A molecule targeting VHL-deficient Renal Cell Carcinoma that induces autophagy (original) (raw)
. Author manuscript; available in PMC: 2010 Feb 10.
Abstract
Renal Cell Carcinomas (RCCs) are refractory to standard therapies. The von Hippel-Lindau (VHL) tumor suppressor gene is inactivated in 75% of RCCs. By screening for small molecules selectively targeting VHL-deficient RCC cells, we identified STF-62247. STF-62247 induces cytotoxicity and reduces tumor growth of VHL-deficient RCC cells compared to genetically matched cells with wild-type VHL. STF-62247-stimulated toxicity occurs in a HIF-independent manner through autophagy. Reduction of protein levels of essential autophagy pathway components reduces sensitivity of VHL-deficient cells to STF-62247. Using the yeast deletion pool, we show that loss of proteins involved in Golgi trafficking increased killing by STF-62247. Thus, we found a small molecule that selectively induces cell death in VHL-deficient cells that represents a paradigm shift for targeted therapy.
Keywords: Renal cell carcinoma, von Hippel-Lindau, autophagy, LC3, autophagosomes, lysosomes, cell death
Significance.
Inactivation of the von Hippel-Lindau (VHL) tumor suppressor gene arises in the majority of Renal Cell Carcinoma (RCC) and is associated with a high degree of vascularization and poor prognosis. We evaluated the possibility of targeting VHL-deficient cells through a synthetic lethality approach by screening for small molecules that exhibited toxicity to renal cells lacking VHL. We identified a compound, STF-62247, which was selectively cytotoxic towards VHL-deficient cells in vitro and in vivo. We demonstrate that STF-62247 induces autophagy and inhibition of autophagy significantly reduces sensitivity to STF-62247. Since RCC are refractory to standard cytotoxic chemotherapies, there is a need for additional chemotherapies. STF-62247 is a small molecule that exploits the loss of VHL in RCC.
Introduction
Mutations and/or inactivation of the von Hippel-Lindau (VHL) tumor suppressor gene occur in the majority of Renal Cell Carcinoma (RCC) of the clear cell phenotype. Restoration of VHL function in VHL−/− RCC results in significant inhibition of these cells to form tumors in nude mice demonstrating the tumor suppressor function of VHL (Iliopoulos et al., 1995). Functional studies indicate that pVHL, the protein product of VHL, is an E3 ubiquitin ligase that targets the α-subunit of the hypoxia-inducible factor (HIF) for proteasomal degradation under normoxia. In the presence of oxygen, hydroxylation on proline residues 564 and 402 by prolyl hydroxylases (PHDs) marks HIF-α for recognition and binding with pVHL, leading to degradation of HIF-α. Under hypoxic conditions, activity of the PHDs decrease reducing recognition of HIF-α by pVHL (Chan et al., 2002; Ivan et al., 2001; Jaakkola et al., 2001). In cells that lack VHL, stabilized HIF-α binds constitutively expressed HIF-β to activate the transcription of genes involved in metabolism, angiogenesis, invasion and metastases, and proliferation (Erler et al., 2006; Gnarra et al., 1996; Iliopoulos et al., 1996; Knebelmann et al., 1998; Staller et al., 2003). In addition to its role in HIF regulation, pVHL has been implicated in a variety of HIF-independent processes including extracellular matrix assembly, regulation of microtubule stability, polyubiquitination of atypical PKC family members, regulation of fibronectin, and modulation of RNA polymerase II subunits (Hergovich et al., 2003; Na et al., 2003; Ohh et al., 1998; Okuda et al., 2001). It has also been reported that an acidic domain present in the N-terminal region of pVHL contributes to its tumor suppression function in a HIF-independent manner and may be relevant for the development of VHL-associated malignancies (Lolkema et al., 2005).
Defects in apoptosis that are observed in many solid tumor cells, including RCCs, increase the resistance of tumor cells to chemotherapy, radiotherapy and molecularly targeted therapies. In contrast to apoptosis, autophagy regulates the turnover of organelles and long-lived proteins to ensure homeostasis. Under metabolic stress, autophagy is activated and promotes survival (Mathew et al., 2007). In some cell types, autophagy can also result in cell death under persistent stress conditions. Studies have reported a dysregulated level of autophagy in diverse diseases including neuronal degeneration, infectious disease, and cancer (Kondo et al., 2005). Whether autophagy protects against or causes these diseases is still unclear, but the regulation of autophagy has promising therapeutic implications.
Autophagy occurs in all eukaryotic cells from yeast to mammals. In response to a diverse number of stimuli such as starvation, hypoxia, or high temperature, portions of cytoplasm and organelles are sequestered in a double-membrane vesicle called an autophagosome. These vesicles undergo maturation by fusion with endosomes and/or lysosomes to become autolysosomes where hydrolases degrade their contents (Klionsky and Emr, 2000). Among _a_u_t_ophagy-related _g_enes (ATGs) identified to regulate this process, Beclin 1 (BECN), the mammalian ortholog of yeast Atg6, acts as a tumor suppressor gene in mice and is frequently deleted in human cancers (Aita et al., 1999). In mammalian cells, BECN forms a complex with the class III family of phosphatidylinositol 3-kinase (PI3K) and localizes at the trans-Golgi network (TGN) (Kihara et al., 2001). The elongation of the double-membrane involves two ubiquitin-like conjugation systems: the ATG12-ATG5 complex and the microtubule associated protein-light chain 3 called MAP-LC3 (Atg8 in yeast). In unstressed cells, LC3 is present in the cytoplasm, while the lipidated form of LC3 is associated with double-membrane containing organelles in cells undergoing autophagy (Kabeya et al., 2000). The recruitment of LC3 to the membrane occurs in an ATG5-dependent manner. While ATG5-ATG12 dissociate from the membrane upon completion of autophagosome formation, LC3 remains associated with the membrane (Kabeya et al., 2000; Mizushima et al., 2001). ATG7 acts as an E1 enzyme in the autophagy conjugation system where in combination with an E2 enzyme, conjugtes the phosphatidyl ethanolamine to LC3I to form, LC3II and no lipidation of LC3 occurs in the brain of ATG7-deficient mice (Tanida et al., 2002). On the other hand, ATG9 has been reported to cycle between the trans-Golgi network and endosomes and its disruption impairs formation of autophagosomes (Young et al., 2006).
Cell-based small molecule screening has been used to discover compounds that inhibit specific proteins, such as HIF, or to overcome drug resistance to reduce tumorigenicity (Isaacs et al., 2002; Mabjeesh et al., 2003; Rapisarda et al., 2002; Smukste et al., 2006). In this report, we describe a small molecule, STF-62247, identified from a screen that induces autophagy and selectively induces lethality in RCCs that have lost the VHL tumor suppressor gene. We further used the yeast deletion pool to functionally investigate the targets of STF-62247 that are involved in cell killing.
Results
STF-62247 induces cytotoxicity and reduces tumor growth in VHL-deficient cells in a HIF-independent manner
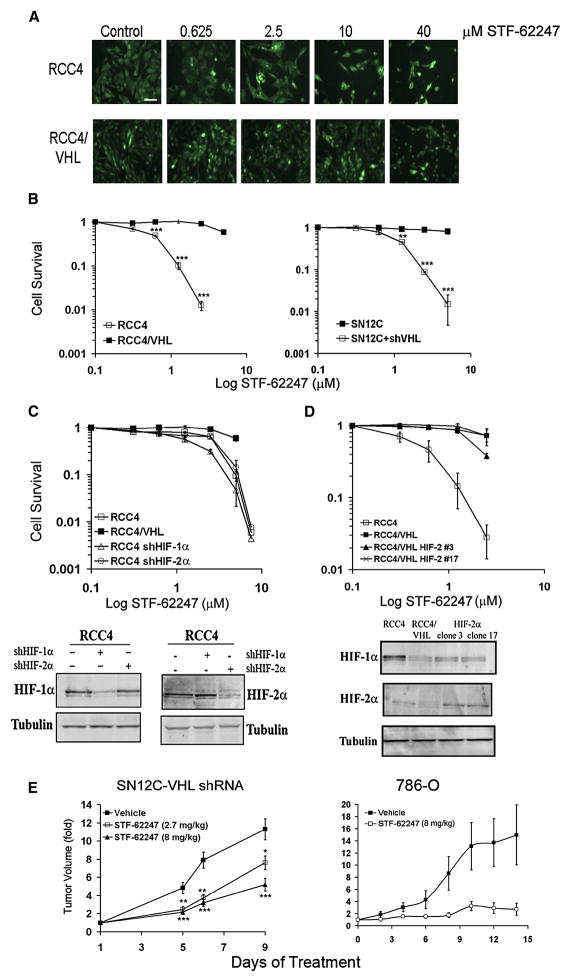
In this study, we evaluated the possibility of selectively targeting VHL-deficient cells using small molecule compounds. We screened 64,000 compounds against wild-type VHL and VHL-deficient RCCs (manuscript in preparation) that were stably transfected with EYFP. The effect of small molecules on each cell type was monitored separately by fluorescence (Figure 1A). The drug STF-62247 was identified in this screen (Figure S1A) and decreases the viability of VHL-deficient cells in a short term assay (Figure S1B). Moreover, STF-62247 specifically results in cytotoxicity in renal cells that have lost VHL as demonstrated using two genetic models: VHL-deficient RCC4 cells and their counterparts that express ectopically introduced wild-type VHL, and VHL-proficient SN12C cells and their counterparts that stably express shRNA targeting VHL (Thomas et al., 2006). Clonogenic assays demonstrate the STF-62247 is selectively toxic to VHL-deficient cells compared to their VHL wild-type counterparts (Figure 1B).
Figure 1. STF-62247 induces cytotoxicity and reduces tumor growth in VHL-deficient cells in HIF-independent manner.
A. RCC4 and RCC4/VHL cells were labeled with EYFP and treated with increasing concentration of STF-62247 for 4 days. Each cell type was monitored by fluorescence separately. Cells are pseudocolored green. Scale bar, 10 μm. B. Clonogenic assay in RCC4, RCC4/VHL, SN12C, SN12C-VHL shRNA cells in the presence of STF-62247 (0–30 μM). Each point of the cell survival is calculated by the average of three different experiments in triplicate as the percent of treated on untreated plate. C. Upper Panel, clonogenic assay in RCC4 infected with HIF-1α or HIF-2α shRNA by retroviral infection in presence of drug in same conditions described above. Bottom panel, HIF-1α or HIF-2α protein expression evaluated by western blot in RCC4 cells with HIF-1α or HIF-2α shRNA. Tubulin is used as loading control. D. Upper Panel, clonogenic assay with STF-62247 in RCC4 where HIF-2α was stably introduced into VHL-positive cells by retroviral infection. Bottom panel, western blot for HIF-1α or HIF-2α in RCC4 VHL-positive cells. E. SN12C-VHL shRNA (2–3 × 106 cells) and 786-O (5 × 106 cells) were injected into SCID mice. SN12C shVHL tumor bearing mice were treated daily with vehicle, 2.7 mg/kg, or 8 mg/kg of STF-62247 by intraperitoneal injection. 786-O tumor bearing mice were treated with vehicle or 8 mg/kg. Five tumors per condition were analyzed where * p<0.05, **p<0.01 and ***p<0.005. All error bars represented the standard error of the mean (SEM).
Since VHL is an important negative regulator of HIF-α through its E3-ligase activity, we used VHL-null RCC4 cells expressing shRNA to HIF-1α or HIF2α to determine whether STF-62247 cytotoxicity is dependent on HIF (Figure 1C bottom panel). The level of HIF target genes is quantified by qRT-PCR verifying functional knockdown of HIF (Figure S1C). Clonogenic assays demonstrate that the reduction of either HIF-1α or HIF-2α in VHL-null cells did not affect cell death induced by STF-62247 (Figure 1C upper panel). Also, decreasing the level of ARNT (HIF-1β) using shRNA did not change the cell sensitivity to STF-62247 (data not shown). Previous studies have indicated that HIF-2 is critical for tumor growth of RCCs (Kondo et al., 2003; Zimmer et al., 2004). To further investigate the effect of HIF-2 in the sensitivity of RCC4 cells to STF-62247, we generated a VHL cell line that stably expressed a normoxically stable, constitutively active HIF-2α mutant P531A N547A. Two individual HIF-2α RCC4/VHL clones were randomly chosen that had elevated HIF-2α expression under normoxic conditions (Figure 1D bottom panel). Results show that cytotoxicity of STF-62247 was unaffected by the presence of HIF-2α expression under normoxic conditions when compared to RCC4/VHL cells. Thus, by using complementary approaches, our results indicate that STF-62247 induces cytotoxicity in VHL-null cells in a HIF-independent manner.
To evaluate the effect of STF-62247 on tumor growth in vivo, we implanted SN12C, SN12C-VHL shRNA or 786-O cells subcutaneously into the flanks of immune-deficient mice. The selective cytotoxicity of STF-62247 for the VHL-deficient cells is also demonstrated in 786-O cells compared to their VHL wild-type counterparts by clonogenic assay in vitro (Figure S1D). Daily treatment with STF-62247 significantly reduced tumor growth of VHL-deficient cells (Figure 1E). This decrease in tumor growth was concentration-dependent. Importantly, drug treatment did not have any effect on the growth of SN12C tumor cells that have wild-type VHL (Figure S1E). Together, these results show that STF-62247 selectively kills RCC cells with a loss of VHL in vitro and also significantly reduces tumor growth in cells deficient in VHL.
Autophagy is induced by STF-62247 treatment
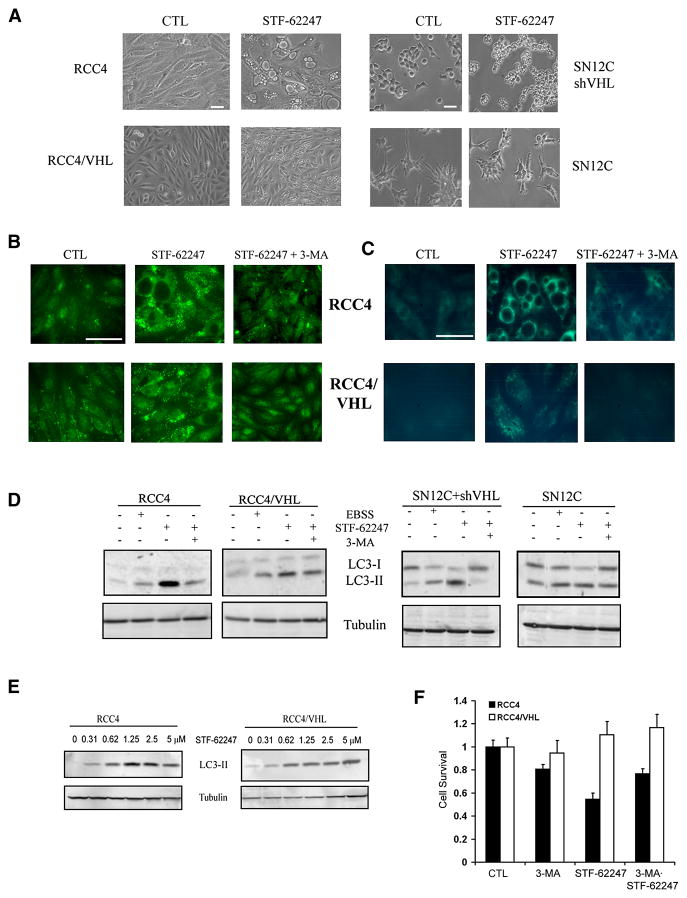
We found that STF-62247 did not induce apoptosis in vitro in VHL-deficient cells as assayed by Hoechst (Figure S1F), Annexin V staining (Figure S1G) or caspase-3 (Figure S1H). These results were further confirmed by immunostaining for caspase-3 in tumor sections (Figure S1I). There was no difference in proliferation as assayed by Ki67 staining in response to STF-62247 in RCC deficient in VHL or with wild-type VHL (Figure S1J). STF-62247 did not induce DNA damage as measured by the comet assay (data not shown). In addition, the phosphorylation of p53 on serine 15 and total p53 levels were unaffected by STF-62247 treatment, further supporting the lack of a DNA damage signal in cell killing induced by STF-62247 (Figure S1K). We observed that STF-62247-treated cells accumulated intracytoplasmic vacuoles characteristic of cells undergoing autophagy. Moreover, these vacuoles were larger in VHL-deficient RCC4 and SN12C-VHL shRNA cells than in wild-type VHL cells (Figure 2A). Vacuole formation in RCC cells with and without VHL was monitored by time-lapse microscopy for 48 hours, which shows that vacuoles appear as soon as 3 hours after adding STF-62247 (Movies S1 and S2). Time-lapse movies also indicate that wild-type VHL prevents the formation of larger vacuoles present in VHL-deficient cells.
Figure 2. Presence of autophagic vacuoles with STF-62247 in RCCs.
A. Phase contrast pictures of VHL-deficient RCC4 and SN12C-VHL shRNA cells and VHL-positive cells RCC4 and SN12C after 20 hrs of 1.25 μM STF-62247. B. Immunostaining for LC3 following STF-62247 treatment at 1.25 μM for 20 hrs with or without 2 mM of 3-MA in RCC4 with and without VHL cells. After treatment, cells were fixed, permeabilized and processed for the detection of punctuate staining by indirect immunofluorescence. C. Autophagic vacuoles stained with monodansylcadaverine (MDC). After STF-62247 treatment in the same conditions, MDC was added for the last 10 minutes. Cells were fixed, washed with PBS and observed directly under microscope. D. Western blot for LC3 in RCC4, RCC4/VHL, SN12C, and SN12C-VHL shRNA cells after STF-62247 treatment with or without 3-MA. Nutrient starvation (EBSS) is used as positive control. E. Western blot for LC3 in RCC4 and RCC4/VHL with different concentrations of STF-62247. Tubulin was used as loading control. F. Cell viability assay in VHL-deficient RCC4 and wild-type VHL cells treated with 1 mM of 3-MA, 1.25 μM of STF-62247 alone or in combination for 24 hrs. Viability is measured by trypan blue exclusion assay and is represented by the average of three different experiments in duplicate as the percent of treated on untreated cells. All error bars represent SEM. All scale bars, 10 μm.
Induction of autophagy was confirmed by immunostaining and Western blotting for LC3. STF-62247 increased punctate staining of LC3, which is associated with processing of LC3 to its lipidated form (Figure 2B). LC3-II, corresponding to the lipidated form found with the formation of double-membranes, was induced by STF-62247 treatment, indicating that STF-62247 stimulates autophagy (Figure 2D). Transmission electron microscopy (TEM) was also used to monitor autophagy in response to STF-62247. There were large numbers of autophagic vacuoles present in STF-62247-treated cells, but not in untreated cells (Figure S2A). The presence of double-membrane containing cellular organelles was observed in treated RCC4 cells at higher magnification (Figure S2B).
The increase of the LC3-II form was abrogated by 3-methyladenine (3-MA), an autophagy inhibitor that inhibits PI3 kinase. It is noteworthy that although the stimulation in LC3 is higher in VHL-deficient cells treated with STF-62247, the increase was also observed in wild-type VHL cells, and therefore the presence of autophagosomes cannot by themselves explain the difference in toxicity observed between RCC cells with and without VHL. We also stained cells with the autofluorescent compound monodansylcadaverine (MDC), which acts as a lysosomotropic agent and labels some of the acidic compartments that are observed after fusion with lysosomes (autolysosomes) (Figure 2C). Staining with MDC in RCC4 cells was primarily observed around the larger vacuoles compared to the punctate staining of LC3, suggesting that these large vacuoles are associated with the acidic components of autolysosomes.
Since autophagy might be an early response during drug treatment, LC3 processing was monitored in cells exposed to increasing concentrations of drug. Results show that the LC3-lipidated form increases in a concentration-dependent manner, indicating that the autophagic process is tightly linked with drug cytotoxicity (Figure 2E). To determine whether autophagy induced by STF-62247 was responsible for the reduced viability observed in VHL-deficient cells, we measured the viability after 24 hours of cells treated with 3-MA to inhibit autophagy. The viability of VHL-deficient cells was partially restored by 3-MA, indicating that STF-62247 induced autophagy leads to cell death in VHL-deficient cells (Figure 2F).
ATG5, ATG7 and ATG9 are involved in autophagosome formation in STF-62247 treated cells
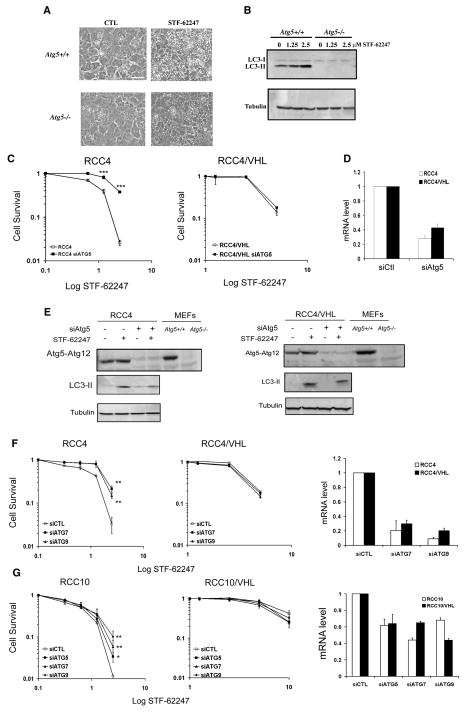
We hypothesized if autophagy was important in STF-62247 cytotoxicity of VHL-deficient cells, Atg5−/− cells would be less sensitive to STF-62247-induced cell death. We found significantly more autophagic vacuoles during STF-62247 treatment in Atg5+/+ cells compared to Atg5−/− cells (Figure 3A). This effect correlates with the LC3 processing that was observed only in Atg5+/+ cells in response to STF-62247 and was concentration dependent (Figure 3B). Importantly, we confirmed that Atg5 is involved in STF-62247-induced toxicity in VHL-deficient RCC cells. Silencing ATG5 in VHL-deficient RCC4 cells by siRNA reduced cell death induced by STF-62247 as assessed by clonogenic assay (Figure 3C). VHL-positive RCC4 cells were unaffected by ATG5 siRNA, suggesting that the cell death observed at higher concentrations in VHL-positive cells is independent of ATG5 and autophagy. A reduction in ATG12-ATG5 levels by siRNA decreases LC3 formation by STF-62247, indicating a critical role for ATG5 in STF-62247 autophagic response in RCC VHL-deficient cells (Figure 3D). Consistent with a role of the autophagy pathway in STF-62247 cytotoxicity, reduction of ATG7 and ATG9 levels by siRNA also reduced the STF-62247 cytotoxicity in VHL-deficient RCC4 cells although to a lesser degree (Figure 3F). Similar results were obtained using VHL-deficient RCC10 and RCC10 cells expressing wild-type VHL (Figure 3G).
Figure 3. Atg5 is involved in autophagic cell death of STF-62247.
A. MEFs Atg5+/+ and Atg5−/− were treated in presence of STF-62247 and examined under light microscopy 20 hrs later. Scale bar, 10 μm. B. LC3 processing evaluated by Western blot following 24 hours of drug treatment in MEFs. C. Cell Survival measured by clonogenic assay with STF-62247 in RCC4 and RCC4/VHL cells transiently transfected with siRNA to ATG5. Colony formation was evaluated as described before. ***p<0.005 represents the difference between RCC4 and RCC4 with siATG5 cells in presence of STF-62247. D. mRNA level of ATG5 quantified by qRT-PCR in RCC4 and RCC4/VHL cells transfected with siRNA to ATG5 for 48 hours. E. Western blot for LC3 and ATG5 in RCC4 and RCC4/VHL cells transiently transfected with siRNA against ATG5. Tubulin was used as loading control. F, G. Left and middle panels, Clonogenic assay with STF-62247 in cells transfected with ATG7 and ATG9 siRNA in VHL-deficient RCC4, RCC10 and wild-type VHL cells. Colony formation was evaluated as described before. **p<0.01 represents the difference between siControl cells and siATG cells in response to STF-62247. Right panels, mRNA levels of ATG7 and ATG9 (and ATG5 in RCC10 cells) quantified by qRT-PCR after 48 hours of transfection using siRNA to ATG7 and ATG9 (and ATG5 in RCC10 cells) in RCC4 and RCC10 cells with or without VHL. All error bars represent SEM.
PI3K and Golgi trafficking are required as initial signals in STF-62247-induced autophagy
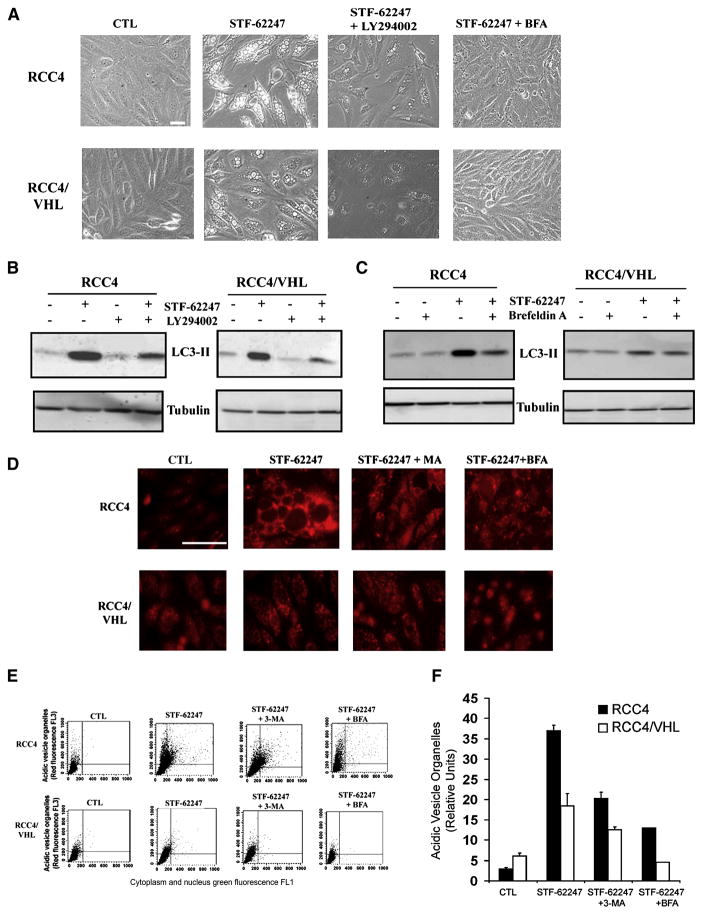
Previous studies have implicated PI3K and ERK signaling as well as Golgi trafficking and endoplasmic reticulum (ER) stress in autophagy (Ogata et al., 2006; Yorimitsu et al., 2006). To evaluate a role for PI3K in response to STF-62247, we compared the effects of the autophagy inhibitor 3-MA to LY294002, another PI3K inhibitor. LY294002 significantly decreased large vacuole formation and the LC3-lipidation form in VHL-deficient cells during STF-62247 treatment to a level similar as 3-MA (Figure 4A, 4B and 2D), indicating a role for PI3K in STF-62247 signaling. In contrast, the MEK inhibitor U0126, which inhibits ERK activation, had little effect on autophagy induced by STF-62247 (Figure S3A, S3B), indicating that MEK signaling is not involved in STF-62247 induced autophagy.
Figure 4. Golgi trafficking and PI3K involved in STF-62247 signaling pathway and acidification of vesicle after STF-62247 treatment.
A. VHL-deficient RCC4 and wild-type VHL were incubated in presence of 10 μM LY294002, or 1 μg/ml Brefeldin A with 1.25 μM of STF-62247 for 20 hrs and the presence of vacuoles was examined under light microscopy. B,C. LC3 processing was evaluated by Western blot after the treatment with Brefeldin A or LY294002 in presence or absence of STF-62247 as described above. Tubulin was used as loading control. D. Cells were treated with 1.25 μM of STF-62247 with or without 2 mM of 3-MA, 1 μg/ml of Brefeldin A for 20 hrs. For the last 15 min, acidic components were stained with 1 μg/ml of acridine orange and visualized under fluorescence microscope. E. FACS analysis to measure the acidic vesicle organelles (AVO) in cells treated in D. y axis represents the concentrated dye in the acidic vesicles, whereas the cytoplasm and the nucleus showed green fluorescence in x axis. F. Quantification of the FACS analysis. The data are calculated as the average of three different experiments as relative units of treated over untreated cells. All error bars represent the SEM. Scale bar, 10 μm.
Furthermore, we found that Brefeldin A (BFA), a blocker of protein transport from the ER to the Golgi apparatus, abolished the formation of autophagic vacuoles and LC3-II stimulated by STF-62247 in VHL-deficient RCC cells. This suggests that Golgi trafficking is involved in the cytotoxicity of STF-62247 in cells that have loss VHL (Figure 4A–C). In contrast, no vacuole or LC3 processing was observed with tunicamycin treatment alone, although it induced BiP (Grp78), a marker of ER stress (Figure S3A, C). Taken together, our results suggest that Golgi trafficking and PI3K are part of the initial signal for the induction of autophagy in RCC cells treated with STF-62247.
STF-62247 increases acidification in VHL-deficient cells
Although autophagosome formation in response to STF-62247 is independent of VHL status, we investigated how STF-62247 specifically kills cells that have lost VHL. A late step in the autophagic cell death process is the fusion of lysosomes with autophagosomes into autolysosomes, which can be detected by measuring their acidification with acridine orange staining. Interestingly, we found acridine orange staining significantly increased in VHL-deficient RCC4 cells compared to their wild-type VHL counterparts (Figure 4D). This observation was quantified and confirmed by FACS analyses and illustrates a significant difference in the acidification of the acidic vesicular organelles (AVOs) in RCC4 and SN12C-VHL shRNA cells (Figure 4E–F and S3D–E) compared to wild-type VHL cells. Blocking autophagy with 3-MA or BFA resulted in a decrease in autolysosomal acidification, demonstrating that acidification induced by STF-62247 is linked to the autophagic pathway. In addition, MDC staining performed using NH4Cl to block vesicle acidification attenuated the functional staining following drug treatment (Figure S3F). However, even if the acidification level in VHL cells increases following STF-62247 treatment, this level of acidification remained significantly lower than in VHL-deficient cells. These results suggest that the fusion of autophagosomes with lysosomes is a key step leading to autophagic cell death in VHL-deficient cells.
Structure-activity studies
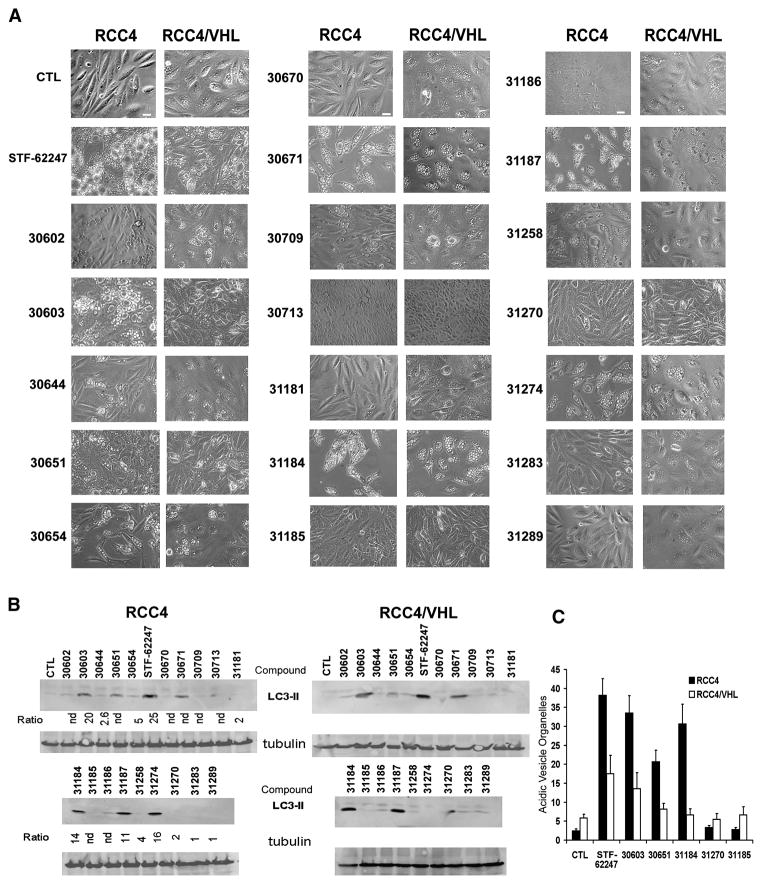
Based on the STF-62247 structure, other compounds were designed and synthesized to evaluate their ability to induce selective cell death in VHL-deficient cells. Analysis of the high throughput screening (HTS) data indicated a specific requirement for a 4-pyridyl unit linked to the 4-position of the thiazole, whereas 3-pyridyl analogs were inactive. The requirement of a thiazole NH suggests an H-bond contact in this region. Substituents at the 2-position on the aryl ring were not tolerated. With these constraints in mind, we prepared a focused set of 19 analogs of STF-62247 to probe structure-activity relationships (SAR) (Table S1 supplementary information). The IC50 for these analogs shows that the STF-62247 has the lowest IC50 in VHL-deficient RCC4 cells and the highest IC50 ratio (VHL/VHL-deficient cells). The compound 30603 with a methyl group in 4-position also has a high ratio but also a higher IC50 for both RCC4 and RCC4/VHL cells. The IC50 could not be determined for some compounds because of poor solubility.
To associate the cytotoxicity of these compounds with autophagy, the vacuole formation and LC3 processing were evaluated (Figure 5A–B). Compounds with an IC50 ratio VHL/VHLdeficient cells >5 correlated positively with the presence of large vacuoles in VHL-deficient cells and the processing of LC3. As observed with STF-62247 treatment, the LC3-lipidated form was present in both VHL wild type and deficient cell lines. To confirm that the cell death observed in VHL-deficient cells is associated with the acidification of autophagic vesicules, the level of AVOs was measured in response to 5 analogs (Figure 5C). The amount of AVOs is consistently higher in VHL-deficient cells treated with compounds 30603, 31184, and 30651, which had relatively low IC50 and induced significant vacuole formation and the LC3-lipidated form. In contrast, AVO staining was not influenced in response to compound 31185 and 31270, which induced neither autophagy nor cytotoxicity. These results confirm that the autophagic cell death observed in VHL-deficient cells is dependent on the acidification of the vacuoles. The strong relationship between autophagic cell death and vacuole acidification might explain why autophagy leads to cell death in VHL-deficient cells and not in wild-type cells.
Figure 5. Evaluation of autophagy in RCC treated with analogs of the STF-62247.
A. RCC4 and RCC4/VHL were treated with 1.25 μM of each 19 analogs for 20 hours and the presence of vacuoles was examined under light microscopy. Scale bar, 10 μm. B. Western blot for LC3 after treatment with each compound. Tubulin was used as loading control. Ratio is the relative from the IC50 in RCC4/VHL cells/IC50 in RCC4 cells. C. Acidic vesicle organelles (AVO) measured by FACS analysis in cells treated with 1.25 μM of STF-62247 or with compounds 30603, 30651, 31184, 31187 and 31270 and stained with acridine orange. All error bars represent SEM.
Use of the yeast deletion pool to identify targets of STF-62247
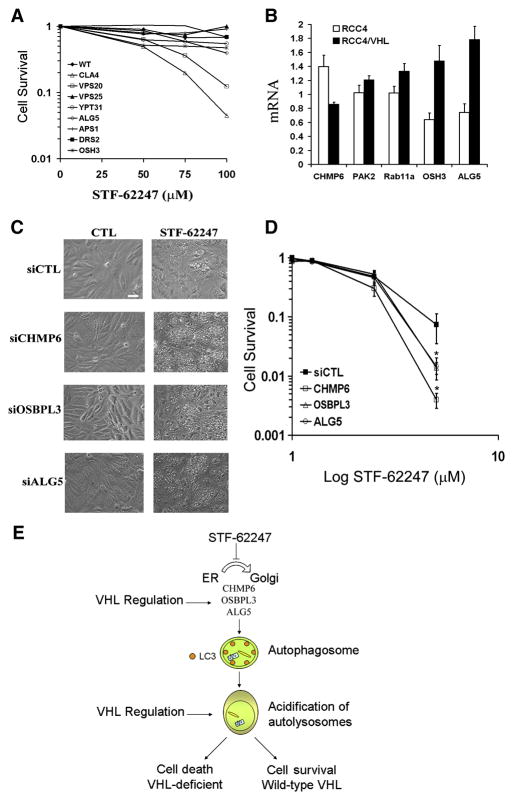
To identify a target for STF-62247, we performed a screen of the yeast deletion pool to determine which genes affect STF-62247 sensitivity. We found that cells lacking proteins involved in Golgi trafficking and in morphogenesis were most sensitive to STF-62247 (Table S2 supplementary information). To validate the data obtained from this screen, we evaluated the sensitivity of 8 deletion yeast strains following STF-62247 treatment (Figure 6A). The killing curves for CLA4, VPS20, ALG5, OSH3, and YPT31 deletion strains show a significant toxicity in response to STF-62247. The Atg8 deletion strain did not exhibit sensitivity to STF-62247 as expected based on our human ATG studies (data not shown). The human orthologs known for CLA4, VPS20, ALG5, OSH3 and YPT31 strains are PAK2, CHMP6, ALG5, OSBPL3 and RAB11A, respectively. In mammalian cells, we found that the expression of ALG5, OSBPL3, and CHMP6, correlated with the VHL status (Figure 6B). These three proteins play a role in coordinating vesicular transport between the ER, the trans-Golgi network (TGN) and lysosomes. Moreover, the knockdown of these proteins using siRNA (Figure S4) increases the vacuole formation in response to STF-62247 (Figure 6C) and cytotoxicity in VHL cells suggesting their loss partially phenocopies the loss of VHL (Figure 6D). Taken together, our results demonstrate that the Trans-Golgi network is a target of the STF-62247 and a drug-selective pathway synthetically lethal in VHL-deficient cells.
Figure 6. Golgi trafficking proteins are sensitive and a target for the STF-62247.
A. Killing curves for 8 deletion yeast strains. Wild-type, ALG5, APS1, CLA4, DRS2, OSH3, VPS20, VPS25, YPT31 were incubated with STF-62247 (0–100 μM) for 1 hour and colony formation was evaluated after two days. Each point of the survival is calculated by the average of two different experiments in triplicate and as the percent of treated on untreated plate. B. Quantitative real time PCR for CHMP6, PAK2, RAB11a, OSBPL3 and ALG5 in RCC4 and RCC4/VHL. Histogram is calculated by the average of three different experiments in triplicate as the percent of RCC4/VHL on RCC4 cells. The error bars are represented as SEM and *p<0.05, **p<0.01. C. VHL cells were transiently transfected with siRNA for CHMP6, OSBPL3 and ALG5 for 72 hours. The last 24 hours, 1.25 μM of STF-62247 was added and the vacuole formation was examined under light microscopy. Scale bar, 10 μm. D. Clonogenic assay with STF-62247 (0–30 μM) in RCC4/VHL cells transiently transfected in the same condition than in C. Colony formation was evaluated as described before. All error bars represent SEM. *p<0.05 represents the difference between VHL cells and VHL with siRNA in presence of 5 μM of STF-62247. E. Representative schema of the effect of STF-62247 in RCC through autophagy. STF-62247 disrupts ER-Golgi trafficking through CHMP6, OSBPL3 and ALG5 and induces autophagy. Lipidation of LC3 associated with the double membrane of autophagosome is observed in an ATG5-dependent manner. A higher acidification of the autolysosomes is associated with autophagic cell death in VHL-deficient cells whereas wild-type VHL cells are protected against acidification and survive. A regulation of CHMP6, OSBPL3 and ALG5 proteins by VHL is also observed and remains to be investigated.
Discussion
Targeting HIF in cancer therapy is ardently being investigated. Different screening approaches have identified drugs, cytotoxin or small molecules as HIF inhibitors. It has been reported that radiation in combination with a HIF inhibitor could improve conventional therapies (Moeller et al., 2005). In RCCs with constitutively elevated levels of HIF, different agents that can disrupt or attenuate the HIF pathway or the protein products of HIF target genes are presently in clinical trials or approved by the U.S. Food and Drug Administration (FDA). For example, temsirolimus (CCI-779) and everolimus (RAD001) inhibit HIF through the targeting of mTOR. In contrast, agents with anti-angiogenic activity that inhibit VEGFR and PDGFR signaling (e.g. sorafenib, sunitinib), the VEGF ligand (bevacizumab), and the EGF ligand (cetuximab) have also shown some effectiveness in the management of renal cell cancer to different degrees (Patel et al., 2006; Sosman et al., 2007). These agents all have been reported to have antitumor effects on kidney tumors, but tumors ultimately progress regardless of these therapies as they are not cytotoxic but cytostatic. In the kidney, the inactivation of the tumor suppressor gene VHL is an early event that promotes tumorigenesis, leading to clear cell-renal cell cancer. We hypothesized that inactivation of VHL in RCC progression can be therapeutically exploited using a screening strategy that targets cells with loss of VHL. While different agents have been used to target the HIF pathway, we investigated an approach of using small molecules to induce selective cell death in RCC that lack a functional VHL gene, the mammalian equivalent of a yeast synthetic lethal screen. Among drugs identified from this screen (manuscript in preparation), we found that STF-62247 is cytotoxic to VHL-deficient cells. Surprisingly, cell death induced by STF-62247 is independent of HIF-1α or HIF-2α and was unaffected by hypoxia (data not shown). Our results support other reports that demonstrate in addition to its well understood role in HIF degradation, VHL has other critical targets that also play important roles in cellular homeostasis (Hergovich et al., 2003; Lolkema et al., 2005; Na et al., 2003; Ohh et al., 1998; Okuda et al., 2001).
It has been proposed that the resistance of RCC to chemotherapy and radiotherapy is due to increased levels of the nuclear factor kB activity and resistance to apoptosis (An et al., 2005; Oya et al., 2001; Qi and Ohh, 2003). Previous studies have reported that the metabolic stress observed in human tumors leads cancer cells to acquire resistance to apoptosis and to stimulate autophagy to maintain energy demand and prevent necrosis (Jin et al., 2007). Increasing evidence implicates a role for autophagy in cancer, but it is still not well understood how cellular and environmental cues drive autophagic cells down survival or death pathways (Jin et al., 2007; Mathew et al., 2007). It has been reported that above a certain threshold which destroys organelles and portions of the cytoplasm, autophagic cell death is induced (Gozuacik and Kimchi, 2007). It is also possible that autophagy can kill by selective degradation of vital proteins in the cell. Anti-cancer therapies using radiation or agents such as tamoxifen, rapamycin, imatinib, arsenic trioxide, resveratrol or soybean B-group have recently been reported to induce autophagic cell death (Bursch et al., 1996; Ellington et al., 2005; Ertmer et al., 2007; Ito et al., 2005; Kanzawa et al., 2003; Opipari et al., 2004; Takeuchi et al., 2005). We provide evidence that STF-62247 induces autophagy and that inhibition of autophagy significantly increases the survival of VHL-deficient cells to STF-62247.
Among different stimuli, signaling cascades or proteins that have been reported to be involved in the initial signal for autophagy, ER stress and Golgi trafficking were identified in the formation of autophagosomes (Ogata et al., 2006; Yorimitsu et al., 2006; Young et al., 2006). Also, the PI3K-beclin-1 (Atg6) complex (Furuya et al., 2005), the PI3K/AKT/mTOR cascade (Takeuchi et al., 2005), the Ras/MEK/ERK pathway (Corcelle et al., 2006) and the p53, p73 family (Crighton et al., 2007; Crighton et al., 2006) have also been found to regulate the autophagic process. Thus, it appears that the upstream signals to induce autophagy are stimuli-and organism-dependent. In our study, we show that the Golgi trafficking and PI3K pathways are involved in the STF-62247 upstream signal for autophagosome formation. More specifically, yeast deletion strains for ALG5, OSH3, and VPS20 are sensitive to STF-62247. In mammalian cells, knockdown of the equivalent protein orthologs (ALG5, OSBPL3, and CHMP6 respectively) that are located in the ER and the trans-Golgi network are found to be lethal in combination with STF-62247 in cells with a functional VHL. While knockdown of ALG5, an enzyme found in the membrane of the ER, sensitizes cell to death in cells with functional VHL, an ER stress induced by tunicamycin has no apparent effect in response to STF-62247. ALG5 catalyzes the transfer of glucose from UDP-glucose to dolichyl phosphate. A deletion of the yeast gene leads to a loss of this activity and an underglycosylation of carboxypeptidase Y (Heesen et al., 1994). However, the ER stress induced by tunicamycin that leads to autophagy increases the unfolded protein response (UPR) and induces the expression of chaperones and proteins involved in the recovery process (Yorimitsu et al., 2006). ER stress induced autophagy has been reported in yeast and in neuroblastoma but is distinct from autophagy induced by STF-62247 (Ogata et al., 2006; Yorimitsu et al., 2006). Our results also show that oxysterol binding protein-like protein 3 (OSBPL3), a protein involved in a variety of processes such as secretory vesicle budding from the Golgi and establishment of cell polarity, the glucosyltransferase ALG5 and CHMP6, a key component of the ESCRT III required for transport of protein into the multivesicular body pathway to the lysosomal/vacuolar lumen are regulated by VHL (Castro et al., 1999; Perry and Ridgway, 2006; Yorikawa et al., 2005). However, the mechanism of regulation of these proteins by VHL is unknown and remains to be investigated. Recently, it was reported that disruption of the ESCRTIII complex induces autophagosome formation and neurodegeneration consistant with our results (Lee et al., 2007). A role for sterol proteins in elongation of the autophagosome membrane has previously been shown (Nazarko et al., 2007). In addition, STF-62247-induced autophagy was blocked in presence of BFA, a compound that causes the trans-Golgi network (TGN) to reassemble into extensive tubular processes and merges with the recycling endosomal system. Our results suggest that certain proteins associated with the trans-Golgi network and the ER are targeted by STF-62247 in VHL-deficient RCC. Furthermore, we show that ATG9, the only transmembrane ATG is involved in autophagic cell death in response to STF-62247. The mammalian ATG9 has been implicated in trafficking between Golgi and endosomes supporting a role for Golgi in the initial signal of autophagy (Young et al., 2006). Recently, a study reported that the transcript of sONE, an antisense of eNOS and a yeast homolog of Atg9, is upregulated in the absence of VHL function through a HIF-independent manner, and may interfere with the HuR/VHL interaction (Fish et al., 2007).
We established that autophagosome formation occurs in both VHL-deficient and VHL wild-type cells after STF-62247 treatment. Interestingly, our analyses indicate that the drug stimulates the maturation of autophagosomes into autolysosomes in VHL-deficient cells and that the size of autolysosomes is larger in VHL-deficient cells. The acidification present after fusion with the lysosomes in VHL-inactivated cells was prevented by the treatment with NH4Cl (Figure S3F). Although the fusion of autophagosomes with lysosomes is known, the maturation steps and proteins implicated remain to be elucidated. It was reported that microtubules (Kochl et al., 2006), GTPase Rab7 (Gutierrez et al., 2004), and the lysosomal protein LAMP1 and 2 (Eskelinen et al., 2002) are essential for the endosome/lysosome fusion process. Disruption or inactivation of these proteins has been shown to block the maturation of autophagic vacuoles likely before the fusion with the lysosomes. Preliminary results indicate that knockdown of LAMP1 or LAMP2 did not influence the toxicity of STF-62247 in VHL-deficient cells or in cells with functional VHL, and therefore cannot explain the difference in acidic vesicle organelles observed. However, our results suggest that VHL-deficient cells are altered in the autolysosomal maturation step increasing the acidification of vesicular organelles.
Many chemotherapeutic agents are limited in their therapeutic index due to cytotoxicity to non-tumor cells. To develop new compounds with higher therapeutic indices, targeted genetic approaches should be considered (Giaccia et al., 2003; Sutphin et al., 2004). Synthetic lethality describes that combination of two nonallelic mutations, which individually are not lethal, result in cell death. This concept has been used successfully in yeast and Drosophila and shows a potential means of achieving selective toxicity based on genotypic differences between tumor cells and normal tissue (Hartman et al., 2001; Kaelin, 2005). In this study, we found that renal cancer cells deficient in VHL were sensitive to the small molecule STF-62247, whereas cells with wild-type VHL were unaffected by STF-62247 treatment, establishing a synthetic lethal situation where both drug treatment and VHL deficiency lead to lethality. We also identified proteins involved in Golgi trafficking as a targeting pathway for STF-62247 and demonstrate that STF-62247 in combination with the loss of VHL, leads to synthetic lethality. This synthetic lethal approach should be considered in the future for the continued development of other molecular targeting agents for the treatment of renal cell cancer as they have the benefit of selectively killing tumor cells based on their genotype.
Experimental Procedures
Cell Culture and Cell Treatments
RCC4 parental and RCC4 with VHL-reintroduced (RCC4/VHL), SN12C and SN12C-CSCG-VHL shRNA (gift from George V. Thomas, UCLA), MEF Atg5+/+ and Atg5−/− (gift from Noboru Mizushima, Japan) were maintained in DMEM supplemented with 10% FCS. Cells were plated at 70% confluency, and treated with 1.25 μM (RCC4) or 3.75 μM (SN12C) of STF-62247 overnight in presence or absence of 2 mM 3-MA, 1 μg/ml Brefeldin A, 15 μM LY294002, 2 μg/ml Tunicamycin, or 10 μM U0126 (Sigma Chemical Co).
Cell Viability and XTT assays
For cell viability, 100,000 cells were plated in a 12-well plate. The following day, 1.25 μM STF-62247 was added in the presence or absence of 1 mM 3-MA for 24 hours at 37°C. Cells were trypsinized and counted by trypan blue exclusion. For XTT assays, 5000 RCC4 with and without VHL cells or 2,500 SN12C with and without VHL shRNA cells were plated in 96-well plates. The following day, vehicle (DMSO), STF-62247 or each of 19 analogs were added to media by serial dilution. Four days later, the media was aspirated and XTT solution containing 0.3 mg/ml of XTT (Sigma Chemical Co) in Phenol Red-free media, 20% FCS and 2.65 mg/ml N-methyl dibenzopyrazine methyl sulfate (PMS) was added to the cells and incubated at 37°C for 1–2 hours. Metabolism of XTT was quantified by measuring the absorbance at 450 nm on a plate reader.
Clonogenic Cell Survival Assay
RCC4, RCC4/VHL, SN12C and SN12C-VHL shRNA were plated at 300 cells per 60 mm tissue culture dish. The following day, vehicle or STF-62247 was added and cells were incubated in the presence of drug for 10 days in RCC4 and RCC4/VHL and 7–10 days for SN12C and SN12C-VHL shRNA. The colonies were stained with crystal violet, and quantified. Each experiment was performed at least three times in triplicate.
Western blot analysis
After treatments, cells were lysed in urea buffer (9M urea, 75mM Tris (pH 7.5), 150mM b-mercaptoethanol), placed on ice for 10 minutes and sonicated briefly (15 seconds). Extracts were centrifugated at 8,000 x g for 10 minutes to eliminate insoluble material. Lysates were quantitated using Bio-Rad protein Assay (Bio-Rad Laboratories) and 20–30 μg of proteins were separated on a 7.5% or 15% bis-acrylamide gels and then transferred onto PVDF membranes. Proteins were visualized using HIF-2α (Novus Biological), HIF-1α, pVHL and BiP (BD Biosciences), LC3 (MBL), Atg5 (gift from Dr. Mizushima), or α-tubulin antibodies.
Quantification of Acidic Vesicule Organelles with Acridine Orange
In acridine orange-stained cells, the cytoplasm and nucleus are bright green and dim red, whereas acidic compartments are bright red. The intensity of the red fluorescence is proportional to the degree of acidity. Following 20 hours of treatment with STF-62247 or analogs, cells were stained with 1 μg/ml of acridine orange for 15 min. Cells were trypsinized and then analyzed by flow cytometry using FACScan cytometer and CellQuest software.
Immunofluorescence staining
Cells were seeded on glass coverslips and at subconfluence, STF-62247 was added at 1.25 μM for RCC4 and 3.75 μM for SN12C. After 20 hours, cells were fixed in 3.7% formaldehyde for 10 min at room temperature and washed in PBS. Then cells were permeabilized in 0.2% Triton X-100 for 5 min at room temperature and washed in PBS. The coverslips were incubated with primary antibody LC3 diluted 1:500 in PBS containing 0.2% BSA for 1 hour at room temperature. Cells were washed three times in PBS-BSA and the bound primary antibody was detected using a fluorescein-conjugated secondary antibody (ALEXA) diluted 1:500 in PBS-BSA and incubated for 1 hour. Coverslips were washed and mounted in mounting medium from Vector Laboratories according to the manufacturer’s instructions. Cells were examined with a confocal microscope
Visualization of Monodansylcadaverine (MDC) vacuoles
Cell were incubated in the presence of drug for 20 hours and labeled with 50 mM of the autofluorescent marker MDC (Sigma Chemical Co) in PBS at 37°C for 10 minutes. Then, cells are fixed with 3.7% formaldehyde for 10 min at room temperature and washed in PBS. MDC was observed by fluorescence microscopy (Nikon Eclipse E800).
In vivo studies
All animal experiments were performed in accordance with institutional and national guidelines and approved by Stanford’s Administrative Panel on Laboratory Animal Care (APLAC). Male scid mice, 4 to 6 weeks old, were obtained from either Stanford University’s Research Animal Facility or Charles River Laboratories. SN12C (2–3 × 106), SN12C-VHL shRNA (2–3 × 106), or 786-O (5 × 106) cells were resuspended in sterile PBS and injected subcutaneously. Mice were randomized into vehicle control group or treatment group and treated daily by intraperitoneal injection. When tumors reached 100 mm3, mice in the treatment group implanted with SN12C or SN12C-VHL shRNA cells were treated with 8 mg/kg or 2.7 mg/kg of STF-62247 whereas 2 weeks after implanted 786-O cells, mice were treated with 8 mg/kg of STF-62247. Tumor growth was measured every 2–3 days after the drug treatment was started.
shRNA constructs, plasmids and retroviral infection
The CMV-EYFP fluorescent protein and the HIF-2α P531A/P847A double mutant were cloned into pBabe-puro vector. The sequence used for HIF-1α and HIF-2α shRNA were gaggaactaccattatat and ggagacggaggtggtctat respectively. Retrovirus were produced by transfecting plasmids into ΦNX-amphotropic cell lines according to manufacturer’s instructions with Lipofectamine Plus (Invitrogen). The next day after transfection, cells were placed at 32°C. Retrovirus was harvested 48 and 72 hours after transfection. Puromycin was used to select for stable retroviral integrants.
Transient transfection of RCC
Cells were transfected with siRNA against ATG5, CHMP6, PAK2, RAB11a, OSBPL3, ALG5, ATG7, ATG9 (SMART pool) and siControl2 non-targeting pool from Dharmacon using Dharmafect Reagent 1 for 48 hours. Cells were split for clonogenic assay, harvested for RNA or protein.
Screen in yeast and killing curves
Genotypes of the parental yeast strain BY4743, construction of the homozygous diploid deletion strains, and construction of the homozygous diploid deletion pool have been described previously (Thomas et al., 2006). A mutant pool of 4728 nonessential homozygous diploid deletion strains was used. The parental diploid strain BY4743 was used as control in survival and complementation assays. For clonogenic survival, equivalent numbers of early log-phase cells parental and deletion strains for ALG5, APS1, OSH3, CLA4, DRS2, DAP1, VPS20, VPS25, FIG4, SPT4 were suspended in yeast extract peptone dextrose (YPD) and treated with STF-62247 (0–100 μM) for 1 h while being shaken at 300 rpm at 30°C. Cells were then pelleted, washed once with buffer (10 mM Tris and 5 mM EDTA), pelleted, and resuspended in the same buffer. Cells were then plated at appropriate dilutions onto YPD solid medium to allow for accurate counting of surviving colonies (range, 50–250). Plates were incubated for 2 days at 30°C before counting colonies. Full survival curves were performed in triplicates for each selected deletion strain.
Quantitative RT-PCR (qRT-PCR)
RNA from cells transfected with siControl or siAtg was prepared using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. The reverse transcription was performed with Moloney Murine Leukemia Virus MMLV reverse transcriptase (Invitrogen) using two μgrams of RNA and 5 μ_M_-random primers (Invitrogen) according to the manufacturer’s instructions. Approximately 0.5% of each RT reaction was used for the quantitative RT (qRT)-PCR reactions containing 5 μl 2× SYBR Green master mix (ABI, Foster City, CA) and 200 n_M_ forward and reverse primers specific of the different genes of interest in a volume of 10 μl. Detection and data analysis was carried out with the ABI PRISM 7900 sequence detection system using TBP primers as an internal control. PCR primer sequences are obtained from the Primer Bank database (http://pga.mgh.harvard.edu/primerbank/) and synthesized in the Stanford Protein and Nucleic Acid Biotechnology Facility.
Supplementary Material
01
02
03
Acknowledgments
This work was supported by a Canadian Institutes of Health Research (CIHR Fellowship (ST); NCI-CA-082566 and by a gift from the Silicon Valley Community Foundation; NCI-CA-123823 (DAC). Authors want to thank Dr. Noboru Mizushima for the MEFs Atg5 cells and the Atg5 antibody and also Dr. George Thomas for SN12C and SN12C-VHL shRNA cells. The authors would also like to thank Dr. Martin Brown, Dr. Jim Brown and Kelly McCann for their technical assistance with the yeast deletion pool and Dr. Trent A. Watkins for help with the time-lapse microscopy.
Abbreviations
ATG
autophagy-related gene
AVO
Acidic vesicle organelles
BECN
Beclin-1
BFA
Brefeldin A
CHMP6
charged multivesicular body protein 6
EGFR
epidermal growth factor receptor
HIF
hypoxia inducible factor
LAMP1 or 2
lysosome-associated membrane protein 1 or 2
MAP-LC3
microtubule associated protein light chain 3
3-MA
3-Methyladenine
NFkB
nuclear factor kB
OSBPL3
oxysterol binding protein-like 3
PDGF
platelet-derived growth factor
PI3K
phosphatidylinositol 3-kinase
RCCs
renal cell carcinomas
TGF-α
transforming growth factor-α
TGN
Trans Golgi network
VEGF
vascular endothelial growth factor
VHL
von Hippel-Lindau
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- An J, Fisher M, Rettig MB. VHL expression in renal cell carcinoma sensitizes to bortezomib (PS-341) through an NF-kappaB-dependent mechanism. Oncogene. 2005;24:1563–1570. doi: 10.1038/sj.onc.1208348. [DOI] [PubMed] [Google Scholar]
- Bursch W, Ellinger A, Kienzl H, Torok L, Pandey S, Sikorska M, Walker R, Hermann RS. Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis. 1996;17:1595–1607. doi: 10.1093/carcin/17.8.1595. [DOI] [PubMed] [Google Scholar]
- Castro O, Chen LY, Parodi AJ, Abeijon C. Uridine diphosphate-glucose transport into the endoplasmic reticulum of Saccharomyces cerevisiae: in vivo and in vitro evidence. Mol Biol Cell. 1999;10:1019–1030. doi: 10.1091/mbc.10.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DA, Sutphin PD, Denko NC, Giaccia AJ. Role of prolyl hydroxylation in oncogenically stabilized hypoxia-inducible factor-1alpha. J Biol Chem. 2002;277:40112–40117. doi: 10.1074/jbc.M206922200. [DOI] [PubMed] [Google Scholar]
- Corcelle E, Nebout M, Bekri S, Gauthier N, Hofman P, Poujeol P, Fenichel P, Mograbi B. Disruption of autophagy at the maturation step by the carcinogen lindane is associated with the sustained mitogen-activated protein kinase/extracellular signal-regulated kinase activity. Cancer Res. 2006;66:6861–6870. doi: 10.1158/0008-5472.CAN-05-3557. [DOI] [PubMed] [Google Scholar]
- Crighton D, O’Prey J, Bell HS, Ryan KM. p73 regulates DRAM-independent autophagy that does not contribute to programmed cell death. Cell Death Differ. 2007;14:1071–1079. doi: 10.1038/sj.cdd.4402108. [DOI] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Ellington AA, Berhow M, Singletary KW. Induction of macroautophagy in human colon cancer cells by soybean B-group triterpenoid saponins. Carcinogenesis. 2005;26:159–167. doi: 10.1093/carcin/bgh297. [DOI] [PubMed] [Google Scholar]
- Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, Chi JT, Jeffrey SS, Giaccia AJ. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- Ertmer A, Huber V, Gilch S, Yoshimori T, Erfle V, Duyster J, Elsasser HP, Schatzl HM. The anticancer drug imatinib induces cellular autophagy. Leukemia. 2007 doi: 10.1038/sj.leu.2404606. [DOI] [PubMed] [Google Scholar]
- Eskelinen EL, Illert AL, Tanaka Y, Schwarzmann G, Blanz J, Von Figura K, Saftig P. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell. 2002;13:3355–3368. doi: 10.1091/mbc.E02-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JE, Matouk CC, Yeboah E, Bevan SC, Khan M, Patil K, Ohh M, Marsden PA. Hypoxia-inducible Expression of a Natural cis-Antisense Transcript Inhibits Endothelial Nitric-oxide Synthase. J Biol Chem. 2007;282:15652–15666. doi: 10.1074/jbc.M608318200. [DOI] [PubMed] [Google Scholar]
- Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- Giaccia A, Siim BG, Johnson RS. HIF-1 as a target for drug development. Nat Rev Drug Discov. 2003;2:803–811. doi: 10.1038/nrd1199. [DOI] [PubMed] [Google Scholar]
- Gnarra JR, Zhou S, Merrill MJ, Wagner JR, Krumm A, Papavassiliou E, Oldfield EH, Klausner RD, Linehan WM. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc Natl Acad Sci U S A. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozuacik D, Kimchi A. Autophagy and cell death. Curr Top Dev Biol. 2007;78:217–245. doi: 10.1016/S0070-2153(06)78006-1. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- Hartman JLt, Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291:1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- Heesen S, Lehle L, Weissmann A, Aebi M. Isolation of the ALG5 locus encoding the UDP-glucose:dolichyl-phosphate glucosyltransferase from Saccharomyces cerevisiae. Eur J Biochem. 1994;224:71–79. doi: 10.1111/j.1432-1033.1994.tb19996.x. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Lisztwan J, Barry R, Ballschmieter P, Krek W. Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat Cell Biol. 2003;5:64–70. doi: 10.1038/ncb899. [DOI] [PubMed] [Google Scholar]
- Iliopoulos O, Kibel A, Gray S, Kaelin WG., Jr Tumour suppression by the human von Hippel-Lindau gene product. Nat Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- Iliopoulos O, Levy AP, Jiang C, Kaelin WG, Jr, Goldberg MA. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc Natl Acad Sci U S A. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs JS, Jung YJ, Mimnaugh EG, Martinez A, Cuttitta F, Neckers LM. Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 alpha-degradative pathway. J Biol Chem. 2002;277:29936–29944. doi: 10.1074/jbc.M204733200. [DOI] [PubMed] [Google Scholar]
- Ito H, Daido S, Kanzawa T, Kondo S, Kondo Y. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int J Oncol. 2005;26:1401–1410. [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Jin S, Dipaola RS, Mathew R, White E. Metabolic catastrophe as a means to cancer cell death. J Cell Sci. 2007;120:379–383. doi: 10.1242/jcs.03349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. Embo J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- Kanzawa T, Kondo Y, Ito H, Kondo S, Germano I. Induction of autophagic cell death in malignant glioma cells by arsenic trioxide. Cancer Res. 2003;63:2103–2108. [PubMed] [Google Scholar]
- Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knebelmann B, Ananth S, Cohen HT, Sukhatme VP. Transforming growth factor alpha is a target for the von Hippel-Lindau tumor suppressor. Cancer Res. 1998;58:226–231. [PubMed] [Google Scholar]
- Kochl R, Hu XW, Chan EY, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 2006;7:129–145. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- Kondo K, Kim WY, Lechpammer M, Kaelin WG., Jr Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003;1:E83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III Dysfunction Causes Autophagosome Accumulation and Neurodegeneration. Curr Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Lolkema MP, Gervais ML, Snijckers CM, Hill RP, Giles RH, Voest EE, Ohh M. Tumor suppression by the von Hippel-Lindau protein requires phosphorylation of the acidic domain. J Biol Chem. 2005;280:22205–22211. doi: 10.1074/jbc.M503220200. [DOI] [PubMed] [Google Scholar]
- Mabjeesh NJ, Escuin D, LaVallee TM, Pribluda VS, Swartz GM, Johnson MS, Willard MT, Zhong H, Simons JW, Giannakakou P. 2ME2 inhibits tumor growth and angiogenesis by disrupting microtubules and dysregulating HIF. Cancer Cell. 2003;3:363–375. doi: 10.1016/s1535-6108(03)00077-1. [DOI] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller BJ, Dreher MR, Rabbani ZN, Schroeder T, Cao Y, Li CY, Dewhirst MW. Pleiotropic effects of HIF-1 blockade on tumor radiosensitivity. Cancer Cell. 2005;8:99–110. doi: 10.1016/j.ccr.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Motzer RJ, Bander NH, Nanus DM. Renal-cell carcinoma. N Engl J Med. 1996;335:865–875. doi: 10.1056/NEJM199609193351207. [DOI] [PubMed] [Google Scholar]
- Na X, Duan HO, Messing EM, Schoen SR, Ryan CK, di Sant’Agnese PA, Golemis EA, Wu G. Identification of the RNA polymerase II subunit hsRPB7 as a novel target of the von Hippel-Lindau protein. Embo J. 2003;22:4249–4259. doi: 10.1093/emboj/cdg410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarko TY, Farre JC, Polupanov AS, Sibirny AA, Subramani S. Autophagy-related pathways and specific role of sterol glucoside in yeasts. Autophagy. 2007;3:263–265. doi: 10.4161/auto.3907. [DOI] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohh M, Yauch RL, Lonergan KM, Whaley JM, Stemmer-Rachamimov AO, Louis DN, Gavin BJ, Kley N, Kaelin WG, Jr, Iliopoulos O. The von Hippel-Lindau tumor suppressor protein is required for proper assembly of an extracellular fibronectin matrix. Mol Cell. 1998;1:959–968. doi: 10.1016/s1097-2765(00)80096-9. [DOI] [PubMed] [Google Scholar]
- Okuda H, Saitoh K, Hirai S, Iwai K, Takaki Y, Baba M, Minato N, Ohno S, Shuin T. The von Hippel-Lindau tumor suppressor protein mediates ubiquitination of activated atypical protein kinase C. J Biol Chem. 2001;276:43611–43617. doi: 10.1074/jbc.M107880200. [DOI] [PubMed] [Google Scholar]
- Opipari AW, Jr, Tan L, Boitano AE, Sorenson DR, Aurora A, Liu JR. Resveratrol-induced autophagocytosis in ovarian cancer cells. Cancer Res. 2004;64:696–703. doi: 10.1158/0008-5472.can-03-2404. [DOI] [PubMed] [Google Scholar]
- Oya M, Ohtsubo M, Takayanagi A, Tachibana M, Shimizu N, Murai M. Constitutive activation of nuclear factor-kappaB prevents TRAIL-induced apoptosis in renal cancer cells. Oncogene. 2001;20:3888–3896. doi: 10.1038/sj.onc.1204525. [DOI] [PubMed] [Google Scholar]
- Patel PH, Chadalavada RS, Chaganti RS, Motzer RJ. Targeting von Hippel-Lindau pathway in renal cell carcinoma. Clin Cancer Res. 2006;12:7215–7220. doi: 10.1158/1078-0432.CCR-06-2254. [DOI] [PubMed] [Google Scholar]
- Perry RJ, Ridgway ND. Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol Biol Cell. 2006;17:2604–2616. doi: 10.1091/mbc.E06-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Ohh M. The von Hippel-Lindau tumor suppressor protein sensitizes renal cell carcinoma cells to tumor necrosis factor-induced cytotoxicity by suppressing the nuclear factor-kappaB-dependent antiapoptotic pathway. Cancer Res. 2003;63:7076–7080. [PubMed] [Google Scholar]
- Rapisarda A, Uranchimeg B, Scudiero DA, Selby M, Sausville EA, Shoemaker RH, Melillo G. Identification of small molecule inhibitors of hypoxia-inducible factor 1 transcriptional activation pathway. Cancer Res. 2002;62:4316–4324. [PubMed] [Google Scholar]
- Smukste I, Bhalala O, Persico M, Stockwell BR. Using small molecules to overcome drug resistance induced by a viral oncogene. Cancer Cell. 2006;9:133–146. doi: 10.1016/j.ccr.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Sosman JA, Puzanov I, Atkins MB. Opportunities and obstacles to combination targeted therapy in renal cell cancer. Clin Cancer Res. 2007;13:764s–769s. doi: 10.1158/1078-0432.CCR-06-1975. [DOI] [PubMed] [Google Scholar]
- Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. 2003;425:307–311. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- Sutphin PD, Chan DA, Giaccia AJ. Dead cells don’t form tumors: HIF-dependent cytotoxins. Cell Cycle. 2004;3:160–163. [PubMed] [Google Scholar]
- Takeuchi H, Kondo Y, Fujiwara K, Kanzawa T, Aoki H, Mills GB, Kondo S. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–3346. doi: 10.1158/0008-5472.CAN-04-3640. [DOI] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem. 2002;277:13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- Thomas GV, Tran C, Mellinghoff IK, Welsbie DS, Chan E, Fueger B, Czernin J, Sawyers CL. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nat Med. 2006;12:122–127. doi: 10.1038/nm1337. [DOI] [PubMed] [Google Scholar]
- Yorikawa C, Shibata H, Waguri S, Hatta K, Horii M, Katoh K, Kobayashi T, Uchiyama Y, Maki M. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem J. 2005;387:17–26. doi: 10.1042/BJ20041227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Zimmer M, Doucette D, Siddiqui N, Iliopoulos O. Inhibition of hypoxia-inducible factor is sufficient for growth suppression of VHL−/− tumors. Mol Cancer Res. 2004;2:89–95. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03