The Effect of Chemical Modification and Nanoparticle Formulation on Stability and Biodistribution of siRNA in Mice (original) (raw)
Abstract
Instability and inadequate biodistribution of double-stranded RNA are major drawbacks to the clinical use of RNA interference. This work compares chemical modification and nanoparticle formulation as strategies to improve the systemic delivery of small interfering RNA (siRNA). Variable levels of chemical modified siRNA, either naked or within nanoparticle, were intravenously injected into mice to study temporal stability and biodistribution detected by direct radioactive labeling or by northern blotting. Naked siRNA showed rapid renal clearance, with circulatory half-life of <5 minutes that could be extended to >30 minutes by cholesterol conjugation. The integrity of the chemically stabilized siRNA was maintained in blood for at least 30 minutes, whereas, unmodified siRNA duplex was degraded within 1 minute. Intact chemically modified siRNA could also be detected in all analyzed organs at 30 minutes but disappeared at 24 hours, except for heavy locked nucleic acid (LNA)-modified and cholesterol-conjugated siRNA in the lungs. Chitosan, liposomal, or JetPEI formulation greatly improved the stability and biodistribution of siRNA. Interestingly, high siRNA accumulation of the chitosan/siRNA formulation within the kidney was observed 24 hours postadministration. This comparative study highlights improvements to siRNA stability and pharmacokinetics, key determinants for development of clinically relevant RNAi therapeutics.
Introduction
Small interfering RNA (siRNA)–mediated gene silencing is a potential therapeutic strategy for human diseases. For clinical use, however, barriers such as instability, renal clearance, and poor cellular uptake associated with systemic administration of naked siRNA need to be overcome.1 Chemical modification of the nucleic acid backbone has been used to improve the stability and alter biodistribution of siRNA. For example, phosphorothioate-modified oligonucleotides can be used to improve cellular uptake and improve circulatory half-life and biodistribution2,3 whilst, incorporation of locked nucleic acid (LNA) molecules that contain methylene bridge connecting the 2′-oxygen with the 4′-carbon has been shown to increase serum stability and reduce off-target gene regulation.4,5 Small internally segmented interfering RNA (sisiRNA) composed of an antisense strand (AS) and two shorter 10–11 nucleotides sense strands is fully functional with the ability to accommodate more heavily modified ASs than standard siRNAs, which is important for in vivo applications.6 Other chemical approaches for siRNA include conjugation of functional groups to improve cell or tissue specific targeting. For instance, cholesterol-conjugated siRNA has been shown to increase serum protein binding, alter the biodistribution and reduce urinary excretion. Silencing endogenous genes with cholesterol-conjugated siRNA demonstrates improved systemic activity through chemical modification strategies.7
Nanoparticle formulation is another strategy to improve the pharmacokinetic properties and maximize the siRNA payload at target sites. Various cationic polymer (polyplexes) and lipid-based (lipoplexes) nanoparticles formed by self-assembly with nucleic acids have been developed.8,9 For example, the use of polyethylenimine (PEI) and chitosan nanoparticles in addition, to lipoplexes have been shown to facilitate siRNA-mediated gene silencing in vivo.10,11,12,13,14,15 In contrast to DNA polyplexes, the circulatory half-life and biodistribution of siRNA polyplexes has not been extensively conducted.
In this study, we compare the effect of chemical modification (LNA-modified siRNA, phosphorothioate, and sisiRNA, or cholesterol-conjugated siRNA), and nanoparticle formulation on siRNA stability, blood clearance, and biodistribution in mice. Additionally experiments were performed to study the effect of siRNA dose. The siRNA was detected directly by radioactive labeling or indirectly by northern blotting to allow a true determination of siRNA integrity.
Results
Chemical modification effects on blood clearance and biodistribution
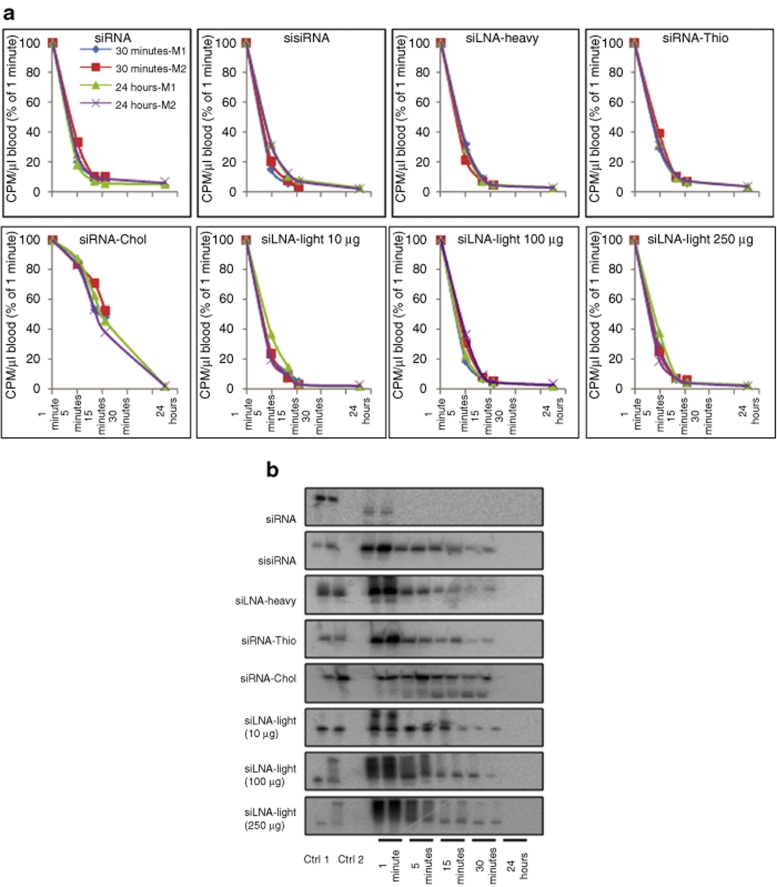
A range of siRNAs with different chemical modifications and conjugations were prepared (refer to Table 1 for nomenclature). siRNAs were intravenously (i.v.) injected into mice (N = 2) at a concentration of 400 µg/kg. Three different detection strategies were used for siRNA analysis postinjection: scintillation count of radiolabeled siRNA or direct gel electrophoresis and northern analysis of total RNA. For the direct labeling method, [γ-32P] adenosine triphosphate 5′-labeling of AS allowed temporal detection of siRNA in whole blood over 1, 5, 15, 30 minutes, and 24 hours, and organs at 30 minutes and 24 hours. Scintillation counts in whole blood samples at different time points are shown in Figure 1a. The lightly LNA–modified siRNA (siLNA-light) and sisiRNA duplexes were rapidly removed from the bloodstream with ~60–80% decline at 5 minutes (relative to 1-minute time point) and ~90 and >95% at 15 and 30 minutes, respectively, with ~1% remaining at 24 hours. In contrast, ~80% of the cholesterol-conjugated siLNA light (siRNA-Chol) remained in the circulation at 5 minutes and decreased to ~50% at 30 minutes.
Table 1.
Oligonucleotide sequences and chemical modifications
Figure 1.
siRNA blood clearance and stability analysis by scintillation counting and gel electrophoresis. (a) Scintillation measurement of blood radioactivity over indicated time course for siRNA, sisiRNA, siLNA-heavy, siRNA-Thio, siRNA-Chol, and siLNA-light (10 µg) after intravenous injection in mice (N = 2). Dose-dependent experiments were performed with doses of 10-, 100-, and 250-µg per mouse, indicated by siLNA-light 10 µg, siLNA-light 100 µg, and siLNA-light 250 µg. The data are obtained from three measurements and presented as average, and normalized with the level at 1 minute. The _x_-axis is a log 5 scale. (b) RNA integrity analysis for blood samples using 15% denaturing polyacrylamide gel electrophoresis. Loading order: lane 1, 0.5-ng 32P-labeled antisense oligos (Ctrl 1), lane 2, 1-ng injected duplexes (Ctrl 2), lane 3, blank, lanes 4–13, time points 1, 5, 15, 30 minutes, and 24 hours blood from mouse 1 and 2, respectively. Total RNA (2 µg) was loaded into each well. CPM, counts per minute; Ctrl, control; LNA, locked nucleic acid; siLNA, LNA-modified siRNA; siLNA-heavy, heavily LNA–modified siRNA; siLNA-light, lightly LNA–modified siRNA; siRNA, small interfering RNA; sisiRNA, small internally segmented interfering RNA; siRNA-Chol, cholesterol-conjugated siLNA-light; siRNA-Thio, phosphorothioate-modified siRNA.
To investigate siRNA integrity, 2 µg of total RNA from whole blood was analyzed on a denaturing polyacrylamide gel and labeled AS strand visualized by autoradiography. As shown in Figure 1b, unmodified siRNA duplex was rapidly and completely degraded 1 minute after injection. In contrast, the modified duplexes, showed a gradual reduction of band intensity in a time-dependent manner with intact siRNA still present after 30 minutes. The absence of degradation products suggests that LNA modifications can significantly reduce the susceptibility to nuclease degradation and increase stability in the bloodstream. A phosphorothioate-modified siRNA (siRNA-Thio) appeared to have a similar blood clearance profile, whereas, circulation of siRNA-Chol was significantly prolonged in the blood with <60% decrease over 30 minutes, although not detectable after 24 hours.
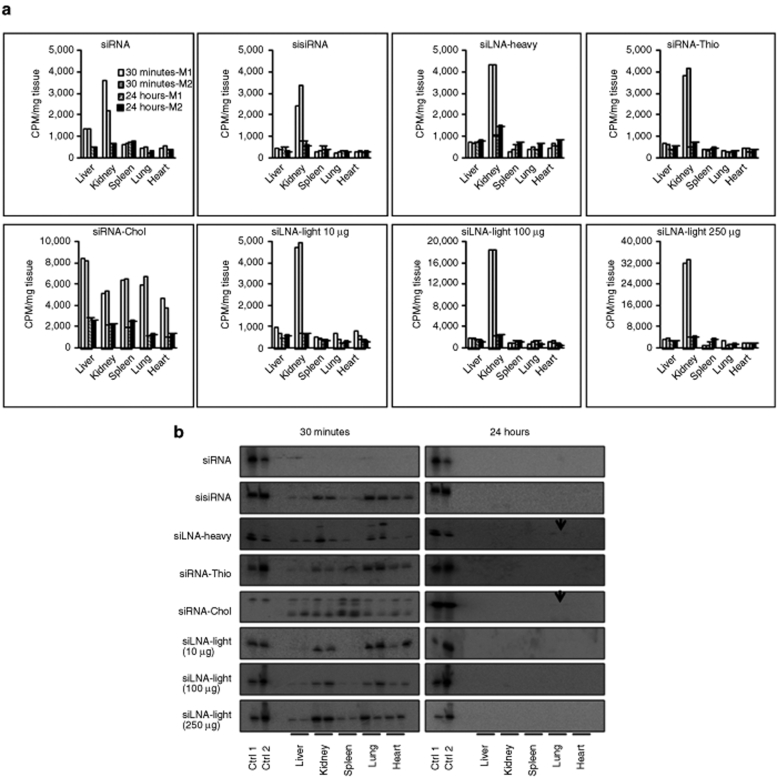
Clearance of naked siRNA from the blood is associated with excretion via the kidney or distribution to systemic organs. The radioactivity level of the siLNA, sisiRNA, and siRNA-Thio reached the highest in the kidney after 30 minutes, 6–8 times higher than in other organs. At 24 hours, the level of radioactivity decreased about five times in the kidney, but was maintained at the same level in other organs. In contrast, siRNA-Chol showed higher levels in all organs, especially the liver both at 30 minutes and 24 hours after injection (Figure 2a).
Figure 2.
siRNA tissue distribution at 30 minutes and 24 hours time points. (a) Scintillation measurement of radioactivity in five organs (expressed as CPM per mg organ). The data were obtained from three measurements and presented as average. (b) RNA integrity of tissue samples using denaturing polyacrylamide gel electrophoresis as above. Loading order: lane 1, 0.05-ng 32P-labeled antisense oligo (Ctrl 1), lane 2, 0.1-ng injected duplexes (Ctrl 2), lane 3, blank, lanes 4–13, organs at 1 and 2 at 30 minutes and 24 hours, respectively. Total RNA (4 µg) was loaded into each well. CPM, counts per minute; Ctrl, control; LNA, locked nucleic acid; siLNA, LNA-modified siRNA; siLNA-heavy, heavily LNA–modified siRNA; siLNA-light, lightly LNA–modified siRNA; siRNA, small interfering RNA; sisiRNA, small internally segmented interfering RNA; siRNA-Chol, cholesterol-conjugated siLNA-light; siRNA-Thio, phosphorothioate-modified siRNA.
Similar to blood, we found that intact siRNA detected by gel analysis in different organs at 30 minutes for the modified duplexes. For the siLNA-light, sisiRNA, and siRNA-Thio, the highest intensity was found in the lung and kidney, followed by the heart with lowest level in liver and spleen. Relatively equal levels were detected in all organs for the siLNA-heavy. For siRNA-Chol, the highest intensity of intact siRNA was found in the spleen at 30 minutes, with relatively equal levels in other organs. The signal was undetectable for unmodified siRNA at any time point, which correlates with the rapid degradation detected in the blood. After 24 hours, only the siLNA-heavy and siRNA-Chol was detected in the lungs (<5% intensity compared to 30 minutes) (Figure 2b).
To investigate the effect of siRNA concentration on clearance rate and biodistribution, dose-dependent experiments were performed with 10-, 100-, or 250-µg siLNA-light per mouse (Figure 1a,b). A similar blood clearance rate and tissue distribution pattern was found for all three doses, although the absolute radioactivity counts were increased in a dose-dependent manner in blood samples and organs (Figures 1a,b and 2a,b).
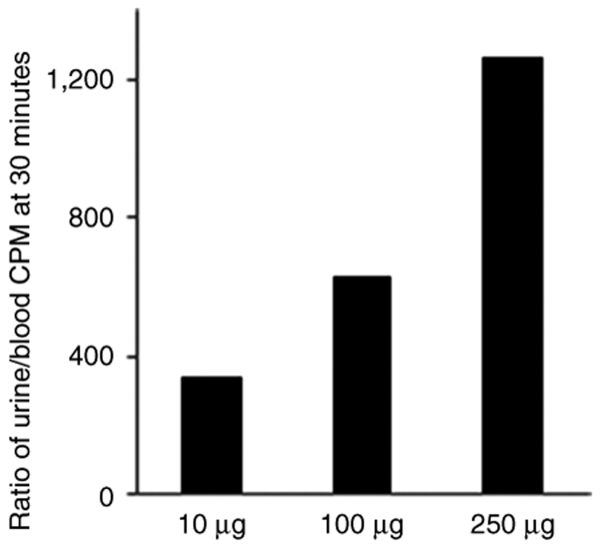
We collected urine samples from one mouse of each group and measured the radioactivity by scintillation counting. The urine/blood (U/B) ratio at 30 minutes showed a dose-dependent increase ~300-, 600-, and 1,200-fold for LNA 10, LNA 100, and LNA 250 µg, respectively, suggesting that increasing the siRNA concentration led to a relatively higher rate of excretion (Figure 3). The observed increase in AS in the kidneys at the highest dosing furthermore adds support for relatively greater siRNA renal excretion at higher siRNA doses.
Figure 3.
Dose-dependent correlation between siRNA dose and urine excretion. The ratio of CPM level in urine to CPM level in blood at 30 minutes at different doses is presented. Data are obtained from one mouse in each group. CPM, counts per minute; siRNA, small interfering RNA.
The drawback to siRNA detection by the means of a 5′-phosphate is possible phosphate removal or exchange with nonradioactive phosphates by phosphatases and kinases. To address this problem, we performed northern blotting on the same filters that were used for blotting of the labeled oligos. The two assays were performed 4 months (eight half-lives) apart to ensure that no detectable radioactivity was remaining on the filters. The result from the northern blotting revealed a very similar pattern to that obtained using the direct detection method, suggesting that a 5′-end labeled phosphate is a robust marker for siRNA biodistribution in mice. A typical example of a northern blot is shown in Figure 4c for siLNA-light.
Figure 4.
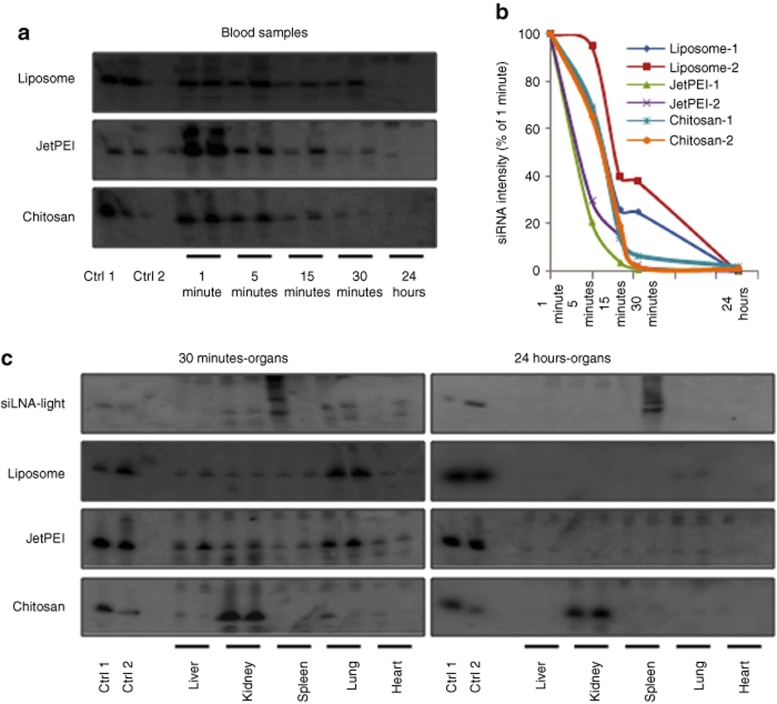
siRNA stability, blood clearance and organ distribution of siRNA formulations using northern analysis. Northern blot analysis over indicated time course for liposomal, chitosan, and JetPEI/siRNA nanoparticles (containing 10 µg siRNA) after 200 µl i.v. injection in mice (N = 2). RNA from blood (2 µg) and from tissues (4 µg) were run on 15% denaturing polyacrylamide gels and northern analysis was performed by probing with [γ-32P] adenosine triphosphate–labeled sense-strand LNA-modified siRNA. Liposome, JetPEI, and chitosan represent nanoparticles complexed with siLNA-light. (a) siRNA stability: Intact siRNA integrity was shown at indicated time course. Loading order: lane 1, 3-ng siRNA duplexes (Ctrl 1), lane 2, 2-ng siRNA/nanoparticle complex (Ctrl 2), lane 3, blank, lanes 4–13, time points 1, 5, 15, 30 minutes, and 24 hours blood from mouse 1 and 2, respectively. (b) siRNA blood clearance: the intensity of the siRNA signals was measured by Quantity ONE programme, and percentages of the intensity at 1 minute and clearance curves are presented as a function of time (log 5 scale). (c) siRNA biodistribution: loading order: lane 1, 1.5-ng duplexes (Ctrl 1), lane 2, 1 ng siRNA/nanoparticle complex (Ctrl 2), lane 3, blank, lanes 4–13, organs at 1 and 2 at 30 minutes and 24 hours, respectively. Comparison of signal intensities reveals ~7–8% of injected siRNA accumulated in the kidneys at 30 minutes with ~1.5–2% still remaining after 24 hours. Ctrl, control; LNA, locked nucleic acid; siLNA, LNA-modified siRNA; siLNA-light, lightly LNA–modified siRNA; siRNA, small interfering RNA.
Pharmacokinetics of nanoparticle-based siRNA vectors
Nanoparticles composed of siLNA-light duplexes in combination with chitosan, JetPEI, or lipids were formulated and evaluated for in vivo stability and biodistribution using northern analysis. Clear differences were found among the different formulations (Figure 4a–c). For liposomal/siRNA, there was slow clearance with 30% remaining in the bloodstream after 30 minutes compared to intensity at 1 minute. No siRNA, however, could be detected in the blood after 24 hours (Figure 4a,b). Liposomal formulation facilitated uptake of siRNA in the lungs five- to tenfold compared to other organs, with 5% of 30-minute levels still present at 24 hours (Figure 4c).
JetPEI/siRNA yielded a strong AS signal at 1 minute but ~80% of this AS was rapidly cleared over the next 4 minutes and then seems more constant later on (Figure 4a,b). The AS could be detected in the blood, even after 24 hours, as a degraded product, at <1% of 1-minute intensity. Similar to the liposomal formulations, greater levels were found in the lung (two- to tenfold) than the other organs, which exhibited similar amounts (Figure 4c). A slight increase was observed in the liver compared to the kidney, similar in effect to cholesterol conjugation (Figure 2b). One surprising observation was that intact siRNA could be detected in all organs at 24 hours, at ~5–10% of the intensity observed at 30 minutes (Figure 4c).
Chitosan prolonged the blood circulation time to 65% after 5 minutes but only ~5% remained at 30 minutes and ~1–2% at 24 hours (Figure 4a,b). Chitosan did, however, alter the organ distribution patterns with pronounced accumulation in the kidney (Figure 4c). At 30 minutes, the intensity of intact siRNA in kidney was much stronger than other organs, showing a tenfold and 100-fold increase compared to lung and spleen, respectively. It was calculated to be ~7–8% of the total amount of injected siRNA based on weight of kidneys, the total RNA obtained and the intensity of samples and the control. The level in the kidney remained very high even after 24 hours, ~1.5–2% of injected amount (~15–20% of the level detected at 30 minutes). Detection of siRNA was also observed in the lungs at 24 hours, (~1–2% measured at 30 minutes) but in much lower level (<1%) than the kidney at 24 hours. This result suggests that chitosan/siRNA complexes preferentially accumulate in the kidneys whilst circumventing rapid glomerular filtration observed for naked siRNA.
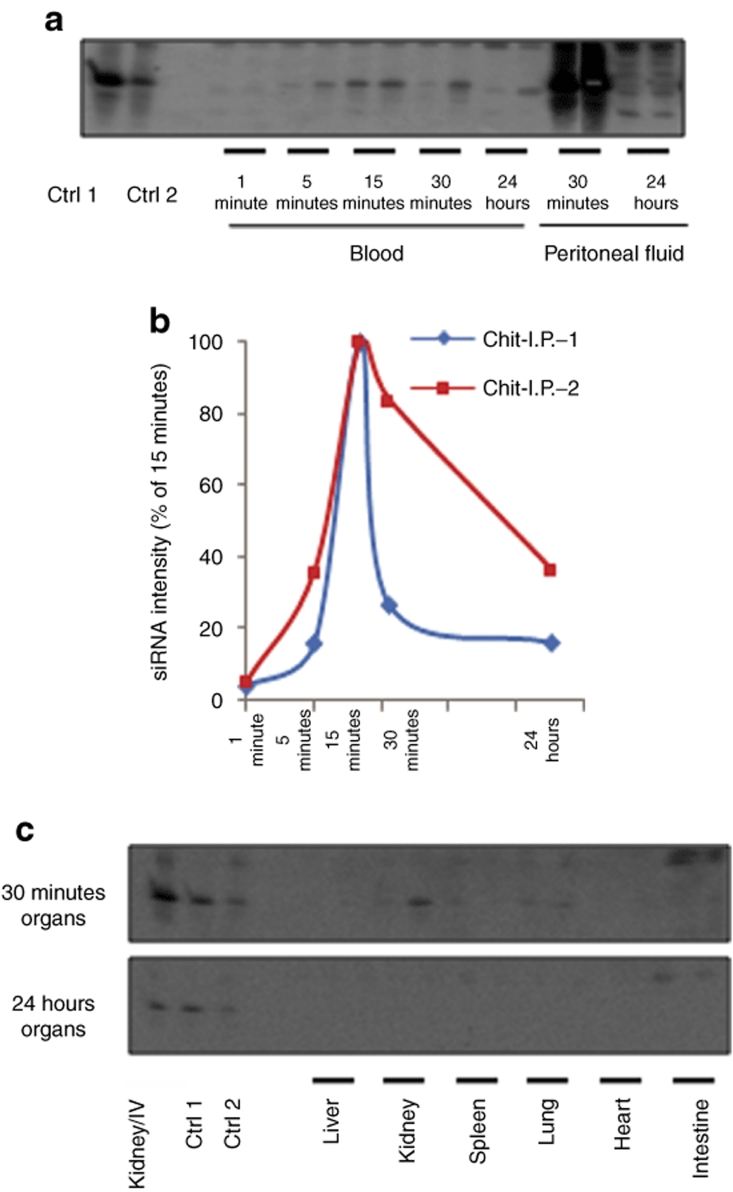
Intraperitoneal (i.p.) administration was used to investigate the effect of administration route on the biodistribution of chitosan/siRNA complexes. siRNA was detected in the blood at 1 minute, and increased at 5 minutes, reaching a maximum at 15 minutes (set as 100% for clearance curve) (Figure 5a,b). The levels thereafter gradually declined and at 24 hours only a degraded product was observed. An extremely high siRNA concentration was present in the peritoneal fluid at 30 minutes, ~150-fold higher than the blood at same time points that suggests retention within the peritoneal cavity. Approximately, a two- to fivefold higher signal was present in the peritoneal fluid than blood at 24 hours (Figure 5a). Intact siRNA was only observed in the kidneys and lungs with weak signals in the intestines up to 30 minutes (Figure 5c), with no signal detectable in any organs at 24 hours. Kidney levels at 30 minutes for the i.v. dose shown in Figure 4c exhibited ~10- to 20-fold increase compared to kidney accumulation after i.p. administration (Figure 5c).
Figure 5.
Northern analysis of siRNA levels in blood and organs after i.p. administration of chitosan/siRNA complexes. Chitosan/siRNA complex containing 10-µg siLNA-light was administered i.p. in 200 µl volume. (a) siRNA stability: RNA integrity was evaluated in blood samples at indicated time course and peritoneal fluid at 30 minutes and 24 hours. Loading order: lane 1, 3-ng siRNA duplex (Ctrl 1), lane 2, injected chitosan/siRNA complex containing 2-ng siRNA (Ctrl 2), lane 3, blank, lanes 4–13, as 1, 5, 15, 30 minutes, and 24 hours blood, 14–17, as peritoneal fluid at 30 minutes and 24 hours from mouse 1 and 2, respectively. (b) siRNA blood clearance: the intensity of the siRNA signals from blood is measured by Quantity ONE programme. Percentages of the intensity at 15 minutes and clearance curves are presented as a function of time (log 5 scale). (c) siRNA biodistribution: RNA integrity is analyzed for tissue samples from six organs at 30 minutes and 24 hours. Loading order is: lane 1, kidney sample from i.v. (Kidney/IV), lane 2, 1.5-ng siRNA duplex (Ctrl 1), lane 3, injected chitosan/siRNA complex containing 1-ng siRNA (Ctrl 2), lane 4, blank, lanes 5–16, six organs as indicated from mouse 1 and 2 at 30 minutes and 24 hours, respectively. Ctrl, control; LNA, locked nucleic acid; siLNA, LNA-modified siRNA; siLNA-light, lightly LNA–modified siRNA; siRNA, small interfering RNA.
Discussion
This work provides a comparative study showing to what extent chemical modification and siRNA design, including siLNA-light, siRNA-Thio, sisiRNA, and siRNA-Chol, as well as nanoparticle delivery can influence siRNA circulatory half-life, biodistribution, and stability.
Chemical modifications have been used previously for optimizing thermal/enzymatic stability and cellular delivery of siRNA molecules.16,17 In vivo factors such as interaction with plasma proteins and degradation by serum ribonucleases are major barriers to systemic stability.2 We found that unmodified siRNA showed degradation in blood 1 minute after i.v. administration, restricting the likelihood of accumulation within peripheral organs. In contrast, all the chemically modified siRNA could be detected in the blood and most organs at 30 minutes postinjection. Reduction in kidney glomerular filtration18 is required in order to improve the circulatory half-life and concomitant siRNA dissemination into the organs.19 Previous work in mice has shown 123I-labeled unmodified siRNA distributed to the kidney and liver 1 minute after i.v. injection peaking at 5 minutes (ref. 2). In another study, siRNA has shown rapid clearance to kidney and excretion into urine in rats.20 Similar observation was also obtained by de Wolf et al.21 In agreement with those reports, we observed six- to eightfold higher radioactivity levels in kidneys than other organs at 30 minutes for the majority of the siRNA duplexes.
We found that the siRNA circulatory half-life could be extended to 30 minutes by cholesterol conjugation. In work by Soutschek,7 the circulatory half-life of siRNA in rats was increased 15-fold by cholesterol conjugation attributed to interaction with human serum albumin. We found widespread tissue distribution of intact Chol-modified siRNA at 30 minutes but at 24 hours only in the lung, in contrast to continued distribution at 24 hours reported by Soutschek. This could reflect the extremely high dose of 50 mg/kg used in the Soutschek study in contrast to the comparatively low amount of 400 µg/kg used in our work. Our findings do, however, support pharmacokinetic improvements at more clinically relevant doses. The effect of LNA modification on tissue distribution could also account for lung accumulation observed in our work.
Increased siLNA-light dose did not significantly affect blood clearance or alter the biodistribution pattern but did increase the relative renal excretion rate. This advocates the use of low siRNA doses to maximize the ratio of retained and excreted siRNA and to minimize possible undesirable kidney damage side effects.19 In our study, three different siRNA detection strategies were used: scintillation counting and direct autoradiography on gels or northern blotting. Our results show that intact siRNA levels obtained by the two latter methods do not correlate with the total 32P-radioactivity levels. For example, no significant differences were observed for blood clearance and biodistribution pattern between unmodified and chemically modified siRNA by scintillation counting. However, a rapid degradation of unmodified siRNA was seen at 1 minute postinjection and chemically modified siRNAs could be clearly detected by gel electrophoresis in blood and organs at 30 minutes postinjection. Although, at 30 minutes, duplexes showed highest radioactivity concentration in kidneys, the strongest intensity of intact siRNA was found in lungs. In addition, there was a stronger radioactive count in purified RNA from liver and spleen than other organs at 24 hours (data not shown), but intact siRNA could only be seen in the lung on the gel (Figure 2b). To address the possibility of end label removal by phosphatases in vivo, we performed northern blotting on filters after 32P disintegration. A clear correlation was observed in the northern both in terms of specificity and overall intensity that promotes the use of northern blotting rather than the direct radioactivity labeling method widely used in nucleic acid pharmacokinetic studies. Other available methods may be also considered in the future studies, such as high-performance liquid chromatography and positron emission tomography imaging.22,23 For quantification of siRNA, Q-RT PCR has become a popular method.24 However, an LNA modification at 3′-end of the siRNA inhibits polyadenylation, which generally is the initial step in this approach (S. Gao and J. Kjems, unpublished data). Hence, the Q-RT-PCR approach is not suitable for this study.
In this study, we compare the cationic polymers, chitosan and JetPEI, and a polyethylene glycolated (PEGylated) liposomal formulation. We found that JetPEI/siRNA formulations showed a rapid blood clearance 1–5 minutes after injection, although, some degradation products at 24 hours were observed. This rapid removal from the circulation for nonmodified polyplexes agrees with previous studies.25,26 Postmodification, with hydrophilic polymers is the common approach used to improve the circulatory half-life by making the polyplex “stealth-like” and avoid hepatic clearance by reducing phagocytic capture by cellular components of the mononuclear phagocyte system.25 In our experiments, however, JetPEI facilitated equal distribution of intact siRNA throughout all organs even at 24 hours, albeit at the low levels. The ability to retain siRNA integrity through chemical modification could, circumvent the necessity for high levels for biological silencing effects. Effective gene silencing has been shown previously for nonmodified particles27,28 and i.p. administered JetPEI.29,30 The slightly higher accumulation in the lung at 30 minutes may reflect the exposed cationic charged-surface that can result in protein-mediated nanoparticle aggregation26 and subsequent physical capture within pulmonary capillary beds. Changing the physical properties of polycations such as charge and molecular weight and NP ratio may also be used to alter biodistribution. In a study by Thomas et al.,31 a fully deacylated linear PEI exhibited more accumulation into the lung attributed to increased polyplex stability resulting from greater ionic interactions with polymers possessing high cationic amino charge density.
We have previously utilized the mucoadhesive properties of chitosan for nanoparticle-based siRNA pulmonary delivery and gene silencing.9,13,32 In this work, we found that chitosan extended the circulatory half-life of siRNA, somewhat surprising given the high cationic charged-surface that can interact with serum proteins,33 cellular surfaces, and connective tissue. Chitosan-coated polyisohexylcyanoacrylate nanoparticles, however, have been shown previously to reduce subcutaneous tumor growth after i.v. administration.34 We show an increase in siRNA stability and a very pronounced accumulation at 24 hours in the kidney after i.v. injection. Previous reports have shown kidney accumulation of chitosan35 and chitosan/DNA complexes33 but this is the first report to our knowledge of siRNA delivery to the kidney using a nanoparticle system. This accumulation could reflect the mucoadhesion of chitosan to the mucosal epithelium lining the kidney and could be exploited for RNAi-based treatments for renal diseases. This is in contrast to renal clearance of siRNA shown with other vectors that may be a consequence of particle instability.23,36 As an alternative to i.v., the i.p. route showed accumulation within the peritoneal cavity. The retention of this chitosan formulation within a serum-free environment allows nanoparticles to enter resident peritoneal macrophages in an intact form and has been exploited in our lab for delivery of antitumor necrosis factor-α siRNA into macrophages as a targeted anti-inflammatory therapy.37
Lipoplexes have previously been shown to increase stability and improve biodistribution of siRNA14,15 and have been utilized in vivo to induce interference in the mouse vascular endothelium within different organs including lung, liver, and heart.38 A PEGylated liposome developed by Regulon (Athens, Greece)39 has been used in our studies. This liposome formulation showed an improved circulatory half-life within the first 30 minutes, however, not as pronounced as the JetPEI or chitosan over 24 hours, surprising considering the PEGylated nature of the liposomal formulation. This may reflect the degree of PEGylation and stability of the liposomes compared to the polycationic formulations. An increased uptake into the lung indicates an insufficient PEG coat resulting in serum-induced aggregation leading to pulmonary capillary bed38,40 or macrophage41 capture shown previously for liposomes. Additionally, the larger hydrodynamic radius of the liposome formulation (>500 nm) in comparison to the polycation systems (~300 nm) could contribute to lung deposition.
Our comprehensive investigation compares the effects of chemical modification and nanoparticle formulation on siRNA stability, blood circulation, and tissue distribution. This work demonstrates the noncorrelation between siRNA integrity and radioactivity promoting the application of our nonradioactive method for sensitive detection of intact siRNA in blood and organs. We found a requirement for LNA, phosphorothioate, and sisiRNA modifications for improved stability of siRNA to avoid rapid degradation observed with the unmodified siRNA. Furthermore, accumulation of intact siRNA was only found for siLNA-heavy and siRNA-Chol in the lungs 24 hours postinjection. Relevant to clinical applications, relatively low doses of modified siRNA allow biodistribution modulation. Nanoparticle formulations improved blood stability and biodistribution compared to naked siRNA. Additionally, in this work we have reported for the first time, use of chitosan/siRNA nanoparticles for extended siRNA accumulation in the kidney that may have potential for treatment of renal diseases with RNAi therapeutics.
Materials and Methods
siRNA duplex preparation. For radioactive labeling siRNA the AS were labeled at the 5′-end in a 100-µl reaction containing ~30 µg single-strand siRNA, 10 µl [γ-32P] (7,000 Ci/mmol) adenosine triphosphate, 5 µl T4 polynucleotide kinase, and 10 µl polynucleotide kinase reaction buffer (New England Biolabs, Beverly, MA) at 37 °C for 0.5 hours. The reaction was stopped by adding 1-µl 0.5 mol/l EDTA (pH 8.0) and run through a G50 spin column (Roche, Basel, Switzerland) to remove unincorporated [γ-32P] adenosine triphosphate, prior to annealing. siRNA duplexes were prepared by annealing equimolar concentrations (20 µmol/l) of the sense and antisense siRNA in 5× annealing buffer (150 mmol/l HEPES, pH 7.6, 500 mmol/l KCl, 0.05 mmol/l MgCl2) at 95 °C for 1 minute and at 37 °C for 1 hour. The duplexes were purified on 15% native polyacrylamide gel run at 4 °C, excised by radiography or UV-shadowing, eluted in 0.3 mol/l NaOAc (pH 6.0) overnight at 4 °C, phenol and chloroform extracted, and precipitated with 2½ vol. ethanol. The siRNA was diluted to 10 µg in a final volume of 200-µl 1× phosphate-buffered saline per mouse before injection. A small fraction was kept for further analysis.
Nanoparticle formulation. Chitosan (MW 160 kd, 80% deacetylation; Bioneer, Hørsholm, Denmark) and JetPEI (Polyplus-transfection, Illkirch, France), and PEGylated liposomes (Regulon)39 were used for the formulations containing siLNA-light siRNA. Preparation of liposomal and JetPEI/siRNA (NP 8) was performed according to the suppliers instructions and chitosan/siRNA complexes (NP 10) reported previously.32 Briefly, liposomes: 5 µl siRNA (10 µg) was added to 195 µl liposome solution and incubated for 5 minutes at 37 °C. JetPEI: 5 µl siRNA (10 µg) in 45 µl water and mixed with 50-µl 10% glucose (solution A) and 2 µl in vivo JetPEI diluted in 48 µl water and mixed with 50-µl 10% glucose (solution B) were mixed, left for 15 minutes at room temperature, and vortexed. Chitosan: 25 µl of siRNA (siLNA-light) (50 µg) was added whilst stirring to 975 µl chitosan (250 µg) in 0.2 mol/l sodium acetate buffer pH 5.5 and left for 1 hour at room temperature. The hydrodynamic radius was measured by photon correlation spectroscopy using Zetasizer Nano ZS (Malvern Instruments, Malvern, UK) prior to injection.
Evaluation of blood clearance and biodistribution. Pharmacokinetic studies were performed in Balb/c mice (~25 g) in duplicates. For i.v. administration, 10-, 100-, or 250 µg siRNA duplex in 200 µl volumes (corresponding to ~400 µg/kg, 4 mg/kg, and 10 mg/kg body weight) either naked or within nanoparticle formulations were administered by tail vein injection. Blood samples (30–40 µl) obtained by orbital puncture were collected into EDTA tubes, at 1, 5, 15, 30 minutes, and 24 hours time points. A volume of 10 µl was transferred to another tube containing 400-µl 10 mol/l NaOH from which 40 µl was added to 5 ml scintillation liquid for radioactivity count and the remaining volume immediately immersed into liquid nitrogen for RNA purification. Kidney, liver, heart, spleen, and lung tissue were collected at 30 minutes and 24 hours and aliquoted. One aliquot (~5 × 5 × 5 mm corresponding to 50–100 mg) was transferred into 1.0–1.5 ml RNAlater (Qiagen, Copenhagen, Denmark) and another aliquot immersed immediately in liquid nitrogen. Tissues (15 mg) were homogenized in 1.5-ml 10 mol/l NaOH from which 200 µl was added to 5 ml scintillation liquid for radioactivity counting using a 32P 5-minute-counting interval with a liquid scintillation counter (Beckman LS 6000; Beckman, Fullerton, CA) in count per min. Noninjected duplex (0.5 ng) was used as a control. The numbers were converted into CPM per µl blood at the various time points. To evaluate the blood clearance of the duplexes, the values were normalized to the CPM measured 1 minute after injection. This is done to avoid systemic errors from the injection procedures, where some of the samples occasionally leaked out after injection. Similarly, 2 µl urine samples were also collected at 30 minutes and 24 hours from one mouse from each group. The radioactivity of urine was measured and presented as CPM per µl urine at indicated time point. The radioactivity in the organs was expressed as CPM per mg tissue. Data were obtained from three measurements and presented as the average. For i.p. administration, chitosan/siRNA nanoparticles using the same amount and volume of siRNA as the i.v. experiment were injected into intraperitoneal cavity of mice with blood and organs collected as above. In addition, the small intestines were collected and the peritoneal fluid was obtained by flushing the peritoneal cavity with phosphate-buffered saline.
Gel electrophoresis and northern blotting determination of siRNA integrity. Total RNA was purified by Trizol reagent (Invitrogen, Carlsbad, CA) from frozen blood samples and weighed tissue in RNAlater. RNA from blood (2 µg) and from tissues (4 µg) was run on 15% denaturing polyacrylamide gels and transferred onto Hybond-N+ membrane (Amersham Biosciences, Uppsala, Sweden). After UV crosslinking, the membranes were analyzed on phophorimager to visualize the 32P labeled AS of the siRNA. To perform northern blotting, the same filters were stored for 4 months (eight half-lives of 32P) and probed with [γ-32P] adenosine triphosphate–labeled sense-strand LNA-modified siRNA according to standard procedures. For mice injected with the siRNA nanoparticles, nonlabeled siRNA was used and northern blotting was performed.
Acknowledgments
This work was supported by Danish Technical Research Council, the Danish Strategic Research Council, and the EU-FP6 RIGHT project (LSHB-CT-2004-005276). We thank Claus Bus and Rita Rosendahl for their excellent technical assistance, and Jesper Bramsen for siRNA selection.
REFERENCES
- Pouton CW., and , Seymour LW. Key issues in non-viral gene delivery. Adv Drug Deliv Rev. 2001;46:187–203. doi: 10.1016/s0169-409x(00)00133-2. [DOI] [PubMed] [Google Scholar]
- Braasch DA, Paroo Z, Constantinescu A, Ren G, Oz OK, Mason RP, et al. Biodistribution of phosphodiester and phosphorothioate siRNA. Bioorg Med Chem Lett. 2004;14:1139–1143. doi: 10.1016/j.bmcl.2003.12.074. [DOI] [PubMed] [Google Scholar]
- Geary RS, Yu RZ., and , Levin AA. Pharmacokinetics of phosphorothioate antisense oligodeoxynucleotides. Curr Opin Investig Drugs. 2001;2:562–573. [PubMed] [Google Scholar]
- Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mook OR, Baas F, de Wissel MB., and , Fluiter K. Evaluation of locked nucleic acid-modified small interfering RNA in vitro and in vivo. Mol Cancer Ther. 2007;6:833–843. doi: 10.1158/1535-7163.MCT-06-0195. [DOI] [PubMed] [Google Scholar]
- Bramsen JB, Laursen MB, Damgaard CK, Lena SW, Babu BR, Wengel J, et al. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res. 2007;35:5886–5897. doi: 10.1093/nar/gkm548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Elouahabi A., and , Ruysschaert JM. Formation and intracellular trafficking of lipoplexes and polyplexes. Mol Ther. 2005;11:336–347. doi: 10.1016/j.ymthe.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Howard KA., and , Kjems J. Polycation-based nanoparticle delivery for improved RNA interference therapeutics. Expert Opin Biol Ther. 2007;7:1811–1822. doi: 10.1517/14712598.7.12.1811. [DOI] [PubMed] [Google Scholar]
- Aigner A. Gene silencing through RNA interference (RNAi) in vivo: strategies based on the direct application of siRNAs. J Biotechnol. 2006;124:12–25. doi: 10.1016/j.jbiotec.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Aigner A. Applications of RNA interference: current state and prospects for siRNA-based strategies in vivo. Appl Microbiol Biotechnol. 2007;76:9–21. doi: 10.1007/s00253-007-0984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard KA, Dong M, Oupicky D, Bisht HS, Buss C, Besenbacher F, et al. Nanocarrier stimuli-activated gene delivery. Small. 2007;3:54–57. doi: 10.1002/smll.200600328. [DOI] [PubMed] [Google Scholar]
- Howard KA, Rahbek UL, Liu X, Damgaard CK, Glud SZ, Andersen MO, et al. RNA interference in vitro and in vivo using a novel chitosan/siRNA nanoparticle system. Mol Ther. 2006;14:476–484. doi: 10.1016/j.ymthe.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Aigner A. Tumor-targeting nanosystems for the delivery of siRNA. Nanomed. 2007;2:569–572. doi: 10.2217/17435889.2.4.569. [DOI] [PubMed] [Google Scholar]
- Landen CN, Jr, Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- Dorsett Y., and , Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr Opin Chem Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Venturoli D., and , Rippe B. Ficoll and dextran vs. globular proteins as probes for testing glomerular permselectivity: effects of molecular size, shape, charge, and deformability. Am J Physiol Renal Physiol. 2005;288:F605–F613. doi: 10.1152/ajprenal.00171.2004. [DOI] [PubMed] [Google Scholar]
- Lu PY, Xie F., and , Woodle MC. In vivo application of RNA interference: from functional genomics to therapeutics. Adv Genet. 2005;54:117–142. doi: 10.1016/S0065-2660(05)54006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Water FM, Boerman OC, Wouterse AC, Peters JG, Russel FG., and , Masereeuw R. Intravenously administered short interfering RNA accumulates in the kidney and selectively suppresses gene function in renal proximal tubules. Drug Metab Dispos. 2006;34:1393–1397. doi: 10.1124/dmd.106.009555. [DOI] [PubMed] [Google Scholar]
- de Wolf HK, Snel CJ, Verbaan FJ, Schiffelers RM, Hennink WE., and , Storm G. Effect of cationic carriers on the pharmacokinetics and tumor localization of nucleic acids after intravenous administration. Int J Pharm. 2007;331:167–175. doi: 10.1016/j.ijpharm.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Viel T, Boisgard R, Kuhnast B, Jego B, Siquier-Pernet K, Hinnen F, et al. Molecular imaging study on in vivo distribution and pharmacokinetics of modified small interfering RNAs (siRNAs) Oligonucleotides. 2008;18:201–212. doi: 10.1089/oli.2008.0133. [DOI] [PubMed] [Google Scholar]
- Bartlett DW, Su H, Hildebrandt IJ, Weber WA., and , Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci USA. 2007;104:15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratford S, Stec S, Jadhav V, Seitzer J, Abrams M., and , Beverly M. Examination of real-time polymerase chain reaction methods for the detection and quantification of modified siRNA. Anal Biochem. 2008;379:96–104. doi: 10.1016/j.ab.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Oupicky D, Ogris M, Howard KA, Dash PR, Ulbrich K., and , Seymour LW. Importance of lateral and steric stabilization of polyelectrolyte gene delivery vectors for extended systemic circulation. Mol Ther. 2002;5:463–472. doi: 10.1006/mthe.2002.0568. [DOI] [PubMed] [Google Scholar]
- Dash PR, Read ML, Fisher KD, Howard KA, Wolfert M, Oupicky D, et al. Decreased binding to proteins and cells of polymeric gene delivery vectors surface modified with a multivalent hydrophilic polymer and retargeting through attachment of transferrin. J Biol Chem. 2000;275:3793–3802. doi: 10.1074/jbc.275.6.3793. [DOI] [PubMed] [Google Scholar]
- Takeshita F, Minakuchi Y, Nagahara S, Honma K, Sasaki H, Hirai K, et al. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc Natl Acad Sci USA. 2005;102:12177–12182. doi: 10.1073/pnas.0501753102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- Urban-Klein B, Werth S, Abuharbeid S, Czubayko F., and , Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005;12:461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- Grzelinski M, Urban-Klein B, Martens T, Lamszus K, Bakowsky U, Hobel S, et al. RNA interference-mediated gene silencing of pleiotrophin through polyethylenimine-complexed small interfering RNAs in vivo exerts antitumoral effects in glioblastoma xenografts. Hum Gene Ther. 2006;17:751–766. doi: 10.1089/hum.2006.17.751. [DOI] [PubMed] [Google Scholar]
- Thomas M, Lu JJ, Ge Q, Zhang C, Chen J., and , Klibanov AM. Full deacylation of polyethylenimine dramatically boosts its gene delivery efficiency and specificity to mouse lung. Proc Natl Acad Sci USA. 2005;102:5679–5684. doi: 10.1073/pnas.0502067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Howard KA, Dong M, Andersen MO, Rahbek UL, Johnsen MG, et al. The influence of polymeric properties on chitosan/siRNA nanoparticle formulation and gene silencing. Biomaterials. 2007;28:1280–1288. doi: 10.1016/j.biomaterials.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Mao HQ, Roy K, Troung L, Janes KA, Lin KY, Wang Y, et al. Chitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiency. J Control Release. 2001;70:399–421. doi: 10.1016/s0168-3659(00)00361-8. [DOI] [PubMed] [Google Scholar]
- Pille JY, Li H, Blot E, Bertrand JR, Pritchard LL, Opolon P, et al. Intravenous delivery of anti-RhoA small interfering RNA loaded in nanoparticles of chitosan in mice: safety and efficacy in xenografted aggressive breast cancer. Hum Gene Ther. 2006;17:1019–1026. doi: 10.1089/hum.2006.17.1019. [DOI] [PubMed] [Google Scholar]
- Hwang HY, Kim IS, Kwon IC., and , Kim YH. Tumor targetability and antitumor effect of docetaxel-loaded hydrophobically modified glycol chitosan nanoparticles. J Control Release. 2008;128:23–31. doi: 10.1016/j.jconrel.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Heidel JD, Yu Z, Liu JY, Rele SM, Liang Y, Zeidan RK, et al. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc Natl Acad Sci USA. 2007;104:5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard KA, Paludan SR, Behlke MA, Besenbacher F, Deleuran B, et al. Chitosan/siRNA nanoparticle-mediated TNF-α knockdown in peritoneal macrophages for anti-inflammatory treatment in a murine arthritis model. Mol Ther. 2009;17:162–168. doi: 10.1038/mt.2008.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Aleku M, Keil O, Endruschat J, Esche V, Fisch G, et al. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 2006;13:1222–1234. doi: 10.1038/sj.gt.3302777. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Low toxicity and anticancer activity of a novel liposomal cisplatin (Lipoplatin) in mouse xenografts. Oncol Rep. 2004;12:3–12. [PubMed] [Google Scholar]
- Aigner A. Nonviral in vivo delivery of therapeutic small interfering RNAs. Curr Opin Mol Ther. 2007;9:345–352. [PubMed] [Google Scholar]
- Poste G, Bucana C, Raz A, Bugelski P, Kirsh R., and , Fidler IJ. Analysis of the fate of systemically administered liposomes and implications for their use in drug delivery. Cancer Res. 1982;42:1412–1422. [PubMed] [Google Scholar]