Specificity of Human Thymine DNA Glycosylase Depends on N-Glycosidic Bond Stability (original) (raw)
. Author manuscript; available in PMC: 2010 Jan 20.
Published in final edited form as: J Am Chem Soc. 2006 Sep 27;128(38):12510–12519. doi: 10.1021/ja0634829
Abstract
Initiating the DNA base excision repair pathway, DNA glycosylases find and hydrolytically excise damaged bases from DNA. While some DNA glycosylases exhibit narrow specificity, others remove multiple forms of damage. Human thymine DNA glycosylase (hTDG) cleaves thymine from mutagenic G·T mispairs and recognizes many additional lesions, and has a strong preference for nucleobases paired with guanine rather than adenine. Yet, hTDG avoids cytosine, despite the million-fold excess of normal G·C pairs over G·T mispairs. The mechanism of this remarkable and essential specificity has remained obscure. Here, we examine the possibility that hTDG specificity depends on the stability of the scissile base-sugar bond by determining the maximal activity (_k_max) against a series of nucleobases with varying leaving group ability. We find that hTDG removes 5-fluorouracil 78-fold faster than uracil and 5-chlorouracil 572-fold faster than thymine, differences that can be attributed predominantly to leaving group ability. Moreover, hTDG readily excises cytosine analogues with improved leaving ability, including 5-fluorocytosine, 5-bromocytosine, and 5-hydroxycytosine, indicating that cytosine has access to the active site. A plot of log(_k_max) versus leaving group p_K_a reveals a Brønsted-type linear free energy relationship with a large negative slope of βlg = −1.6 ± 0.2, consistent with a highly dissociative reaction mechanism. Further, we find that the hydrophobic active site of hTDG contributes to its specificity by enhancing the inherent differences in substrate reactivity. Thus, hTDG specificity depends on _N_-glycosidic bond stability, and the discrimination against cytosine is due largely to its very poor leaving ability rather than its exclusion from the active site.
Introduction
The chemically reactive bases in DNA are continuously modified by agents of cellular metabolism and from exogenous sources, producing lesions that threaten genetic integrity and play a role in ageing and diseases including cancer.1 Counteracting this inevitable damage is the base excision repair (BER) pathway, initiated by a damage-specific DNA glycosylase. Using a base-flipping mechanism, these enzymes recognize damaged bases and remove them by catalyzing the hydrolysis of the _N_-glycosidic bond connecting the nucleobase to the sugar, producing an abasic (AP) site in the DNA. While some DNA glycosylases possess significant catalytic power, they are perhaps more impressive in their specificity for certain damaged bases and against normal bases, in keeping with the threat to genomic integrity posed by the aberrant removal of normal bases from DNA. However, the challenge of distinguishing damaged from normal is daunting, given that many damaged bases differ modestly from the normal counterpart and that the lesions are hidden within the vast excess of normal DNA.
To obtain specificity, some DNA glycosylases employ a highly discriminating active site that accommodates only certain lesions. For example, uracil DNA glycosylase (UDG) uses steric exclusion and specific electrostatic interactions to recognize uracil and prevent other bases from docking in its active site.2 Other enzymes with a highly selective active site include human 8-oxoguanine (OG) DNA glycosylase (hOGG1), which removes OG from OG·C pairs, but not G from G·C pairs,3 and Escherichia coli 3-methyladenine DNA glycosylase I (eTAG), which binds 3-methyladenine but not adenine.4,5 In contrast, many DNA glycosylases allow a broad range of damaged and even some normal bases access to the active site, as exemplified by 3-methyladenine DNA glycosylase II from Escherichia coli (AlkA) and mammalian alkyladenine DNA glycosylase (AAG).6–8 For these more promiscuous enzymes, certain properties of the damaged base contribute to specificity. For example, alkylated bases form a more labile _N_-glycosidic bond and are more easily removed compared to the normal counterpart.6,7 Additionally, some damaged bases have a diminished capacity for H bonding and/or stacking interactions. As a result, such bases are more likely to flip out of the DNA duplex, which can lead to tighter binding to a DNA glycosylase and faster base excision.7,8
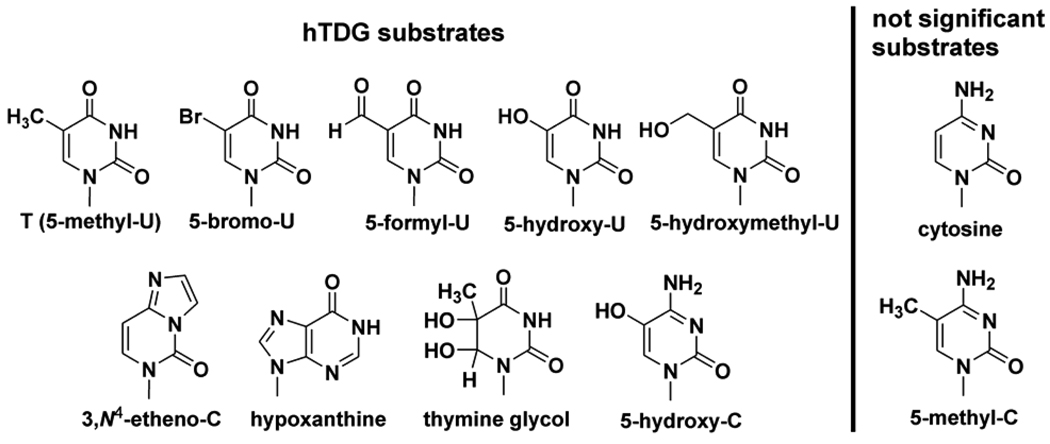
Another enzyme with broad specificity is human thymine DNA glycosylase (hTDG), which excises thymine from G·T mispairs, and removes many additional lesions, with a strong preference for bases that are paired with guanine and located at CpG sites (i.e., 5´-CpG/5´-XpG, where X is the target base).9 In vertebrates, cytosine methylation (m5C) at CpG serves as a mark for transcriptional silencing and is central to many important cellular processes. CpG sites are also associated with disease; improper CpG methylation plays a role in carcinogenesis,10 and the deamination of m5C to T produces G·T mispairs, contributing to the high mutational frequencies observed at CpG sites in humans.11,12 Although hTDG is highly specific for removing bases that are paired with guanine as opposed to adenine, it avoids the removal of cytosine despite the million-fold excess of G·C pairs over G·T mispairs. The challenge of attaining such remarkable specificity would appear to be compounded by the broad range of bases that are excised by hTDG (Figure 1).
Figure 1.
Known hTDG substrates are shown.13–17 Cytosine and 5-methyl-C are not significant substrates.13,18 We report several new hTDG substrates here, including 5-chloro-U, 5-iodo-U, 5-fluoro-C, and 5-bromo-C.
One proposal holds that specificity occurs at the base-flipping step, whereby the higher stability of G·C pairs relative to G·T mispairs would preclude the enzymatic flipping of cytosine into the active site.19 Another possibility is that hTDG uses selective steric and electrostatic interactions to recognize multiple substrates while excluding cytosine. However, examination of the various bases removed by hTDG (Figure 1) indicates no obvious handle that would allow for substrate recognition and cytosine rejection. Moreover, the related mismatch-specific uracil DNA glycosylase from Escherichia coli (eMUG) has a permissive and non-specific active site which provides no hydrogen bonds that account for its specificity against cytosine,20 and a recent structure of the hTDG catalytic core, conjugated to the ubiquitin-like modifier SUMO-1, reveals strong similarity to eMUG.21 Another possibility is that specificity is obtained at the chemical step of the enzymatic reaction, cleavage of the _N_-glycosidic bond connecting the nucleobase to the sugar. Thus, hTDG could potentially flip a broad range of bases, including cytosine, into its active site, with specificity determined predominantly by the heterolytic stability of the scissile C-N bond.
Here, we examine the dependence of hTDG specificity on _N_-glycosidic bond stability. Our strategy stems from previous studies which found that non-enzymatic pyrimidine hydrolyses proceed through a highly dissociative mechanism, where the reaction rate, hence glycosidic bond stability, depends on the leaving ability of the nucleobase, as indicated by the acidity of the glycosidic nitrogen (N1) in aqueous solution (p_K_aN1).22,23 Importantly, these studies found that 2'-deoxycytidine (dC) is significantly more stable than 2'-deoxyuridine (dU) and 2'-deoxythymidine (dT) at pH 7.4,24 raising the possibility that hTDG specificity depends on substrate reactivity. To test this idea directly, we determined the maximal activity (_k_max) of hTDG against a series of 5-substituted uracil and cytosine bases which varied in leaving group ability. We show quantitatively that hTDG specificity is determined largely by _N_-glycosidic bond stability, and we find that a hydrophobic active site enhances the inherent differences in substrate reactivity. Thus, specificity against cytosine can be largely explained by its poor leaving ability rather than its exclusion from the hTDG active site.
Materials and Methods
Duplex DNA substrates
The 19 bp duplex DNA substrates were made by mixing the target strand (5’-CACTGCTCAxGTACAGAGC,x = substrate base) and 5% excess of the complementary strand (5’-GCTCTGTACGTGAGCAGTG) in 0.01 M Tris pH 8.0, 0.1 M NaCl, and 0.1 mM EDTA, heating to 80 °C, and slowly (> 3 hrs) cooling to 22 °C. The oligonucleotides were synthesized at the W.M. Keck Biotechnology Resource Laboratory of Yale University and at the Biopolymer Genomics Core Facility, University of Maryland Baltimore. Nucleoside phosphoramidites with modified bases were purchased from Glen Research (Sterling, VA), and were incorporated using standard phosphoramidite chemistry and the deprotection method recommended by the manufacturer. The phosphoramidite for 5-chloro-dU was generously provided by Dr. Yu Lin Jiang (Johns Hopkins University). The oligonucleotides were purified by anion exchange HPLC using a Zorbax Oligo column (Agilent Technologies), desalted by gel filtration, and stored at −20 °C. The purity was verified by analytical anion-exchange HPLC under denaturing conditions (pH 12) using a DNAPac PA200 column (Dionex Corp.). The molecular weight was verified by ESI mass spectrometry (Integrated DNA Technologies); the observed and calculated masses differed by <0.03%. The possibility that the 5-halogenated cytosine bases had spontaneously deaminated to the 5-halouracil base during purification or storage was eliminated using anion-exchange HPLC at pH 12, which allowed the oligonucleotides (i.e., FC versus FU) to be fully resolved (not shown). The oligonucleotides were quantified by absorbance at 260 nm using pairwise extinction coefficients.25
Expression and purification of hTDG
A pET-28 based expression plasmid for hTDG26 was transformed into BL21(DE3) “Rosetta” cells (Novagen). The cells were grown in LB (Luria Broth) at 37 °C to an OD600 = 0.8, the temperature was reduced to 15 °C, and expression of hTDG was induced with 0.25 mM IPTG (isopropyl β-D-thiogalactoside) and continued for about 15 hrs. The cells were harvested and stored at −80 °C. The pellet was thawed and suspended in lysis buffer (0.05 M sodium phosphate pH 8.0, 0.3 M NaCl, 0.02 M imidazole, 0.01 M beta-mercaptoethanol), incubated with 1 mg/ml lysozyme and DNase (Novagen) for 30 min on ice, and then sonicated. The lysate was cleared by centrifugation, incubated with 4 ml Ni-NTA resin (Qiagen) for 1 hr at 4 °C, and the mixture was added to an empty column with the flow-through collected. The bound hTDG was washed using 30 ml lysis buffer with 20 mM imidazole and 1M NaCl, washed again using 30 ml lysis buffer with 20 mM imidazole and 0.3M NaCl, and eluted using 3 × 5 ml volumes of lysis buffer with 0.15 M imidazole. Purification was continued using a Q sepharose HP column (Amersham) with buffers IE-A (25 mM Tris pH 7.5, 75 mM NaCl, 1 mM DTT, 0.2 mM EDTA, 1% glycerol) and IE-B (IE-A with 1 M NaCl) and a gradient of 0–50% IE-B over 60 min at 2.5 ml/min. hTDG was purified further using a SP sepharose HP column (Amersham) with the same buffers and a gradient of 0–100% IE-B over 60 min at 2.5 ml/min. The purity of hTDG was >99% as judged by a Coomassie stained gel, and its molecular weight was confirmed by mass spectrometry. The yield of hTDG was typically >5 mg per L of culture. Purified hTDG was dialyzed overnight in 1 L storage buffer (20 mM HEPES 7.5, 0.1 M NaCl, 1 mM DTT, 0.5 mM EDTA, and 1% glycerol), concentrated to about 0.1 mM, flash frozen, and stored at −80 °C. The concentration of hTDG was determined by absorbance using ε280 = 31.5 mM−1cm−1.27
Single turnover kinetics
Because hTDG is strongly inhibited by its abasic DNA product,9 we used single turnover kinetics and saturating enzyme conditions. To ensure that maximal rate constants were obtained (i.e., _k_obs = _k_max), the experiments were collected with a large excess of enzyme, at least 100-fold greater than the reported _K_D = 41 nM for hTDG binding to DNA containing a G·T mispair.28 Saturating enzyme conditions were verified by collecting the experiments with at least two hTDG concentrations, typically 5 and 10 µM, which yielded rate constants that were equivalent within experimental uncertainty (± 10%). The DNA substrate concentrations were typically 500 nM, although equivalent _k_max values were obtained for 250 and 1000 nM substrate concentrations (not shown). The reactions were performed either manually or using a three-syringe rapid chemical quenched-flow instrument (RQF-3, Kintek Corp.). For some substrates, both methods were used, yielding data that were in very good agreement. Samples taken at specific time points were quenched with 50 % (v:v) quench solution (0.3 M NaOH, 0.03 M EDTA), incubated at 85 °C for 15 min to induce alkaline cleavage of the DNA backbone at abasic sites, and then analyzed by HPLC to determine the reaction progress (see below). For the DNA substrates containing 5-hydroxyuracil (hoU), 5-hydroxycytosine (hoC), and 5-hydroxymethyluracil (hmU), the reactions were quenched using 0.1 M piperidine with 0.03 M EDTA (final concentration) and heated at 85 °C for 15 min, because a small amount of DNA backbone cleavage was observed (in the absence of hTDG) at the site of the modified base when the NaOH quench was used. The single turnover reactions proceeded to full completion for all substrates (except G·C19). Rate constants were determined by fitting the data to a single exponential equation using non-linear regression with Grafit 5 29. The reactions were conducted at 22 °C in HEMN.1 buffer (20 mM HEPES pH 7.50, 0.2 mM EDTA, 2.5 mM MgCl2, 0.1 M NaCl) with 0.1 mg/ml bovine serum albumin.
HPLC assay for monitoring the hTDG reaction
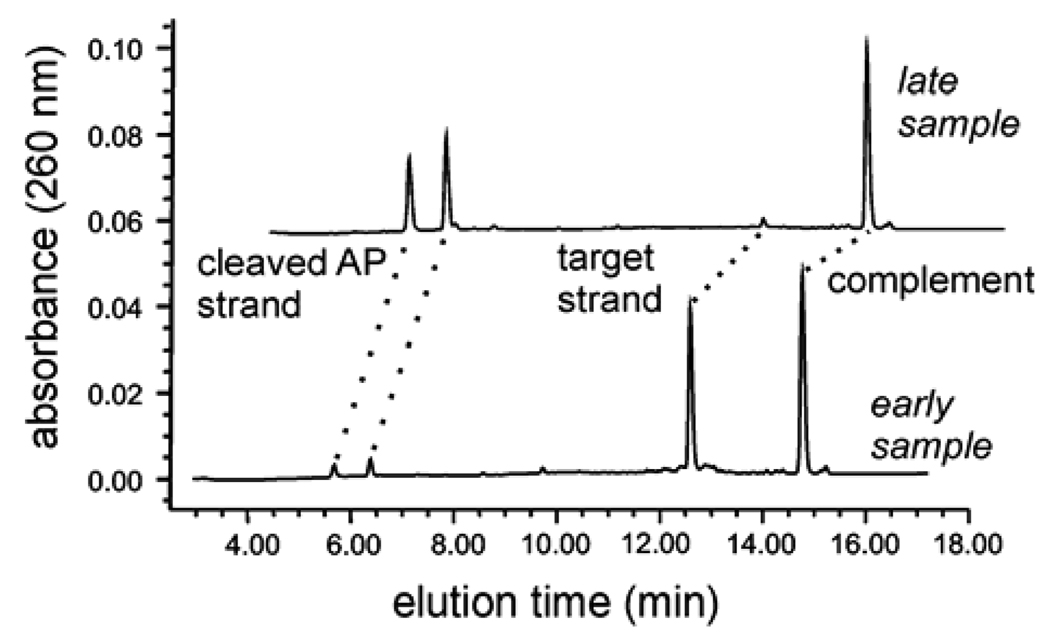
We have developed a HPLC assay for monitoring the kinetics of the hTDG-catalyzed reaction. Samples taken from the kinetic reactions at various time points contained a mixture of four oligonucleotides: the full-length (19mer) target strand, its complement, and the two smaller product strands resulting from alkaline cleavage of the abasic strand (produced by hTDG activity). As shown in Figure 2, these four strands are fully resolved by anion exchange HPLC using denaturing (pH 12.0) conditions with a DNAPac PA200 column (Dionex Corp.). The alkaline conditions serve to suppress hybridization and to increase the resolution, because thymine and guanine are negatively charged at pH 12. The elution buffer was 0.02 M sodium phosphate pH 12.0 containing either (A) 0.03 M NaClO4 or (B) 0.50 M NaClO4. The protocol employed: 8% B for 1 min, linear gradient of 8–25% B over 14 min, 100% B for 2 min, 8% B for 6 min, and a flow rate of 1.2 ml/min throughout. The oligonucleotides were detected by absorbance (260 nm). Assignment of the elution peaks was confirmed by running individually the oligonucleotides corresponding to the target and product strands. The fraction product was determined from the integrated peak areas for the target strand (AS) and the two product strands (AP1 and AP2) using eq 1:
| F=fraction product=(AP1+AP2)/(AS+AP1+AP2) | (1) |
|---|
Using multiple injections of an identical sample, we found that the fraction product was reproducible to within 1% (not shown). This HPLC assay is amenable to automation, as we routinely analyze dozens of samples overnight using an auto-sampling device. We anticipate that this approach will be applicable to other DNA glycosylases, and other enzymes that act upon DNA. Indeed, we have used it to monitor the kinetics of human AP endonuclease (not shown).
Figure 2.
HPLC assay for monitoring DNA glycosylase reactions. Shown are chromatograms for samples taken from a single-turnover reaction at an “early” time point where there was little product formation, and a “late” time point where the reaction was nearly complete. The ion-exchange HPLC under denaturing (pH 12) conditions gives excellent resolution of the target strand, its complement, and the two shorter product strands resulting from alkaline-induced cleavage of the nascent abasic strand produced by hTDG activity.
Theoretical Calculations
To obtain model structures, vibrational frequencies, and energetics for the neutral and N1 deprotonated nucleobases, theoretical calculations were performed using Gaussian 03.30 For the systems excluding 5-iodouracil, geometry optimizations and vibrational analyses were performed at the MP2(full)/6-31G* level. When used to calculate thermal energy corrections, the MP2(full)/6-31G* vibrational frequencies are scaled by a factor of 0.9646.31 Single point energy calculations and electrostatic potential surfaces were performed at the MP2(full)/6-311+G(2d,2p) level using the MP2(full)/6-31G* geometries. Parameters for iodine are not available in the above basis sets; therefore, calculations for the 5-iodouracil systems were performed using the modified LANL2DZ32,33 effective core potentials (ECPs) and valence basis sets for I, while the standard basis sets described above were used for all other atoms. To obtain accurate deprotonation enthalpies and free energies, zero-point energy (ZPE) and basis set superposition error (BSSE) corrections34,35 were included in the determination of these values.
To determine the relative size of uracil and cytosine and the influence of the 5-substituents upon the size of these nucleobases, we calculated molar volumes with Gaussian 03 at the MP2(full)/6-311+G(2d,2p) level using the MP2(full)/6-31G* geometries. To determine accurate volumes, the MP2 density was used and numerical integration was employed as the default Monte Carlo algorithm produced spurious and irreproducible results.
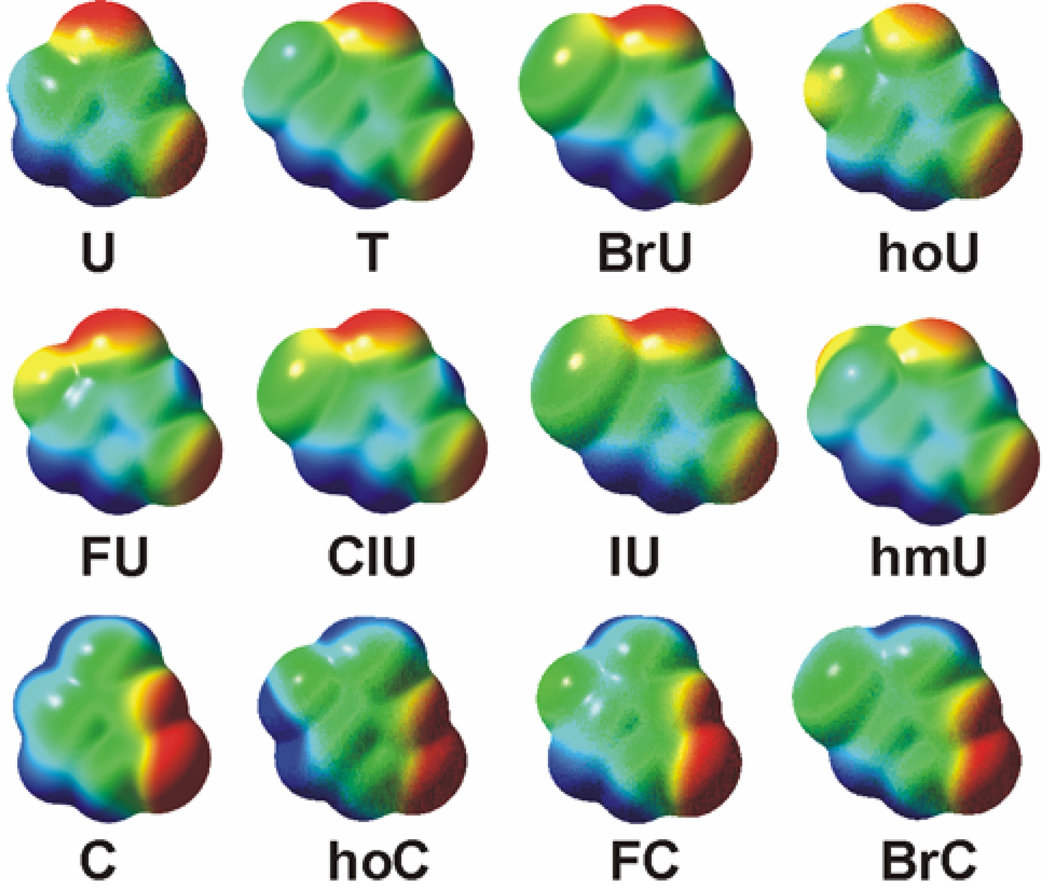
To more clearly visualize the influence of the 5-substituents upon the electronic properties of the nucleobases, we calculated electrostatic potential maps for the neutral species. The process involves calculation of the interaction of a +1 probe charge and every part of the electron density cloud of these species calculated at the MP2(full)/6-311+G(2d,2p) level. The electrostatic potential was then mapped onto an isosurface of 0.002 electrons/Å3 of the total SCF electron density for the species of interest. The electrostatic potential maps were then color-coded according to their potential with the regions of negative electrostatic potential shown in red, neutral regions shown in green, and positive regions shown in blue. The electrostatic potential range used for the maps generated in this work varied from –31 to +31 kcal/mol.
Results
Single turnover kinetics
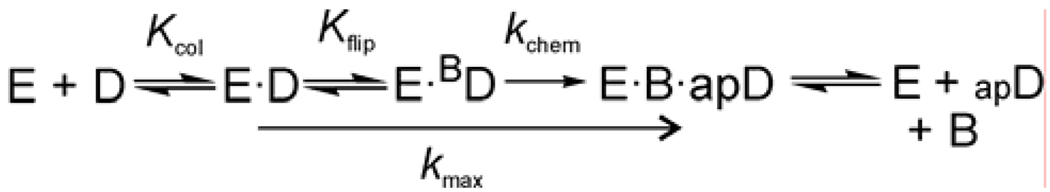
Like many DNA glycosylases, hTDG binds with high affinity to its reaction product, abasic DNA.36–38 For hTDG, the resulting product inhibition is so potent that it exhibits almost no turnover in vitro. We therefore used single turnover kinetics and saturating enzyme conditions to obtain rate constants (_k_max) that were not impacted by product release or the bimolecular association of enzyme and substrate, and thus correspond to the maximal catalytic activity of hTDG (Figure 3).39
Figure 3.
A minimal kinetic mechanism for the hTDG-catalyzed reaction. The initial association of hTDG (E) and substrate DNA (D) produces the collision complex (E·D) followed by a base-flipping step to form the reactive complex (E·BD, where BD is DNA with an extrahelical base). Cleavage of the base-sugar bond (_k_chem) yields the ternary product complex (E·B·apD). Release of the base (B) likely precedes very slow release of AP DNA (apD).36,38 The _k_max values report on the reaction steps from the initial hTDG·DNA collision complex to the ternary product complex.
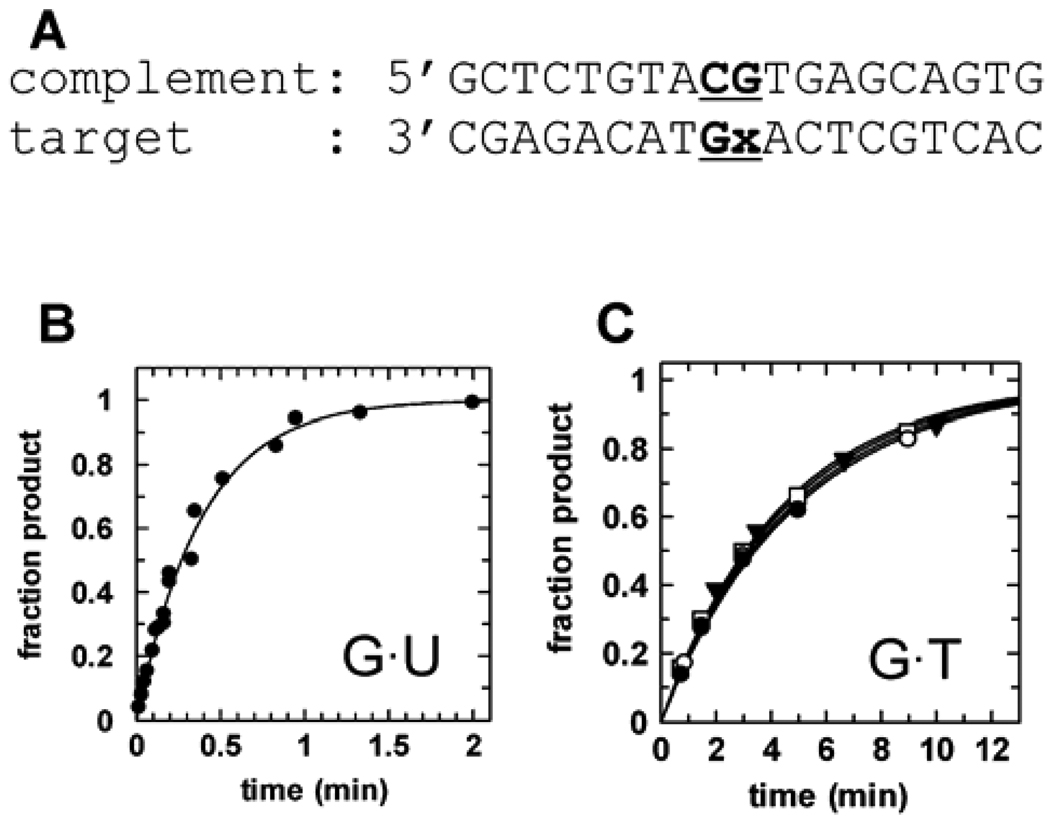
The kinetics experiments were conducted with 19 bp duplex DNA substrates that varied only in the identity of the target base (Figure 4A). Because hTDG is specific for bases that are paired with guanine and located in a CpG site,18,41 the target base was placed in this context for all substrates. Representative data for the single turnover activity of hTDG against the G·U19 substrate is shown in Figure 4B. To confirm saturating enzyme conditions, the single turnover experiments were collected with multiple hTDG concentrations. The _k_max values obtained for G·T19 are constant (within experimental error) for hTDG concentrations ranging from 1 to 10 µM (Figure 4C). Similar results are observed for the U·G19 substrate over the same range of concentrations of hTDG (data not shown). For all the other substrates examined here, data were collected at 5 and 10 µM concentrations of hTDG and gave equivalent _k_max values (within experimental error). The results of these kinetic experiments are summarized in Table 1.
Figure 4.
Single turnover kinetics for hTDG. (A) Structure of the 19 bp DNA substrate used for the kinetics experiments, where the target base (x) is located at a CpG site (bold and underlined). A substrate in which x = uracil is referred to as G·U19 (B) Representative data for the maximal activity of hTDG (5 µM) against G·U19 (500 nM) gives a rate constant of _k_max = 2.6 min−1. Data for G·U19 were collected using manual sampling and a rapid chemical quench-flow instrument (for t <8 s). (C) Confirming the saturating enzyme conditions, _k_max values for G·T19 are independent of enzyme for hTDG concentrations ≥ 1.0 µM: _k_obs = 0.20 min−1 for 1.0 µM hTDG (●); _k_obs = 0.21 min−1 for 2.5 µM hTDG (○); _k_obs = 0.23 min−1 for 5 µM hTDG (▼); _k_obs = 0.22 min−1 for 10 µM hTDG (□).
Table 1.
Kinetic parameters for hTDG
| Substrate | _k_max (min−1) a | Relative to G·U | Relative to G·T | Relative to G·C | _k_non (min−1) b | Rate enhancement e |
|---|---|---|---|---|---|---|
| G·U | 2.6 ± 0.3 | (1) | 11.8 | 2.4 × 10−9 | 109.0 | |
| G·T | 0.22 ± 0.04 | 0.08 | (1) | 7.5 × 10−10 | 108.5 | |
| G·FU | 202 ± 16 | 78 | 918 | 2.9 × 10−8 | 109.9 | |
| G·ClU | 126 ± 16 | 48 | 572 | 5.0 × 10−8 | 109.4 | |
| G·BrU | 11.6 ± 1.0 | 4.5 | 53 | 4.2× 10−8c | 108.4 | |
| G·IU | 0.06 ± 0.006 | 0.023 | 0.27 | 9.5 × 10−9 | 106.8 | |
| G·hoU | 2.2 ± 0.2 | 0.85 | 10 | |||
| G·hmU | 1.9 ± 0.2 | 0.73 | 8.6 | |||
| G·C | 1.2 × 10−5 ± 0.5 × 10−5 | 10−5.3 | 10−4.3 | (1) | 5 × 10−12 d | 106.4 |
| G·FC | 0.035 ± 0.004 | 103.5 | ||||
| G·BrC | 0.008 ± 0.0007 | 102.8 | ||||
| G·hoC | 0.010 ± 0.001 | 102.9 |
Theoretical results
Theoretical structures for the neutral and N1 deprotonated nucleobases were calculated as described in the Theoretical calculations section. The calculated deprotonation enthalpies and free energies are summarized in the Supporting Information (Table S1). Independent zero point energy, basis set superposition error, and thermal corrections are provided for all nucleobases listed. In previous work, we also calculated the N1 and N3 acidities of uracil at the CBS-Q level of theory.43 Those calculations suggested that the MP2(full)/6-311+G(2d,2p)//MP2(full)/6-31G* level of theory used here somewhat overestimates the acidities, but showed that the relative acidities are accurately reproduced. Thus the trends in the MP2 acidities should be a good descriptor of the influence of the 5-substituent on the acidity of the nucleobase. Our calculated deprotonation enthalpy for uracil N1 (Table S1, Supporting Information) is in good agreement with previously reported experimental44,45 and calculated44,46–48 values. Likewise, good absolute and excellent relative agreement between our calculations and previous studies is obtained for N1 acidities of thymine,47,49,50 cytosine,47,51 5-flurouracil (FU)50,52 5-chlorouracil (ClU),52 and 5-hydroxyuracil (hoU).50 It was also previously established that the use of ECPs for the halogen substituent produced only a modest effect on the calculated acidities of BrU43 and thus it is expected that the calculated acidity of IU is also reasonably accurate.
The calculated molar volumes of the isolated nucleobases are summarized in Table 2 along with other steric and electrostatic properties. The calculated electrostatic potential maps are shown in Figure 5.
Table 2.
Parameters for the 5-substituted uracil and cytosine bases
| Base | C5 substituent | p_K_aN1a | Electronic substituent constant (σm)b | hydrophobic substituent constant (π) c | projection along C5-X axis (Å) d | Volume of base (Å3) |
|---|---|---|---|---|---|---|
| U | H | 9.76 | (0) | (0) | 2.28 | 127.6 |
| T | CH3 | 10.19 | −0.07 | 0.56 | 150.5 | |
| FU | F | 8.43 | 0.34 | 0.14 | 2.81 | 132.7 |
| ClU | Cl | 8.14 | 0.37 | 0.71 | 3.49 | 149.9 |
| BrU | Br | 8.24 | 0.39 | 0.86 | 3.80 | 155.4 |
| IU | I | 8.44 | 0.35 | 1.12 | 4.01 | 166.7 |
| hoU | OH | 9.34 | 0.12 | −0.67 | 137.3 | |
| hmU | CH2OH | 9.82 | 0.00 | −1.03 | 160.2 | |
| C | H | 12.20 | (0) | (0) | 2.28 | 133.7 |
| FC | F | 10.87 | 0.34 | 0.14 | 2.83 | 138.6 |
| BrC | Br | 10.33 | 0.39 | 0.86 | 3.81 | 160.7 |
| hoC | OH | 11.66 | 0.12 | −0.67 | 143.7 |
Figure 5.
Electrostatic potential maps of the 5-substituted uracil and cytosine bases used in this work. The range of electrostatic potential varies between −31 kcal/mol (red) to +31 kcal/mol (blue), on an isosurface of 0.002 electrons/Å3 of the total SCF electron density.
Structure-activity correlations
Previous studies indicate that non-enzymatic pyrimidine hydrolysis reactions are dissociative,22,23 where the rate depends on the leaving ability of the nucleobase, as indicated by its N1 acidity in aqueous solution (p_K_aN1). This suggested that we could test the role of _N_-glycosidic bond stability in the specificity of hTDG by determining its maximal activity against substrates with varying leaving group ability. Such structure-activity correlations often require substrates with significantly different steric and electrostatic properties, and are therefore not feasible for many enzymes, including probably most DNA glycosylases. However, hTDG is a good candidate for this approach, because it has a relatively accommodating and non-specific active site.20,21 The substrates examined here differ significantly in leaving group ability, where the p_K_aN1 for a 5-substituted uracil or cytosine base depends on the electronic effect (σm) of the C5 substituent. The C5 substituent also affects the steric and electrostatic properties of a nucleobase, which could potentially impact its interaction with the active site and its rate of excision by hTDG. These properties are given in Table 2.
hTDG activity against 5-substituted uracils
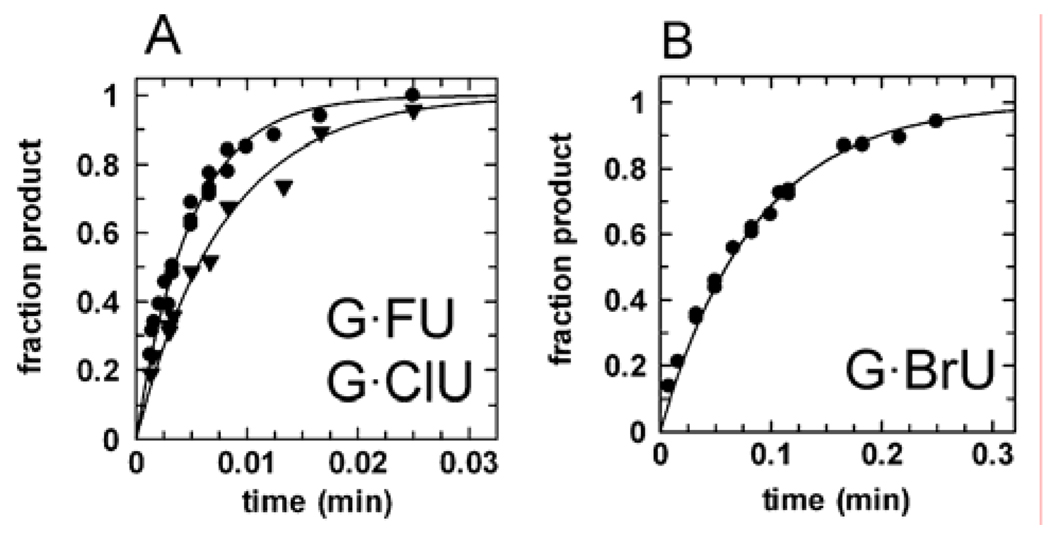
We found that 5-halogen substituents dramatically alter the excision rate of uracil by hTDG. Indeed, the activity against G·FU pairs, _k_max = 202 min−1 (Fig. 6A), is 78 times faster than for G·U pairs (Table 1). Thus, the small increase in size of FU over U (Table 2, Figure 5) does not appear to significantly impact its access to the hTDG active site. The propensity of FU to form electrostatic interactions is probably similar to that of U, because the change in polarity due to the 5-F substitution is relatively small (π, Table 2), and it has been shown that C-F groups in aromatic systems are poor hydrogen bond acceptors60 and poor halogen bond donors.61 Indeed, the enthalpy of base pairing for isolated A·U and A·FU pairs was calculated to be 12.2 and 12.6 kcal/mol at the MP2(full)/6-311+G(2d,2p)//B3LYP/6-31G* level of theory.43 Thus, the greater hTDG activity against FU can be largely attributed to the superior leaving ability of FU over U (Δp_K_aN1 = −1.33), and is consistent with the higher intrinsic reactivity of FdU compared to dU (_k_non, Table 1).
Figure 6.
Representative data for the activity of hTDG against 5-halouracils. (A) hTDG rapidly excises FU, _k_max = 202 min−1 (●), and ClU, _k_max = 126 min−1 (▼). (B) The removal of BrU is significantly slower, _k_max = 11.6 min−1, likely due to steric effects with the active site (see text).
We also discovered that hTDG is highly active against G·ClU pairs, _k_max = 126 min−1 (Figure 6A), corresponding to a 48-fold and 572-fold increase over G·U and G·T pairs, respectively (Table 1).62 This is remarkable considering that ClU is much bulkier than U and nearly isosteric with T (Figure 5, Table 2). The H bonding properties of ClU and T are probably not substantially different, because the 5-Cl and 5-CH3 substituents have a similar hydrophobic effect (π, Table 2), and previous studies have shown that the hydrophobicity of uracil was increased to the same extent by halogen (5-Br) and methyl (5-CH3) substituents.65,66 Moreover, the enthalpy of base pairing for the isolated A·T and A·ClU pairs was calculated to be 12.1 and 12.8 kcal/mol at the MP2(full)/6-311+G(2d,2p)//B3LYP/6-31G* level of theory.43,67 Thus, the 572-fold greater activity of hTDG against ClU over T can be most readily explained by the superior leaving ability of ClU compared to T (Δp_K_aN1 = −2.05), and is consistent with the higher intrinsic reactivity (_k_non) of CldU over dT (Table 1).
In contrast with FU and ClU, the limits of the hTDG active site appear to be tested by BrU and IU. The activity against G·BrU pairs, _k_max = 11.6 min−1 (Figure 6B), is significantly lower than for G·FU and G·ClU pairs, although still 53 times greater than for G·T pairs (Table 1). Given that BrU and ClU are nearly equivalent in leaving ability and have similar inherent reactivity (_k_non), the 11-fold lower hTDG activity against BrU indicates that its access to the active site is limited, presumably by steric effects (see “projection along C5-X axis”, Table 2). Consistent with this, hTDG activity against the bulky IU is sharply reduced, _k_max = 0.06 min−1 (data not shown), and is about 4-fold slower than for G·T pairs. The leaving ability of IU is similar to the other 5-halouracils, suggesting that its low reactivity is due to steric hindrance in the active site (Table 2). Indeed, IU is removed 10−3.5 times more slowly than FU, even though FU and IU have the same p_K_aN1 values, and FdU and IdU exhibit similar non-enzymatic rates (Table 1).
We also examined the activity against 5-hydroxyuracil (hoU) and 5-hydroxymethyluracil (hmU), which are similar to U in leaving ability but differ significantly in their steric and electrostatic properties (Table 2, Figure 5). The activity against G·hmU pairs, _k_max = 1.9 s−1, and G·hoU pairs, _k_max = 2.2 s−1 (data not shown) is about the same as for G·U pairs.68 This finding is consistent with indications from the results above that hTDG activity depends on glycosidic bond stability.
Cytosine analogues with improved leaving ability are substrates of hTDG
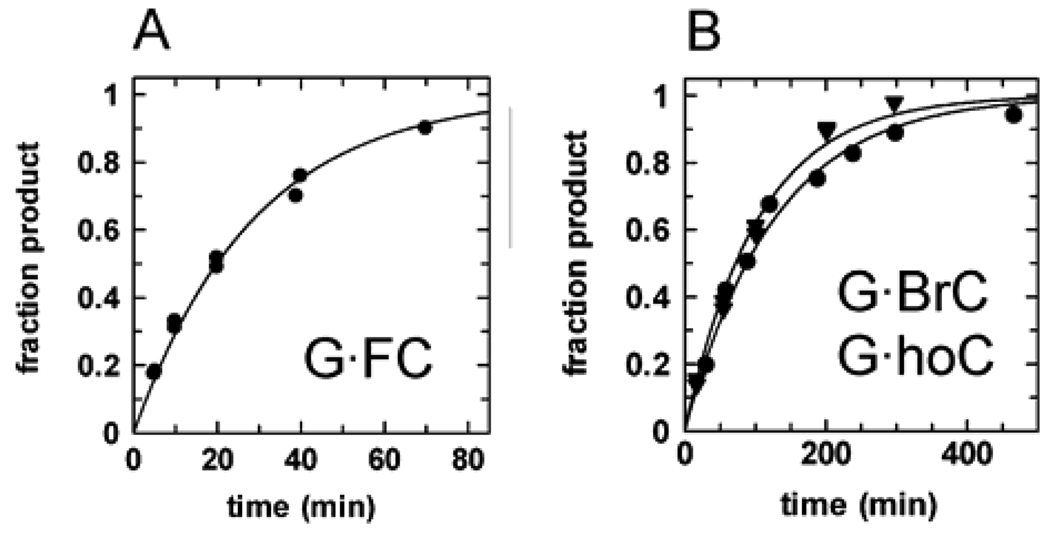
The strong specificity of hTDG for G·T over A·T pairs 9 likely arises from H bond interactions that are compatible with the Watson-Crick base pairing groups of guanine but not adenine.19 Given its specificity for removing bases that are paired with guanine, hTDG must employ a stringent mechanism to avoid the excision of cytosine from the million-fold excess of G·C pairs over G·T mispairs. Accordingly, hTDG activity against G·C pairs has not previously been observed.13,18 Nevertheless, under the saturating enzyme conditions used in this work, we were able to measure an exceedingly weak activity against G·C pairs, _k_obs = 1.2 × 10−5 min−1 (Figure S1, Supporting Information). Given the strong hTDG activity against FU and ClU (Table 1), we suspected that improving the leaving ability of cytosine (p_K_aN1 = 12.2) could substantially increase its rate of excision by hTDG. Indeed, hTDG exhibits significant activity against 5-fluorocytosine (FC), _k_max = 0.035 min−1, 5-bromocytosine (BrC), _k_max = 0.008 min−1, and 5-hydroxycytosine (hoC), _k_max = 0.010 min−1 (Figure 7). Remarkably, these cytosine analogues are removed 102.8- to 103.5-fold faster than cytosine (Table 1), consistent with their increased N1 acidities (Table 2). Although the leaving ability of BrC is better than FC and nearly the same as T, BrC is excised 4-fold and 28-fold slower than FC and T, respectively, suggesting that the access of BrC to the active site is limited, probably by steric effects (Table 2, Figure 5). The observation that hTDG excises cytosine, albeit very slowly, and readily removes cytosine analogues that are bulkier than cytosine indicates that cytosine can flip into the hTDG active site.
Figure 7.
hTDG excises cytosine analogs with improved leaving group ability. Shown are representative data from single turnover experiments for (A) the removal of FC from G·FC pairs, _k_max = 0.035 min−1, and (B) the removal of BrC from G·BrC pairs, _k_max = 0.008 min−1 (●), and hoC from G·hoC pairs, _k_max = 0.010 min−1 (▼).
As an important control, we sought to determine whether the halogen substitutions perturbed the stability of G·U and G·C pairs in the duplex substrates, because previous studies indicate that a target base which is more prone to flipping out of the duplex may be more rapidly removed by a DNA glycosylase.8,70 This could lead to an increase in _k_max unrelated to glycosidic bond stability, i.e., due to a change in _K_flip rather than _k_chem (Figure 3). To address this question, we conducted melting studies on the DNA substrates. We find that the 5-halogen substitutions do not alter the stability of the G·U or G·C base pairs in the substrates examined here (Supporting Information).
Linear free energy relationships for _N_-glycosidic bond hydrolysis
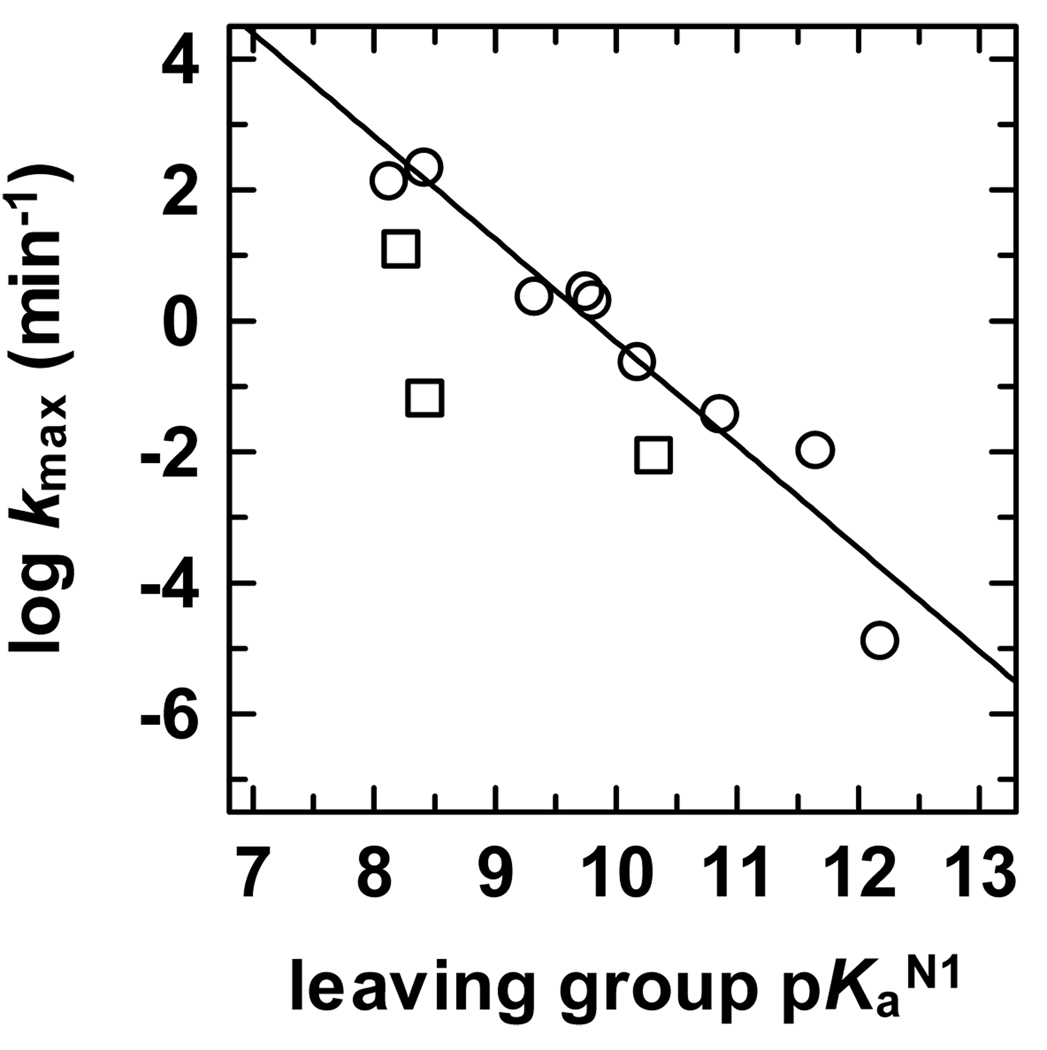
Taken together, our observations indicate a strong dependence of hTDG activity on the leaving ability of the target base, suggesting that a quantitative analysis would be informative. Indeed, a plot of log (_k_max) versus leaving group p_K_aN1 reveals a Brønsted-type linear free energy relationship (LFER) with a large negative slope of βlg = −1.6 ± 0.2 (Figure 8). The large negative slope of the LFER indicates that negative charge accumulates on the departing base in the transition state of the hTDG reaction and that the _N_-glycosidic bond is largely or perhaps fully ruptured such that the rate of C-N bond cleavage is highly sensitive to the influence of the 5-substituent, where electron-withdrawing substituents (e.g., Cl) increase the rate by stabilizing the transition state, and electron-donating groups (e.g., CH3) destabilize the transition state and slow the reaction. Thus, our findings suggest the hTDG-catalyzed reaction follows a highly dissociative or perhaps a stepwise mechanism, consistent with previous work showing that nearly all enzymatic and non-enzymatic _O_- and _N_-glycoside hydrolyses proceed through a highly dissociative ANDN (SN2) or a stepwise DN*AN (SN1) mechanism.24
Figure 8.
Brønsted-type linear free energy relationship (LFER) for the hTDG reaction. The LFER has a good correlation coefficient (r = 0.96) and a large negative slope of βlg = −1.6 ± 0.2 (Brønsted coefficient). The LFER includes data for U, T, FU, ClU, hoU, hmU, FC, hoC, and C (○) and covers a range in leaving group p_K_aN1 of >4 log units. A determination of the LFER using only the 5-substituted uracils gives essentially the same result, βlg = −1.4 ± 0.2, r = 0.96. Data for BrU, IU, and BrC are shown (□) but were not included in the LFER because the _k_max values for these bases suggests limited access to the active site (see text).
The Brønsted-type LFER shows that hTDG activity (k_max) is highly correlated with the N1 acidity of the target base in aqueous solution. However, structural studies indicate that the hTDG active site is hydrophobic,20,21 and nucleobase acidity varies with the polarity of the environment.44,56 Indeed, the N1 and N3 acidities of uracil differ by 14 kcal/mol in the gas phase,44,46,56 whereas they are nearly equivalent in aqueous solution.22 Thus, as indicated by previous studies, gas-phase acidities can be informative regarding the leaving ability of a nucleobase within a hydrophobic active site, and may be relevant to the mechanism of substrate discrimination.44,71 We therefore examined the dependence of hTDG activity on N1 acidity in a non-polar environment by calculating the gas phase N1 acidities of the nucleobases at 298 K (Δ_G_acid,g, Table S1, Supporting Information). (For comparison with other studies, the more commonly reported deprotonation enthalpies at 0 and 298 K are also given). A plot of (2.3_RT)log(_k_max) versus Δ_G_acid,g gives a LFER with a good correlation coefficient (r = 0.94) and a slope of m = −0.42 ± 0.06 (Figure S2, Supporting Information). The strong correlation of log(_k_max) with the nucleobase N1 acidities in both the gas phase (Δ_G_acid,g) and in aqueous solution (p_K_aN1) is explained by the finding that Δ_G_acid,g is highly correlated with p_K_aN1 (r = 0.98, Figure S2, Supporting Information), which is perhaps not surprising for acids of similar structure. The slope of this correlation (m = 3.7 ± 0.2) is similar to that observed previously for substituted carboxylic acids,72,73 and indicates that differences in N1 acidities are nearly four-fold greater in the gas phase than in aqueous solution. The enhanced acidity in the gas phase as compared to aqueous solution is relevant because it predicts that p_K_aN1 differences should be enhanced in a hydrophobic active site, leading to greater differences in _k_max between various substrates than would be expected for an aqueous environment. As discussed below, our findings indicate that a hydrophobic active site increases the specificity of hTDG for G·T over G·C pairs.
Discussion
Implications of the Brønsted-type LFER for the mechanism of hTDG
We found that the hTDG reaction is remarkably amenable to Brønsted-type LFER analysis (Figure 8), to our knowledge, the first such study for a DNA glycosylase. Indeed, this approach is probably not feasible for most DNA glycosylases, because a selective active site may not accommodate the perturbations in substrate size and/or polarity that can accompany changes in leaving group ability. Even the closely related eMUG is probably too restrictive for such studies, because its activity is exceedingly slow for uracil analogues with modestly sized 5-substituents such as T (5-CH3) and hmU (5-CH2OH).74 In contrast, T, hmU, and ClU are well fit by the LFERs for hTDG (Figure 8 and Figure S2, Supporting Information). Moreover, the Brønsted LFER for hTDG includes nine substrates, covers a large range in p_K_aN1 (> 4 log units), and exhibits a good correlation coefficient (r = 0.96), indicating that the βlg = −1.6 ± 0.2 accurately reflects the dependence of _k_max on the leaving ability of the target base (p_K_aN1).
It is informative to compare the results for hTDG with the non-enzymatic reaction. A plot of log(_k_non) versus p_K_aN1 for the spontaneous hydrolysis of dU, dT, and 5-Br-dU at pH 6.5 and 75 °C has a slope of βlg = −0.86 ± 0.03,22,75 and the same value (βlg = −0.86 ± 0.05) is obtained using _k_non values extrapolated to 22 °C for dU, dT, 5-F-dU, 5-Cl-dU, and 5-Br-dU (Figure S3, Supporting Information). These βlg values, together with the observed low and positive activation entropies,22 are consistent with a highly dissociative transition state for the non-enzymatic reaction,22,23,76 although it was concluded from computational studies that the mechanism is concerted (ANDN) rather than stepwise (DN*AN).77 The more negative βlg = −1.6 observed for hTDG indicates that the transition state (TS) of the enzymatic reaction is more sensitive to the development of negative charge on N1 of the leaving group base than is the TS of the non-enzymatic reaction. Thus, the βlg = −1.6 may reflect a more dissociative TS for hTDG compared to the non-enzymatic reaction. The difference in βlg may also be attributable to the hydrophobic active site of hTDG.20,21 As noted previously,78,79 a more negative βlg can be expected for a reaction occurring in an environment that is less polar than aqueous solution, where charge development in the TS of the latter is stabilized to a greater extent by solvent. Nevertheless, the large negative βlg = −1.6 implies that the hTDG reaction proceeds through a highly dissociative, perhaps stepwise mechanism. Accordingly, hTDG does not appear to have an active-site group that could serve as a general base catalyst, which would likely be needed to activate (deprotonate) the water nucleophile in a more associative reaction. Thus, our findings are consistent with previous work showing that nearly all enzymatic and non-enzymatic _O_- and _N_-glycoside hydrolyses follow a highly dissociative ANDN (SN2) or stepwise DN*AN (SN1) mechanism.24
It is of interest to consider how hTDG could stabilize a highly dissociative transition state. The precedent for enzymatic _N_-glycosidic bond hydrolysis in DNA is the reaction catalyzed by uracil DNA glycosylase (UDG), which proceeds through a stepwise mechanism involving an oxacarbenium cation-uracil anion intermediate.77,80–82 For UDG, two conserved residues each contribute about 5 kcal/mol of TS stabilization; an Asp side-chain serves to stabilize the oxacarbenium cation and activate (or position) the water nucleophile, and a His side-chain stabilizes the uracil anion leaving group via formation of a strong H bond.80,83–85 Although these catalytic groups are absent in hTDG, the enzyme still provides over 12 kcal/mol of TS stabilization for a dU substrate (Table 2) as compared to 17 kcal/mol for UDG.83 Structural studies of eMUG suggest that it and hTDG may provide some stabilization to an anionic nucleobase leaving group via H bond interactions involving backbone amides and an active-site water molecule.20 To stabilize the oxacarbenium ion, hTDG may use tactics employed by UDG, including electrostatic interactions with the anionic leaving group and with the negatively charged DNA backbone.76,77,82,86
Role of a hydrophobic active site
Our findings indicate that the hydrophobic active site of hTDG20,21 enhances the inherent differences in substrate reactivity, thereby increasing the specificity against cytosine. The potential impact of a hydrophobic active site is indicated by the highly linear plot of Δ_G_acid,g versus (2.3_RT_)p_K_aN1 (Figure S2, Supporting Information), which shows that the differences in nucleobase N1 acidity are four-fold larger in the non-polar extreme of the gas phase as compared to aqueous solution. This relationship indicates that differences in nucleobase leaving ability (p_K_aN1) should be enhanced in the hydrophobic hTDG active site, thereby leading to larger differences in _k_max. Consistent with this prediction, the Brønsted LFER for hTDG (Figure 8) shows that the dependence of log(_k_max) on p_K_aN1 is steeper (βlg = −1.6) than would be expected for an active-site environment that mimics aqueous solution (βlg = −0.86). Thus, the hydrophobic active site of hTDG increases its specificity for G·T over G·C pairs by enhancing the difference in the inherent reactivity between dT and dC. Indeed, hTDG exhibits a 104.3-fold difference in _k_max for G·T relative to G·C pairs, much larger than the 102.2-fold difference in the non-enzymatic rates (_k_non) for dT and dC (Table 1). The observed 104.3-fold _k_max difference also exceeds the 103.2-fold difference that would be predicted by the Brønsted LFER (Δp_K_aN1 = 2.0, βlg = −1.6), as indicated by the relatively poor fit of cytosine to the LFER (Figure 8). The origin of this additional specificity against C is presently unknown, although, as discussed below, our findings indicate that cytosine can flip into the active site. Given that hTDG is a promiscuous enzyme, the enhanced (20,000-fold) difference in hTDG activity for G·T versus G·C pairs, as compared to the 150-fold difference in intrinsic reactivity between dT and dC, may be essential for avoiding the danger of either excessive activity against normal G·C pairs or insufficient activity against G·T mispairs.
Our finding that a hydrophobic active site contributes to hTDG specificity by enhancing the differences in nucleobase N1 acidity is reminiscent of previous studies which found that the difference in uracil N1 and N3 acidities is much greater in the gas phase than in aqueous solution, suggesting this difference in acidities may be observed in a hydrophobic active site.44,46 Although a hydrophobic active site may be expected to slow the reaction because charge development becomes less favorable, it has been shown that the N1 site of uracil is as acidic as HCl in the gas phase. In addition, it has previously been suggested that N1-deprotonated U may be a relatively good leaving group in a non-polar active site.44,87 A similar argument can be made for thymine, which is merely 1.5 kcal/mol less acidic than U (Table S1, Supporting Information). Moreover, computational studies indicate that the gas-phase N1 acidities of U, T, FU, and hoU can be significantly increased by H bond interactions,48,50 and structural studies indicate that such interactions are provided by backbone amides and/or a water molecule in the eMUG and hTDG active sites.20
It is of interest to compare our findings with recent studies of another promiscuous DNA glycosylase, AlkA, for which the base excision rate was also found to depend on substrate reactivity.7 These studies showed that AlkA exhibits the same rate enhancement (_k_max/_k_non) for several damaged and normal purine bases.7 In contrast, we find that hTDG exhibits different rate enhancements for the various pyrimidine substrates examined here, where the rate enhancement decreases with diminishing leaving ability of the nucleobase (Table 2). This observation can be largely attributed to the hydrophobic active site of hTDG, which enhances the inherent differences in substrate reactivity.
Role of base-flipping in the specificity against cytosine
It was previously suggested that the specificity of eMUG and hTDG against cytosine may be attributable to the greater stability of G·C pairs over G·U (and G·T) mispairs, such that the enzymes are unable to flip cytosine into the active site.19 However, our observation that hTDG removes cytosine (albeit very slowly) and readily excises the larger cytosine analogues FC, BrC, and hoC, indicates that cytosine has access to the active site. Nevertheless, given that the equilibrium constant for the spontaneous opening of G·C base pairs is 3–4 orders of magnitude smaller than for G·T mispairs,76, it could be argued that greater G·C stability decreases the equilibrium for enzymatic base flipping (_K_flip, Figure 3), thereby decreasing the lifetime of cytosine in the active site. However, our observations suggest otherwise. If _K_flip was diminished for C, FC, BrC, and hoC compared to the 5-xU bases, then the _k_max values would be lower (because _k_max = _k_chem(_K_flip/1+_K_flip)) and would not provide a good fit to the LFERs (Figure 8 and Figure S2 of the Supporting Information). On the contrary, the LFERs provide no indication that the _k_max values are uniformly lower for C, FC, BrC, and hoC relative to the uracil analogues. Thus, our findings indicate that cytosine can flip into the hTDG active site, but the enzyme simply lacks the necessary catalytic power to remove cytosine at a significant rate. In marked contrast, uracil DNA glycosylase (UDG) employs steric hindrance and selective electrostatic interactions to preclude cytosine from docking in its active site2. Moreover, UDG possesses enough catalytic power to remove cytosine, as a UDG variant (N123D) designed to form H bonds with cytosine can remove cytosine at a significant rate (_k_cat = 0.02 min−1).88,89 Thus, by excluding cytosine from its active site, UDG can attain high catalytic power while avoiding the danger of aberrant cytosine excision.
Mechanism of hTDG-catalyzed excision of C and 5-xC analogues
Our results indicate that the mechanism employed by hTDG in the excision of C and 5-xC analogues differs from that of the non-enzymatic and UDG-catalyzed reactions. The non-enzymatic dC and 5BrdC hydrolyses are acid catalyzed through N3 protonation.23 The bromine substituent increases the acidity of the N1 and N3 sites, lowering the concentration of the N3-protonated species.58 Thus, dC is hydrolyzed as rapidly as 5BrdC (at 95 °C, pH 5) even though BrC has a much lower p_K_aN1 than C (Δp_K_aN1 = 1.9).23 Likewise, studies of a UDG variant (N123D) designed to remove cytosine concluded that the reaction involves protonation of cytosine N3 prior to glycosidic bond cleavage.88 In contrast, our findings indicate that the hTDG-catalyzed excision of C, FC, BrC, and hoC is not acid catalyzed. Indeed, hTDG removes FC 103.5-fold faster than C even though the population of protonated FC is expected to be 60-fold lower than protonated C (Δp_K_aN3 = 1.8).58 Similarly, hTDG removes BrC and hoC much more rapidly than C, despite the lower population of protonated BrC and hoC compared to protonated C. Consistent with our findings, hTDG does not appear to have an active-site side chain that could protonate cytosine N3.20,21 Thus, we conclude that the excision of C, FC, BrC, and hoC by hTDG is not acid catalyzed, and the base departs as the monoanion rather than the neutral species, such that the rate depends largely on p_K_aN1.
Biological relevance of hTDG activity against ClU and BrU
The observation of strong hTDG activity against ClU and BrU may have biological implications, because these lesions arise in DNA due to oxidative processes associated with inflammation.90,91 ClU and BrU can be incorporated opposite G92 to give lesions that are mutagenic, genotoxic, and cytotoxic,93,94 consistent with the correlation between chronic inflammation and carcinogenesis.95,96 Our findings raise the possibility that hTDG provides some protection against ClU and BrU lesions, and previous studies indicate that such protection is not offered by other pyrimidine-specific enzymes. Indeed, human UDG and SMUG1 (single-stranded specific monofunctional uracil-DNA glycosylase) are inactive against ClU and BrU,90,97,98 due likely to their restrictive active sites. Human MBD4 (methyl binding domain IV) reportedly removes ClU and U with similar efficiency, and is much less effective against BrU than U.99 The activity of hTDG will likely be lower for A·BrU and A·ClU pairs, given the reported 102.4-fold decrease in hTDG activity for A·U versus G·U pairs.9 Although the activity will also be somewhat lower for G·ClU and G·BrU lesions located outside of CpG sites, the activity may still be biologically relevant given the potent activity against ClU and BrU observed here (Table 2). The activity of hTDG against ClU, BrU, and other lesions located in DNA contexts other than CpG sites is under investigation.
Supplementary Material
1
Acknowledgement
We thank Dr. Yu Lin Jiang for converting 5-chloro-2'-deoxyuridine to its corresponding phosphoramidite, Dr. Primo Schär, University of Basel, for generously providing an expression plasmid for hTDG, and the reviewers for their helpful suggestions. This work was supported by grants from the NIH (GM 72711) to A.C.D and the NSF (CHE-0518262) to M.T.R, and by the University of Maryland Greenebaum Cancer Center.
Footnotes
Supporting Information Available: A figure showing the hTDG activity against G·C base pairs; experimental methods and results for melting temperature studies on the DNA substrates; a figure showing a plot of (2.3_RT_)log(k_max) versus nucleobase N1 acidity in the gas phase (Δ_G_acid,g) and a plot of Δ_G_acid,g versus(2.3_RT)p_K_aN1; figures showing Brønsted-type LFERs for non-enzymatic pyrimidine hydrolyses at 75 °C and at 22 °C; and the complete reference for Frisch, M. J., et al., Gaussian, Inc.: Pittsburgh, PA, 2003. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lindahl T. Nature. 1993 362;:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackburn GM, Tainer JA. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee A, Yang W, Karplus M, Verdine GL. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 4.Drohat AC, Kwon K, Krosky DJ, Stivers JT. Nat. Struct. Biol. 2002;9:659–664. doi: 10.1038/nsb829. [DOI] [PubMed] [Google Scholar]
- 5.Cao C, Kwon K, Jiang YL, Drohat AC, Stivers JT. J. Biol. Chem. 2003;278:48012–48020. doi: 10.1074/jbc.M307500200. [DOI] [PubMed] [Google Scholar]
- 6.Berdal KG, Johansen RF, Seeberg E. Embo. J. 1998;17:363–367. doi: 10.1093/emboj/17.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Brien PJ, Ellenberger T. J. Biol. Chem. 2004;279:26876–26884. doi: 10.1074/jbc.M403860200. [DOI] [PubMed] [Google Scholar]
- 8.O'Brien PJ, Ellenberger T. J. Biol. Chem. 2004:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 9.Waters TR, Swann PF. J. Biol. Chem. 1998;273:20007–20014. doi: 10.1074/jbc.273.32.20007. [DOI] [PubMed] [Google Scholar]
- 10.Jones PA, Baylin SB. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 11.Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- 12.Rideout WM, 3rd, Coetzee GA, Olumi AF, Jones PA. Science. 1990;249:1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- 13.Hardeland U, Bentele M, Jiricny J, Schar P. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neddermann P, Jiricny J. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu P, Burdzy A, Sowers LC. DNA Repair (Amst) 2003;2:199–210. doi: 10.1016/s1568-7864(02)00198-2. [DOI] [PubMed] [Google Scholar]
- 16.Saparbaev M, Laval J. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8508–8513. doi: 10.1073/pnas.95.15.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon JH, Iwai S, O'Connor TR, Pfeifer GP. Nucleic Acids Res. 2003;31:5399–5404. doi: 10.1093/nar/gkg730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sibghat U, Gallinari P, Xu YZ, Goodman MF, Bloom LB, Jiricny J, Day RS., 3rd Biochemistry. 1996;35:12926–12932. doi: 10.1021/bi961022u. [DOI] [PubMed] [Google Scholar]
- 19.Barrett TE, Savva R, Panayotou G, Barlow T, Brown T, Jiricny J, Pearl LH. Cell. 1998;92:117–129. doi: 10.1016/s0092-8674(00)80904-6. [DOI] [PubMed] [Google Scholar]
- 20.Barrett TE, Scharer OD, Savva R, Brown T, Jiricny J, Verdine GL, Pearl LH. Embo. J. 1999;18:6599–6609. doi: 10.1093/emboj/18.23.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baba D, Maita N, Jee J-G, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M. Nature. 2005;435:979–982. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- 22.Shapiro R, Kang S. Biochemistry. 1969;8:1806–1810. doi: 10.1021/bi00833a004. [DOI] [PubMed] [Google Scholar]
- 23.Shapiro R, Danzig M. Biochemistry. 1972;11:23–29. doi: 10.1021/bi00751a005. [DOI] [PubMed] [Google Scholar]
- 24.Berti PJ, McCann JA. Chem. Rev. 2006;106:506–555. doi: 10.1021/cr040461t. [DOI] [PubMed] [Google Scholar]
- 25.Fasman G. CRC Handbook of Biochemistry and Molecular Biology, 3rd ed. Boca Ratton, FL: CRC Press; 1975. [Google Scholar]
- 26.Hardeland U, Steinacher R, Jiricny J, Schar P. Embo. J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gill SC, von Hippel PH. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 28.Abu M, Waters TR. J. Biol. Chem. 2003;278:8739–8744. doi: 10.1074/jbc.M211084200. [DOI] [PubMed] [Google Scholar]
- 29.Leatherbarrow RJ. Staines, U.K.: Erithacus Software Ltd.; 1998. [Google Scholar]
- 30.Frisch MJ, et al. Pittsburgh, PA: Gaussian, Inc.; 2003. [Google Scholar]
- 31.Foresman J, Frisch A. Exploring Chemistry with Electronic Structure Methods: A Guide to Using Gaussian. 2nd ed. Pittsburgh: Gaussian; 1996. [Google Scholar]
- 32.Basis sets were obtained from the Extensible Computational Chemistry Environment Basis Set Database, Version 10/21/03, as developed and distributed by the Molecular Science Computing Facility, Environmental and Molecular Sciences Laboratory which is part of the Pacific Northwest Laboratory, P.O. Box 999, Richland, Washington 99352, USA, and funded by the U.S. Department of Energy. The Pacific Northwest Laboratory is a multi-program laboratory operated by Battelle Memorial Institute for the U.S. Department of Energy under contract DE-AC06-76RLO 1830.
- 33.Check CE, Faust TO, Bailey JM, Wright BJ, Gilbert TM, Sunderlin LS. J. Phys. Chem. A. 2001;105:8111–8116. [Google Scholar]
- 34.Boys SF, Bernardi R. Mol. Phys. 1979;19:553. [Google Scholar]
- 35.Van Duijneveldt FB, van Duijneveldt-van de Rijdt JGCM, van Lenthe JH. Chem. Rev. 1994;94:1873–1885. [Google Scholar]
- 36.Waters TR, Gallinari P, Jiricny J, Swann PF. J. Biol. Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
- 37.Noll DM, Gogos A, Granek JA, Clarke ND. Biochemistry. 1999;38:6374–6379. doi: 10.1021/bi990335x. [DOI] [PubMed] [Google Scholar]
- 38.McCann JA, Berti PJ. J Biol Chem. 2003;278:29587–29592. doi: 10.1074/jbc.M212474200. [DOI] [PubMed] [Google Scholar]
- 39.For the single turnover experiments under saturating enzyme conditions, _k_max = _k_chem(_K_flip/1+_K_flip),8 where _K_flip is the equilibrium constant for the base-flipping step. Our findings indicate that _k_max depends on _k_chem for most of the substrates examined, except for those with bulky 5-substituents such as BrU, IU, and BrC (see text). A direct determination of _k_chem would require stopped-flow experiments with substrates containing a fluorophore that reports on base-flipping (i.e., 2-aminopurine),40 which is beyond the scope of this work.
- 40.Stivers JT, Pankiewicz KW, Watanabe KA. Biochemistry. 1999;38:952–963. doi: 10.1021/bi9818669. [DOI] [PubMed] [Google Scholar]
- 41.Waters TR, Swann PF. Mutat. Res. 2000;462:137–147. doi: 10.1016/s1383-5742(00)00031-4. [DOI] [PubMed] [Google Scholar]
- 42.Van Schepdael A, Ossembe N, Herdewijn P, Roets E, Hoogmartens J. J. Pharm. Biomed. Anal. 1993;11:345–351. doi: 10.1016/0731-7085(93)80027-x. [DOI] [PubMed] [Google Scholar]
- 43.Yang Z, Rodgers MT. J. Am. Chem. Soc. 2004;126:16217–16226. doi: 10.1021/ja045375p. [DOI] [PubMed] [Google Scholar]
- 44.Kurinovich MA, Lee JK. J. Am. Chem. Soc. 2000;122:6258–6262. [Google Scholar]
- 45.Miller TM, Arnold ST, Viggiano AA, Miller AES. J. Phys. Chem. A. 2004;108:3439–3446. [Google Scholar]
- 46.Nguyen MT, Chandra AK, Zeegers-Huyskens T. J. Chem. Soc., Faraday Trans. 1998;94:1277–1280. [Google Scholar]
- 47.Huang YQ, Kenttamaa H. J. Phys. Chem. A. 2003;107:4893–4897. [Google Scholar]
- 48.Di Laudo M, Whittleton SR, Wetmore SD. J. Phys. Chem. A. 2003;107:10406–10413. [Google Scholar]
- 49.Chandra AK, Nguyen MT, Zeegers-Huyskens T. J. Phys. Chem. A. 1998;102:6010–6016. [Google Scholar]
- 50.Whittleton SR, Hunter KC, Wetmore SD. J. Phys. Chem. A. 2004;108:7709–7718. [Google Scholar]
- 51.Chandra AK, Nguyen MT, Zeegers-Huyskens T. J. Mol. Struct. 2000;519:1–11. [Google Scholar]
- 52.Chandra AK, Uchimaru T, Zeegers-Huyskens T. J. Mol. Struct. 2002;605:213–220. [Google Scholar]
- 53.Nakanishi K, Suzuki N, Yamazaki F. Bull. Chem. Soc. Jpn. 1961;34:53–57. [Google Scholar]
- 54.Berens K, Shugar D. Acta Biochim. Pol. 1963;10:25–48. [PubMed] [Google Scholar]
- 55.Wempen I, Fox JJ. J. Am. Chem. Soc. 1964;86:2474–2477. [Google Scholar]
- 56.Wierzchowski KL, Litonska E, Shugar D. J. Am. Chem. Soc. 1965;87:4621–4629. doi: 10.1021/ja00948a039. [DOI] [PubMed] [Google Scholar]
- 57.Hansch C, Leo A, Unger SH, Kim KH, Nikaitani D, Lien EJ. J. Med.Chem. 1973;16:1207–1216. doi: 10.1021/jm00269a003. [DOI] [PubMed] [Google Scholar]
- 58.Wempen I, Fox JJ. J. Med. Chem. 1963;122:688–693. doi: 10.1021/jm00342a013. [DOI] [PubMed] [Google Scholar]
- 59.Bondi A. J. Am. Chem. Soc. 1964;68:441–451. [Google Scholar]
- 60.Kool ET. Acc. Chem. Res. 2002;35:936–943. doi: 10.1021/ar000183u. [DOI] [PubMed] [Google Scholar]
- 61.Auffinger P, Hays FA, Westhof E, Ho PS. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16789–16794. doi: 10.1073/pnas.0407607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Halogen substitutions lower the p_K_a for both the N1 and N3 sites of uracil, raising the possibility that the 5-halouracils could be partially ionized at pH 7.5, which could lead to decreased _k_max values because the ionized base is a poor substrate. However, previous studies reported that for a G·BrU pair in a 7 bp duplex, BrU ionizes with p_K_aN3 = 8.6 and is neutral at pH 7.5.63 Similarly, for a G·FU pair in a 7 bp duplex, FU ionizes with p_K_aN3 = 8.3 and is predominantly neutral at pH 7.5.64 Nevertheless, we repeated the single turnover experiments at pH 7.0 and found that the _k_max values are the same within error (not shown). As an important control, _k_max is the same for G·U pairs at pH 7.0 and pH 7.5, where uracil is known to be neutral (p_K_aN3 = 9.7).
- 63.Sowers LC, Goodman MF, Eritja R, Kaplan B, Fazakerley GV. J. Mol. Biol. 1989;205:437–447. doi: 10.1016/0022-2836(89)90353-7. [DOI] [PubMed] [Google Scholar]
- 64.Sowers L, Eritja R, Kaplan B, Goodman M, Fazakerly G. J. Biol. Chem. 1988;263:14794–14801. [PubMed] [Google Scholar]
- 65.Shih P, Pedersen LG, Gibbs PR, Wolfenden R. J. Mol. Biol. 1998;280:421–430. doi: 10.1006/jmbi.1998.1880. [DOI] [PubMed] [Google Scholar]
- 66.Giesen DJ, Chambers CC, Cramer CJ, Truhlar DG. J. Phys. Chem. B. 1997;101:5084–5088. [Google Scholar]
- 67.Yang Z, Rodgers MT. Int. J. Mass Spectrom. 2005;241:225–242. [Google Scholar]
- 68.Ionization of the hydroxyl group of hoU, hmU, and hoC must be considered, because this would significantly increase p_K_aN1 and therefore lower _k_max. This is not a concern for the G·hmU substrate, because a p_K_aOH = 9.33 was reported for 5-hm-dU.69 The ionization of hoC in DNA can be predicted from the p_K_aOH = 8.5 reported for 5-hydroxy-dC-5'-monophosphate, and a similar value is likely for hoU in DNA.69 Nevertheless, for the hoU and hoC substrates, we collected the single turnover data at pH 6.5 to ensure that the _k_max values were not impacted by 5-OH ionization, and found the same _k_max values within experimental error.
- 69.La Francois CJ, Jang YH, Cagin T, Goddard WA, Sowers LC. Chem. Res. Toxicol. 2000;13:462–470. doi: 10.1021/tx990209u. [DOI] [PubMed] [Google Scholar]
- 70.Krosky DJ, Schwarz FP, Stivers JT. Biochemistry. 2004;43:4188–4195. doi: 10.1021/bi036303y. [DOI] [PubMed] [Google Scholar]
- 71.Sharma S, Lee JK. J. Org. Chem. 2002;67:8360–8365. doi: 10.1021/jo0204303. [DOI] [PubMed] [Google Scholar]
- 72.Wilson B, Georgiadis R, Bartmess JE. J. Am. Chem. Soc. 1991;113:1762–1766. [Google Scholar]
- 73.Kumar GA, McAllister MA. J. Am. Chem. Soc. 1998;120:3159–3165. [Google Scholar]
- 74.O'Neill RJ, Vorob'eva OV, Shahbakhti H, Zmuda E, Bhagwat AS, Baldwin GS. J. Biol. Chem. 2003 doi: 10.1074/jbc.M210860200. [DOI] [PubMed] [Google Scholar]
- 75.Shapiro and Kang measured the non-enzymatic rates (_k_non) for the hydrolysis of dU, dT, and 5-Br-dU and showed that log(_k_non) versus p_K_aN1 was highly linear.22 Although they did not report a slope, one obtains βlg = −0.99 ± 0.01 using their data, which included p_K_aN1 = 8.49 for BrU, a value they calculated based on 36% N1 deprotonated species (N1−) and 64% N3 deprotonated species (N3−), citing Wempen and Fox.55 However, Wempen and Fox actually report the opposite: 64% N1− and 36% N3− for the BrU monoanion,55 from which one obtains p_K_aN1 = 8.24 for BrU and βlg = −0.86 ± 0.03 for the non-enzymatic reaction (Supporting Information).
- 76.Stivers JT, Jiang YL. Chem. Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 77.Dinner AR, Blackburn GM, Karplus M. Nature. 2001;413:752–755. doi: 10.1038/35099587. [DOI] [PubMed] [Google Scholar]
- 78.Jencks WP. Cold Spring. Harb. Symp. Quant. Biol. 1972;36:1–11. doi: 10.1101/sqb.1972.036.01.004. [DOI] [PubMed] [Google Scholar]
- 79.Hollfelder F, Herschlag D. Biochemistry. 1995;34:12255–12264. doi: 10.1021/bi00038a021. [DOI] [PubMed] [Google Scholar]
- 80.Werner RM, Stivers JT. Biochemistry. 2000;39:14054–14064. doi: 10.1021/bi0018178. [DOI] [PubMed] [Google Scholar]
- 81.Drohat AC, Stivers JT. J. Am. Chem. Soc. 2000;122:1840–1841. [Google Scholar]
- 82.Jiang YL, Drohat AC, Ichikawa Y, Stivers JT. J. Biol. Chem. 2002;277:15385–15392. doi: 10.1074/jbc.M200634200. [DOI] [PubMed] [Google Scholar]
- 83.Drohat AC, Jagadeesh J, Ferguson E, Stivers JT. Biochemistry. 1999;38:11866–11875. doi: 10.1021/bi9910878. [DOI] [PubMed] [Google Scholar]
- 84.Drohat AC, Stivers JT. Biochemistry. 2000;39:11865–11875. doi: 10.1021/bi000922e. [DOI] [PubMed] [Google Scholar]
- 85.Dong J, Drohat AC, Stivers JT, Pankiewicz KW, Carey PR. Biochemistry. 2000;39:13241–13250. doi: 10.1021/bi001437m. [DOI] [PubMed] [Google Scholar]
- 86.Jiang YL, Ichikawa Y, Song F, Stivers JT. Biochemistry. 2003;42:1922–1929. doi: 10.1021/bi027014x. [DOI] [PubMed] [Google Scholar]
- 87.Kurinovich MA, Lee JK. J. Am. Soc. Mass Spectrom. 2002;13:985–995. doi: 10.1016/S1044-0305(02)00410-5. [DOI] [PubMed] [Google Scholar]
- 88.Kwon K, Jiang YL, Stivers JT. Chem. Biol. 2003;10:351–359. doi: 10.1016/s1074-5521(03)00077-2. [DOI] [PubMed] [Google Scholar]
- 89.Kavli B, Slupphaug G, Mol CD, Arvai AS, Peterson SB, Tainer JA, Krokan HE. Embo. J. 1996;15:3442–3447. [PMC free article] [PubMed] [Google Scholar]
- 90.Jiang Q, Blount BC, Ames BN. J. Biol. Chem. 2003;278:32834–32840. doi: 10.1074/jbc.M304021200. [DOI] [PubMed] [Google Scholar]
- 91.Henderson JP, Byun J, Takeshita J, Heinecke JW. J. Biol. Chem. 2003;278:23522–23528. doi: 10.1074/jbc.M303928200. [DOI] [PubMed] [Google Scholar]
- 92.Yu H, Eritja R, Bloom L, Goodman M. J. Biol. Chem. 1993;268:15935–15943. [PubMed] [Google Scholar]
- 93.Morris SM. Mutat. Res. 1991;258:161–188. doi: 10.1016/0165-1110(91)90007-i. [DOI] [PubMed] [Google Scholar]
- 94.Morris SM. Mutat. Res. 1993;297:39–51. doi: 10.1016/0165-1110(93)90006-9. [DOI] [PubMed] [Google Scholar]
- 95.Ames BN, Gold LS, Willett WC. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5258–5265. doi: 10.1073/pnas.92.12.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ohshima H, Tatemichi M, Sawa T. Arch. Biochem. Biophys. 2003;417:3–11. doi: 10.1016/s0003-9861(03)00283-2. [DOI] [PubMed] [Google Scholar]
- 97.Brandon ML, Mi L-J, Chaung W, Teebor G, Boorstein RJ. Mutat. Res. 2000;459:161–169. doi: 10.1016/s0921-8777(99)00061-0. [DOI] [PubMed] [Google Scholar]
- 98.Kubareva EA, Volkov EM, Vinogradova NL, Kanevsky IA, Oretskaya TS, Kuznetsova SA, Brevnov MG, Gromova ES, Nevinsky GA, Shabarova ZA. Gene. 1995;157:167–171. doi: 10.1016/0378-1119(94)00771-j. [DOI] [PubMed] [Google Scholar]
- 99.Valinluck V, Liu P, Kang JI, Jr, Burdzy A, Sowers LC. Nucl. Acids Res. 2005;33:3057–3064. doi: 10.1093/nar/gki612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1