Altered MicroRNA Expression following Traumatic Spinal Cord Injury (original) (raw)
. Author manuscript; available in PMC: 2010 Oct 1.
Abstract
MicroRNAs (miRNAs) are a novel class of small non-coding RNAs that negatively regulate gene expression at the posttranscriptional level by binding to the 3’ untranslated region of target mRNAs leading to their translational inhibition or sometimes degradation. We uncovered a previously unknown alteration in temporal expression of a large set of miRNAs following a contusive spinal cord injury (SCI) in adult rats using microarray analysis. These altered miRNAs can be classified into 3 categories: (1) up-regulation, (2) down-regulation and (3) an early up-regulation at 4 hours followed by down-regulation at 1 and 7 days post-SCI. The bioinformatics analysis indicates that the potential targets for miRNAs altered after SCI include genes encoding components that are involved in the inflammation, oxidation, and apoptosis that are known to play important roles in the pathogenesis of SCI. These findings suggest that abnormal expression of miRNAs may contribute to the pathogenesis of SCI and are potential targets for therapeutic interventions following SCI.
Keywords: spinal cord injury, microRNA, inflammation, oxidation, apoptosis, gene expression
Introduction
There are two mechanisms of damage after acute spinal cord injury (SCI): a primary mechanical injury and a secondary injury induced by multiple biological processes including extensive temporal changes in gene expression (Bareyre and Schwab 2003; Di Giovanni et al. 2003; Nesic et al. 2002). Alteration in expression of many genes has been shown to play important roles in the pathogenesis of secondary SCI (Bareyre and Schwab 2003; Di Giovanni et al. 2003; Nesic et al. 2002). Much less insight, however, has been gained into the regulatory network that establishes such altered gene expression. miRNAs are attractive candidates as upstream regulators of the secondary SCI progression because miRNAs can post-transcriptionally regulate the entire set of genes (Chan et al. 2005; Lim et al. 2005). miRNAs are endogenous, non-coding ~22nt RNA molecules that have been discovered recently as fundamental and posttranscriptional regulators of gene expression (Kosik 2006; Krichevsky 2007). Recent evidence suggests that expression of at least 20–30% of human protein-coding genes is modulated by miRNAs (Krichevsky 2007). A number of miRNAs were found in the mammalian CNS, including the brain and spinal cord, where they play key roles in neurodevelopment and are likely to be important mediators of plasticity (Bak et al. 2008; Kosik 2006; Krichevsky 2007; Miska et al. 2004). Some miRNAs have been implicated in several neurological diseases such as Tourette’s syndrome and Fragile × syndrome (Kosik 2006). Recently, several studies suggested the possibility of miRNA involvement in neurodegeneration (Bilen et al. 2006; Kim et al. 2007; Schaefer et al. 2007). To date, however, no reports are available on the expression of miRNAs after SCI. In the present study, we examined temporal expression of miRNAs in the spinal cord following contusive SCI in adult rats using microarray analysis and confirmed these results with real-time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). Our findings provide the first evidence of an altered miRNA expressional profile in the spinal cord following injury. The bioinformatics analysis suggests that the altered expression of miRNAs may contribute to the pathogenesis of secondary SCI.
Materials and Methods
Spinal cord contusion injury
Adult female SD rats (200–230g) underwent a T10 contusive spinal cord injury using an NYU impactor (10 g, 12.5 mm), as described previously (Liu et al. 2004). Control animals received laminectomy only. The rats were sacrificed at 4 hours, 1 day, or 7 days post-SCI, and a 10 mm spinal cord segment containing the injury epicenter was removed quickly for microarray and qRT-PCR analyses. All surgical interventions and postoperative animal care were performed in accordance with the Guide for the Care and Use of Laboratory Animals (National Research Council, 1996) and the Guidelines of the Indiana Institutional Animal Care and Use Committee.
Total RNA extraction
Total RNA from 10 mm-long spinal cord segments containing the injury epicenter was extracted with miRNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. RNA purity was examined by spectrophotometric determination at 260/280 and 260/230 nm.
miRNA microarray assay and analysis
Microarray assay for mature miRNAs was performed by the LC Sciences Microarray Service (LC Sciences, Houston, TX). Briefly, total RNA (5 µg) was size fractionated (<300 nucleotides) by using a YM-100 Microcon centrifugal filter (Millipore) and the small RNAs (< 300 nt) isolated were 3’-extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was then ligated to the poly(A) tail for later fluorescent dye staining. Dye switching was done for duplicates to eliminate potential dye bias.
Hybridization was performed overnight on a µParaflo microfluidic chip using a microcirculation pump (Atactic Technologies). On the microfluidic chip, each detection probe consisted of a chemically modified nucleotide coding segment complementary to target all 350 mature rat miRNAs sequences from miRBase version 11.0 (Sanger Institute, Cambridge, U.K.; http://microrna.sanger.ac.uk/sequences). After RNA hybridization, tag-conjugating Cy3 and Cy5 dyes were circulated through the microfluidic chip for dye staining. Fluorescence images were collected using a laser scanner (GenePix 4000B, Molecular Device) and digitized using Array-Pro image analysis software (Media Cybernetics).
Data were analyzed by first subtracting the background and then normalizing the signals using a LOWESS filter (Locally-Weighted Regression) for two color experiments. Transcripts were determined as detectable if their signal intensity was higher than 3 times the background standard deviation, spot CV (standard deviation/signal intensity) was <0.5, and transcripts had at least 50% of replicate probe signals registering above the detection level. _p_-values of the ANOVA were calculated. The miRNAs with _p_ values <0.05 and signal intensity > 500 were selected for cluster analysis according to a hierarchical method, which was performed with average linkage and Euclidean distance metrics. The clustering plot was generated using TIGR MeV (Multiple Experimental Viewer) software from the Institute for Genomic Research.
Rea-time qRT-PCR
Real-time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) was performed by the LC Sciences Service (LC Sciences, Houston, TX). Briefly, 10 ng of total RNA from each sample was reverse-transcribed to cDNA using TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) and miRNA-specific primers (Applied Biosystems) for four down-regulated (miR-137, miR-181a, miR-219-2-3p and miR-7a) and one up-regulated miRNA species (miR-21). Each group had five samples: three from the microarray studies and two from additional samples not used in the microarray studies. U87 and 4.5S were used as endogenous controls. A no-template control was used as a negative control. Critical threshold (CT) values were determined using ABI Prism 7000 SDS Software (Applied Biosystems). The relative quantity of each miRNA to U87 was described using 2−ΔCT, where ΔCT=(CT miRNA-CT U87).
Statistical analysis
All data are presented as mean ± SD. One-way ANOVA with Tukey HSD post hoc t-tests were used to determine levels of statistical significance. A p value of < 0.05 was considered statistically significant.
Results
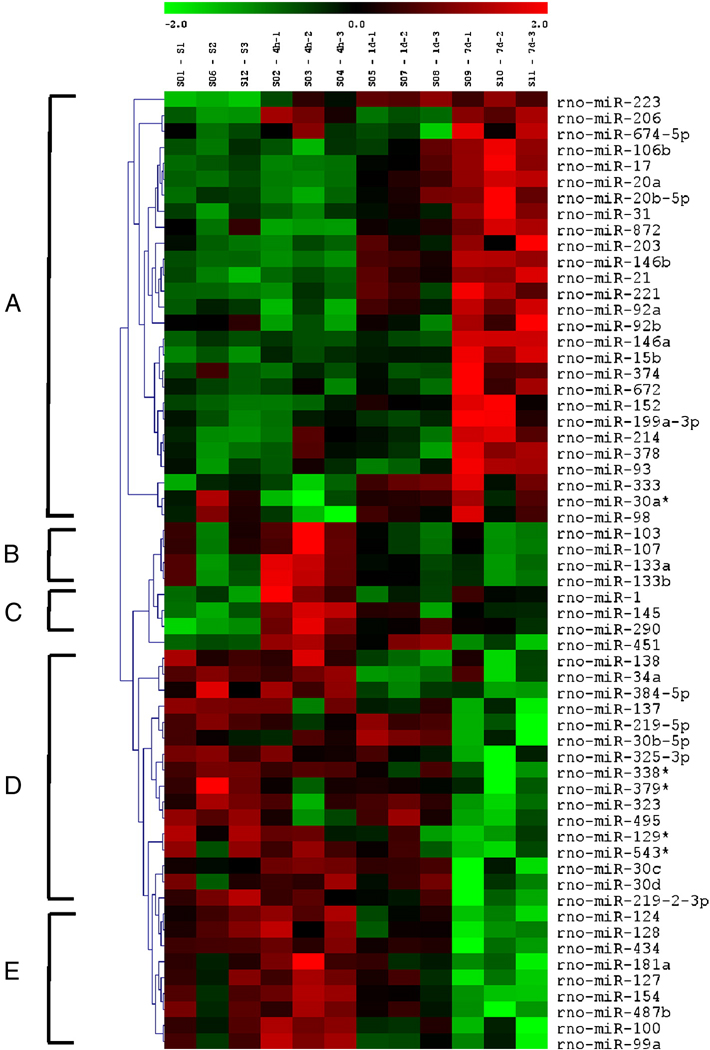
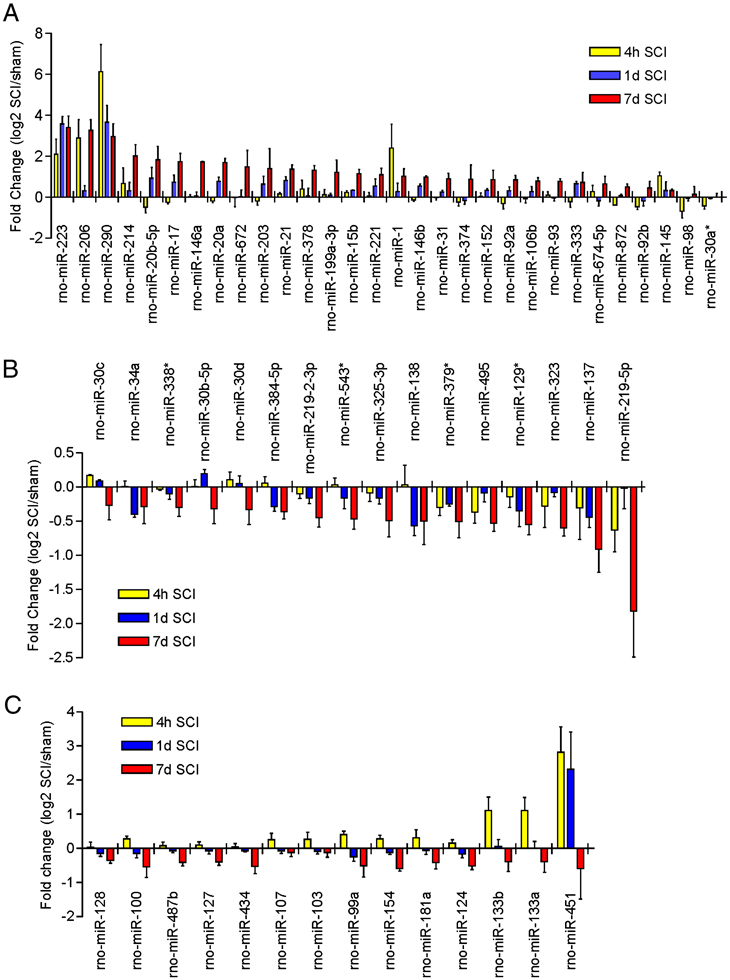
We examined expression of 350 _Rattus norvegicus_-miRNAs based on Version 11.0 of the Sanger miRBase (Sanger Institute, Cambridge, U.K.; http://microrna.sanger.ac.uk/sequences) in the injured spinal cords at 4 hours, 1 and 7 days after acute SCI. Microarray analysis revealed that 269 of 350 miRNAs were detected in the adult rat spinal cord. Based on signal intensity of the miRNA expression, levels of miRNAs expressed in adult rat spinal cords were divided into four categories: low level (intensity<500), moderate level (500–4999), high level (5000–9999) and the highest level (>1000). Among all expressed miRNAs, 36 were expressed at the highest level in the normal spinal cord (Supplementary Table 1). After SCI, 97 of 269 miRNAs showed significant expressional changes (ANOVA, p<0.05). Of those, 60 miRNAs were expressed greater than the low level, i.e. between the moderate and highest levels (Supplementary Table 2). These miRNAs can be classified into five distinct clusters by hierarchical clustering analysis (A–E; Fig. 1). Clusters A and C contain up-regulated miRNAs (Fig. 2A), cluster D contains down-regulated miRNAs (Fig. 2B), and clusters B and E contain miRNAs that were up-regulated at 4 hour post-SCI and then down-regulated at 1 and 7 days (Fig. 2C). The remaining 37 miRNAs were expressed at low level and were significantly deregulated after SCI (Supplementary Table 3). The expression of 89 miRNAs above the low level and 83 miRNAs at the low level were unaffected by SCI (Supplementary Table 4 and 5).
Figure 1.
Heat map shows significant expressional changes of 60 miRNAs with levels of intensity >500 in rats that received a contusive SCI and were sacrificed at 4 hours, 1 day, and 7 days post-injury as compared to the sham-operated control (n=3/group; S1–3, sham; 4h 1–3, 4h post-SCI; 1d 1–3, 1d post-SCI; 7d 1–3, 7d post-SCI). The color green indicates down-regulation and red up-regulation. The contusive SCI model, the microarray assay and analysis are provided in the supplementary methods online.
Figure 2.
Bar graphs show significant expressional changes of 60 miRNAs at 4 hour, 1 day and 7 days post-SCI with levels of intensity >500 (ANOVA, p<0.05; n=3/group). A. miRNAs significantly up-regulated following SCI. B. miRNAs significantly down-regulated following SCI. C. miRNAs significantly up-regulated at 4 hours and then down-regulated at 1 and 7 days post-SCI.
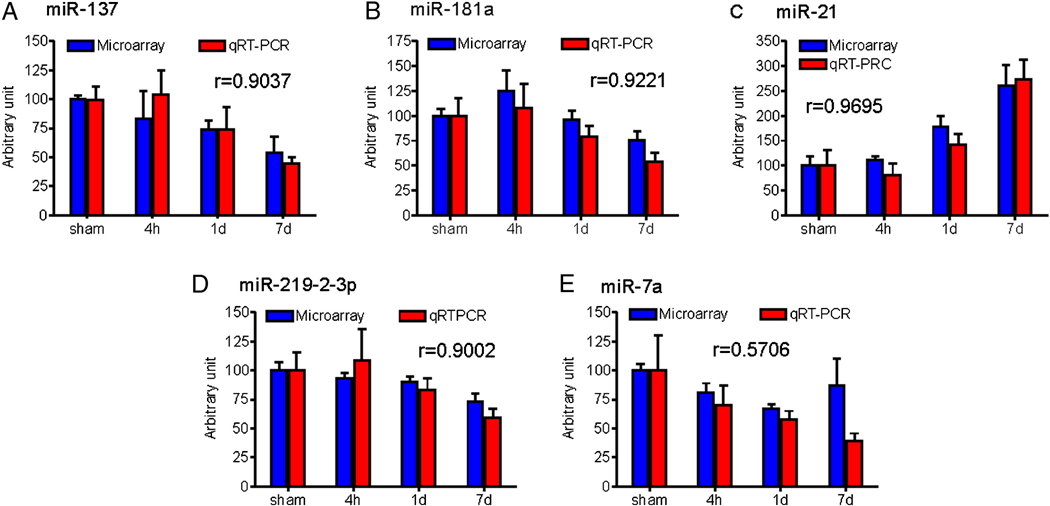
To validate the microarray platform, we assessed the expression of a subset of miRNAs including four down-regulated (miR-137, miR-181a, miR-219-2-3p and miR-7a) and one up-regulated (miR-21) miRNAs by real time qRT-PCR. A strong correlation between our microarray profiling and real-time RT-PCR data was found (Fig. 3), suggesting that the microarray data were reliable to warrant further analysis.
Figure 3.
Real time qRT-PCR validation shows a strong correlation between the microarray and real-time qRT-PCR data. r = correlation coefficient (n=3/group for microarray, n=5/group for qRT-PCR).
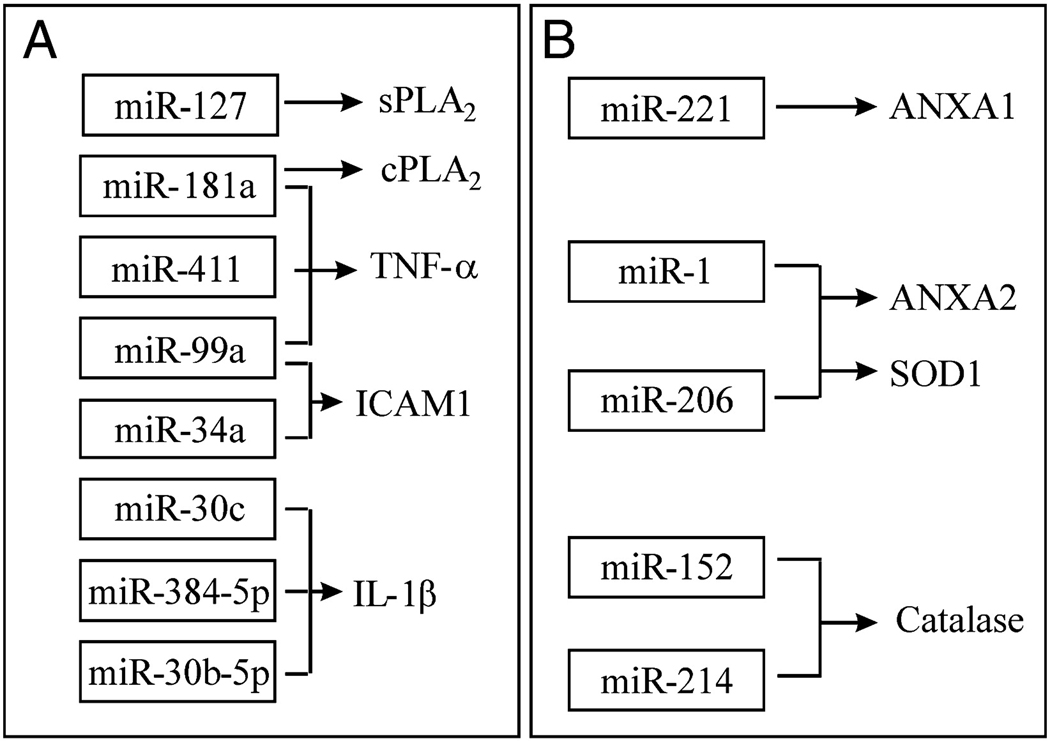
To analyze the role of miRNAs following SCI, potential downstream targets for miRNAs altered after SCI, predicted by miRanda, were retrieved from the Sanger miR Database. The bioinformatics analysis revealed that some inflammatory mediator mRNAs such as tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β) and intercellular adhesion molecule 1 (ICAM1) mRNAs were potential targets of miR-181a, miR-411, miR-99a, miR-34a, miR-30c, miR-384-5p and miR-30b-5p which were down-regulated after SCI. Conversely, some anti-inflammatory mRNAs such as annexin A1 and annexin A2 mRNAs were potential targets of miR-221 and miR-1 respectively, which were up-regulated after SCI (Fig. 4A & B). Cytosolic phospholipases A2 (cPLA2) and secretory PLA2 (sPLA2) mRNAs were potential targets of miR-181a and miR-127, respectively, which were down-regulated after SCI (Fig. 4A). Recent evidence showed that SCI-induced PLA2 activation may play an important role in the pathogenesis of secondary SCI (Liu et al. 2006). Several up-regulated miRNAs after SCI, such as miR-1, miR-206, miR-152 and miR-214, are upstream to some anti-oxidant genes such as SOD1 and catalase genes (Fig. 4B).
Figure 4.
The potential targets of altered miRNAs following SCI. A, Several inflammatory genes are potential targets of down-regulated miRNAs after SCI. sPLA2: secretary phospholipase A2; cPLA2: cytosolic phospholipase A2; TNF-α: Tumor necrosis factor-α; IL-1β: Interleukin-1β; ICAM1: Intercellular adhesion molecule 1. B, Several anti-inflammatory or anti-oxidative genes are potential targets of up-regulated miRNAs after SCI. ANXA1: annexin A1; ANXA2: annexin A2; SOD1: Superoxide dismutase 1.
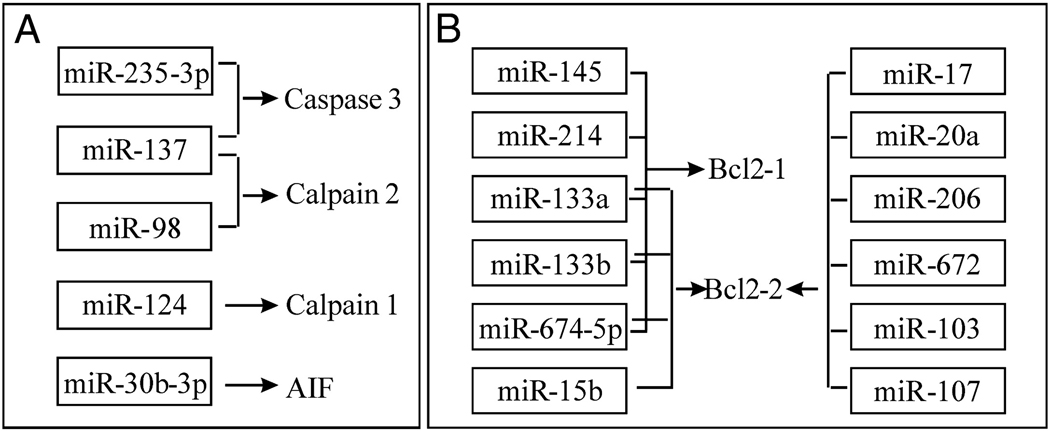
In recent years, apoptosis has been identified as an important mechanism of cell death in many neurological disorders including SCI (Beattie et al. 2000; Crowe et al. 1997; Liu et al. 1997). Apoptotic genes such as caspase-3, calpain 1, calpain 2 and apoptosis inducing factor (AIF) were potential targets of several miRNAs such as miR-235-3p, miR-137, miR-98, miR-124 and miR-30b-3p, which were down-regulated after SCI. However, some anti-apoptotic genes such as Bcl2-1 and Bcl2-2 were potential targets of several miRNAs such as miR-145, miR-214, miR-133a, miR-133b, miR-674-5p, miR-15b, miR-17, miR-20a, miR206, miR-672, miR-103 and miR-107, which were up-regulated after SCI (Fig. 5A & B).
Figure 5.
The potential targets of altered miRNAs following SCI. A, Several apoptotic genes are potential targets of down-regulated miRNAs after SCI. AIF: Apoptosis inducing factor. B, Several anti-apoptotic genes are potential targets of up-regulated miRNAs after SCI.
Discussion
To our knowledge, this is the first study demonstrating the miRNA expression profile after traumatic SCI in adult rats. Real time qRT-PCR analysis verified the results of the microarray study and showed that the microarray data were consistent and reliable. We demonstrated that ≈77% of the identified rat mature miRNAs were expressed in the adult rat spinal cord, suggesting that the rat spinal cord is a rich source of miRNA expression, which is consistent with a previous report showing the expression of a large number of miRNAs in the spinal cord of adult mice (Bak et al. 2008). We then demonstrated that a large set of miRNAs were significantly deregulated after SCI. Among the 60 miRNAs that showed expressional changes greater than the low level, 30 were up-regulated, 16 down-regulated, and 14 up-regulated at 4 hours post-SCI and then subsequently down-regulated at 1 and 7 days (Fig. 1 & 2). These altered miRNAs with a diversity of functions may affect a large number of neuronal genes (Kim et al. 2004; Krichevsky et al. 2003; Lagos-Quintana et al. 2002).
Demonstrating a large number of miRNA expressional changes following acute spinal cord injury is an important first step in understanding the underlying molecular mechanism of injury. However, the pathophysiological relevance of deregulation of these miRNAs after traumatic SCI remains to be determined. Using a bioinformatics approach, we analyzed potential targets for miRNAs that were altered after SCI. The bioinformatics analysis indicates that the potential targets for these miRNAs include genes encoding components that are involved in many pathophysiological processes such as inflammation, oxidation and apoptosis following SCI. For example, several miRNAs down-regulated after SCI are upstream to several inflammatory (Fig. 4A) and apoptotic (Fig. 5A) genes whereas other miRNAs up-regulated after SCI are upstream to several anti-inflammatory, anti-oxidative and anti-apoptotic genes (Fig. 4B & 5B). These findings suggest that abnormal expression of miRNAs following SCI may contribute to the pathogenesis of secondary SCI, and could be potential targets for therapeutic intervention following SCI.
Although the roles of miRNAs in human diseases including that of the SCI are to be elucidated, increasing evidence suggests that these miRNAs represent a new class of drug target (Esau and Monia 2007; Mirnezami et al. 2009; Weiler et al. 2006). miRNAs reduce steady state protein levels for the targeted genes by posttranscriptional regulation (Kosik 2006; Krichevsky 2007). Inhibition of a particular miRNA linked to a particular disease may remove the blockade against expression of a therapeutic protein. Conversely, administration of a miRNA mimetic may boost an endogenous miRNA population which in turn represses a detrimental gene. Taking advantage of their small size and the current knowledge of miRNA biogenesis, modified RNAs can be transiently delivered as a synthetic, pre-processed miRNA or anti-miRNA oligonucleotides (Medina and Slack 2009). Several studies have shown the potential of customized miRNA inhibitors to target specific pathologic miRNAs in vitro, and more importantly, in vivo (Medina and Slack 2009; Mirnezami et al. 2009).
In conclusion, our study has provided the first evidence of altered miRNA expression following SCI. To elucidate the role of miRNAs in SCI, additional studies are required to investigate the function and targets of these miRNAs. Experiments along these lines are currently in progress in our laboratory.
Supplementary Material
1
ACKNOWLEDGEMENTS
This work was supported by NIH NS36350, NS52290, NS50243, Mari Hulman George Endowment Funds and the State of Indiana (Grant # 91910 and 91913).
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version.
References
- Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14(3):432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bareyre FM, Schwab ME. Inflammation, degeneration and regeneration in the injured spinal cord: insights from DNA microarrays. Trends Neurosci. 2003;26(10):555–563. doi: 10.1016/j.tins.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Beattie MS, Farooqui AA, Bresnahan JC. Review of current evidence for apoptosis after spinal cord injury. J Neurotrauma. 2000;17(10):915–925. doi: 10.1089/neu.2000.17.915. [DOI] [PubMed] [Google Scholar]
- Bilen J, Liu N, Burnett BG, Pittman RN, Bonini NM. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol Cell. 2006;24(1):157–163. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- Chan CS, Elemento O, Tavazoie S. Revealing posttranscriptional regulatory elements through network-level conservation. PLoS Comput Biol. 2005;1(7):e69. doi: 10.1371/journal.pcbi.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe MJ, Bresnahan JC, Shuman SL, Masters JN, Beattie MS. Apoptosis and delayed degeneration after spinal cord injury in rats and monkeys. Nat Med. 1997;3(1):73–76. doi: 10.1038/nm0197-73. [DOI] [PubMed] [Google Scholar]
- Di Giovanni S, Knoblach SM, Brandoli C, Aden SA, Hoffman EP, Faden AI. Gene profiling in spinal cord injury shows role of cell cycle in neuronal death. Ann Neurol. 2003;53(4):454–468. doi: 10.1002/ana.10472. [DOI] [PubMed] [Google Scholar]
- Esau CC, Monia BP. Therapeutic potential for microRNAs. Adv Drug Deliv Rev. 2007;59(2–3):101–114. doi: 10.1016/j.addr.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317(5842):1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci U S A. 2004;101(1):360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7(12):911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM. MicroRNA profiling: from dark matter to white matter, or identifying new players in neurobiology. ScientificWorldJournal. 2007;7:155–166. doi: 10.1100/tsw.2007.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9(10):1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu N, Han S, Lu PH, Xu XM. Upregulation of annexins I, II, and V after traumatic spinal cord injury in adult rats. J Neurosci Res. 2004;77(3):391–401. doi: 10.1002/jnr.20167. [DOI] [PubMed] [Google Scholar]
- Liu NK, Zhang YP, Titsworth WL, Jiang X, Han S, Lu PH, Shields CB, Xu XM. A novel role of phospholipase A2 in mediating spinal cord secondary injury. Ann Neurol. 2006;59(4):606–619. doi: 10.1002/ana.20798. [DOI] [PubMed] [Google Scholar]
- Liu XZ, Xu XM, Hu R, Du C, Zhang SX, McDonald JW, Dong HX, Wu YJ, Fan GS, Jacquin MF, Hsu CY, Choi DW. Neuronal and glial apoptosis after traumatic spinal cord injury. J Neurosci. 1997;17(14):5395–5406. doi: 10.1523/JNEUROSCI.17-14-05395.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina PP, Slack FJ. Inhibiting microRNA function in vivo. Nat Methods. 2009;6(1):37–38. doi: 10.1038/nmeth0109-37. [DOI] [PubMed] [Google Scholar]
- Mirnezami AH, Pickard K, Zhang L, Primrose JN, Packham G. MicroRNAs: key players in carcinogenesis and novel therapeutic targets. Eur J Surg Oncol. 2009;35(4):339–347. doi: 10.1016/j.ejso.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5(9):R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic O, Svrakic NM, Xu GY, McAdoo D, Westlund KN, Hulsebosch CE, Ye Z, Galante A, Soteropoulos P, Tolias P, Young W, Hart RP, Perez-Polo JR. DNA microarray analysis of the contused spinal cord: effect of NMDA receptor inhibition. J Neurosci Res. 2002;68(4):406–423. doi: 10.1002/jnr.10171. [DOI] [PubMed] [Google Scholar]
- Schaefer A, O'Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204(7):1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13(6):496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1