In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy (original) (raw)
. Author manuscript; available in PMC: 2010 Apr 28.
Published in final edited form as: Curr Biol. 2009 Apr 2;19(8):694–699. doi: 10.1016/j.cub.2009.02.056
Summary
The kinetochore is a macromolecular protein machine [1] that links centromeric chromatin to the plus-ends of one or more microtubules (MT), and segregates chromosomes during cell division. Its core structure consists of eight multi-component protein complexes, most of which are conserved in all eukaryotes. We use an in vivo two-color fluorescence microscopy technique to determine, for the first time, the location of these proteins along the budding yeast kinetochore axis at nanometer resolution. Together with kinetochore protein counts [2, 3], these localizations predict the 3-D protein architecture of a kinetochore-microtubule attachment, and provide new functional insights. We also find that the kinetochore becomes much shorter in anaphase as metaphase tension is lost. Shortening is mainly due to a decrease in the length of the Ndc80 complex, which may result either from intra-molecular bending of the Ndc80 complex at the kink within the stalk region of the Ndc80/Nuf2 dimer [4, 5] or from a change in its orientation relative to the microtubule axis. Conformational changes within the Ndc80 and Mtw1 complexes may serve as mechanical cues for tension-dependent regulation of MT attachment and the spindle assembly checkpoint. The geometry of the core structure of the budding yeast kinetochore reported here is remarkably similar to that found in mammalian kinetochores, indicating that kinetochore structure is conserved in eukaryotes with either point or regional centromeres.
Results and Discussion
The budding yeast kinetochore is nucleated by one centromeric nucleosome containing the centromere-specific histone H3 variant Cse4 [6]. The centromere also binds the DNA binding protein Mif2p and the CBF3 complex. Genetic, structural, and biochemical studies show that this assembly is stably linked to one microtubule (MT) plus-end by a network of protein complexes comprised of the Ctf19 complex [6], the Mtw1 complex [7, 8], the Spc105-Ydr532c complex [8], and the MT-binding Ndc80 complex [9, 10]. The MT-associated protein complex Dam1-DASH [11, 12] is also necessary for MT attachment. With the exception of the CBF3 and Dam1-DASH complex, these protein complexes are conserved in all eukaryotes [1, 13]. We have previously shown that the single MT attachment at the point centromere in budding yeast contains a specific number of each core structural protein complex [2]. Kinetochores at regional centromeres with 2–3 MT attachments in fission yeast also have nearly identical protein numbers per MT attachment (with the exception of the Dam1-DASH complex, ref. [3]), indicating that the protein architecture of individual MT attachment sites at these complex kinetochores is also conserved. The next critical task is to determine the organization of these structural protein complexes within a kinetochore-MT attachment in living cells, which remains poorly understood because of poor visibility by electron microscopy methods [14].
We have used a two-color, in vivo fluorescence microscopy technique to determine the relative position of budding yeast kinetochore proteins along the kinetochore axis with 10 nm resolution. Measurements are made pair-wise, with one protein fused to EGFP (a green florescent protein) and the other fused to tdTomato (a red fluorescent protein, [15]). Our technique is largely based on the in vitro method of Single molecule High REsolution Colocalization(SHREC, [16]), and extends its scope to measurements in vivo. The ability to fuse fluorescent protein genes at the C-terminus of budding yeast genes through homologous recombination–a technique not generally available in vertebrates, is critical for obtaining accurate localizations. The well-defined structure of the budding yeast mitotic spindle is also crucial. In a metaphase spindle, sister kinetochores on each chromosome are attached to MT plus ends from opposite poles and stretch their interconnecting chromatin apart by ~ 800 nm across of the spindle equator [17]. The kinetochores from all sixteen sister chromosome pairs form two well separated clusters on opposite sides of the spindle equator that appear as nearly diffraction-limited spots when imaged with wide-field fluorescence microscopy (Fig. 1a). After spindle elongation in anaphase, the sister kinetochore clusters become separated by > 4 μm (average spindle length in our mid- to late anaphase measurements was 5–6μm; Supplemental Data 1). In both metaphase and anaphase, kinetochores within the same cluster face the same pole (Fig. 1a). At metaphase, opposing pulling forces produced by each pair of sister kinetochores stretch the chromatin between sisters, and thus align the kinetochores and the axes of their attached MTs closely with the central spindle axis (Supplemental Data 1). In mid- to late anaphase, the kinetochore axes can be expected to be roughly perpendicular to the face of the spindle pole body to which they are connected by very short (~ 60 nm) MTs [17].
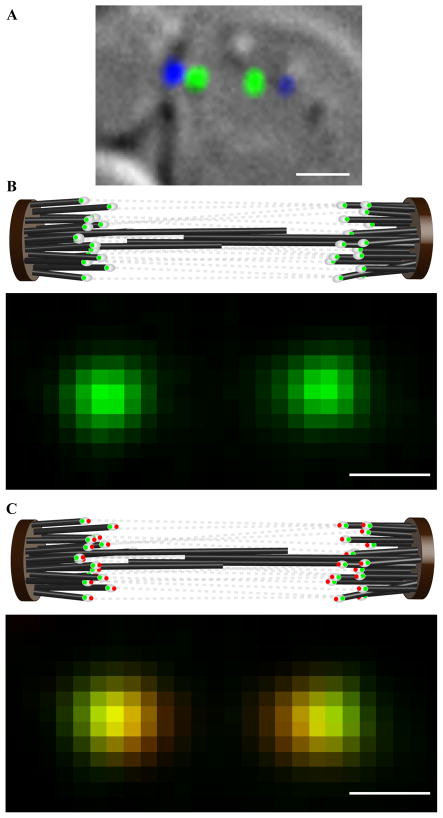
Figure 1. Measuring the distance separating two kinetochore proteins in a budding yeast metaphase spindle.
(a) A budding yeast cell in metaphase (DIC) with fluorescently labeled kinetochores (green) and spindle pole body (blue). (b) The cartoon depicts arrangement of kinetochores tagged with EGFP (white ovals with green dots) within the metaphase spindle. Tense chromatin connections (gray dotted lines) between sister kinetochores align them closely with the spindle axis. When such a cell is visualized with wide-field fluorescence microscopy, the two kinetochore clusters (each containing 16 kinetochores) appear as nearly diffraction-limited spots. (c) Metaphase spindle in a strain that has two kinetochore proteins, one protein fused to EGFP (green dots) and the other with tdTomato (red dots). When such cells are imaged simultaneously in the EGFP and tdTomato channels (lower panel), the offset between the centroids of the EGFP and tdTomato images of a kinetochore cluster can be used to determine the average distance separating the ends of the two proteins accurately. (scale bar ~ 1μm in a, ~ 500 nm in b and c; 1 pixel ~ 107 nm in b and c).
We simultaneously recorded red and green images of kinetochore clusters in cells expressing a selected pair of fluorescently labeled kinetochore proteins (Experimental Procedures, Fig. 1b). After red-green image registration, the distance separating the centroids of each pair of EGFP and tdTomato spots reflect the average distance separating the labeled kinetochore proteins within a cluster, even if the kinetochores themselves were staggered as much as 150 nm along the spindle axis (Supplemental Data 2). The centroids of the EGFP and tdTomato spots were determined within the in-focus plane with accuracy better than 10 nm by fitting the intensity distribution with a 2-D Gaussian function (Supplemental Data 2, [16, 18]). Residual error after red-green image registration was 6 nm or less (Supplemental Data 3). Image registration and the random orientation of spindle axes within the image plane within each data-set suppressed any bias due to chromatic aberrations to negligible levels (Supplemental Data 4). Measured distances were also corrected for the tilt of the spindle axis along the optical axis, which projects actual distances in the image plane, and thus underestimates the actual centroids separations (Supplemental Data 5). It has been previously established that the separation between the peaks of two normally distributed probability density functions is most accurately obtained using maximum likelihood estimation (Supplemental Data 6, ref. [19]). An additional source of error was incomplete maturation of the EGFP and tdTomato labels within each kinetochore cluster. However, this error only increases measurement variance, and does not introduce any systematic error leaving measurement accuracy unaffected. The minimum separation distance that could be directly measured by our technique was ~ 10 nm (Supplemental Data 7). As discussed below, the 68% confidence interval for measured separations > 10 nm was less than ± 3 nm. The average contributions are expected to be much smaller than the maximum size contribution of 2 nm for EGFP and 2–4 nm for tdTomato [20]. We have neglected the physical size of the fluorescent proteins from our analysis, because both EGFP and tdTomato are linked to a protein of interest via a flexible linker. The two monomers within tdTomato are also connected by a flexible linker [15]. These flexible linkers should allow the fluorescent proteins to freely rotate in space about the kinetochore protein end, thus significantly reducing their contribution to our distance measurements.
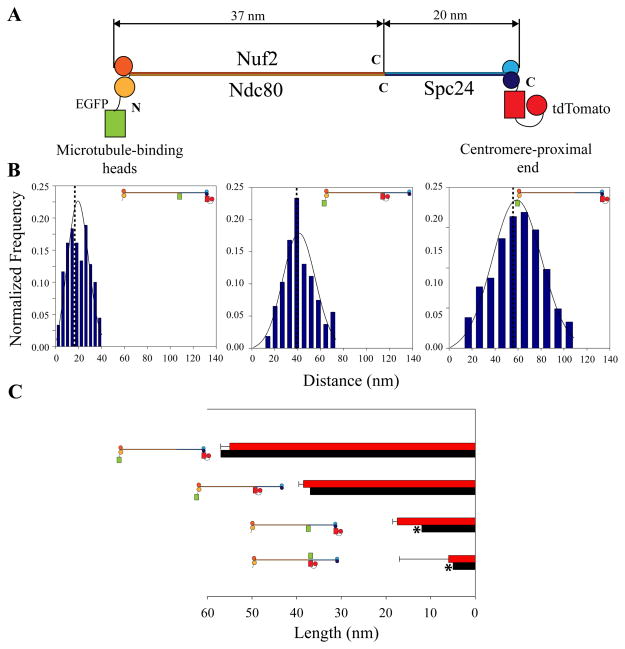
The NDC80 complex (Fig. 2a) is a 56 nm long, rod-shaped molecule with globular domains at both ends that are separated by a long alpha-helical coiled coil rod domain [5, 21, 22]. Because of its well-characterized shape, we used the Ndc80 complex as an in vivo ruler to test the accuracy of our technique. The N-terminal heads of Ndc80/Nuf2 bind the microtubule lattice while C-terminal globular domains of Spc24/Spc25 link the Ndc80 complex to the inner kinetochore [21]. To characterize the in vivo structure and orientation of the Ndc80 complex within the kinetochore, we constructed four strains through combinations of N- and C-terminal tagging for Ndc80p, Nuf2p, and Spc24p. We found that the overall length of the complex was 55 nm, which is almost equal to its full length. In contrast, if the complex were bound to the MT lattice at a 40 angle as observed in vitro [22], its end to end distance projected in the image plane would be 44 nm. Independent measurements of the lengths of the two sections of the complex yielded lengths of 38.5 nm for the Nuf2p-Ndc80p dimer, and 17 nm for the Spc24p-Spc25p dimer (Fig. 2b). The sum of these lengths is also close to the 55 nm measured for the total length. These results support the idea of an extended orientation for the Ndc80 complex along the MT axis in metaphase. They also demonstrate the accuracy of our measurement method.
Figure 2. Ndc80 complex as an in vivo molecular ruler.
(a) Structure of purified NDC80 complex [21]. An 80 amino acid long tail at the Ncd80p C-terminus separates it from the C-terminus of Nuf2p. Due to its unspecified structure, the exact distance separating the C-termini of Ndc80p and Nuf2p is unknown. (b) Distance measurements from four strains (N-Ndc80-C:Spc24-C, N-Ndc80:Nuf2-C, Ndc80-C:Spc24-C, and Ndc80-C:Nuf2-C) determine the dimension and orientation of the NDC80 complex in vivo. The non-Gaussian probability distribution fits (Supplemental Data 6) for three strains from the above list are shown [19]. Plots on the left hand side display histograms of measurements and the maximum likelihood fits for the data (solid lines). Dotted lines represent the true distance value predicted by maximum likelihood estimation. (c) Comparison of experimental distances (red bars) with the expected distances (black bars). The error bars represent the standard deviation estimated from the maximum likelihood estimation. The exact location of the C-terminus of Ndc80p is unknown. Therefore, the expected distance between the Ndc80-C: Nuf2-C and Ndc80-C:Spc24-C domains is a close estimate (marked by stars). The graphed measurement was the distance separating these two domains projected along the spindle axis (Supplemental Data 7).
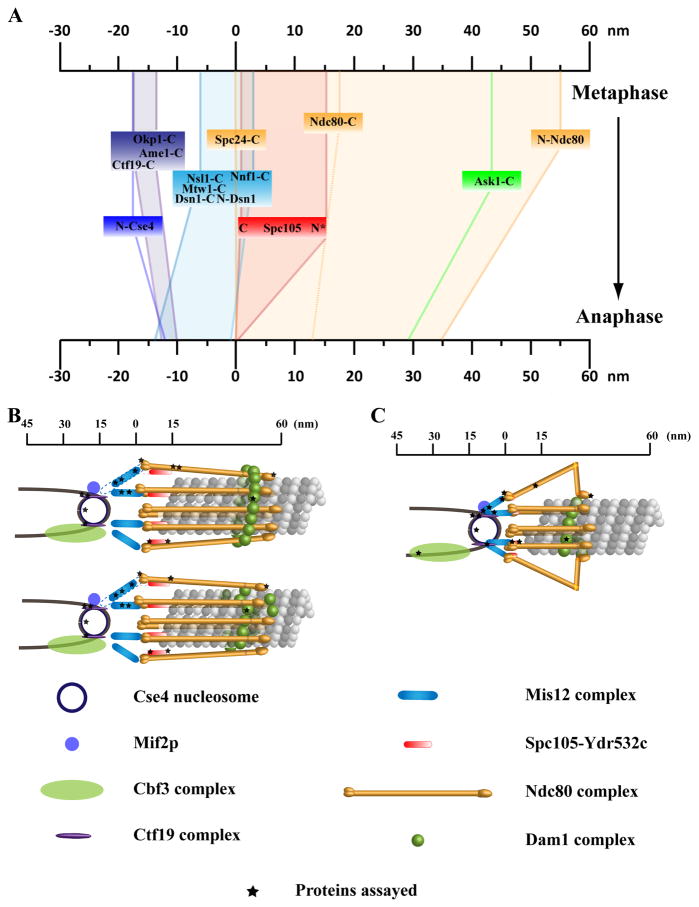
We next measured the average location of other kinetochore protein complexes in metaphase cells relative to the C-terminus of Ndc80p (Supplemental Data 8). We display the results with respect to the C-terminus of Spc24p for ease of interpretation (Fig. 3a), which is 17 nm inside (towards the centromere) of the C-terminus of Ndc80p. Relative to the C-terminus of Spc24, three members of the Mtw1 complex were inside (towards the centromere). The C-termini of Mtw1p and Nsl1p were localized 5 nm inside. For Dsn1p, the N-terminus was 2 nm and the C-terminus was 6 nm inside. Biochemical data links members of the Mtw1 complex with the centromere-proximal end of the NDC80 complex as well as with the DNA-binding protein Mif2p [7, 23]. Structural studies measure a length of 30 nm for reconstituted Mtw1 complex molecules in vitro (Dr. Eva Nogales, personal communication). Our measurements are consistent with both these data. The Mtw1complex is also linked to the centromere through the Ctf19 complex [23, 24]. We localized the C-termini of three members of the Ctf19 complex [7] Ctf19p, Ame1p, and Okp1p. Both Ame1p and Okp1p colocalized 13 nm inside the Spc24 C-terminus, while Ctf19p was located a distance of 16 nm. These proteins were close to the N-terminus of centromeric histone Cse4p, which was 17 nm inside Spc24 C-terminus.
Figure 3. Protein architecture of a kinetochore-microtubule attachment.
(a) The average location of kinetochore proteins along the axis of the kinetochore-microtubule attachment in metaphase and late anaphase. Each colored box represents a protein complex within the kinetochore. 68% confidence intervals on the mean position for all the measurements are less than 3 nm. The exception is Spc105p-C (indicated by stars), which could not be localized using maximum likelihood estimation. The positions in this case reflect the average offset along the spindle axis, which is likely an underestimate of the actual distance. For the Mtw1 and Ctf19 complexes, we only show the spans as measured by the positions of the respective member proteins. (b) 3-D visualization of the metaphase budding yeast kinetochore-microtubule attachment as predicted by the protein localization data assuming a symmetric arrangement of kinetochore protein complexes around the cylindrical microtubule lattice. Black stars indicate the positions of fluorescent labels used in distance measurements. The configuration of the Dam1/DASH complex suggests two possibilities–a kinetochore that contains an oligomeric ring of the Dam1 complex (top), and a kinetochore that employs Dam1/DASH patches or incomplete rings (bottom). Dashed lines indicate established biochemical interactions between two protein complexes. (c) Loss of centromeric tension and changes induced the cell-cycle regulation result in a shorter kinetochore in late anaphase. A striking change occurs in the Ndc80 complex as the Nuf2p-Ndc80p dimer shows a length reduction that is 40% larger than the reduction predicted by an overall change in the orientation of the molecule with respect to the MT. The model displays a possible mechanism that relies on bending of the Ndc80-Nuf2p dimer at the kink as observed in vitro.
In addition to the Ndc80 complex, MT attachment within the kinetochore outer domain depends on Spc105p and the Dam1/DASH complex. The C-terminus of Spc105p, a large 100 kDa protein, colocalized with Spc24p C-terminus (1 nm inside; Fig. 3a). The position of the N-terminus of Spc105p could not be accurately determined using the maximum likelihood method, but we estimate it to be 16 nm outside (towards the MT) the Spc24p C-terminus (Supplemental Data 8). This indicates that the protein likely extends outward along the microtubule axis. Surprisingly, we found that Ask1p, a key component of the Dam1/DASH complex was 12 nm inside of the microtubule-binding head domains of Ndc80 complex (Fig. 3a).
There were significant changes in the relative positions of kinetochore proteins from metaphase to anaphase. We found that the overall kinetochore length in late anaphase cells was reduced by 25 nm (Fig. 3a, Supplemental Data 9 and 10). The end-to-end length of the Ndc80 complex decreased from 55 nm in metaphase to 34 nm. Within the Ndc80 complex, the separation between the two ends of the Ndc80-Nuf2 dimer was reduced by 13 nm, whereas the Spc24-Spc25 dimer showed a smaller decrease of 5 nm. Also important was the movement of the Spc24/Spc25 end of the Ndc80 complex 5 nm closer to the centromeric nucleosome (Fig. 3a). Components of the Mtw1 complex also showed a significant redistribution; notably Nsl1p moved closer to the centromeric nucleosome. On the other hand, the position of the Ctf19 complex with respect to the N-terminus of Cse4p did not change significantly suggesting a rigid coupling between this complex and the centromeric nucleosome. It is known that the CBF3 complex binds to the CDE III region [26] of the centromeric DNA via Ndc10p and Cep3p in metaphase. We found that the C-terminus of Ndc10p was 35 nm inside N-terminus of Cse4p in anaphase. This large distance is likely due to the anaphase dislocation of Ndc10p from the kinetochore [25]. Finally, the position of the Dam1-DASH complex does not change significantly with respect to the Ndc80 head domain. It should be noted that the average number of Dam1-DASH complex molecules per budding yeast kinetochore decreases from 16–20 in metaphase to 9 in anaphase, which is insufficient to form Dam1-DASH rings around the late anaphase MTs [2].
This study assembles the first in vivo, high resolution map of kinetochore protein localization along the axis of a kinetochore-microtubule attachment (Fig. 3a, ref. [27]). It should be noted that these locations reflect average positions of kinetochore proteins. Furthermore, our technique can measure distances in the image plane, and it is insensitive to distance changes that may occur either along the optical axis or perpendicular to the spindle axis. Therefore, positional changes that take place within complexes of unknown shape (such as the Mtw1, Spc105-YDR532c, and Ctf19 complexes) along these directions could not be detected. The position of the MT plus-end within the kinetochore could not be determined with our technique. The location of the MT-associated Dam1-DASH complex suggests that the MT plus-end extends at least 10 nm beyond the contact point between the MT and Nuf2p/Ndc80p head domains [28]. KNL-1, the C. elegans homolog of Spc105p KNL-1 [22], and the N-terminal domain of Spc7, the S. pombe homolog show MT-binding activity [29]. These data suggest that the MT plus-end may extend up to the Spc24/Spc25 end of the Ndc80 complex.
The localization data can be combined with protein numbers [2] and existing structural information to predict a 3-D visualization of kinetochore-MT attachment assuming a symmetric distribution of proteins around the cylindrical MT lattice (Fig. 3b). The end-to-end measurement of the metaphase length of the Ndc80 complex shows that it binds the MT lattice while making a small angle with the MT axis, rather than the 40 angle of unbound Ndc80 complexes observed in vitro. This alignment of the Ndc80 complex and MT axes can be expected, given that the Ndc80 complex is one of the primary force generators at the kinetochore, and that this force acts along the MT axis. Available biochemical data suggests that the contact between the Ndc80 complex with the inner kinetochore is achieved through interactions of Spc24/Spc25 globular domains with the Mtw1 and Spc105 complexes [22]. Additional points of contact would be necessary to resist the pulling forces tending to align the Ndc80 complex along the MT axis, and maintain its tilted orientation (with respect to the MT axis) in metaphase. The possible conformations [11, 30] and functional mechanisms [31, 32] of the Dam1-DASH complex in vivo are critical questions that remain unanswered. The Dam1-DASH complex can form oligomeric rings (containing 16–23 copies) around the MT lattice in vitro that have an inner diameter of 35 nm and an outer diameter of 45–54 nm [11, 12]. Structural and theoretical studies also show that in this configuration, individual subunits within the ring interact with the MT lattice via projections that span the ~ 5 nm gap between the MT lattice and the inner surface of the Dam1-DASH ring [31, 33]. There are 16–20 DAM1-DASH complex molecules per budding yeast kinetochore in vivo, enough to build one ring [2]. If a persistent Dam1-DASH ring structure exists in vivo, its location within the kinetochore would require the Ndc80 complex molecules to attach the MT lattice at angles of 50–60 to accommodate the Dam1-DASH ring underneath. These large angles are inconsistent with the measured end-to-end length of 55 nm for the Ndc80 complex. Therefore, the Dam1-DASH ring will have to encircle both the MT lattice and the rod domains of Ndc80 complex molecules (Fig. 3b). Individual Dam1-DASH monomers can still interact with the MT lattice via the projections spanning the gap between the inner surface of the ring and the MT lattice. This configuration may also promote rapid rebinding of Ndc80 heads to the MT lattice by limiting their diffusion. Alternatively, the Dam1-DASH complex may not form a single ring structure at the kinetochore. Instead, spiral oligomers that incompletely surround the microtubule lattice at several locations along the microtubule axis may assemble (Fig. 3b). In this configuration, direct binding between the Dam1-DASH monomers and the Ndc80 complex becomes necessary for their stable association with the kinetochore. Although a direct biochemical link between the Dam1-DASH complex and other kinetochore complexes has not been established, such a linkage is necessary for transmitting the force generated through interactions between the Dam1-DASH complex and the MT lattice to the rest of the kinetochore for the participation of either configuration in force generation [31, 34].
The anaphase measurements reveal tension and/or cell-cycle dependent changes within the kinetochore (Fig. 3c). The reduction in the end-to-end length of the Ndc80 complex in late anaphase indicates that the Ndc80 complex directly participates in force generation, and transmits this force to the inner kinetochore components through the Mtw1 complex. The observed decrease may be explained either through intra-molecular bending at the kink domain within the complex observed in vitro (depicted in Fig. 3c) or a re-orientation of the entire complex, so that it makes an angle of 45–50 with the axis of the MT. The later configuration requires a large extension (~ 40 nm) of the inner kinetochore complexes perpendicular to the MT axis to stably link the Ndc80 complex back to the inner kinetochore and the centromere. The elongated shapes of the Mtw1 complex and Ctf19 complex may facilitate such an alignment of the Ndc80 complex in anaphase. The total length of such a linkage in anaphase would predict a much longer distance between the centromere and the Spc24/Spc25 end of the Ndc80 complex under the metaphase pulling forces acting along the axis of the MT. We therefore show the simpler configuration of the anaphase kinetochore that relies on intra-molecular bending of the Ndc80 complex.
Many of the structural proteins and protein complexes are conserved in all eukaryotes [1], although the complex architecture of the regional centromeres likely necessitates significant modifications, especially to the centromere-proximal proteins [35–37]. Architecture of the kinetochore-microtubule attachment site built on either the point or regional centromere foundation however, is likely conserved in all eukaryotes as evidenced by the conserved stoichiometry of kinetochore proteins between point and regional centromeres [2, 3]. Indeed, kinetochore protein localizations obtained by antibody labeling in fixed HeLa cells show a strikingly similar pattern (submitted). This conservation of kinetochore protein structure and the protein architecture of the kinetochore-MT attachment demonstrate that the core structure of the kinetochore, along with its basic functional mechanisms in force generation and spindle assembly checkpoint signaling, are conserved throughout eukaryotic phylogeny.
Experimental Procedures
Strains & Growing conditions
Strains (Supplemental Data 12) were grown in either YPD or YPG at 32°C. Proteins were tagged with either EGFP or tdTomato through homologous recombination, mostly at the C-terminus, using PCR-amplified cassettes. Cells from mid-log phase cultures were re-suspended in synthetic media, and immobilized on concanavaline A (cat. # 7275, Sigma, St. Louis, MO) coated coverslips for imaging.
Imaging
Cells were imaged at room temperature on a Nikon TE-2000E (Nikon Instruments Inc., Melville, NY) inverted microscope equipped with a 1.4 NA, 100x DIC objective and 1.5x optovar lens (1 pixel ~ 107 nm). A dual-excitation filter set (FITC/TRITC ET set# 59004, Chroma Technology, Rockingham, VT) was used for simultaneous excitation of both, EGFP and tdTomato. Images were acquired with a DV-887B iXon camera (Andor Technology, South Windsor, CT) using the conventional acquisition mode mounted on the bottom port of the microscope. The Dual-View attachment (MAG Bio-systems, Pleasanton, CA) was used for simultaneous acquisition of images at both wavelengths with pre-mounted dichroic and emission filters for EGFP and tdTomato. Prior to each experiment 100 nm TetraSpek (cat. # T-7279, Invitrogen, Carlsbad, CA) bead images were acquired for image registration (Supplemental Data 4). For each cell, 10 image slices were obtained by moving the piezoelectric Z-stage (MadCity Labs, Madison, WI) through 200 nm steps, and a 300x300 pixel wide, centrally located region was recorded in each image. The exposure time was set at 800 ms per image, to maintain a high signal-to-noise ratio with minimal bleaching during image acquisition. The imaging and image acquisition hardware was run by Metamorph 7 (Molecular Devices, Sunnyvale, CA).
Image Analysis
Image analysis was carried out using custom software written in MatLAB 7 (MathWorks Inc., Natick, Md). The tdTomato image stack was registered with the EGFP image stack (described in detail in Supplemental Data 4). For centroid determination, the area of interest for centroid localization was determined by placing an 8×8 pixel region (for metaphase measurements) on an EGFP image such that the cumulative intensity within the centrally located 2×2 pixel square was maximized. The corresponding region from the registered tdTomato image was then extracted for centroid localization. A similarly selected 10×10 pixel region was used for analyzing anaphase cells.
Supplementary Material
Supplementary Data
Acknowledgments
We thank Stirling Churchman for suggestions and for sharing software and Arshad Desai for reading the manuscript. A.P.J. holds a Career Award at the Scientific Interface from the Burroughs-Wellcome Fund. The research is supported by grants to K.S.B. and E.D.S. from NIH and NIGMS.
References
- 1.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 2.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon ED, Bloom KS. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008 doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang HW, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E. Architecture and Flexibility of the Yeast Ndc80 Kinetochore Complex. J Mol Biol. 2008 doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ortiz J, Stemmann O, Rank S, Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nekrasov VS, Smith MA, Peak-Chew S, Kilmartin JV. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:4931–4946. doi: 10.1091/mbc.E03-06-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janke C, Ortiz J, Lechner J, Shevchenko A, Magiera MM, Schramm C, Schiebel E. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. Embo J. 2001;20:777–791. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wigge PA, Kilmartin JV. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol. 2001;152:349–360. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 12.Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006 doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- 13.Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature biotechnology. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 16.Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc Natl Acad Sci U S A. 2005;102:1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Churchman LS, Flyvbjerg H, Spudich JA. A non-gaussian distribution quantifies distances measured with fluorescence localization techniques. Biophys J. 2006;90:668–671. doi: 10.1529/biophysj.105.065599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang F, Moss LG, Phillips GN. The molecular structure of green fluorescent protein. Nat Biotech. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 21.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci U S A. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 23.Euskirchen GM. Nnf1p, Dsn1p, Mtw1p, and Nsl1p: a new group of proteins important for chromosome segregation in Saccharomyces cerevisiae. Eukaryot Cell. 2002;1:229–240. doi: 10.1128/EC.1.2.229-240.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westermann S, Cheeseman IM, Anderson S, Yates JR, 3rd, Drubin DG, Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouck DC, Bloom KS. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc Natl Acad Sci U S A. 2005;102:5408–5413. doi: 10.1073/pnas.0405925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Espelin CW, Kaplan KB, Sorger PK. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schittenhelm R, Heeger S, Althoff F, Walter A, Heidmann S, Mechtler K, Lehner C. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma. 2007;116:385–402. doi: 10.1007/s00412-007-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Bachant J, Alcasabas AA, Wang Y, Qin J, Elledge SJ. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerres A, Jakopec V, Fleig U. The conserved Spc7 protein is required for spindle integrity and links kinetochore complexes in fission yeast. Mol Biol Cell. 2007;18:2441–2454. doi: 10.1091/mbc.E06-08-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- 31.Grishchuk EL, Spiridonov IS, Volkov VA, Efremov A, Westermann S, Drubin D, Barnes G, Ataullakhanov FI, McIntosh JR. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc Natl Acad Sci U S A. 2008;105:6918–6923. doi: 10.1073/pnas.0801811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gestaut DR, Graczyk B, Cooper J, Widlund PO, Zelter A, Wordeman L, Asbury CL, Davis TN. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat Cell Biol. 2008;10:407–414. doi: 10.1038/ncb1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang HW, Ramey VH, Westermann S, Leschziner AE, Welburn JP, Nakajima Y, Drubin DG, Barnes G, Nogales E. Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat Struct Mol Biol. 2007;14:721–726. doi: 10.1038/nsmb1274. [DOI] [PubMed] [Google Scholar]
- 34.Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci U S A. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 36.Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, et al. CCAN Makes Multiple Contacts with Centromeric DNA to Provide Distinct Pathways to the Outer Kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 37.Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, 3rd, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data