Fig. S3 shows functional analysis of splenic T cells and thymocytes in _Nfil3_−/− mice. Table S1 shows comparison of numbers of NK and NK T cells in various organs of WT and _Nfil3_−/− mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20092176/DC1.
Acknowledgments
The authors thank Carlos Lopez Otin, Adolfo Ferrando, and Nobuyuki Ono for helpful suggestions and comments, Rakash Nayyar and Frances Kiti Tong for cell sorting, Roderick Bronson for pathology analysis, and Mary Saunders for scientific editing.
Financial support for this project was provided by grants from the National Institutes of Health), Canadian Institute of Health Research, the National Cancer Institute of Canada, and the International Research Fund of the Kyushu University School of Medicine Alumni, Japan.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
BMT
BM transfer
cDNA
complementary DNA
E4bp4
E4-binding protein 4
ES
embryonic stem
iNK
immature NK
mFlt3L
murine FMS-like tyrosine kinase 3 ligand
mNK
mature NK
mRNA
messenger RNA
mSlf
murine steel factor
Nfil3
nuclear factor IL-3
NKP
NK progenitor
Par
proline- and acidic amino acid–rich
RBC
red blood cell
References
- Barton K., Muthusamy N., Fischer C., Ting C.N., Walunas T.L., Lanier L.L., Leiden J.M. 1998. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 9:555–563 10.1016/S1074-7613(00)80638-X [DOI] [PubMed] [Google Scholar]
- Boos M.D., Yokota Y., Eberl G., Kee B.L. 2007. Mature natural killer cell and lymphoid tissue–inducing cell development requires Id2-mediated suppression of E protein activity. J. Exp. Med. 204:1119–1130 10.1084/jem.20061959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos M.D., Ramirez K., Kee B.L. 2008. Extrinsic and intrinsic regulation of early natural killer cell development. Immunol. Res. 40:193–207 10.1007/s12026-007-8006-9 [DOI] [PubMed] [Google Scholar]
- Brown D.D., Wang Z., Furlow J.D., Kanamori A., Schwartzman R.A., Remo B.F., Pinder A. 1996. The thyroid hormone-induced tail resorption program during Xenopus laevis metamorphosis. Proc. Natl. Acad. Sci. USA. 93:1924–1929 10.1073/pnas.93.5.1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell I.G., Skinner A., Hurst H.C. 1992. Transcriptional repression by a novel member of the bZIP family of transcription factors. Mol. Cell. Biol. 12:3070–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Santo J.P. 2006. Natural killer cell developmental pathways: a question of balance. Annu. Rev. Immunol. 24:257–286 10.1146/annurev.immunol.24.021605.090700 [DOI] [PubMed] [Google Scholar]
- Drolet D.W., Scully K.M., Simmons D.M., Wegner M., Chu K.T., Swanson L.W., Rosenfeld M.G. 1991. TEF, a transcription factor expressed specifically in the anterior pituitary during embryogenesis, defines a new class of leucine zipper proteins. Genes Dev. 5:1739–1753 10.1101/gad.5.10.1739 [DOI] [PubMed] [Google Scholar]
- Furlow J.D., Brown D.D. 1999. In vitro and in vivo analysis of the regulation of a transcription factor gene by thyroid hormone during Xenopus laevis metamorphosis. Mol. Endocrinol. 13:2076–2089 10.1210/me.13.12.2076 [DOI] [PubMed] [Google Scholar]
- Gascoyne D.M., Long E., Veiga-Fernandes H., de Boer J., Williams O., Seddon B., Coles M., Kioussis D., Brady H.J.M. 2009. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 10:1118–1124 10.1038/ni.1787 [DOI] [PubMed] [Google Scholar]
- Guerra N., Tan Y.X., Joncker N.T., Choy A., Gallardo F., Xiong N., Knoblaugh S., Cado D., Greenberg N.M., Greenberg N.R., Raulet D.H. 2008. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 28:571–580 10.1016/j.immuni.2008.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington N.D., Vosshenrich C.A., Di Santo J.P. 2007. Developmental pathways that generate natural-killer-cell diversity in mice and humans. Nat. Rev. Immunol. 7:703–714 10.1038/nri2154 [DOI] [PubMed] [Google Scholar]
- Ikushima S., Inukai T., Inaba T., Nimer S.D., Cleveland J.L., Look A.T. 1997. Pivotal role for the NFIL3/E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Proc. Natl. Acad. Sci. USA. 94:2609–2614 10.1073/pnas.94.6.2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T., Roberts W.M., Shapiro L.H., Jolly K.W., Raimondi S.C., Smith S.D., Look A.T. 1992. Fusion of the leucine zipper gene HLF to the E2A gene in human acute B-lineage leukemia. Science. 257:531–534 10.1126/science.1386162 [DOI] [PubMed] [Google Scholar]
- Ishida H., Ueda K., Ohkawa K., Kanazawa Y., Hosui A., Nakanishi F., Mita E., Kasahara A., Sasaki Y., Hori M., Hayashi N. 2000. Identification of multiple transcription factors, HLF, FTF, and E4BP4, controlling hepatitis B virus enhancer II. J. Virol. 74:1241–1251 10.1128/JVI.74.3.1241-1251.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Iizuka K., Aguila H.L., Weissman I.L., Yokoyama W.M. 2000. In vivo natural killer cell activities revealed by natural killer cell-deficient mice. Proc. Natl. Acad. Sci. USA. 97:2731–2736 10.1073/pnas.050588297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Iizuka K., Kang H.S., Dokun A., French A.R., Greco S., Yokoyama W.M. 2002. In vivo developmental stages in murine natural killer cell maturation. Nat. Immunol. 3:523–528 10.1038/ni796 [DOI] [PubMed] [Google Scholar]
- Kondo M., Weissman I.L., Akashi K. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672 10.1016/S0092-8674(00)80453-5 [DOI] [PubMed] [Google Scholar]
- Kuribara R., Kinoshita T., Miyajima A., Shinjyo T., Yoshihara T., Inukai T., Ozawa K., Look A.T., Inaba T. 1999. Two distinct interleukin-3-mediated signal pathways, Ras-NFIL3 (E4BP4) and Bcl-xL, regulate the survival of murine pro-B lymphocytes. Mol. Cell. Biol. 19:2754–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzstein M.M., Hengartner M.O., Tsung N., Ellis R.E., Horvitz H.R. 1996. Transcriptional regulator of programmed cell death encoded by Caenorhabditis elegans gene ces-2. Nature. 382:545–547 10.1038/382545a0 [DOI] [PubMed] [Google Scholar]
- Mitsui S., Yamaguchi S., Matsuo T., Ishida Y., Okamura H. 2001. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 15:995–1006 10.1101/gad.873501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C.R., Maire P., Schibler U. 1990. DBP, a liver-enriched transcriptional activator, is expressed late in ontogeny and its tissue specificity is determined posttranscriptionally. Cell. 61:279–291 10.1016/0092-8674(90)90808-R [DOI] [PubMed] [Google Scholar]
- Murakami M., Watanabe H., Niikura Y., Kameda T., Saitoh K., Yamamoto M., Yokouchi Y., Kuroiwa A., Mizumoto K., Iba H. 1997. High-level expression of exogenous genes by replication-competent retrovirus vectors with an internal ribosomal entry site. Gene. 202:23–29 10.1016/S0378-1119(97)00468-X [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Tanaka T. 2001. Calcium-dependent activation of nuclear factor regulated by interleukin 3/adenovirus E4 promoter-binding protein gene expression by calcineurin/nuclear factor of activated T cells and calcium/calmodulin-dependent protein kinase signaling. J. Biol. Chem. 276:19921–19928 10.1074/jbc.M010332200 [DOI] [PubMed] [Google Scholar]
- Ohno T., Onishi Y., Ishida N. 2007. A novel E4BP4 element drives circadian expression of mPeriod2. Nucleic Acids Res. 35:648–655 10.1093/nar/gkl868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., Saijo K., Takahashi T., Osawa M., Arase H., Hirayama N., Miyake K., Nakauchi H., Shirasawa T., Saito T. 1995. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 3:771–782 10.1016/1074-7613(95)90066-7 [DOI] [PubMed] [Google Scholar]
- Pellegrini M., Bath S., Marsden V.S., Huang D.C., Metcalf D., Harris A.W., Strasser A. 2005. FADD and caspase-8 are required for cytokine-induced proliferation of hemopoietic progenitor cells. Blood. 106:1581–1589 10.1182/blood-2005-01-0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prussin C., Metcalfe D.D. 1995. Detection of intracytoplasmic cytokine using flow cytometry and directly conjugated anti-cytokine antibodies. J. Immunol. Methods. 188:117–128 10.1016/0022-1759(95)00209-X [DOI] [PubMed] [Google Scholar]
- Samson S.I., Richard O., Tavian M., Ranson T., Vosshenrich C.A., Colucci F., Buer J., Grosveld F., Godin I., Di Santo J.P. 2003. GATA-3 promotes maturation, IFN-gamma production, and liver-specific homing of NK cells. Immunity. 19:701–711 10.1016/S1074-7613(03)00294-2 [DOI] [PubMed] [Google Scholar]
- Shimozato O., Ortaldo J.R., Komschlies K.L., Young H.A. 2002. Impaired NK cell development in an IFN-gamma transgenic mouse: aberrantly expressed IFN-gamma enhances hematopoietic stem cell apoptosis and affects NK cell differentiation. J. Immunol. 168:1746–1752 [DOI] [PubMed] [Google Scholar]
- Souza S.S., Castro F.A., Mendonça H.C., Palma P.V., Morais F.R., Ferriani R.A., Voltarelli J.C. 2001. Influence of menstrual cycle on NK activity. J. Reprod. Immunol. 50:151–159 10.1016/S0165-0378(00)00091-7 [DOI] [PubMed] [Google Scholar]
- Vosshenrich C.A., Samson-Villéger S.I., Di Santo J.P. 2005. Distinguishing features of developing natural killer cells. Curr. Opin. Immunol. 17:151–158 10.1016/j.coi.2005.01.005 [DOI] [PubMed] [Google Scholar]
- Wallace A.D., Wheeler T.T., Young D.A. 1997. Inducibility of E4BP4 suggests a novel mechanism of negative gene regulation by glucocorticoids. Biochem. Biophys. Res. Commun. 232:403–406 10.1006/bbrc.1997.6206 [DOI] [PubMed] [Google Scholar]
- Zhang W., Zhang J., Kornuc M., Kwan K., Frank R., Nimer S.D. 1995. Molecular cloning and characterization of NF-IL3A, a transcriptional activator of the human interleukin-3 promoter. Mol. Cell. Biol. 15:6055–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]