Model Systems for Examining Effects of Leukemia-Associated Oncogenes in Primary Human CD34+ Cells via Retroviral Transduction (original) (raw)
. Author manuscript; available in PMC: 2010 Feb 22.
Summary
The use of primary human cells to model cancer initiation and progression is now within the grasp of investigators. It has been nearly a decade since the first defined genetic elements were introduced into primary human epithelial and fibroblast cells to model oncogenesis. This approach has now been extended to the hematopoietic system, with the first described experimental transformation of primary human hematopoietic cells. Human cell model systems will lead to a better understanding of the species and cell type specific signals necessary for oncogenic initiation and progression, and will allow investigators to interrogate the cancer stem cell hypothesis using a well-defined hierarchical system that has been studied for decades. The molecular and biochemical link between self-renewal and differentiation can now be experimentally approached using primary human cells. In addition, the models that result from these experiments are likely to generate highly relevant systems for use in identification and validation of potential therapeutic targets as well as testing of small molecule therapeutics. We describe here the methodologies and reagents that are used to examine the effects of leukemia fusion protein expression on primary human hematopoietic cells, both in vitro and in vivo.
Keywords: Human HSPC, CD34, Retroviral transduction, Leukemia fusion genes, Differentiation, NOD/SCID, Xenograft
1. Introduction
Many different systems are now available for modeling leukemia, but by far the most popular model is the mouse, using either genetically engineered mice or a bone marrow transduction and transplantation system (1)(refer to Chapter “Retroviral/Lentiviral Transduction and Transformation Assay”). However, it is becoming increasingly clear that there are important differences between murine and human cells with respect to cellular transformation (2–4). Species-specific differences in ras signaling as well as in telomerase and telomere length regulation limit the conclusions that can be reached when using murine cells to model human cancer (5–7). The inbred nature of the mouse strains used for these studies further complicates the extrapolation of results to humans. For these reasons, some labs are now using human cells to study oncogenesis, and the controlled transformation of primary human fibroblasts and epithelial cells has been previously reported (8, 9). Similar studies have been pursued using hematopoietic stem and progenitor cells (HSPCs) to model preleukemia (10–14), and recently the experimental transformation of a primary human HSPC has been accomplished in the laboratory (15).
The strategy that is used in these studies with primary human HSPC necessarily involves the introduction of oncogenes using retroviral or lentiviral delivery systems. The specific HSPC that is used, whether the cord blood, bone marrow or mobilized CD34+ (or lineage negative) cell, may alter the results that are obtained, since it is now recognized that these cells differ significantly in their properties (16). In addition, many delivery options are available using different viral promoters, marker genes, and viral envelopes. These options give great flexibility to the investigator for pursuing different experimental approaches as well as in using combinations of genes together in the same experiment. The in vitro and in vivo systems in which to examine the phenotype upon oncogene expression are also expanding. As we learn more about the specific signals that are important in self-renewal divisions of the human HSPC, we strive to mimic these signals in vitro, to expand normal or preleukemic cells for analysis and therapeutic purposes. The strains of immunodeficient mice are continually improving, allowing greater sensitivity for xenograft analysis and ultimately for in vivo drug treatment studies. With the defined transformation of primary human cells of hematopoietic origin now in hand, we can use these systems to study the cell type specific signals involved in oncogenesis and identify the critical pathways that can be therapeutically targeted in these cancers.
2. Materials
2.1. Isolation of Human CD34+ Cells
- Human umbilical cord blood (UCB) derived CD34+ cells. Selected, cryopreserved cells are available commercially from multiple vendors. Acquiring umbilical cord blood from donors is a cost effective alternative (see Note 1).
- Dulbecco’s Phosphate Buffered Saline (DPBS), without calcium and magnesium (Mediatech). Ca2+ and Mg2+ aid in cell-to-cell adhesion and clumping and thus should be avoided.
- Ficoll-Paque PLUS (GE Healthcare).
- Selection buffer: DPBS, 0.5% BSA, 2 mM ethylenediamine tetraacetic acid (EDTA), 50 U/mL each penicillin and streptomycin (antibiotics). Filter-sterilize and store at 4°C.
- CD34+ selection kit. Either EasySep human CD34 Positive Selection kit (StemCell Technologies) or human CD34 MicroBead Kit (Miltenyi Biotech) works well. Both kits utilize antibodies to CD34 that are directly or indirectly linked to magnetic particles. Use of either kit requires a specialized magnet, available separately from the manufacturers.
- Counting solution: Trypan blue dye solution, 3% acetic acid.
- Hetastarch freezing media solutions (Store at 4°C). Hetastarch solution 1: 50% Hetastarch solution (6% stock solution in 0.9% NaCl)(Baxter Healthcare Corp, Deerfield IL), 30% Iscove’s Modified Dulbecco’s Eagle’s Medium (IMDM), and 20% BSA fraction V solution (25% stock solution). Hetastarch solution 2: 10% DMSO, 50% hetastarch solution (6% stock solution in 0.9% NaCl), 20% IMDM, and 20% BSA fraction V solution (25% solution) (see Note 2).
2.2. Virus Preparation
- Producer cells. These are generally 293T cells (ATCC) or derivatives.
- 293T Media: Dulbecco’s Modified Eagle’s Medium (DMEM), 10% fetal bovine serum (FBS), and antibiotics.
- Trypsin-EDTA: Hank’s balanced salt solution (without calcium and magnesium), 0.05% trypsin, 0.5 mM EDTA. Store at 4°C, or at −20°C for long-term storage.
- Poly-l-lysine: 0.1 mg/mL solution of poly-l-lysine is prepared in water and stored at 4°C.
- Calcium phosphate precipitation reagents: Kits are commercially available; however the components are easily made. Three solutions are required: (1) Sterile, nuclease-free water. (2) 2 M CaCl2. (3) 2× HEPES buffered saline (2× HBS): 50 mM HEPES, 280 mM NaCl, 1.5 mM Na2HPO4, pH 7.10. A large batch can be prepared and aliquots can be stored long-term at −20°C. The pH of the 2× HBS solution is critical. Each batch of reagent should be tested prior to use.
- Virus collection media: IMDM, 10% FBS, antibiotics. Alternatively, FBS can be replaced with BIT (BSA, Insulin, Transferrin) serum substitute (StemCell Technologies) at a final concentration of 20% (see Note 3).
- Large syringes (10–60 mL).
- Syringe filters, 0.45 µm.
- Tubes for concentration of virus. These are protein purification columns with a 100-kD molecular weight cutoff (Centricon Plus concentrators, Millipore). Viral particles are retained when the supernatant is spun at 2,000 × g in these columns.
- HT1080 cells (ATCC).
2.3. Transduction of Human CD34+ Cells
- Prestimulation media: IMDM, 10% FBS (see Note 4), 10−4 M β-mercaptoethanol (BME)(see Note 5), antibiotics, and 100 ng/mL each of the human cytokines stem cell factor (SCF), megakaryocyte growth and differentiation factor (MGDF), and FMS-like tyrosine kinase-3 ligand (Flt3L). All cytokines used in these procedures are available for purchase (Peprotech, Rocky Hill, NJ).
- RetroNectin (TaKaRa): Prepare a 24 µg/mL solution by dissolving RetroNectin into water. Aliquot and store at −20°C. Six-milliliter aliquots will be sufficient for coating an entire six-well nontissue culture treated plate.
- DPBS containing 2% BSA. Sterilize by vacuum filtration with a low protein binding filter such as SFCA. Store the solution at 4°C.
- Hank’s balanced salt solution (HBSS) containing 2.5% (v/v) 1 M HEPES. Ensure sterility by vacuum filtration. Store at room temperature.
- Polybrene (hexadimethrine bromide). Prepare an 8-mg/mL solution in water. Store at 4°C or −20°C for long-term storage.
- Six-well nontissue culture treated plate.
- Non-enzymatic cell dissociation buffer (Gibco Invitrogen).
2.4. In Vitro Culture of Transduced Cells
- Myeloid culture media. This is the same media as that used for prestimulation prior to transduction with the exception that cytokines (10 ng/mL) are SCF, MDGF, Flt3L, interleukin-3 (IL-3), and interleukin-6 (IL-6).
- B-cell culture media: Minimum essential medium α (MEMα), 10% FBS, antibiotics, and 10 ng/mL of each of the human cytokines SCF, Flt3L, Interleukin-7 (IL-7).
- MS-5 mouse stroma cell line.
- MS-5 media: MEMα, 10% FBS, antibiotics.
- Methylcellulose media: 40 mL Base Methylcult H4100 (Stem-Cell Technologies), 20% FBS (20 mL), 2 mM l-glutamine, 10−4 M BME, antibiotics. IMDM (~20 mL) is added to bring the volume to 80 mL. All calculations should be based on a final volume of 100 mL, which includes the cell suspension (20% of the final volume) when performing the assay.
- Methylcellulose cytokine cocktail. Make a 20× concentrated cytokine cocktail by adding each human cytokine into IMDM. Our typical cytokines and final concentrations are granulocyte colony stimulating factor (G-CSF), SCF, IL-3, and IL-6 (all at 10 ng/mL), and erythropoietin (EPO, 6 U/mL).
- 16-gauge needles.
- 3-mL syringes.
- 35-mm tissue culture plate with grid lines.
2.5. Injection of Transduced Cells into Immunodeficient Mice
- A strain of immunodeficient NOD/SCID mice (Jackson Laboratories, Bar Harbor, ME).
- Doxycycline-treated chow (Purina prolab RMH 1500 with 0.0625% doxycycline, which provides approximately 2–3 mg of doxycycline per adult mouse per day; Harlan Teklad, Indianapolis, IN).
- Bactrim-treated chow (Teklab 2018 with 0.373% Bactrim; Harlan Teklad).
- Isoflurane.
- Anesthesia machine.
- Buprenex.
- Betadine (povidone–iodine, 10%; Purdue Pharma).
- Alcohol wipes.
- 25-gauge needles, 5/8 in.
- Insulin syringe (28 gauge, ½ in.).
- Ammonium chloride solution: 150 mM NH4 Cl, 100 nM KHCO3, 10 nM Na4 EDTA, pH 7.3 +/− 0.1. Generally, a 10× concentrated solution is prepared that is diluted with water before use. Store at 4°C.
- Murine anti-Fc receptor γ antibody clone 2.4G2 (BD Fc Block™, Pharmingen).
3. Methods
We typically use cord blood CD34+ cells in our protocols, as these cells have been shown to give the most robust engraftment in immunodeficient mice (17). However, whether these fetal/infant cells are representative of the bulk of human leukemia, which occurs in the adult, remains an open question, and it will be necessary to perform comparative experiments to answer these questions. Our system is focused on retroviral constructs rather than lentiviral, and it remains to be determined whether the strong viral promoters that are present in these constructs are contributing to the phenotypes obtained, presumably through retroviral insertional activation of endogenous oncogenes. It will also be interesting to examine whether lentiviral transduction of quiescent, noncycled human HSPC will increase the transduction frequency of the most primitive cells. The optimal viral envelope for use in human CD34+ experiments is also a variable that has been analyzed in some studies, with varying and sometimes contradictory results (18–20). As more labs become proficient in the growth, transduction, and transplantation of human CD34+ cells, it is likely that these questions will be answered.
The specific conditions that are used for the transduction of the human CD34+ cells depend upon the ultimate use of the cells upon transduction. If the cells are to be propagated in vitro, and will not be injected into immunodeficient mice, it is less critical that the cytokines IL-3 and IL-6 are excluded from the prestimulation mix. Including these cytokines will increase cell yield and transduction efficiency, and typically does not negatively impact on the overall expansion and proliferation of the cells in vitro. The choice of immunodeficient mouse, and the route of delivery as well as the age at which to transplant, depends upon the availability of the strains and the expertise of the lab. Intravenous injection of 6- to 8-week-old NOD/SCID or NOG mice (both commercially available strains) is the most popular approach, but the use of newborn pups, cranial facial vein injection, intrafemoral injection, and the newer strains of immunodeficient mice (e.g., NOD/SCID-SGM3 mice for myeloid biased grafts) are gaining popularity, and only time will tell which approach will be superior for studying normal human hematopoiesis as well as leukemogenesis in the mouse.
3.1. Isolation of Human CD34+ Cells
- Mix the whole blood in an equal volume of sterile PBS in a 50-mL tube, layer slowly onto 0.5 volumes of Ficoll-Paque PLUS solution, spin at 250 × g for 30 min to 1 h at 18–20°C. The majority of red blood cells and granulocytes will be pelleted while mononuclear cells including CD34+ cells will form a white interface just above the ficoll. Serum is left above the interface.
- Harvest the mononuclear cells (MNC) from the interface, transfer to a 50-mL tube, wash cells in at least 3 volumes of selection buffer, centrifuge at 500 × g for 15 min at 18–20°C.
- Resuspend the pellet in 5 mL of selection buffer and transfer to a 15-mL tube. Rinse the original 50-mL tube with an additional 5 ml of selection buffer and transfer to the 15-mL tube to make 10 mL of total volume.
- Take 10 µL of MNC and mix with 90 µL of counting solution (see Note 6).
- Spin at 700 × g for 8 min, aspirate the supernatant, resuspend the MNC in selection buffer at the density recommended by the manufacturer of the selection kit.
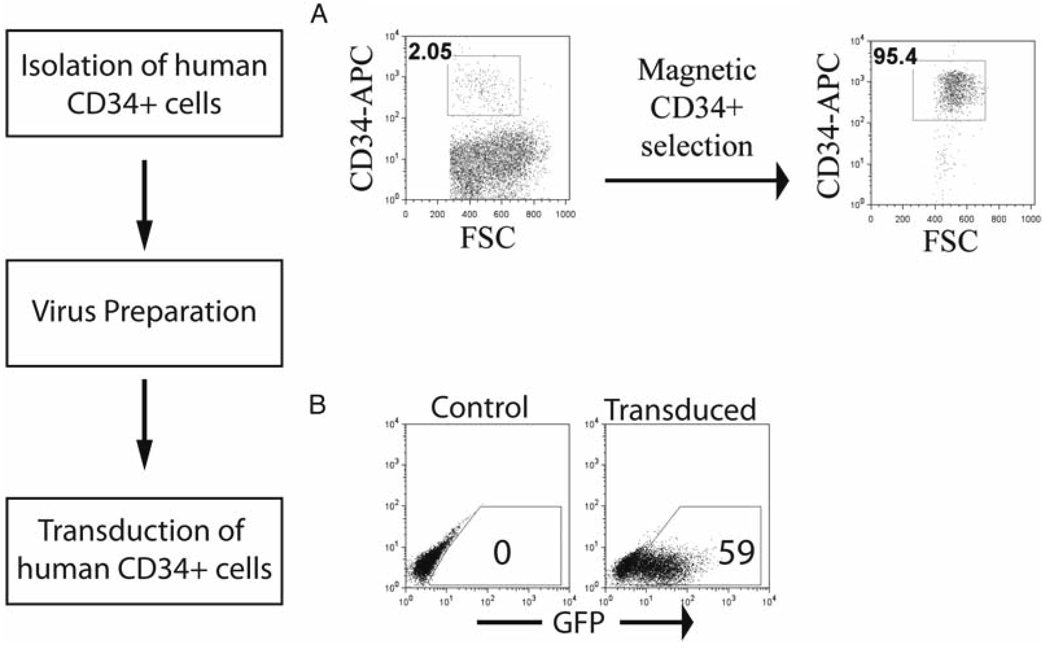
- CD34+ cells are positively selected from this population using either the Miltenyi Macs CD34 Microbead Kit or the Stem Cell Technologies CD34 EasySep procedure. Both protocols are successful and give comparable yields and purity of CD34+ cells (Fig. 1a). Each protocol is followed according to the manufacturer’s recommendations (see Note 7).
- After selection of CD34+ cells, count the cells and centrifuge at 700 × g for 10 min to pellet them. Aspirate the supernatant. Cells can either be used immediately as described in Subheading 3.3 below, or viably frozen by resuspending the pellet in 300–500 µL of hetastarch solution 1 and transferring to a cryovial. An equal volume of hetastarch solution 2 is then added dropwise (105 to 2 × 106 cells per vial).
Fig. 1.
Overview of the model system for retroviral transduction of human CD34+ cells. (a) Human CD34+ cells are purified by magnetic selection, and purity is confirmed by flow cytometric analysis. (b) A transduction of human CD34+ cells is shown, with a non-transduced control. GFP expression is visualized by flow cytometry.
3.2. Virus Preparation and Titer
- Either the 293T or Phoenix cell line is used for transient virus production using a plasmid transfection procedure. The “strain” of Phoenix cell line used depends on the species of the cells that will be targeted (see Note 8). Virus production is best done in batches that are titered and aliquoted for future use. 10-cm tissue culture dishes are coated with poly-l-lysine by adding 3 mL of solution to the dish, swirling to cover the whole bottom, and transferring to subsequent dishes. 5 million cells are then plated to each dish (day 1). To ensure even distribution, do not swirl the media, but rock the dishes twice side-to-side and twice to-and-fro. The dishes are placed in the incubator overnight (37°C, 5% CO2).
- Virus production is initiated by transfecting the cells in the 10-cm dish with three plasmids:
- Retroviral vector 12 µg
- Gag/Pol vector 10 µg
- Envelope vector 3 µg
All three plasmids are used in the transfection, even if a Phoenix cell line already contains the gag/pol or envelope plasmids as stable integrants. The quality of the plasmid used in the transfection is critical for maximal uptake by the cells. Only DNA from a maxiprep should be used for virus production. Different transfection protocols can be used, but one of the most reliable and cost-efficient is the calcium phosphate method (see Note 9).
- Transfect the cells the afternoon of day 2. The cells should be approximately 75% confluent. All solutions should be at room temperature. For calcium phosphate transfection, a final volume of 1-mL transfection mix is needed per 10-cm dish. The volumes can be scaled up if multiple dishes are transfected with the same retroviral vector. Add plasmids to ddH2 O to a final volume of 438 µL and mix by brief pipeting. Next, add 62 µL of 2 M CaCl2 and mix. Then add this solution dropwise to a Falcon 2054 polystyrene tube containing 500 µL of 2× HBS (pH 7.10). The solution should appear somewhat cloudy after this step. Some investigators prefer to vortex while adding the DNA mixture, others prefer to bubble air through the 2× HBS during the addition. Results seem to be comparable using the two techniques in our experience.
- Remove dishes from the incubator and slowly add 1 mL of the DNA–CaPO4 mixture solution directly on top of the cells. Use a 1-mL pipetman with the tip beneath the media to discharge the mixture while moving the tip around, blanketing the entire cell layer. Return the dishes to the incubator. Some investigators prefer to incubate the mixture for 15–45 min before adding to the cells, but we find that this increases the size of the precipitate which decreases the transduction efficiency. Very fine black particles (visible at 20× power under a microscope) are the best precipitate for transfection.
- On the morning of Day 3 (12–16 h after initiating transfection), remove dishes from the incubator and aspirate the media by gently tipping the dish at a 45° angle and touching the tip of the aspirating device to the side of the dish. Slowly add 6–7 mL of warmed collection media to the side of the angled dish, so as not to disturb the cell monolayer. The media that is used depends on the target cells that will be transduced and should be the optimal media for these cells. Ideally, virus-containing supernatant is now collected at 12-h intervals, for a total of 4–5 collections. Fresh warm collection media is replaced each time, gently, to prevent disruption of the cell monolayer. Virus supernatant can be kept overnight on ice at 4°C to allow concentration of the supernatant in one step. Producer cells should be visualized under the fluorescent microscope, if a fluorescent marker protein is used (i.e., EGFP). At least 50% of cells should be transfected to justify proceeding with the collections (see Note 10).
- Combine virus collections, filter through a 0.45- µm filter (0.2 µm can also be used with no loss of virus and less risk of producer cell contamination), and concentrate using a protein purification column with a molecular weight cutoff of 100-kD. The specific fold concentration depends on the needs of the investigator and the transgene used, but can range between 5 and 100× or more. These filters can be re-used for the same virus supernatant. Aliquot and freeze concentrated virus at −80°C. Keep a very small aliquot for determining viral titer. Depending on the envelope used, viral titer will drop by one-half for each freeze-thaw cycle. (see Note 11).
- To titer the virus, we use the HT1080 adherent cell line (for a human-tropic virus). These cells are available from ATCC.
- Remove sub-confluent HT1080 cells from the plate with trypsin–EDTA and plate in six-well plates at 2.5 × 105 cells per well, in DMEM 10% FBS (Day 1).
- The following day, dilute viral supernatant by taking 30 µL of virus and mixing with 3 mL of DMEM 10% FBS. This is a 10−2 dilution. Make tenfold serial dilutions by taking 300 µL of the 10−2 dilution and adding to 2.7 mL of DMEM 10% FBS (10−3), repeating this three more times until a 10−6 dilution is reached. Aspirate the media from the six-well plate of HT1080 cells, add 2 mL of the 10−6 dilution to a single well, and continue for each of the remaining dilutions. Use a new pipet and add 2 mL of DMEM 10% FBS to the final well of the six-well plate (no-virus control well). Add 2 µL of an 8 mg/mL polybrene solution to each well (final concentration is 8 µg/mL). Incubate overnight at 37°C in a 5% CO2 incubator.
- On day 3, aspirate the medium from each well. Add 2 mL of complete media. If the retrovirus contains a drug-selectable cassette, add the working solution of drug in complete media at this time (see Note 12).
- After 10–14 days, fix the colonies of cells that have formed by adding ice-cold methanol for 5–10 min and then rinse once with distilled deionized water. Stain the colonies with an aqueous solution of 0.4% methylene blue for one minute, wash twice with water and dry the plate upside down. Calculate the titer. The best dilution for calculations will be one that gives between 10 and 100 colonies.
- For viral vectors that contain a fluorescent marker, collect cells two days after the end of transduction (day 5) and analyze by flow cytometry.
- Viral particle number is calculated by multiplying the percentage marked cells by the total number of target cells present at the time of incubation with virus (day 2; a replicate well is included, and this is counted at the time of incubation with virus) (It is important that the dilution that is used for the calculation of titer has given a transduction efficiency of less than 37%, to ensure single hit kinetics). Multiplying this number by the dilution factor for that well (e.g., 100 for a 10−2 dilution) will give the titer of the virus per mL. We have found that the viral titer should not be less than 105/mL for successful transduction of human CD34+ cells.
3.3. Transduction of Human CD34+ Cells
- To thaw frozen human CD34+ cells, agitate the vial rapidly in a 37°C water bath until only a small piece of ice remains. Add 2 mL of room temperature HBSS + 2% BSA to the vial, mix, and transfer to a 15-mL tube. Rinse the vial and cap with HBSS + 2% BSA and increase the volume in the 15-mL tube to 10 mL. Centrifuge at 4°C, 500 × g for 10 min, flick the tube with index finger to disperse the cell pellet, and resuspend in prestimulation media to give a final concentration of 106 cells per mL (see Note 13).
- Culture the cells for 1.5–2 days to ensure that a population of cells are replicating (essential for transduction with retrovirus).
- Prepare a RetroNectin-coated plate the day before transduction. Incubate a six-well nontissue culture treated plate with RetroNectin solution (2 h, room temperature), then PBS containing 2% BSA (30 min, room temperature), and finally with HBSS containing 2.5% HEPES (quick wash). Although plates can be stored for months at 4°C, we have found that a fresh treatment is superior to stored plates. The primary solution of RetroNectin can be used in a secondary (and tertiary) treatment, and these plates can be used for transduction of less critical cell lines if desired.
- Count the cells. The cell number should be approximately equal to the number when cells were thawed (see Note 14).
- Precoat the RetroNectin well with 2–4 mL of unconcentrated viral supernatant or 1 mL of concentrated supernatant. Centrifuge the plate for 45 min at 2,000 × g at room temperature. Aspirate the solution and repeat the spin with additional virus supernatant. After the second treatment, add approximately 1 million prestimulated cells in an equal volume of fresh prestimulation media to the viral supernatant in the well, and incubate at 37°C for 4–8 h (during the day), with 8 µg/mL polybrene (see Note 15).
- At the end of the day, carefully aspirate the majority of the media (cells should be predominantly attached to the Retro-Nectin-coated plastic) and add 2 mL of fresh prestimulation media. Allow the cells to recover in the incubator overnight.
- The next morning, add 2–4 mL (unconcentrated) or 1 mL (concentrated) of virus and polybrene. Do not centrifuge. At the end of the day, remove the cells from the plate by collecting the media and detaching the cells from the plate using a non-enzymatic cell dissociation buffer. Centrifuge at 500 × g for 7 min and resuspend in the appropriate media at a density of 106 cells per mL.
- Two days later, cells can be analyzed by flow cytometry (if a fluorescent co-marker was present in the retroviral construct; Fig. 1b) to determine the transduction frequency, or drug selection can begin if a drug-selectable construct was used (see Note 16).
3.4. In Vitro Culture of Transduced Cells
- The choice of culture medium will depend on the specific lineage that is desired and the oncogene that is used in the transduction. For culture under myeloid conditions, a complex mixture of cytokines is most likely to allow the greatest diversity in terms of myeloid potential for the cells. After transduction, cells can be cultured in a five-cytokine cocktail of SCF, MDGF, Flt3L, IL-6, and IL-3. The specific concentrations to use can be determined empirically and may differ depending on the transgene. We have found that 10 ng/mL of each is sufficient for the AML1-ETO, CBFB-SMMHC, and MLL-AF9 fusion protein-expressing long-term cultures (12, 21, 22). Typically the control-transduced CD34+ cells will proliferate for 8–12 weeks under these conditions, while the oncogene-transduced cultures grow for varying lengths of time, depending on the specific oncogene.
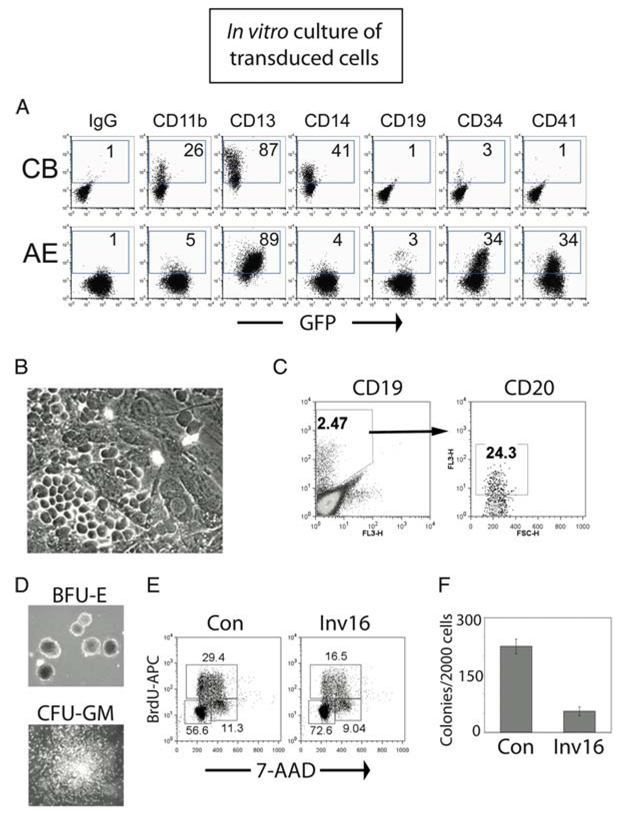
- Count cells weekly and seed at 4 × 105 cells/mL, in a volume that will give the desired number of cells after the 1-week expansion period (see Note 17). During the first 3–4 weeks, cells will expand approximately tenfold each week, and supplementation of the medium may be needed on day 4 or 5 to prevent depletion of the medium. Under these conditions, the control cultures will maintain a population of CD34+ cells of approximately 2–10%, depending on the specific cord blood. The remainders of the cells are at various stages of myelopoiesis. A layer of adherent cells will slowly form over time, and the suspension cells can be moved to a new well each week if desired. Toward the end of the proliferative period (weeks 7–12 depending on the cord blood), cells will double only once or twice per week, and the majority of the cells will be monocyte/macrophage. Expression of the fluorescent marker, if present in the retroviral construct, can be monitored during this time, to determine effects of the transgene on proliferation and/or differentiation. An example of a myeloid culture of cells expressing AML1-ETO in shown in Fig. 2a.
- For expansion of B-lymphoid cells in vitro, the use of a stroma coculture will give the best growth and the most reliable and reproducible results. We use the MS-5 murine stroma cell line (23) (see Note 18). Immediately after transduction, seed 1 × 105 cells onto a monolayer of MS-5 stroma cells formed in a T25 culture flask with 5 mL of B-cell media. Some cells will invade the stroma layer and grow beneath the stroma as “phase-dark” cells, meaning that they will appear as dark, nontranslucent cells through the phase-contrast microscope. Under B-cell growth conditions, these cells will typically form organized areas known as cobblestone areas, and will also appear dispersed throughout the monolayer in addition to growing in loosely organized areas. An example of a typical cobblestone area forming cells is shown in Fig. 2b. The majority of cells will grow as suspension cells or weakly attached to the topside of the stroma. The dilution to make during weekly passage of cells will depend upon the individual capacity of each cord blood preparation, which demonstrates large proliferative variations under B-cell growth conditions (see Note 19). For the first 2–3 weeks of growth, only myeloid cells (CD33+) will be present in the suspension. At weeks 3–4, a small population of CD19+ cells will form, which can become the majority population by 5–6 weeks of culture. Most of the CD19+ B-cells will co-express CD10, and a percentage (10–50%) will co-express CD20, but in our hands the B-cells that develop under these conditions do not express surface Ig (Fig. 2c).
- The methylcellulose assay is a convenient and powerful way to determine the effects of a particular transgene on hematopoietic differentiation and proliferation. For the most quantitative assay, transduced cells should be sorted or drug selected prior to use in a methylcellulose assay. Typically, after transduction, cells are washed with IMDM without cytokines or FBS and counted. Then, 8,000 cells are deposited into a 15-mL tube in a final volume of 800 µL IMDM (enough cells to give triplicate methylcellulose plates with 2,000 cells each, and allowing for pipetting error by calculating for four plates). The cells are mixed with 3.2 mL of methylcellulose media while vortexing at high speed. The methylcellulose is easiest to measure and manipulate using a 3-mL syringe with a 16 gauge needle (fill the syringe with methylcellulose solution from the bottle, discharge to clear the air void, and completely fill the syringe by slowly withdrawing the plunger fully. This will give a volume of approximately 3.2 mL). Rest the syringe and needle in the falcon tube for approximately 5 min to allow the air bubbles to rise.
- Draw up 3 mL of methylcellulose/cell suspension, and add 1 mL to each of triplicate 35-mm dishes that contain 50 µL of 20× cytokine cocktail (see Note 20). Tilt and rotate the dish to distribute the solution over the entire bottom. Incubate the dishes for 2 weeks in a humidified chamber to prevent drying of the methylcellulose media (see Note 21). Minimize disturbance or movement of the dishes during this time.
- After two weeks, count the colonies and score the colony type. We typically will score three types of colonies, including granulocyte/macrophage (GM), burst forming uniterythroid (BFU-E), and granulocyte/erythroid/macrophage/megakaryocyte (GEMM) colonies (Fig. 2d). Transgene expression in human CD34+ cells could affect the size, the number as well as the specific type of colonies present. For example, expression of the CBFB-SMMHC oncogene causes a G0/G1 arrest, and the number of colonies that are present after 2 weeks are severely decreased (Fig. 2e and 2f). The expected number of colonies from 2,000 input cells varies greatly depending on the particular cord blood as well as the timing of the experiments. Numbers could range from 50–400 colonies per dish. The best growth/differentiation will typically occur when colony number is around 100–200 per dish.
Fig. 2.
Overview of the in vitro analysis of retrovirally transduced human CD34+ cells. (A) Representative long-term myeloid cultures (6 weeks) of control CB cells and cells expressing the AML1-ETO oncogene (AE). This phenotype is preserved throughout the 6–8 months of growth. (B) Example of a cobblestone area that forms upon coculture of human CD34+ cells with the MS-5 stroma cell line. (C) B cells can be expanded from the transduced CD34+ cultures upon coculture with MS-5 and the cytokines Flt3L, SCF and IL-7. Shown is a CBFB-SMMHC-transduced culture that was 12 weeks posttransduction. Cells were cultured on MS-5 for an additional 4 weeks. A fraction of the CD19+ cells also express the CD20 B-cell marker. (D) A clonogenic methylcellulose assay is shown, with representative BFU-E (red cell colony) and CFU-GM (myeloid colony) that result from the growth of a single cell. (E, F) Expression of a fusion oncogene frequently leads to loss of clonogenic potential immediately upon transduction, and this effect is due to a G0/G1 cell cycle block for cells expressing the inv16 fusion protein CBFB-SMMHC.
3.5 Injection of Transduced Cells into Immunodeficient Mice
- If cells are going to be used for injection into immunodeficient mice, the culture medium for prestimulation should have only minimal cytokines to preserve the primitive nature of the cells as much as possible. The cytokines SCF, TPO, and Flt3L should be sufficient to promote cell cycle entry and survival; IL-3 should be avoided, since this cytokine has been found to promote differentiation and loss of SCID repopulating potential.
- Cells should be injected into animals as soon after thaw as possible. Our usual approach would be to prestimulate for 1.5 days, transduce cells for 0.5–1.0 days (1–2 incubations with virus, 4–6 h each) and then immediately inject into animals, so that only two or three days pass from initial thaw. A small aliquot can be retained in vitro to check verify transduction and calculate efficiency.
- The best characterized animal model for xenograft of human hematopoietic cells is the NOD/SCID mouse. A number of variants are now available, including the NOD/SCID-β2M−/−, NOD/SCID-IL2Rγc−/− (NOG), and NOD/SCID-SGM3 mice. The latter two strains are just becoming widely used, and limited information is available in the literature for these mice. The NOG mouse should be highly immunocompromised and possess no residual NK activity and may be defective for dendritic cell and macrophage function as well (24). The SGM3 mouse is transgenic for the myeloid-promoting cytokines SCF, GM-CSF, and IL-3 and skews the human graft towards myeloid differentiation, but has also been documented to promote loss of the normal human graft, possibly due to mobilization and differentiation of the primitive human cells in the mouse bone marrow (25, 26).
- The number of human cells that must be injected to ensure a reliable hematopoietic graft will vary depending on the mouse strain and sub-strain that is used. A good rule of thumb for regular NOD/SCID mice, with injection of prestimulated, cycling cells, is to use 300,000 cells per mouse, calculated based on the starting number of the CD34+ population. For example, if 9 × 105 CD34+ cells are prestimulated, and after 4 days have doubled in number to 1.8 × 106, this would be enough cell number for three mice (900,000 starting cells/300,000 = 3). If the cells tripled in this time to 2.7 × 106, this would still be enough cell number for only three mice. (see Note 22).
- Mice (6–8 weeks old on the day of injection) should be prepared for injection by feeding with doxycycline-treated chow for 1 week before irradiation, and continued on this chow for 1 week after irradiation (we have recently found that we now achieve better results using Bactrim-treated chow for 2 weeks post-irradiation, with doxycycline-treated chow for 1 week before irradiation. It is possible that this treatment needs optimization for individual animal facilities, and procedures may need to be altered if mouse loss becomes unacceptable after irradiation). This minimizes the loss of mice due to radiation illness. The dose of radiation needs to be determined empirically for each colony, and the dose may need to be recalculated as the colony ages. We have found that our colony gave very good results using 375 Gy initially, but we now use 300 Gy (2 years after establishing the colony) with similar survival numbers (approximately 10–20% of mice will be lost in each experiment due to radiation illness).
- Mice are irradiated up to 24 h in advance of injection. For intravenous injection, a volume of 300 µL works well (cells in a PBS solution). For intrafemoral injection, a volume of 25 µL or less should be used. (see Note 23).
- To inject cells intravenously, place mice under a heat lamp for several minutes. This will allow easier visualization of the tail vein. Injection is easiest with immobilized mice, either by containment in a mouse restraint device or by anesthetization with isoflurane. After cleaning the tail with an alcohol wipe, inject the cells into the tail vein with an insulin syringe taking special care to avoid injection of any air bubble (this will kill the mouse). You will be able to see the clear PBS-cell mixture travel through the vein. If the needle is not in the vein, the plunger will not easily be depressed and a bump will form in the tail of the mouse. Attempt the initial injection midway down the tail so that if this occurs, another attempt can be made closer to the body of the mouse.
- To inject intrafemorally, anesthetize the mouse with isoflurane. Give the mouse painkiller such as Buprenex (buprenorphine) by injecting under the skin of the back. Place the mouse on its back, and insert the snout of the mouse into a tube for constant delivery of isoflurane during the procedure. Clean the leg with an alcohol wipe and Betadine. Insert a 25 gauge needle (5/8 in.) attached to a 1-mL syringe into the femur (see Note 24). Remove the needle slowly, while keeping the leg steady and remaining fully focused on the location of the hole. Insert the insulin syringe containing the cells and slowly inject the mixture into the hole. Move the mouse back to its cage where it will regain consciousness within 5 min.
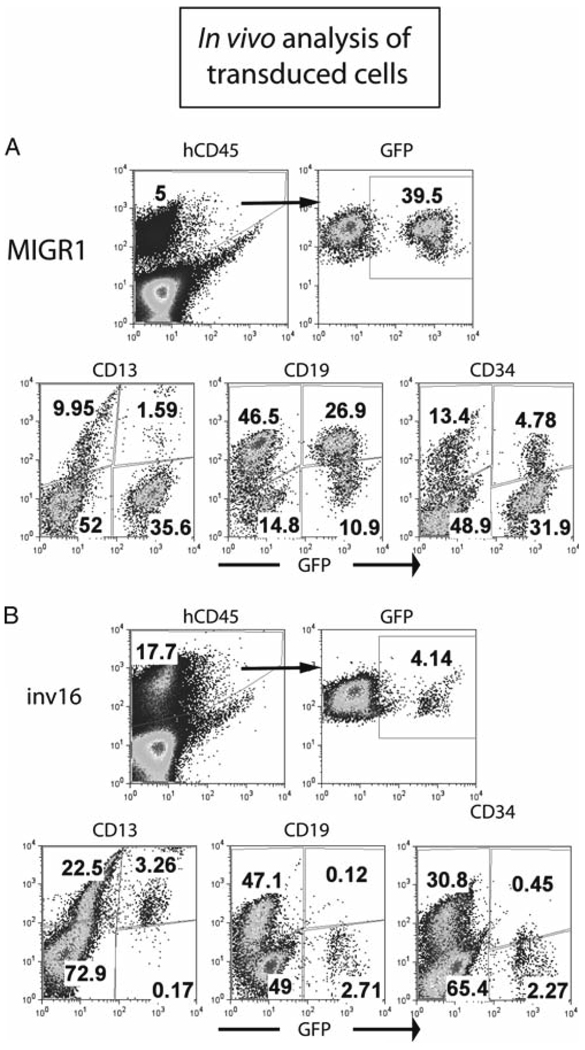
For measurement of a graft due to the most primitive human cells, it is recommended that the mice be monitored at 9–12 weeks post injection. It is often difficult to measure human cells in the peripheral blood in these mice, but depending on the size of the graft it is possible. We use the CD45, CD33 and CD19 antibodies to determine total cell graft, myeloid and B-lymphoid populations respectively. To block non-specific staining of murine hematopoietic cells that express FcR, the use of 1 µL of Fc block is recommended per 1 × 106 cells. For surface staining of PB, red cells should be lysed in an ammonium chloride solution, and the residual red blood cells should be excluded from the viable cell gate during flow to determine an accurate percentage of human white blood cells in the mouse. - When the experiment is to be ended, the bone marrow, spleen and peripheral blood of the animal should be processed and analyzed for human cells. The four long bones of the hind legs are removed, and either crushed using mortar and pestle or flushed using an insulin syringe and IMDM media after cutting the ends of the bones with scissors. Either way, cells are then filtered through a 40 µm filter, and red blood cells are lysed. The spleen or other organs can be crushed through a 40 µm filter, using the flat end of a 3 mL syringe, and red cells are then lysed. Cells are stained for surface markers to determine the lineage composition of the human graft (Fig. 3a and 3b). Expression of a leukemia fusion gene can specifically affect the composition of the graft, as shown by expression of the inv16 oncogene CBFBSMMHC, which leads to a myeloid-dominated graft with very few CD19+ cells and a decreased number of CD34+ cells (Fig. 3a and 3b). Methylcellulose assays using human-specific cytokines can be performed to determine the progenitor activity, using the protocol described earlier. Cells can be viably frozen for later use. To show self-renewal of the human cells using the most stringent criteria, a secondary transplant is performed, following the same procedures as for the primary transplant. Secondary mice are analyzed at 6–12 weeks for the presence of human cells..
Fig. 3.
Overview of the in vivo analysis of retrovirally transduced human CD34+ cells. (A, B) Analysis of the bone marrow of a NOD/SCID-β2M−/− mouse that was injected with control transduced (MIGR1) and CBFB-SMMHC-transduced (inv16) cells. 8.5 weeks after injection, the human graft in the bone marrow of the mouse was readily detectable, and GFP+ cells were present in both mice. Most of the human cells in the control mouse were CD19+ B-cells, as is typical, but for the CBFB-SMMHC-expressing cells, there is a loss of the B-cell fraction, and essentially all of the cells express the myeloid marker CD13.
Footnotes
1
The quality of the whole blood sample should be considered prior to proceeding with the selection protocol. We have found that a significant drop in yield occurs at volumes smaller than 80 mL and we generally do not proceed with these samples. Additionally, the procedure should begin within 48 h of collection. The samples can be stored at room temperature in the presence of anticoagulants. Samples with obvious clumps or a very dark color should be avoided.
2
Solutions should be prepared without including the BSA. Solutions can then be stored at 4°C for months. Withdraw an aliquot of each solution and add BSA to each immediately before use.
3
The choice of virus collection media will depend on which culture condition is preferred (serum or serum-free) for culture of the CD34+ cells after transduction. Phoenix-GP producer cells produce adequate viral particles in either virus collection media.
4
The response of CD34+ cells to FBS culture conditions can vary significantly depending on the lot of serum. For this reason, it is important to carefully test several lots before purchase. We generally perform methylcellulose colony assays of UCB CD34+ cells with several lots of serum before purchasing a bulk quantity of a satisfactory lot (most colonies, with good growth and multilineage differentiation, with performance at least as good as the current or past lots).
5
A typical stock BME solution is 14.4 M. To prepare aliquots of 500× concentrated BME, dilute 35 µL of stock BME into 10 mL water and sterilize with a 0.2- µm syringe filter. This yields a 10−2 M solution (500×, add 1 mL to a 500-mL bottle IMDM) that can be aliquoted and stored at −20°C. Solutions are generally stable for 1 month at 4°C, wrapped in foil.
6
The acetic acid/trypan blue solution will allow easy counting of nucleated cells. The red blood cells will be lysed in this solution. Depending on the number of cells in the preparation, a larger or smaller dilution of the suspension may be needed.
7
The yield and quality of cells from these procedures can vary significantly, due to sample differences. From an average UCB of 100 mL, the expected yield of CD34+ cells should be between 0.5 and 1.5 million cells. Occasionally far more CD34+ cells can be recovered, but typical recovery is approximately 50% of the CD34+ cells present in the MNC fraction. The purity of the selected cells should be confirmed by flow cytometry and the cell counts adjusted to represent the actual number of CD34+ cells. We routinely achieve purities of greater than 90% using these methods (Fig. 1a). A small aliquot of the selected cells can be tested for growth and viability by culturing in myeloid media and monitoring cell counts or by performing methylcellulose assays.
8
The Phoenix cells, based on the 293T cell line, are available through ATCC in an agreement with Dr. Garry Nolan. The cells can be obtained with only the gag and pol genes (Phoenix-GP), or additionally with the ecotropic or amphotropic envelopes (Phoenix-E or Phoenix-A). We typically use Phoenix-GP and transfect additional gag/pol helper plasmid, as this has been shown to be the limiting construct in viral preparations. Full details on these cell lines are available upon purchase from ATCC.
9
Many methods of transfection exist. The amounts of DNA required by these methods vary. The DNA amounts presented here are optimized for the calcium phosphate precipitation method. In using different methods of transfection we have found that the ratio of the different plasmids is an important factor and should be preserved if the total amount of DNA is altered.
10
The fluorescent intensity of EGFP may not be impressive on the first day after transfection, but by day 2 it should be readily visible, especially from the control cells transfected with empty vector. It is possible that the transgene in the viral construct will severely diminish the intensity of the EGFP. We have found that this is transgene-specific but often occurs when large genes or oncogenes are used. In this case, the intensity of the EGFP will not correlate with the viral titer.
11
The rule of thumb is that approximately 50% of viral titer is lost for each freeze/thaw cycle when using amphotropic or ecotropic envelopes. We have not formally tested this. When using the feline endogenous virus (RD118) or the vesicular stomatitus virus envelope (VSV-G), there is little to no loss of virus titer. These two envelopes are also reported to allow concentration of virus particles by ultracentrifugation without loss of viral titer.
12
For HT1080, use the following concentrations for commonly used drugs. G418 for neomycin resistance, 800 µg/mL, (10–14 days, with a media change at day 5); hygromycin B, 500 µg/mL, (7–10 days); puromycin, 1 µg/mL (4–7 days).
13
Media is IMDM with 10% heat-inactivated FBS, 50 U/mL penicillin, 50 U/mL streptomycin, 2 mM l-glutamine, 10−4 M β-mercaptoethanol. Alternatively, if serum-free conditions are preferred, the FBS is substituted with the BIT supplement from Stem Cell Technologies, supplied as a 5× concentrate. If the transduced cells will be used in vivo (in immunodeficient mice), the human cytokines SCF, TPO (MDGF), and FLT3L are included at 100 ng/mL. For in vitro applications only, the addition of human IL-3 at 20 ng/mL and IL-6 at 100 ng/mL will increase growth and transduction efficiency but will also promote differentiation.
14
Some cells will die during the prestimulation period, and some cells will begin to divide. This typically results in approximately an equal viable population relative to the starting number on Day 0. If the number is lower than 50% of the viable cell number on Day 0 it is often not worth proceeding with the transduction, since the cells that do recover are usually less hardy and long-lived and may not represent the expected population of human CD34+ cells.
15
Ensure that cells have adhered to the RetroNectin-coated plastic after approximately 15 min of incubation. This is essential for good transduction by all pseudotypes of virus with the exception of VSV-G; if the virus is pseudotyped with VSV-G, no RetroNectin is used for transduction (27, 28).
16
Working concentrations for human CD34+ cells for some common drugs are: G418 (for neomycin), 800 µg/mL (will take approximately 1.5 weeks; fresh drug should be added at day 4 or 5); hygromycin B, 300 µg/mL (will take approximately 1 week); puromycin, 0.5 µg/mL (will take 3–5 days for selection). Nontransduced cells should be incubated with drug, and selection is complete when all of these cells are dead. As a note of caution, it is possible that some transgenes will have deleterious effects on human CD34+ cells, and selection with drug resistance may be impossible. A fluorescent marker is more useful in this case; cells can be sorted after transduction and analyzed for these effects.
17
We find the cells grow best when we use six-well plates, for reasons we have not determined. Cultures can reach a density of 2 million cells/mL without significant loss of viability, and a volume of up to 8 mL can be used for each well of the plate.
18
The MS-5 cell line is grown in α-MEM medium with 10–20% FBS. Cells are never allowed to become confluent. A large number of viably frozen vials should be made to ensure a reliable stock of cells that are at an early passage. Over time in vitro, the phenotype/morphology of the cells can become more fibroblastoid, and these cultures are less able to support hematopoiesis and also tend to lose contact inhibition. The MS-5 cells do not need to be irradiated for use as a feeder layer. If cells will not be used for experiments within 2–3 weeks, it is better to discontinue the culture and thaw a new vial rather than to continue passage of cells during this time.
19
A typical cell split would include a gentle agitation of the flask to loosen the hematopoietic cells that are weakly attached to stroma cells, followed by removal of 50% of the volume with replacement by fresh media. This procedure is frequently referred to as demi-depopulation. Care must be taken to ensure that the stroma does not loosen, which becomes more likely with each passing week.
20
The cytokines to use in a methylcellulose assay that will permit the broadest range of colony types includes 10 ng/mL G-CSF, 20 ng/mL each of SCF, IL-3 and IL-6, and 6 U/mL of Epo.
21
Large 500 cm2 square tissue culture dishes can be used to create a humidified chamber. Fill both halves of two 60-mm culture plates with sterile water and place one piece into each corner of the dish (use each lid and the bottom separately). This chamber can hold approximately twenty 35-mm methylcellulose cultures. The chambers are sterilized and reused.
22
There is great variability in the engraftment of different cord blood preparations and also large variability mouse to mouse from the same cord blood. This biological variability is impossible to control at this time, and means that a large number of mice must be used for experiments.
23
We have found that the intrafemoral route, although significantly more time-consuming and requiring more practice, results in a significantly superior graft. It has also been shown that the cell number can be markedly reduced given the superiority of this route of injection (29, 30). It is likely that with the newly available NOG mice, and the intrafemoral route of injection, the number of CD34+ cells that are needed for each mouse will be less than tenfold what is currently recommended.
24
While looking at the mouse knee, locate the highest point (just above the white connective tissue of the joint; it is easier for some investigators if the hair at the knee joint is removed, by simply plucking it off with gloved index finger and thumb). Insert the needle just above that point and slightly to the outside of the leg. The inside of the bone will have a gritty texture that can be sensed by moving the needle up and down slightly. Before attempting this procedure on live mice, it is best to obtain the skill by practicing with sacrificed mice. Try inserting a colored dye into the femur; e.g., trypan blue. Dissection will confirm injection and allow a certain amount of trial and error that is required to successfully and consistently perform the procedure.
References
- 1.Sharpless NE, Depinho RA. The mighty mouse: Genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5(9):741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 2.Rangarajan A, Weinberg RA. Opinion: Comparative biology of mouse versus human cells: Modelling human cancer in mice. Nat Rev Cancer. 2003;3(12):952–959. doi: 10.1038/nrc1235. [DOI] [PubMed] [Google Scholar]
- 3.Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6(2):171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Drayton S, Peters G. Immortalisation and transformation revisited. Curr Opin Genet Dev. 2002;12(1):98–104. doi: 10.1016/s0959-437x(01)00271-4. [DOI] [PubMed] [Google Scholar]
- 5.Smogorzewska A, de Lange T. Different telomere damage signaling pathways in human and mouse cells. Embo J. 2002;21(16):4338–4348. doi: 10.1093/emboj/cdf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamad NM, Elconin JH, Karnoub AE, et al. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 2002;16(16):2045–2057. doi: 10.1101/gad.993902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim KH, Baines AT, Fiordalisi JJ, et al. Activation of RalA is critical for Rasinduced tumorigenesis of human cells. Cancer Cell. 2005;7(6):533–545. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 8.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400(6743):464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 9.Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002;347(20):1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- 10.Pereira DS, Dorrell C, Ito CY, et al. Retroviral transduction of TLS-ERG initiates a leukemogenic program in normal human hematopoietic cells. Proc Natl Acad Sci U S A. 1998;95(14):8239–8244. doi: 10.1073/pnas.95.14.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grignani F, Valtieri M, Gabbianelli M, et al. PML/RAR alpha fusion protein expression in normal human hematopoietic progenitors dictates myeloid commitment and the promyelocytic phenotype. Blood. 2000;96(4):1531–1537. [PubMed] [Google Scholar]
- 12.Mulloy JC, Cammenga J, MacKenzie KL, Berguido FJ, Moore MA, Nimer SD. The AML1-ETO fusion protein promotes the expansion of human hematopoietic stem cells. Blood. 2002;99(1):15–23. doi: 10.1182/blood.v99.1.15. [DOI] [PubMed] [Google Scholar]
- 13.Buske C, Feuring-Buske M, Antonchuk J, et al. Overexpression of HOXA10 perturbs human lymphomyelopoiesis in vitro and in vivo. Blood. 2001;97(8):2286–2292. doi: 10.1182/blood.v97.8.2286. [DOI] [PubMed] [Google Scholar]
- 14.Daga A, Podesta M, Capra MC, Piaggio G, Frassoni F, Corte G. The retroviral transduction of HOXC4 into human CD34(+) cells induces an in vitro expansion of clonogenic and early progenitors. Exp Hematol. 2002;28(5):569–574. doi: 10.1016/s0301-472x(00)00135-1. [DOI] [PubMed] [Google Scholar]
- 15.Barabe F, Kennedy JA, Hope KJ, Dick JE. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316(5824):600–604. doi: 10.1126/science.1139851. [DOI] [PubMed] [Google Scholar]
- 16.Bowie MB, Kent DG, Dykstra B, et al. Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proc Natl Acad Sci U S A. 2007;104(14):5878–5882. doi: 10.1073/pnas.0700460104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holyoake TL, Nicolini FE, Eaves CJ. Functional differences between transplantable human hematopoietic stem cells from fetal liver, cord blood, and adult marrow. Exp Hematol. 1999;27(9):1418–1427. doi: 10.1016/s0301-472x(99)00078-8. [DOI] [PubMed] [Google Scholar]
- 18.Kelly PF, Carrington J, Nathwani A, Vanin EF. RD114-pseudotyped oncoretroviral vectors. Biological and physical properties. Ann N Y Acad Sci. 2001;938:262–276. discussion 76–7. [PubMed] [Google Scholar]
- 19.Kelly PF, Vandergriff J, Nathwani A, Nienhuis AW, Vanin EF. Highly efficient gene transfer into cord blood nonobese diabetic/severe combined immunodeficiency repopulating cells by oncoretroviral vector particles pseudotyped with the feline endogenous retrovirus (RD114) envelope protein. Blood. 2000;96(4):1206–1214. [PubMed] [Google Scholar]
- 20.Hanawa H, Kelly PF, Nathwani AC, et al. Comparison of various envelope proteins for their ability to pseudotype lentiviral vectors and transduce primitive hematopoietic cells from human blood. Mol Ther. 2002;5(3):242–251. doi: 10.1006/mthe.2002.0549. [DOI] [PubMed] [Google Scholar]
- 21.Wunderlich M, Krejci O, Wei J, Mulloy JC. Human CD34+ cells expressing the inv(16) fusion protein exhibit a myelomonocytic phenotype with greatly enhanced proliferative ability. Blood. 2006;108(5):1690–1697. doi: 10.1182/blood-2005-12-012773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulloy JC, Cammenga J, Berguido FJ, et al. Maintaining the self-renewal and differentiation potential of human CD34+ hematopoietic cells using a single genetic element. Blood. 2003;102(13):4369–4376. doi: 10.1182/blood-2003-05-1762. [DOI] [PubMed] [Google Scholar]
- 23.Itoh K, Tezuka H, Sakoda H, et al. Reproducible establishment of hemopoietic supportive stromal cell lines from murine bone marrow. Exp Hematol. 1989;17(2):145–153. [PubMed] [Google Scholar]
- 24.Ito M, Hiramatsu H, Kobayashi K, et al. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood. 2002;100(9):3175–3182. doi: 10.1182/blood-2001-12-0207. [DOI] [PubMed] [Google Scholar]
- 25.Nicolini FE, Cashman JD, Hogge DE, Humphries RK, Eaves CJ. NOD/SCID mice engineered to express human IL-3, GMCSF and Steel factor constitutively mobilize engrafted human progenitors and compromise human stem cell regeneration. Leukemia. 2004;18(2):341–347. doi: 10.1038/sj.leu.2403222. [DOI] [PubMed] [Google Scholar]
- 26.Feuring-Buske M, Gerhard B, Cashman J, Humphries RK, Eaves CJ, Hogge DE. Improved engraftment of human acute myeloid leukemia progenitor cells in beta 2-microglobulin-deficient NOD/SCID mice and in NOD/SCID mice transgenic for human growth factors. Leukemia. 2003;17(4):760–763. doi: 10.1038/sj.leu.2402882. [DOI] [PubMed] [Google Scholar]
- 27.Haas DL, Case SS, Crooks GM, Kohn DB. Critical factors influencing stable transduction of human CD34(+) cells with HIV-1-derived lentiviral vectors. Mol Ther. 2000;2(1):71–80. doi: 10.1006/mthe.2000.0094. [DOI] [PubMed] [Google Scholar]
- 28.Sandrin V, Boson B, Salmon P, et al. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood. 2002;100(3):823–832. doi: 10.1182/blood-2001-11-0042. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Kimura T, Asada R, et al. SCID-repopulating cell activity of human cord blood-derived CD34− cells assured by intra-bone marrow injection. Blood. 2003;101(8):2924–2931. doi: 10.1182/blood-2002-09-2782. [DOI] [PubMed] [Google Scholar]
- 30.Yahata T, Ando K, Sato T, et al. A highly sensitive strategy for SCID-repopulating cell assay by direct injection of primitive human hematopoietic cells into NOD/SCID mice bone marrow. Blood. 2003;101(8):2905–2913. doi: 10.1182/blood-2002-07-1995. [DOI] [PubMed] [Google Scholar]