Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection (original) (raw)
Abstract
In mammals, microRNAs (miRNAs) can play diverse roles in viral infection through their capacity to regulate both host and viral genes. Recent reports have demonstrated that specific miRNAs change in expression level upon infection and can impact viral production and infectivity. It is clear that miRNAs are an integral component of viral–host interactions, and it is likely that both host and virus contain mechanisms to regulate miRNA expression and/or activity. To date, little is known about the mechanisms by which miRNAs are regulated in viral infection. Here we report the rapid down-regulation of miR-27a in multiple mouse cell lines as well as primary macrophages upon infection with the murine cytomegalovirus. Down-regulation of miR-27a occurs independently from two other miRNAs, miR-23a and miR-24, located within the same genomic cluster, and analysis of pri-miRNA levels suggest that regulation occurs post-transcriptionally. miR-27b, a close homolog of miR-27a (20/21 nucleotide identity), also decreases upon infection, and we demonstrate that both miR-27a and miR-27b exert an antiviral function upon over-expression. Drug sensitivity experiments suggest that virus entry is not sufficient to induce the down-regulation of miR-27 and that the mechanism requires synthesis of RNA. Altogether, our findings indicate that miR-27a and miR-27b have antiviral activity against MCMV, and that either the virus or the host encodes molecule(s) for regulating miR-27 accumulation, most likely by inducing the rapid decay of the mature species.
Keywords: microRNA, RNA silencing, small RNA profiling, herpesvirus, mouse cytomegalovirus, regulation, RNA processing
INTRODUCTION
RNA silencing is a widespread phenomenon conserved in all eukaryotes from the unicellular fungi to human. Small noncoding RNAs, 19–30 nucleotides (nt) in size, are invariably found at the core of all RNA silencing pathways (Farazi et al. 2008). They exert their activity by directing protein complexes to the nucleic acid targets, enabling various modes of regulation. In mammals, miRNAs constitute an important and diverse class of noncoding RNA (Ambros 2004; Bartel 2004) with more than 700 and 540 miRNA genes identified in human and mouse, respectively (Griffiths-Jones et al. 2008). Their biogenesis involves several processing steps, starting with a large primary transcript and ending with a ∼22-nt double-stranded RNA; the functional strand from this duplex is then incorporated into an RNA-induced silencing complex (RISC) that invariably contains a member of the Argonaute protein family (for review, see Bartel 2004; Meister and Tuschl 2004; Du and Zamore 2005; Kim 2005; Okamura et al. 2008). Once loaded, the mature miRNA remains remarkably stable with a miRNA half-life in the range of days (Gatfield et al. 2009). The other strand, also known as the star sequence, or “miRNA*,” is usually degraded. The active RISC is directed toward its messenger RNA target to regulate, predominantly negatively, translation of the message (for review, see Pillai et al. 2007; Eulalio et al. 2008).
In addition to their regulatory roles in diverse biological pathways, miRNAs have also been associated with viral infections. The most striking association between viruses and miRNAs is the usurpation of miRNAs themselves by some mammalian viruses, such as herpesviruses or polyomaviruses (Cullen 2006; Pfeffer 2008). In this case, miRNAs of viral origin are beneficial for the viruses and contribute, in a nonimmunogenic manner, to the establishment of ideal conditions for the infection to occur. At the other range of the spectrum, unbeneficial interactions between cellular miRNAs and viruses have also been reported. Here, miRNAs of cellular origin can negatively impact viral infection either by targeting the viral genome itself, or by regulating cellular genes essential for the replication cycle. Thus, the primate foamy retrovirus is negatively regulated by miR-32 (Lecellier et al. 2005), vesicular stomatitis virus (VSV) is targeted by miR-24 and miR-93 (Otsuka et al. 2007), and HIV is targeted by miR-29 (Ahluwalia et al. 2008; Nathans et al. 2009). This antiviral activity of miRNAs can be perhaps paralleled to the more general antiviral role of RNA silencing in other phyla such as plant and insect organisms (Ding and Voinnet 2007). On the other hand, the down-regulation of viral genes by host miRNAs may in fact be a strategy employed by viruses in order to evade detection by the host and establish persistence (Pedersen et al. 2007; Mahajan et al. 2009).
Although extensive focus has been placed on uncovering the mechanisms by which miRNAs regulate their mRNA targets, there is a contrasting lack of information on mechanisms by which miRNAs themselves are regulated. This regulation can take place at several different steps (for review, see Winter et al. 2009): transcription of the primary transcript, processing of the pri- and pre-miRNAs, or stability of the miRNA. Hence, some miRNA primary transcripts are known to be under the control of classic transcription factors such as E2F1 (Woods et al. 2007) or p53 (He et al. 2007). Other miRNAs require auxiliary factors for their biogenesis, and many miRNAs have evolutionary conserved loops that might dictate their regulation (Michlewski et al. 2008). In line with this, the precursor of Let-7 has been shown to recruit the Lin-28 protein through interactions with the Let-7 loop, leading to the pre-miRNA polyuridylation, which in turn prevents its efficient processing (Heo et al. 2008; Rybak et al. 2008). Additionally, differential tissue-specific accumulation of pre- and mature miR-138, a brain-specific miRNA, suggests that processing of this miRNA may be inhibited under certain conditions (Obernosterer et al. 2006). It is likely that many more regulatory mechanisms will emerge. For example, recent reports suggest that exoribonucleases modulate miRNA turnover in Arabidopsis (Ramachandran and Chen 2008) and in C. elegans (Chatterjee and Grosshans 2009). To our knowledge there are no reports so far demonstrating regulation at the level of mature miRNA stability in mammalian systems.
We previously showed that mouse cytomegalovirus (MCMV) encodes a total of 18 pre-miRNAs that are abundantly expressed during lytic infection (Buck et al. 2007; Dölken et al. 2007). Here, we report on the modulation of cellular miRNA profiles by lytic MCMV infection. Despite the high level of expression of viral miRNAs during lytic infection, the levels of most cellular miRNAs are only modestly altered. In contrast, we identified one miRNA, miR-27a, which is rapidly down-regulated. MiR-23a and miR-24, two miRNAs clustered within 400 bp of miR-27a, are not affected by MCMV infection, and analysis of the pri-miRNA(s) suggests that regulation of mir-27a occurs post-transcriptionally. miR-27b, which differs from miR-27a by only one nucleotide, is similarly down-regulated and overexpression of either miR-27a or miR-27b results in a log-fold defect in viral replication, suggesting an antiviral function of these miRNAs. Drug sensitivity experiments further show a dependency on RNA synthesis for miR-27 down-regulation. The results presented here suggest that either the virus or the host encodes molecule(s) for regulating miR-27 accumulation, most likely at the level of mature miRNA stability.
RESULTS AND DISCUSSION
Down-regulation of miR-27a in murine cytomegalovirus infection
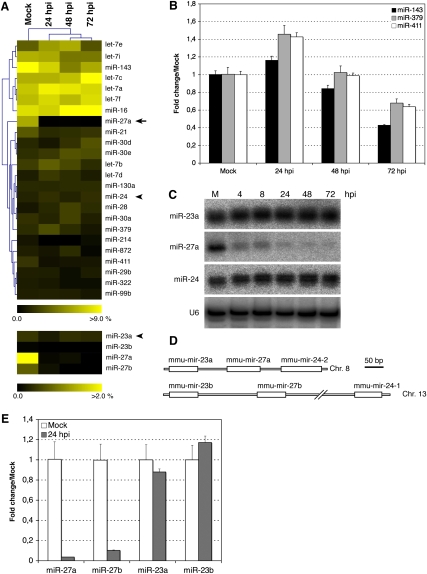
Many studies have demonstrated changes in host miRNA expression levels in response to viral infection (for review, see Gottwein and Cullen 2008; Ghosh et al. 2009). Depending on the kinetics of viral replication, the miRNA composition of infected cells could influence the ability of a virus to replicate or spread, since many pathways (cell cycle, apoptosis, immune response) are modulated by miRNAs. Manipulation of the host miRNome could therefore be used by either virus or host as part of their evolutionary strategies for survival (Obbard et al. 2009). In the lytic stage of MCMV infection, abundant viral-encoded miRNAs are expressed that could theoretically alter, or saturate, the host miRNA composition. We previously reported that virus-encoded miRNAs dominate the small RNA content of NIH-3T3 fibroblasts infected with murine cytomegalovirus (35% of clones at 24 hours post-infection [hpi] and 60% of clones at 72 hpi) (Dölken et al. 2007). If viral-encoded miRNAs were to saturate components of the miRNA machinery, this could lead to a global impact on host miRNA content. Supplemental Table 1 is provided, which details all changes in host miRNAs upon MCMV infection of NIH-3T3 cells. Analysis of the top 25 most abundant mouse miRNAs in these cells (which altogether represent 80% of cloned miRNAs) indicates that the global profile of cellular miRNAs is not dramatically altered upon MCMV infection (Fig. 1A, top panel). Quantitative real-time PCR (qRT-PCR) analysis confirms that some of the miRNAs display subtle changes and fluctuations over the time course of the infection (Fig. 1B). Qualitatively, the results obtained from cloning versus qRT-PCR display the same trends (given the technical aspects of both approaches and the low number of clones analyzed, we would expect a certain level of variability). In addition to the observed fluctuations, one glaring observation is the down-regulation of miR-27a, which represents 6% of all cloned miRNAs in mock-infected cells, but only 0.1% after 24 h of infection (Fig. 1A, arrow). MiR-27a is located on chromosome 8 and is proximal to two other miRNAs, miR-23a and miR-24-2. Neither miR-24 nor miR-23a clone numbers display such a substantial drop-off upon infection (Fig. 1A, arrowheads; Supplemental Table 1). To confirm the specific down-regulation of miR-27a, we analyzed the expression levels of miR-23a, miR-27a, and miR-24 at various times after infection using Northern blot. MiR-27a expression levels drop rapidly by 4 h and 8 h post-infection (hpi) to become barely detectable after 24 hpi, while the levels of miR-24 and miR-23a do not significantly change (Fig. 1C). Another locus on chromosome 13 encodes homologs of the three miRNAs: miR-23b, miR-27b, and miR-24-1 (Fig. 1D). There is a 1-nt difference in the 3′ end of the mature “a” and “b” forms of miR-27 and miR-23 that allows them to be distinguishable by cloning (the mature miR-24 sequence is identical in both clusters). As shown in the bottom panel of Figure 1A and in Supplemental Table 1, miR-27b and miR-23b were cloned in fewer numbers than the miR-27a and miR-23a counterparts in these cells, yet the pattern of down-regulation of the miR-27 species is preserved. Since the cloning numbers in this study are not sufficient to enable statistical conclusions, we validated this observation with qRT-PCR. Using synthetic oligonucleotides, qRT-PCR conditions were optimized to distinguish the “a” and “b” forms of miR-27 and miR-23 (Materials and Methods). Figure 1E demonstrates the down-regulation of both miR-27 species upon infection, while both miR-23 species are unaltered. Based on the above results, we would postulate that down-regulation of miR-27 occurs after the production of the primary transcript. However, we cannot rule out the possibility that miR-27 derives from a pri-miRNA that is regulated independently of miR-23 and miR-24. Indeed, Sun et al. (2009) have characterized the primary transcript for miR-23b, miR-27b, and miR-24-1 and reported that three different pri-miRNAs could co-exist: one containing all three miRNAs, one containing only miR-27b, and one with only miR-24-1. However, there is no evidence for independent pri-miRNAs in the miR-23a–27a–24-2 cluster, and the miRNAs are arranged in closer proximity to one another compared with the miR-23b–27b–24-1 cluster (Fig. 1D). It seems unlikely that miR-27a is expressed from its own primary transcript, since no potential poly(A) binding site could be identified inbetween miR-27a and miR-24-2, while one was found roughly 700-nt downstream from miR-24-2 (data not shown). It also seems probable that regulation occurs at the level of the mature miRNA species, since both miR-27a and miR-27b are impacted (explored further below). In the remainder of this work we will use miR-27 to refer to both miR-27a and miR-27b.
FIGURE 1.
miR-27a and miR-27b are strongly down-regulated upon MCMV infection in NIH-3T3 cells. (A) (Top) Top 25 host miRNAs cloned in mock and infected NIH-3T3 cells, (bottom) miR-23 and miR-27 isoforms. For each miRNA, the percentage from total cellular miRNAs is displayed as a heat-map, miR-27a is indicated by an arrow, and miR-24 and miR-23b are indicated by an arrowhead. (B) Real-time PCR quantification of three miRNAs whose expression levels are modestly impacted by MCMV infection. The fold changes relative to mock-infected cells are displayed. (C) Northern blot analysis of miR-23a, miR-27a, and miR-24 accumulation during MCMV infection (MOI = 1 with centrifugal enhancement), (M) mock infected, (hpi) hours post-infection. (D) Schematic representation of miR-23a/27a/24-2 and miR-23b/27b/24-1 clusters on chromosomes 8 and 13; the position of the predicted pre-miRNAs are indicated by white rectangles. (E) Quantitative real-time PCR analysis of miR-27a and miR-27b and miR-23a and miR-23b in mock and infected NIH-3T3 fibroblast cells (n = 3).
Functional implications of miR-27 down-regulation
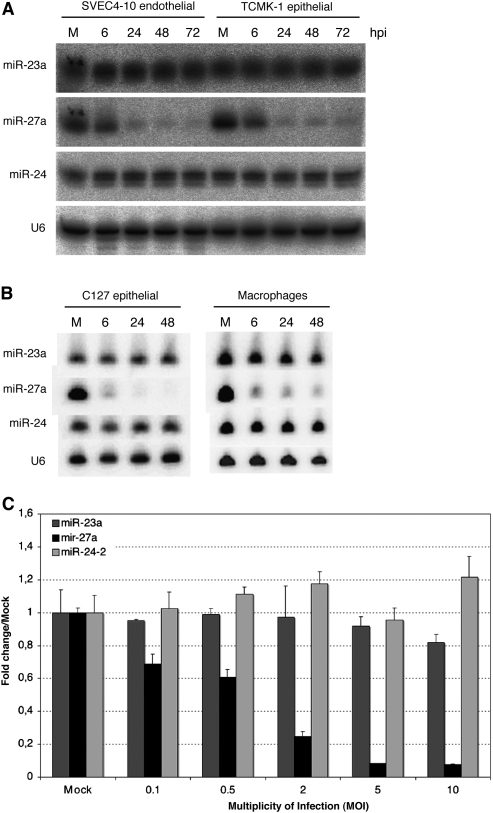
In the context of viral infection, host cells can make use of miRNAs as another arm of the innate immune response. For example, miR-155 is up-regulated within 2 h after treatment with poly (I:C) (O'Connell et al. 2007) or upon infection with Epstein-Barr virus (Cameron et al. 2008; Gatto et al. 2008). Both miR-155 and miR-146 contain NF-_k_B binding sites in their promoter regions, and their regulation is therefore tied to larger transcriptional networks and a coordinated host response. However, there is limited information on the elements involved in post-transcriptional regulation of miRNAs. It is therefore unclear if down-regulation of miR-27 is part of a host response or is a viral strategy that might involve viral factors. To date, down-regulation of miR-27 during viral infection has not been reported. Depending on the mechanism of miR-27 down-regulation, our observations could be specific to MCMV infection and could also be exclusive to particular cell types. MCMV has a wide cell tropism in vivo, infecting many cell types such as fibroblast, epithelial, endothelial, smooth muscle, neuronal and macrophage cells. Therefore, we analyzed miR-27a levels in a range of cell types upon MCMV infection. In addition to the results shown for NIH-3T3 cells (Fig. 1), miR-27a was specifically down-regulated in the endothelial cell line SVEC4-10, epithelial cell lines TCMK-1 and C127, as well as in primary bone marrow-derived macrophages (all of the cells examined) (Fig. 2A,B). Down-regulation of miR-27a in fibroblasts displays a clear dependence on the multiplicity of infection (MOI), with an inverse relationship between mature miR-27a levels and the applied MOI (Fig. 2C). The effect is qualitatively different in some cell types and is partly attributable to MOIs used in experiments (as indicated in the figure legends), which were conducted independently in separate laboratories. These observations indicate that miR-27a regulation is not cell-type-specific and is either mediated by a factor of viral origin or by a ubiquitously expressed cellular factor whose activity is altered upon MCMV infection. Further work is required to establish the generality of our observations. Preliminary data suggest that miR-27a is not altered upon infection with human cytomegalovirus or mouse gammaherpesvirus 68 (data not shown). However, this is not necessarily surprising, as the evolutionary distance between MCMV and HCMV is estimated at ∼80 million years (McGeoch et al. 1995), and there are a number of viral mechanisms for modifying host cells that are not conserved between the two viruses. Related to this, a previous study reported down-regulation of miR-100 and miR-101 upon infection of fibroblasts with human cytomegalovirus (Wang et al. 2008); we do not observe the same down-regulation upon MCMV infection of murine fibroblast cell lines (AH Buck, unpubl.). If modulation of miRNAs is indeed a viral strategy (rather than a host response), it will be interesting to further explore this parameter in the context of viral evolution.
FIGURE 2.
Down-regulation of miR-27a occurs in multiple cell types and displays a clear dependence on the multiplicity of infection. (A) Northern blot analysis of total RNA isolated from endothelial SVEC cells and epithelial TMCK cells infected with MCMV at MOI = 1 with centrifugal enhancement. (B) Northern blot analysis of total RNA isolated from primary bone marrow macrophages and epithelial C127 cells infected with MCMV at MOI = 3 with centrifugal enhancement. (C) qRT-PCR analysis of total RNA isolated from NIH-3T3 cells infected with an increasing number of infectious particles (MOI of 0.1–10.0), 24 hpi.
Antiviral effects of miR-27
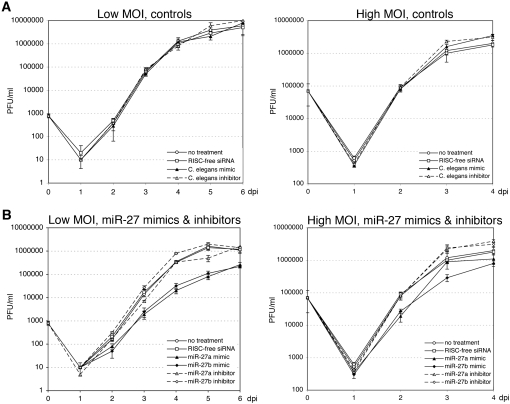
Based on the dramatic and rapid alteration in miR-27 levels upon infection, we might postulate that this miRNA plays some role in MCMV infection. Indeed, HCMV infection alters the expression of miR-100 and miR-101, and these miRNAs were shown to influence viral replication (Wang et al. 2008). To examine this hypothesis, we measured viral growth upon inhibition or overexpression of either miR-27a or miR-27b. These experiments were carried out by transfecting cells with synthetic miRNA inhibitors or mimics. Since transfection of small RNAs has been shown to induce immune activation in various cell types (Kariko et al. 2004; Reynolds et al. 2006), we first examined the effect of transfection on viral replication. Transfection of C. elegans miR-67, which is not encoded in mouse cells, and RISC-free siRNA, which is chemically modified to prevent incorporation into RISC, had no effect on the viral growth rate of MCMV at either low or high multiplicities of infection (Fig. 3A). In contrast, transfection with miR-27a or miR-27b mimics results in greater than a log-fold decrease in viral titer under conditions of low MOI, and an approximately fivefold decrease in viral titer at 2 days post-infection (dpi) at high MOI, consistent with an antiviral function (Fig. 3B). Under these transfection conditions, we confirmed that miR-27a levels increased by ∼50- to 100-fold based on qRT-PCR, and that these conditions did not alter cell viability (data not shown). Theoretically, transfection of the miR-27 inhibitors should show the opposite effect compared with the miR-27 mimics, resulting in an increase in viral growth rate. However, interpretation of results with inhibitors should take into account that miR-27 is naturally inhibited in the infected cells (Figs. 1, 2). Indeed, we observed little effect of inhibitors on viral growth: the miR-27b inhibitor results in an approximately twofold increase in virus titer by day 4 at low MOI (P < 0.0005), but this is not observed with the miR-27a inhibitor.
FIGURE 3.
miR-27 has antiviral properties. MCMV growth curve in NIH-3T3 cells transfected with 25 nmolar C. elegans miR-67 mimic or inhibitor and RISC-free (nontargeting) siRNA (A), or 25 nmolar miR-27a or miR-27b mimic and 50 nmolar inhibitors (B). Cells were infected at MOI of 0.01 and 3.0. (PFU) Plaque forming units; (dpi) days post-infection.
Further analysis of miR-27 targets will shed light on the mechanism underlying the antiviral activity; this could be based on direct targeting of viral genes or based on targeting cellular factors essential for MCMV replication. For example, the antiviral effect of miR-100 and miR-101 during HCMV infection could be partly explained by their targeting of the mTOR pathway (Wang et al. 2008). Similarly, the miRNA cluster miR-17–92 has an inhibitory effect on HIV that is linked to the targeting of the histone acetylase PCAF by miR-17 and miR-20 (Triboulet et al. 2007). A few cellular targets for miR-27a or miR-27b have been published; in particular, miR-27a has been identified as an oncogene that can impact cell cycle regulation, proliferation, apoptosis, megakaryopoiesis, and lipid metabolism (Mertens-Talcott et al. 2007; Guttilla and White 2009; Ji et al. 2009; Liu et al. 2009). As discussed above, miRNAs have also been shown to directly target viral genes. Several putative binding sites for miR-27 can be found in the MCMV genome (data not shown), but based on the high probability of finding complementarities in such a large (∼230 kb) viral genome, this requires extensive validation and analysis before drawing further conclusions.
Mechanism for miR-27 regulation
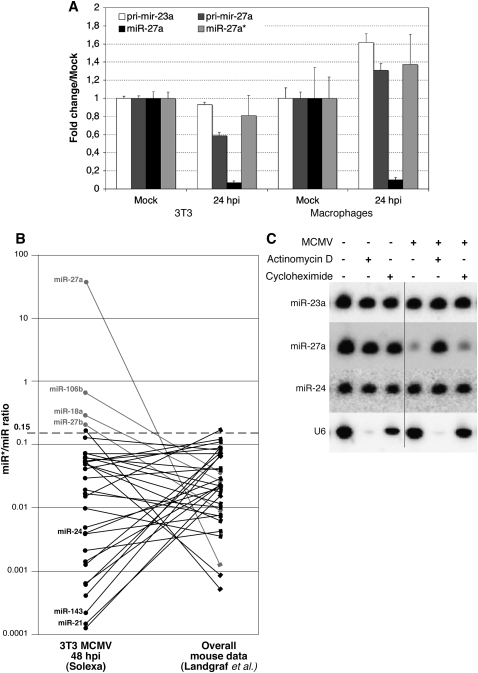
It seems likely that the regulation of miR-27 involves post-transcriptional mechanisms, based on the lack of change in miR-24 and miR-23a expression levels upon infection (Figs. 1, 2). However, it is necessary to consider the possibility that miR-27a derives from a pri-miRNA that is regulated independently of miR-23a and miR-24-2. In order to examine whether regulation might occur at the level of transcription (and/or at the Drosha processing step), we used qRT-PCR to examine changes in pri-miR-27a and pri-miR-23a levels upon infection. Primers were designed within the loop and just 5′ to the respective hairpins. The location of these primers should ensure that pri-miR-27a is detected, regardless of whether it is within a larger pri-miRNA containing miR-23a and miR-24-2. As shown in Figure 4A, the levels of pri-miR-27a and pri-miR-23a are only moderately altered (less than approximately twofold) upon infection. Given the similarity in modest changes between pri-miR-27a and pri-miR-23a upon infection (slightly down in fibroblasts and slightly up in macrophages), it is unlikely that the rapid down-regulation of mature miR-27a in multiple cell types is due to specific regulation of the primary transcript. These data are consistent with regulation of miR-27a occurring after the production of the primary transcript.
FIGURE 4.
miR-27a regulation occurs post-transcriptionally and requires RNA synthesis. (A) Relative pri-miRNA and mature miRNA levels in uninfected versus infected cells, quantified using qRT-PCR (n = 3) normalized to GAPDH or U6 RNA (respectively). Total RNA was isolated from NIH-3T3 cells or bone marrow-derived macrophages 24 hpi with MCMV (MOI = 10 for NIH-3T3 cells and MOI = 5 with centrifugal enhancement for bone-marrow-derived macrophage cells). (B) Large-scale Solexa sequencing of MCMV-infected NIH-3T3 cells reveals that the ratio of miR-27a*/miR-27a is significantly higher than expected from previous analyses in uninfected cells (Landgraf et al. 2007), whereas most of the other miR*/miR ratios stay below 0.15. (C) Northern blot analysis of total RNA isolated from NIH-3T3 cells treated with actinomycin D or cycloheximide (as indicated) in the absence or presence of MCMV (MOI = 10). Though the juxtaposed lanes are not contiguous, all of them are from a single gel (indicated by the dotted line).
Regulation of miR-27a could occur at several steps in the processing pathway: pri-miRNA processing by Drosha, export of pre-miRNA, precursor processing by Dicer, or degradation/turnover of the mature miRNA. Different approaches can be used to tease apart regulation at each step. For example, quantification of the pre-miRNA and/or the miRNA* sequence would shed light on whether regulation occurs during processing of the pre-miRNA. If the pre-miRNA accumulates, but the mature miRNA is not detected, then regulation is likely occurring at processing of pre-miRNA (and the miRNA* levels should also decrease). We were not able to detect pre-miR-27a by Northern blot with a number of probe designs (data not shown), most likely because it is processed very efficiently. The passenger strand, miR-27a*, could be detected using qRT-PCR and does not significantly change in expression level upon infection (Fig. 4A). These data suggest a stark difference in the effect of MCMV infection on mature miR-27a compared with the passenger strand, miR-27a*. To further confirm the difference observed for miR-27a and miR-27a*, we made use of Solexa sequencing data of small RNAs isolated from MCMV-infected (48 hpi) NIH-3T3 cells. In this data set, a total of 7,840,619 small RNAs were sequenced, of which 5,374,302 are annotated as miRNAs (the detailed analysis will be published elsewhere). The balance between a miRNA and its star sequence, as per definition, is lower than a ratio of 0.15 to 1 of star vs. mature miRNA (Landgraf et al. 2007). When the ratio is higher, both strands of the miRNA are defined as functional and the 5p/3p nomenclature is used. Reported ratios for miR-27a*/miR-27a are far below 0.15, and these miRNAs have been cloned in numerous cell lines and tissues (Landgraf et al. 2007). However, in our large-scale sequencing data set of MCMV-infected cells, we sequenced more clones of miR-27a* than miR-27a (for a ratio of 35.5 miR-27a*/miR-27a, compared with 0.0014 in the Landgraf et al. data set) (Fig. 4B). Similarly, the ratio of miR-27b*/miR-27b was inverted in MCMV-infected 3T3 compared with the normal situation, although not as strongly as for miR-27a. Beside miR-27a and miR-27b, only two other miRNAs, miR-106b and miR-18a, showed a similar trend, and all of the other miRNA*/miRNA pairs that we cloned (including miR-24) were below the ratio of 0.15 to 1. The qRT-PCR and cloning results suggest that miR-27a* is not down-regulated upon MCMV infection and, thus, processing of pre-miR-27a occurs normally. Hence, we hypothesize that the regulation of miR-27 occurs at the level of the stability of the mature miRNA. This suggests that the recognition element for down-regulation of miR-27 is contained within the mature 21-nt sequence, consistent with the down-regulation of both miR-27a and miR-27b (Fig. 1E; Supplemental Table 1), which differ from one another by only 1 nt at the 3′end. Regulation at the level of the mature miRNA would involve mechanisms that are distinct from other reported post-transcriptional mechanisms, where elements in the miRNA loop are central to recognition (discussed above). In line with this, the loop of miR-27 does not display obvious conservation and is highly variable between miR-27a and miR-27b (data not shown). Drug-sensitivity experiments demonstrate that virus entry is not sufficient to induce down-regulation of miR-27a, as it can be blocked by actinomycin D treatment (Fig. 4C). Treatment with cycloheximide, which blocks protein synthesis, is not as effective at blocking down-regulation (Fig. 4C). The most consistent model to describe all of the data presented here is that an RNA molecule, which is synthesized upon infection, is involved in inducing the decay or inhibition of mature miR-27. Given that the recognition element for this mechanism appears to be contained within the 21-nt mature miR-27 sequence, it is probable that another RNA molecule, potentially a viral transcript, is involved in the regulatory mechanism, enabling specificity through base pairing. Further work is required to pin down this mechanism and the breadth of miR-27 regulation in other viral infections. Given the ubiquity of miRNAs in signaling and immune responses, it is anticipated that miRNA expression would be controlled at multiple levels and with varying kinetic parameters. Depending on the mechanism, regulation of mature miRNA stability might enable a rapid host response, or provide a rapid means by which pathogens could inhibit miRNA function.
MATERIALS AND METHODS
Cell lines and viruses
NIH-3T3, SVEC4-10, C127, and TCMK-1 cells were grown in DMEM supplemented with 10% FBS and penicillin/streptomycin. Bone marrow-derived macrophages were prepared as described in Buck et al. (2007). Wild-type virus (Smith strain) was generated from bacterial artificial chromosome clone pSM3fr described in Wagner et al. (1999). For experiments in NIH-3T3, SVEC4-10, and TCMK-1 (Figs. 1, 2), virus stocks were prepared on M2-10B4 cells and titers determined as described in Dölken et al. (2007). For experiments in NIH-3T3 (Figs. 3, 4), C127 and bone marrow-derived macrophages (Figs. 2, 4), virus stocks were prepared on NIH-3T3 cells as described by Buck et al. (2007). The MOI for each experiment is indicated in the figure legends. To obtain a high level of infection, centrifugal enhancement was employed during the incubation with virus where indicated, as described by Dölken et al. (2007). This results in an up to 10-fold higher viral load compared with incubations without enhancement (data not shown).
RNA preparation and Northern blotting
RNA was extracted using Trizol reagent (Invitrogen) and Northern blotting was performed on 5–10 μg of total RNA as described before (Pfeffer et al. 2005; Dölken et al. 2007), except that we used 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC)-mediated, chemical cross-linking (Pall and Hamilton 2008) instead of UV cross-linking. Probes were 5′ 32P-radiolabeled oligodeoxynucleotides perfectly complementary to the miRNA sequence or to part of the U6 snRNA sequence. Blots were analyzed and quantified by phosphorimaging using an FLA5100 scanner from Fuji.
miRNA and pri-miRNA quantification by quantitative real-time PCR
For experiments with NIH-3T3 cells in Figure 1, quantitative real-time PCRs were established using a Roche LightCycler 480 Real-Time PCR System. For mature miRNAs, total RNA was polyadenylated and reverse transcribed using the miScript Reverse Transcription kit (QIAGEN) as per the manufacturer's instructions with 1 μg of total RNA per reaction. The obtained cDNA (1 μL of a 1/5 dilution) was then PCR-amplified using the miScript SYBR Green PCR kit (QIAGEN) in 10 μL reaction mixtures consisting of Quantitect SYBR Green PCR master mix and 0.625 μM of the miRNA-specific primer and the miScript universal primer. PCRs were subjected to 15 min of 95°C hot-start enzyme activation, and 40 cycles of 95°C denaturation for 15 sec, 55°C annealing for 30 sec, and 72°C elongation for 30 sec, including subsequent melting curve analysis. The data was analyzed using the ΔΔCt method (Bookout et al. 2006) using U6 snRNA as an endogenous reference, and the mock-infected sample as a calibrator. The amplification efficiency as determined with the LinReg software (Ramakers et al. 2003) was used to calculate fold changes relative to mock. Primers were the mature miRNA sequences for the forward primers, and the universal miScript primer provided by QIAGEN for the reverse primer. The primer specific for miR-27b could discriminate miR-27b from miR-27a with a cycle difference of 7.3 (i.e., a 168.9-fold difference), and the primer for miR-23a could discriminate miR-23a from miR-23b with a cycle difference of 6.5 (a 90.5-fold difference). Similar results were obtained with primers specific for miR-27a and miR-23b, and the use of synthetic standards also allowed us to calculate that there is sixfold more miR-27a than miR-27b in NIH-3T3 fibroblasts. For detection of pri-miRNAs, cDNA was prepared using ThermoScript reverse transcriptase (Invitrogen) according to the manufacturer's protocol (at a temperature of 55°C) with gene-specific primers for pri-miR-23a and pri-miR-27a and the oligo dT primer for GAPDH (similar results were obtained when comparing pri-miR-23a and pri-miR-27a in uninfected and infected cells using cDNA prepared with either the gene-specific primers or the oligo dT primer). cDNAs were amplified using SYBR green SuperMix (Invitrogen) with the following primers:
- pri-miR-27a forward, 5′-CTATCATGACAACTGGCCTGAG-3′;
- pri-miR-27a reverse, 5′-GACTTTGCTGTGGACCTTGC-3′;
- pri-miR-23a forward, 5′-CTACACTCCGCTCCCACC-3′;
- pri-miR-23a reverse, 5′-TTGTGACTGGCATCAAATCC-3′;
- GAPDH forward, 5′-AATGTGTCCGTCGTGGATCT-3′; and
- GAPDH reverse, 5′-CCTGCTTCACCACCTTCTTG-3′.
Detection of miR-27a and miR-27a* in Figure 4A was performed using miRCURY LNA qRT-PCR probes (Exiqon) according to the manufacturer's protocols (using 100 ng input RNA); standard curves were performed with synthetic RNAs (Eurogentec).
Small RNA cloning and analysis
The generation of small RNA libraries used in the initial analysis has been described before (Dölken et al. 2007). For the generation of Solexa-compatible libraries, the protocol was modified according to Hafner et al. (2008), and 3 pmol of the final PCR product were used for sequencing on one lane of an Illumina flow cell using the DGE-Small RNA Cluster Generation Kit—GA II and the 36-cycle sequencing kit V. Adaptor sequences were removed and sequences annotated as described before (Dölken et al. 2007), except that the databases used were miRbase release 13.0, GenBank release 169, and the mm9 version of the mouse genome, with the Illumina pipeline scripts v.1.3.2. The hierarchical clustering analysis described in Figure 1A was done using the MultiExperiment Viewer version 4.4 from the TM4 software suite (Saeed et al. 2003).
Viral growth assays and microRNA transfections
NIH-3T3 cells were reverse transfected with miRNA mimics (25 nM) or inhibitors (50 nM) (Dharmacon) using 0.4% Dharmafect 1 in a 96-well plate format. Cells were seeded at 1.5 × 104 cells/well in antibiotic-free media and were incubated 48 h at 37_°_ C and 5% CO2 prior to infection with MCMV. Supernatant from the infected cells was collected over a period of 6 dpi, and the viral titer measured by standard plaque assay on mouse embryonic fibroblast p53 cell line.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by Agence Nationale pour la Recherche (ANR-07-MIME-012-01), and an ATIP grant from CNRS (S.P.); Wellcome Trust CIIE grant 082611/Z/07/Z and Biotechnology and Biological Science Research Council grant BB/G01552X/1 (A.H.B.); and the German Federal Ministry of Education and Research, NGFNplus grant 01GS0801 (L.D.). We thank Peter Ghazal, Alexei Kazantsev, Eric Westhof, and members of the Pfeffer laboratory for critically reading the manuscript.
Footnotes
REFERENCES
- Ahluwalia JK, Khan SZ, Soni K, Rawat P, Gupta A, Hariharan M, Scaria V, Lalwani M, Pillai B, Mitra D, et al. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology. 2008;5:117. doi: 10.1186/1742-4690-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, Kramer MF. High-throughput real-time quantitative reverse transcription PCR. Curr Protoc Mol Biol. 2006 doi: 10.1002/0471142727.mb1508s73. [DOI] [PubMed] [Google Scholar]
- Buck AH, Santoyo-Lopez J, Robertson KA, Kumar DS, Reczko M, Ghazal P. Discrete clusters of virus-encoded micrornas are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J Virol. 2007;81:13761–13770. doi: 10.1128/JVI.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron JE, Yin Q, Fewell C, Lacey M, McBride J, Wang X, Lin Z, Schaefer BC, Flemington EK. Epstein-Barr virus latent membrane protein 1 induces cellular MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol. 2008;82:1946–1958. doi: 10.1128/JVI.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38(Suppl 1):S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölken L, Perot J, Cognat V, Alioua A, John M, Soutschek J, Ruzsics Z, Koszinowski U, Voinnet O, Pfeffer S. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J Virol. 2007;81:13771–13782. doi: 10.1128/JVI.01313-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Zamore PD. microPrimer: The biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepaa AL, Oresic M, Esau CC, Zdobnov EM, et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes & Dev. 2009;23:1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, Mallardo M. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-κB pathway. Nucleic Acids Res. 2008;36:6608–6619. doi: 10.1093/nar/gkn666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh Z, Mallick B, Chakrabarti J. Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res. 2009;37:1035–1048. doi: 10.1093/nar/gkn1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284:23204–23216. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Ji J, Zhang J, Huang G, Qian J, Wang X, Mei S. Overexpressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 2009;583:759–766. doi: 10.1016/j.febslet.2009.01.034. [DOI] [PubMed] [Google Scholar]
- Kariko K, Bhuyan P, Capodici J, Weissman D. Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through toll-like receptor 3. J Immunol. 2004;172:6545–6549. doi: 10.4049/jimmunol.172.11.6545. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Silva A, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- Liu T, Tang H, Lang Y, Liu M, Li X. MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett. 2009;273:233–242. doi: 10.1016/j.canlet.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Mahajan VS, Drake A, Chen J. Virus-specific host miRNAs: Antiviral defenses or promoters of persistent infection? Trends Immunol. 2009;30:1–7. doi: 10.1016/j.it.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ, Cook S, Dolan A, Jamieson FE, Telford EA. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol. 1995;247:443–458. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001–11011. doi: 10.1158/0008-5472.CAN-07-2416. [DOI] [PubMed] [Google Scholar]
- Michlewski G, Guil S, Semple CA, Caceres JF. Post-transcriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Gordon KH, Buck AH, Jiggins FM. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci. 2009;364:99–115. doi: 10.1098/rstb.2008.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Lai EC. The long and short of inverted repeat genes in animals: microRNAs, mirtrons, and hairpin RNAs. Cell Cycle. 2008;7:2840–2845. doi: 10.4161/cc.7.18.6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S. Viral miRNAs: Tiny but mighty helpers for large and small DNA viruses. Future Virol. 2008;3:291–298. [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Anderson EM, Vermeulen A, Fedorov Y, Robinson K, Leake D, Karpilow J, Marshall WS, Khvorova A. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. RNA. 2006;12:988–993. doi: 10.1261/rna.2340906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Sun F, Wang J, Pan Q, Yu Y, Zhang Y, Wan Y, Wang J, Li X, Hong A. Characterization of function and regulation of miR-24-1 and miR-31. Biochem Biophys Res Commun. 2009;380:660–665. doi: 10.1016/j.bbrc.2009.01.161. [DOI] [PubMed] [Google Scholar]
- Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, et al. Suppression of MicroRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- Wagner M, Jonjic S, Koszinowski UH, Messerle M. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J Virol. 1999;73:7056–7060. doi: 10.1128/jvi.73.8.7056-7060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FZ, Weber F, Croce C, Liu CG, Liao X, Pellett PE. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J Virol. 2008;82:9065–9074. doi: 10.1128/JVI.00961-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]