Drosophila Parkin requires PINK1 for mitochondrial translocation and ubiquitinates Mitofusin (original) (raw)
Abstract
Loss of the E3 ubiquitin ligase Parkin causes early onset Parkinson's disease, a neurodegenerative disorder of unknown etiology. Parkin has been linked to multiple cellular processes including protein degradation, mitochondrial homeostasis, and autophagy; however, its precise role in pathogenesis is unclear. Recent evidence suggests that Parkin is recruited to damaged mitochondria, possibly affecting mitochondrial fission and/or fusion, to mediate their autophagic turnover. The precise mechanism of recruitment and the ubiquitination target are unclear. Here we show in Drosophila cells that PINK1 is required to recruit Parkin to dysfunctional mitochondria and promote their degradation. Furthermore, PINK1 and Parkin mediate the ubiquitination of the profusion factor Mfn on the outer surface of mitochondria. Loss of Drosophila PINK1 or parkin causes an increase in Mfn abundance in vivo and concomitant elongation of mitochondria. These findings provide a molecular mechanism by which the PINK1/Parkin pathway affects mitochondrial fission/fusion as suggested by previous genetic interaction studies. We hypothesize that Mfn ubiquitination may provide a mechanism by which terminally damaged mitochondria are labeled and sequestered for degradation by autophagy.
Keywords: mitophagy, neurodegeneration, Parkinson's disease, quality control
Parkinson's disease (PD) is a common neurodegenerative disorder principally affecting the degeneration of nigral dopaminergic neurons. The pathogenic mechanisms are unknown, but valuable insight has been gained from identifying gene mutations causative for familial forms of PD (1). Loss-of-function mutations in PINK1 and parkin are the major cause of autosomal recessive, early onset PD. PINK1 encodes a mitochondria-targeted kinase (2) whereas parkin encodes an E3 ubiquitin ligase (3), a class of enzymes that conjugate ubiquitin to target substrates. This modification is usually considered in the context of substrate degradation by the proteasome, but ubiquitination also serves many other cellular functions. Consequently, much emphasis has been put on elucidating a link between Parkin dysfunction and protein aggregation. Despite the identification of numerous putative Parkin substrates, an unequivocal causative link between substrate aggregation and pathogenesis remains debatable.
There is strong evidence, however, that supports an important role for Parkin in regulating mitochondrial homeostasis (4). Studies have revealed a conserved function of Parkin acting downstream of PINK1 to protect mitochondrial integrity and prevent oxidative stress-induced apoptosis (5–8). Recently, we and others have reported that Drosophila parkin and PINK1 genetically interact with components of the mitochondrial fission and fusion machinery (9–12), suggesting that loss of PINK1/parkin function may lead to excess mitochondrial fusion. Consistent with this, mitochondrial elongation has been reported in cells derived from PD patients with parkin mutations (13). However, the effects of parkin or PINK1 deficiency in mammalian cells remain unresolved because additional reports describe inconsistent phenotypes in _PINK1_- and _parkin_-deficient cells (6, 14–17). The reasons for this discrepancy are unclear but warrant further clarification.
Important insight into the mechanism by which Parkin regulates mitochondrial homeostasis has revealed that upon mitochondrial depolarization Parkin translocates from the cytoplasm to accumulate on a subset of mitochondria and promotes their degradation by autophagy (18). It has also been proposed that regulated mitochondrial fission/fusion helps sort out damaged mitochondria for degradation (19–21). These findings raise the possibility that Parkin receives some signal identifying dysfunctional or damaged mitochondria and translocates and interacts with an unknown factor to effect mitochondrial fission and/or fusion and promote mitophagy (22).
Here we show that PINK1 is required for the recruitment of Parkin to damaged mitochondria and subsequent mitophagy, consistent with recent reports (23–25). Furthermore, we show that PINK1 and Parkin promote the ubiquitination of the profusion factor Mfn. These findings suggest that Mfn ubiquitination may provide a mechanism by which terminally damaged mitochondria are recognized for degradation by autophagy.
Results
PINK1 and Parkin Knockdown Cause Mitochondrial Elongation.
Our previous genetic interaction studies in vivo, using both loss- and gain-of-function assays, indicate that Drosophila PINK1 and Parkin act to promote fission and/or inhibit fusion of mitochondria. To extend this study, we quantified mitochondrial morphology upon loss or gain of function of parkin and PINK1, along with key regulators of mitochondrial morphology, in Drosophila S2R+ cells.
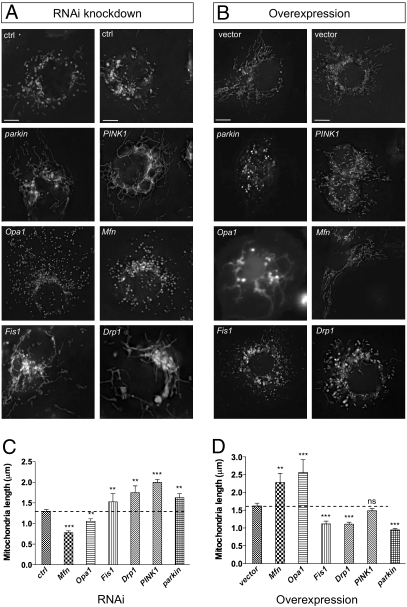
RNA interference (RNAi) can be robustly achieved in Drosophila cells by incubation of long double-stranded RNA molecules complementary to the target gene (26). We performed targeted RNAi against PINK1, parkin, and the Drosophila homologs of Drp1, Fis1, Opa1, and Mfn1/2 (here called Mfn). RNAi of these targets typically caused ∼90% knockdown of the endogenous message after 3 days (Fig. S1). We found that sample fixation grossly altered the mitochondrial morphology (Fig. S2_A_), so for this study cells were imaged live using the selective mitochondrial dye rhodamine 123 (Fig. 1 A and C). As expected, we found that knockdown of the profusion genes Drp1 and Fis1 resulted in elongated mitochondria whereas knockdown of the profusion genes Opa1 and Mfn (also known as Marf) resulted in fragmented mitochondria. We found that knockdown of PINK1 or parkin caused a significant elongation of mitochondrial length. Elongated mitochondria were also seen when transfected mitoGFP was used to label mitochondria (Fig. S2_B_). Conversely, overexpression of Opa1 and Mfn caused a hyper-fused network whereas overexpression of Drp1 and Fis1 caused fragmentation (Fig. 1 B and D). Overexpression of parkin also caused fragmentation; however, the effect of PINK1 was not significant, suggesting that PINK1 may not be limiting or may need to be activated itself to ectopically promote fragmentation.
Fig. 1.
Drosophila PINK1 and parkin activity promotes mitochondrial fission and/or inhibits fusion. (A) S2R+ cells were treated with the indicated RNAi probe for 3 days. Rhodamine 123 was used to visualize mitochondrial morphology in live cells. (B) S2R+ cells were transfected with plasmids to express indicated genes and cotransfected with mitoGFP to visualize mitochondria. (C and D) Quantification of mitochondrial length as imaged in A and B, respectively. Two control (ctrl) RNAi and vector-only treatments are shown to demonstrate phenotype variability. (Scale bar: 5 μm.) The bar graphs represent the mean and sem for at least three independent experiments. Significance was determined by one-way ANOVA with Bonferroni correction (**P < 0.01, ***P < 0.001).
The relative levels of key mitochondrial morphology factor transcripts were unchanged by parkin or PINK1 knockdown (Fig. S3_A_), indicating that the morphology changes do not occur from a transcriptional response altering the level of fission or fusion gene expression. Also, in contrast to a previous report (8), we find that Parkin protein levels are not changed in a PINK1 mutant (Fig. S3_B_). Together these observations are consistent with the PINK1/Parkin pathway modulating the action of mitochondrial fission/fusion proteins to alter morphology.
PINK1 Is Required for Parkin Translocation and Parkin-Mediated Mitophagy.
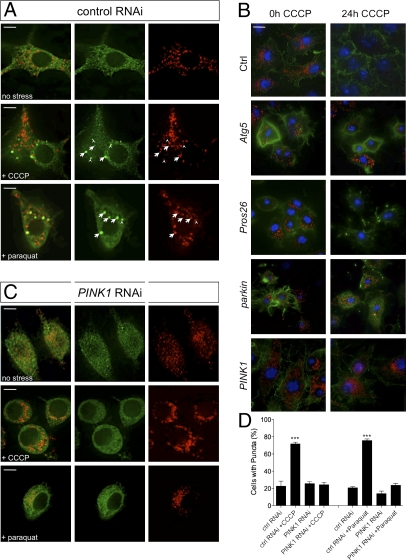
The recent study by Narendra et al. (18) revealed an important insight into the function of Parkin, reporting that it translocates to dysfunctional mitochondria and promotes mitophagy. We sought to determine whether this was a conserved function. In wild-type Drosophila cells, Parkin-GFP typically showed a diffuse localization throughout the cytoplasm with occasional accumulations at mitochondria (Fig. 2_A_ and Fig. S4). Upon treatment with the mitochondrial uncoupling agent carbonyl cyanide _m_-chlorophenylhydrazone (CCCP) or the oxidative stressor paraquat, a large proportion of cells showed Parkin-GFP accumulated at or near mitochondria (Fig. 2_A_ and Fig. S5), whereas overall Parkin-GFP abundance remained constant (Fig. S3_C_). Prolonged exposure to CCCP also led to the autophagy-dependent, but proteasome-independent, loss of mitochondria (Fig. 2_B_ and Fig. S6). Consistent with previous reports, Parkin was also required for CCCP-induced mitophagy in Drosophila cells (Fig. 2_B_).
Fig. 2.
PINK1 is required for Parkin translocation and mitophagy. (A) S2R+ cells cotransfected with parkin-GFP and mitoDsRed, treated with control dsRNA and then treated with CCCP or paraquat and fixed for imaging. Most Parkin-GFP puncta colocalize with (arrow) or abut (arrowhead) mitochondria. (B) Cells were treated with indicated dsRNAs and exposed to CCCP for 24 h. Samples were fixed and stained to label mitochondria (anti-Complex Vα, red), actin (phalloidin-488, green) or nuclei (Hoechst, blue). (C) S2R+ cells transfected as in A and treated with PINK1 dsRNA before exposure to CCCP or paraquat. Comparison of Parkin-GFP distribution under different image analysis methods is shown in Fig. S5. (D) Percentage of cells with Parkin-GFP puncta. (Scale bar: 5 μm.) The bar graphs represent the mean and SEM of at least four independent tests. Significance was determined by one-way ANOVA with Bonferroni correction (***P < 0.001).
To extend this observation, we addressed whether PINK1, which acts upstream of Parkin, may affect the recruitment of Parkin to mitochondria and their autophagic degradation. Following PINK1 RNAi knockdown, CCCP- or paraquat-induced Parkin-GFP recruitment to mitochondria was prevented (Fig. 2 C and D). In addition, loss of PINK1 also abrogated mitophagy (Fig. 2_B_). These data are consistent with PINK1 acting upstream to promote Parkin recruitment to dysfunctional mitochondria, which in turn promotes their degradation.
Drosophila Mitofusin Is Ubiquitinated by PINK1/Parkin and Accumulates in Mutants.
From the previous observations, we hypothesized that Parkin translocates to damaged mitochondria to alter their fission/fusion ability and to promote their degradation, presumably via its ubiquitin ligase activity. Thus, we sought to determine if any of the key mitochondrial morphology factors may be modified by ubiquitin and whether such modifications are Parkin dependent.
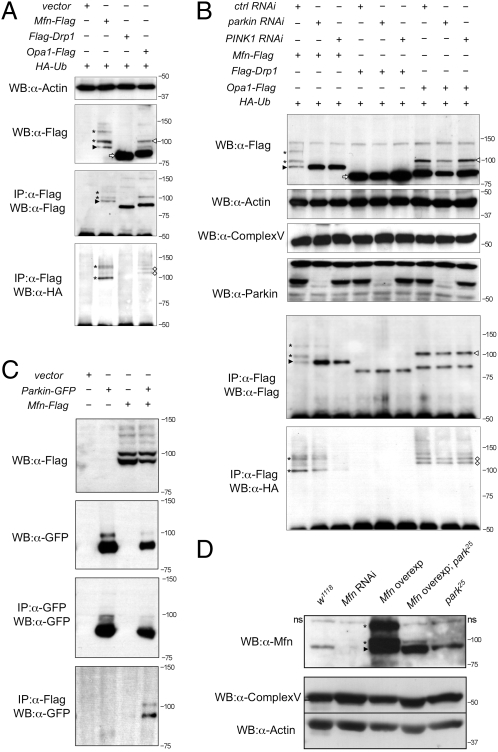
Flag-tagged forms of Drp1, Opa1, and Mfn were coexpressed with hemagglutinin (HA)-tagged ubiquitin (HA-Ub) in Drosophila cells. Western blot analysis of these proteins revealed a single major band of the predicted size for Drp1 (Fig. 3_A_, white arrow). Opa1 was present in a small amount of the full-length form but predominantly in the processed form (Fig. 3_A_, white arrowhead), as previously reported (27). However, Mfn was detected as a number of high-molecular-weight isoforms (asterisks) in addition to the expected full-length form (Fig. 3_A_, black arrowhead). The smallest Mfn isoform at ∼91 kDa is consistent with the predicted size of the full-length, unmodified form of Drosophila Mfn.
Fig. 3.
Mfn is ubiquitinated in a PINK1/Parkin-dependent manner in vitro and in vivo. (A) S2R+ cells cotransfected with HA-Ub plus empty vector or Flag-tagged Mfn, Drp1, or Opa1 plasmids as indicated were immunoprecipitated and subjected to Western blot analysis. (B) S2R+ cells were treated with control, parkin, and PINK1 RNAi before being transfected as indicated. Cells were harvested and subjected to Western blot analysis as shown. Samples were also subjected to immunoprecipitation, and Western bots were probed with antibodies against Flag and HA. (C) Immunoprecipitates of S2R+ cells expressing combinations of empty vector, parkin-GFP, and Mfn-Flag as shown. (D) Western blot analysis of Drosophila Mfn levels in vivo. A nonspecific band seen in all samples is denoted “ns.” Complex Vα and actin are used as loading controls. Genotypes: w1118, wild type; Mfn RNAi, da-GAL4,UAS-Mfn-RNAi; Mfn overexpression, da-GAL4,UAS-Mfn; Mfn overexpression park25, da-GAL4,UAS-Mfn,park25/park25; park25, park25/park25. In all panels, black arrowheads indicate full-length Mfn, asterisks are ubiquitinated Mfn, white arrowheads denote full-length Opa1, white diamonds are ubiquitinated Opa1, and white arrows show full-length Drp1.
To assess whether these isoforms are ubiquitin modified, we performed coimmunoprecipitation experiments against HA-Ub. Immunoprecipitates of Flag-tagged Drp1, Opa1, and Mfn were prepared and Western blots were probed with an antibody against HA. HA-positive bands were detected in the Mfn (asterisks) and Opa1 (white diamonds) samples but not in Drp1 (Fig. 3_A_). Comparison with Western blots probed against Flag showed bands of equivalent size for Mfn-Flag (Fig. 3_A_, asterisks), consistent with these isoforms being ubiquitinated. The smallest Mfn isoform was not HA-Ub–positive (Fig. 3_A_, black arrowhead), which is consistent with this isoform being unmodified. Larger ubiquitinated isoforms of Opa1 are not detected by anti-Flag immunoblotting, suggesting that these are in very low abundance.
We next tested whether this ubiquitination is Parkin dependent by performing the same analysis again following RNAi knockdown of parkin. We also addressed whether attenuating the pathway more generally may affect Mfn ubiquitination, so PINK1 RNAi knockdown was also performed. Western blots of whole-cell lysates from parkin and PINK1 knockdown cells revealed a striking loss of the ubiquitinated forms of Mfn (Mfn-Ub) (Fig. 3_B_, asterisks; Fig. S7_A_). Interestingly, this appeared to be accompanied by an accumulation of nonubiquitinated Mfn. Following enrichment by immunoprecipitation, low levels of Mfn-Ub were detected in parkin knockdown cells that were undetectable in whole-cell lysates, likely reflecting incomplete RNAi knockdown. However, in PINK1 knockdown cells, no ubiquitinated Mfn could be detected even upon immunoprecipitation (Fig. 3_B_). In contrast, however, ubiquitinated forms of Opa1 were unchanged by the loss of parkin or PINK1 (Fig. 3_B_, white diamonds), suggesting that this modification is not PINK1/Parkin dependent. Again, no modification of Drp1 was observed or its abundance changed by parkin or PINK1 RNAi.
To gain further evidence that Mfn ubiquitination may be mediated by Parkin, we tested whether they may physically interact. Immunoprecipitation assays revealed that upon coexpression Parkin-GFP is detected in precipitates of Mfn-Flag (Fig. 3_C_). Together these data strongly support that Parkin mediates the ubiquitination of Mfn.
We sought to extend this observation in vivo. Using antibodies against Drosophila Mfn, we could detect full-length Mfn and high-molecular-weight Mfn-Ub (Fig. 3_D_), consistent with that seen in vitro. These bands were absent upon Mfn RNAi knockdown in vivo (Fig. 3_D_), indicating the specificity of the antiserum. Consistent with our in vitro results, Mfn-Ub was greatly reduced in a parkin mutant background (Fig. 3_D_).
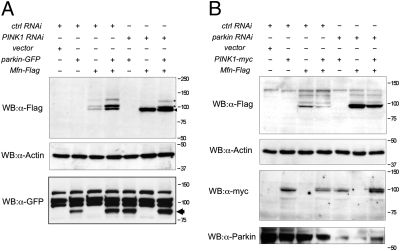
We have shown that PINK1 is required for Parkin recruitment to mitochondria; however, previous genetic experiments in vivo have shown that overexpression of parkin can compensate for loss of PINK1. Therefore, we addressed whether overexpressing parkin may restore Mfn ubiquitination in the absence of PINK1. In PINK1 RNAi knockdown cells only unmodified Mfn was seen (Fig. 4_A_); however, when Parkin was overexpressed in PINK1 RNAi cells, the Mfn-Ub isoforms were again detected (Fig. 4_A_). Conversely, if PINK1 was overexpressed in parkin RNAi cells, Mfn-Ub isoforms were not restored (Fig. 4_B_), consistent with genetic experiments showing that PINK1 overexpression cannot compensate for loss of parkin in vivo.
Fig. 4.
Parkin overexpression can cause Mfn ubiquitination in the absence of PINK1. (A) S2R+ cells were treated with control or PINK1 RNAi probe and transfected with combinations of empty vector, parkin-GFP, and Mfn-Flag, as shown. (B) S2R+ cells were treated with control or parkin RNAi probe and transfected with combinations of empty vector, PINK1-myc, and Mfn-Flag, as shown. Cells were harvested and subjected to Western blot analysis using the specific antibodies indicated.
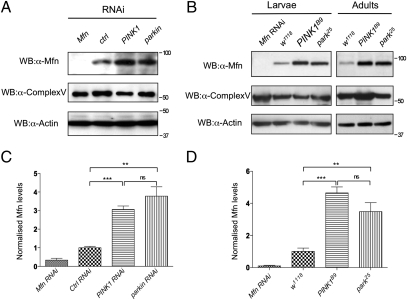
Ubiquitination is a common signal for the degradation of proteins and organelles. Because evidence indicates that Parkin and PINK1 promote mitophagy, we next assessed whether Mfn abundance is altered by loss of parkin and PINK1. The steady-state levels of endogenous Drosophila Mfn were determined in vitro and in vivo. Western blot of control treated cells and wild-type flies showed a band of the predicted full-length Mfn, which was absent in Mfn RNAi knockdown cells or flies (Fig. 5). parkin knockdown cells or park25 null mutant flies showed a significant increase in Mfn abundance (Fig. 5). Mfn was also significantly more abundant in PINK1 knockdown or PINK1B9 null mutant flies (Fig. 5). Interestingly, the level of Complex Vα did not appreciably increase (Fig. S7_B_); however, this technique is not sensitive to small changes. These data indicate that Mfn levels accumulate upon loss of PINK1/parkin, consistent with their role in mitophagy, but likely reflects turnover of a small proportion of the entire mitochondrial content.
Fig. 5.
Mfn accumulates in the absence of PINK1/Parkin. (A) S2R+ cells were treated with RNAi probe as indicated, harvested, and subjected to Western blot analysis by using antibodies against Drosophila Mfn. Antibodies against Complex Vα and actin are used as loading controls. (B) Wild-type and mutant animals were collected and subjected to Western blot analysis. Genotypes: w1118, wild type; Mfn RNAi, da-GAL4,UAS-Mfn-RNAi; park25, park25/park25; PINK1B9, PINK1B9/Y. Quantification of Mfn levels relative to the Complex Vα loading control in S2R+ cells (C) and wild-type and mutant animals (D). Charts represent mean and SEM for three independent experiments. Significance was determined by one-way ANOVA with Bonferroni correction (**P < 0.01, ***P < 0.001).
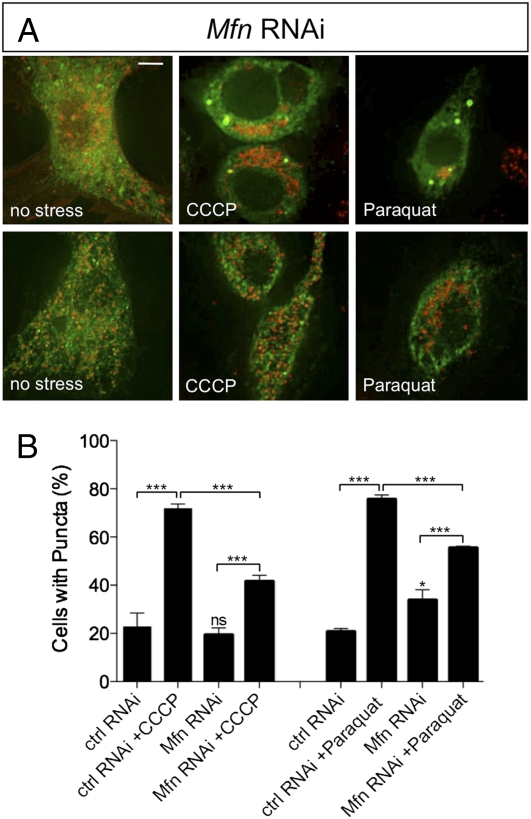
Because our evidence suggests that Parkin is recruited to mitochondria where it may ubiquitinate Mfn, we addressed whether Mfn is required for Parkin translocation. Parkin-GFP localization upon CCCP and paraquat treatment was monitored as before and compared with Mfn RNAi knockdown cells. Under control conditions we saw Parkin-GFP translocation comparable to previous results; however, in the absence of Mfn, Parkin-GFP translocation was not eliminated although the prevalence was markedly reduced (Fig. 6). As before, the overall abundance of Parkin-GFP was unaffected by treatments in Mfn RNAi cells (Fig. S3_C_). These results indicate that Mfn is not an absolute requirement for Parkin translocation.
Fig. 6.
Loss of Mfn partially impairs Parkin translocation to mitochondria. S2R+ cells were cotransfected with plasmids expressing parkin-GFP and mitoDsRed and treated with control RNAi probe or Mfn RNAi probe before being treated with CCCP or paraquat and analyzed for Parkin-GFP accumulation. Control RNAi-treated cells are represented in Fig. 2. (A) Mfn RNAi-treated cells exposed to CCCP, paraquat, or no stress. Two representative images are shown for each treatment to show variability. (Scale bar: 5 μm.) (B) Percentage of cells with green puncta. The chart shows the mean and SEM for counting of at least four independent well fields containing 70–90 cells/field. Significance was determined by one-way ANOVA with Bonferroni correction (*P < 0.05, ***P < 0.001).
Discussion
Maintenance of mitochondrial homeostasis appears to be an important function of the PINK1/Parkin pathway in multiple model systems and is likely a key factor in mediating neurodegeneration. Recent studies have begun to shed light on the potential mechanism by which this pathway maintains a healthy mitochondrial population. Emerging evidence indicates that PINK1 is required to recruit Parkin to damaged or dysfunctional mitochondria, whereupon it promotes mitophagy (23–25). Regulated mitochondrial fission and fusion events are thought to contribute to a quality control mechanism to help “sort out” terminally damaged mitochondria for degradation (19–21). Importantly, PINK1 and parkin have previously been shown to genetically interact with components of the mitochondrial fission/fusion machinery and to affect mitochondrial morphology (9–12); however, the molecular mechanisms are not known. Here we provide further evidence that PINK1 is required for Parkin translocation to damaged mitochondria and that this pathway affects mitochondrial morphology. We also provide evidence that the PINK1/Parkin pathway promotes the ubiquitination and regulates the levels of the profusion protein Mfn, thus providing a potential molecular mechanism by which PINK1/Parkin may modulate mitochondrial dynamics.
Consistent with recent reports (23–25), we find that the translocation of Parkin to damaged mitochondria and their subsequent autophagy is dependent on PINK1. However, the molecular mechanisms that promote Parkin's recruitment to mitochondria are still unclear. PINK1’s kinase activity, but not mitochondrial localization, appears to be necessary for Parkin translocation (23–25). Because PINK1 can be found extramitochondrially (27–30) and may directly phosphorylate Parkin (31, 32), this may be a mechanism to stimulate its translocation. Alternatively, it may phosphorylate a Parkin substrate, e.g., Mfn, and thereby provide a recruitment signal. Interestingly, we find that loss of Mfn reduces but does not eliminate Parkin translocation. Recent evidence indicates that Parkin also ubiqutinates VDAC on the outer mitochondrial surface (23), suggesting that there may be multiple recruitment substrates. Although further work is required to elucidate these mechanisms, these findings suggest a molecular basis for the genetic hierarchy in which PINK1 acts upstream of Parkin (5–8).
To understand the role of Parkin translocation, we took a candidate approach to identifying putative substrates. Because the function of Parkin and PINK1 has been linked with mitochondrial dynamics, we surveyed key components of the mitochondrial fission and fusion machinery for ubiquitin modification. We found that Mfn, which localizes to the outer surface of mitochondria, is ubiquitinated in a PINK1/Parkin-dependent manner and accumulates upon loss of PINK1 or parkin. Interestingly, the ubiquitinated isoforms do not show a typical ubiquitination “ladder” but instead appear to reflect a pattern of one and three or four ubiquitin adducts. Although it remains to be shown that Parkin directly mediates this ubiquitination, there is evidence that Parkin can mediate monoubiquitination (33–37) and K27 (23) and K63 linkages (38, 39). These modes of ubiquitination are not typically linked to proteasome degradation, and there is growing speculation that important pathogenic functions of Parkin may be proteasome independent (reviewed in ref. 40).
Numerous elegant studies have demonstrated that the mitochondrial network is extremely dynamic and responds rapidly and reversibly to many physiological changes including potentially toxic challenges such as oxidative stress and calcium flux (reviewed in refs. 41 and 42). Although mitochondrial remodeling can contribute to promoting cell death, it can also act in a protective manner by contributing to a quality control process that likely involves degradation by autophagy/lysosomes. Recent work has reported observations that, following a fission event, regulated fusion of daughter mitochondria can determine whether they rejoin the network or are sequestered for degradation (19). Refusion appears to be dependent upon the recovery of mitochondrial membrane potential after division and likely represents a mechanism to sort out terminally dysfunctional mitochondria (20). Because Mitofusins mediate the tethering and fusion of mitochondria via homo- and heterotypic interaction of their HR2 domains (43), we hypothesize that Parkin-mediated Mfn ubiquitination may interfere with intermolecular interactions preventing fusion. Alternatively, Mfn ubiquitination may lead to a selective removal of Mfn from damaged mitochondria and thus reduce the refusion capacity of those mitochondria. Consistent with this, we find that loss of parkin or PINK1, and hence loss of ubiquitination, leads to increased Mfn levels and mitochondrial elongation, presumably due to excess fusion. Thus, Mfn ubiquitination may provide a signal that simultaneously prevents the refusion of terminally damaged mitochondria and labels them for safe degradation by autophagy (22).
It is reasonable to suppose that under normal conditions the majority of mitochondria are relatively healthy, and thus mitochondrial turnover is an infrequent event. This is supported by the observation that Complex Vα levels are not significantly altered by decreased mitophagy. However, this rationale implies that Mfn accumulates and is selectively ubiquitinated on mitochondria targeted for degradation although this remains to be shown. Interestingly, our findings provide a molecular mechanism that can explain the previously reported genetic interactions between PINK1 and parkin and the fission/fusion factors—in particular, that promoting mitochondrial fragmentation by overexpression of Drp1 or by reduction of Mfn and Opa1 is able to partially suppress the locomotor deficits, muscle degeneration, and mitochondrial abnormalities (9–12). Together these findings suggest that aberrant accumulation of Mfn may mediate the loss of mitochondrial homeostasis caused by loss of PINK1 or parkin. Although further work will be needed to determine whether this contributes to PD pathogenesis, our results support the emerging hypothesis that the PINK1/Parkin pathway acts to regulate the safe degradation of terminally damaged mitochondria as a quality control mechanism (22, 44).
Materials and Methods
Cell Culture and Transfection.
S2R+ cells were cultured in Schneider's medium (Invitrogen) supplemented with 5% FCS (Sigma) and 1% penicillin–streptomycin (Invitrogen-Gibco). Cells were transfected using Effectene reagent (Qiagen) following the manufacturer's instructions and collected after 24–48 h. Copper sulfate solution (500 μM) was added to the cells to induce plasmid expression when required. Where indicated, cells were treated with 10 μM CCCP for 2 h or 10 mM paraquat for 6 h.
RNAi Treatment and Quantification.
Double-stranded RNAs (dsRNAs) were prepared using the MEGA script kit (Ambion). Primers used to generate dsRNAs are described in SI Materials and Methods. A total of 1.2 million cells were plated on a six-well plate and treated with 15 μg dsRNA probe in serum-free medium. Two hours after probe treatment, complete medium was added to the wells, and cells were cultured for 2 days before being transfected.
For mRNA quantification, total RNA was extracted using TRI reagent (Sigma) or an RNeasy mini kit (Qiagen) following the manufacturer's instruction. Total RNA (1.5 μg) was reverse-transcribed by using a random decamer primer (RETROscript kit; Ambion). Quantitative real-time PCR was performed using the SYBR Green Master Mix method (Sigma) with a Bio-Rad MyiQ system. Full details are in SI Materials and Methods.
Immunoblotting and Immunoprecipitation.
Standard protocols were used for Western blotting. The following commercial antibodies were used: anti-Complex Vα (1:5,000; MitoSciences), anti-Flag (1:1,000; Cell Signaling), anti-HA (1:1,000; Cell Signaling), anti-actin (1:10,000; Chemicon), anti-α-tubulin (1:10,000; Sigma), anti-GFP (1:2,000; Abcam), and anti-Myc (1:1,000; Cell Signaling). Anti-Parkin (1:3,000) has been described before. Anti-Mfn (1:2,000) was raised in rabbit against an N-terminal peptide, DTVDKSGPGSPLSRF. Detection was done using HRP-conjugated secondary antibodies and ECL chemiluminescence. For immunoprecipitation, cells were lysed in a standard lysis buffer (see SI Materials and Methods for details). Anti-Flag (1:50) or anti-GFP (1:200) antibodies were conjugated to Protein A agarose beads, incubated with 1 mg of whole-cell extract overnight, and extracted with 4× Laemmli buffer.
Cell Imaging.
Cells were plated on imaging dishes and treated as indicated. For mitochondrial morphology analysis, live cells were incubated with 200 μM rhodamine 123 and imaged live in growing medium. Quantification of mitochondria length was performed by using ImageJ software as previously described (45). For transfected cells, cells were fixed with paraformaldehyde and prepared by standard protocols for immunocytochemistry following treatment: Hoechst (2 μg/mL), phalloidin Alexa-Fluor488 (2 units), and anti-Complex Vα (1:1,000). All images were acquired on a DeltaVision DV microscope using standard epifluorescence. Unless stated otherwise, images were deconvolved after acquisition to improve image clarity and sharpness.
Drosophila Stocks and Procedures.
Drosophila were raised under standard conditions at 25 °C. park25 and PINK1B9 mutants and UAS-parkin have been described before (7, 46). w1118 and da-GAL4 strains were obtained from the Bloomington Drosophila Stock Center and UAS-Mfn-RNAi from the Vienna Drosophila Research Centre. _UAS-Mfn_3 was constructed by cloning the entire Drosophila Mfn ORF from cDNA (RE04414) into pUAST vector, which was injected into w1118 embryos for germline transformation (BestGene). Multiple independent lines were isolated and assessed.
Supplementary Material
Supporting Information
Acknowledgments
We thank D. Strutt, E. Smythe, and L. Pallanck for critical reading of the manuscript. This work is supported by grants from the Wellcome Trust (Grant 081987) and the Parkinson's Disease Society (Grant G-0713) to A.J.W. The Wellcome Trust is acknowledged for support of the Light Microscopy Facility (Grant GR077544AIA). This work is also supported by Grant G070091 from The Medical Research Council Centre for Developmental and Biomedical Genetics.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Gasser T. Molecular pathogenesis of Parkinson disease: Insights from genetic studies. Expert Rev Mol Med. 2009;11:e22. doi: 10.1017/S1462399409001148. [DOI] [PubMed] [Google Scholar]
- 2.Silvestri L, et al. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum Mol Genet. 2005;14:3477–3492. doi: 10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- 3.Shimura H, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 4.Büeler H. Impaired mitochondrial dynamics and function in the pathogenesis of Parkinson's disease. Exp Neurol. 2009;218:235–246. doi: 10.1016/j.expneurol.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Clark IE, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 6.Exner N, et al. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J Neurosci. 2007;27:12413–12418. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park J, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y, et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci USA. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poole AC, et al. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci USA. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng H, Dodson MW, Huang H, Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci USA. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park J, Lee G, Chung J. The PINK1-Parkin pathway is involved in the regulation of mitochondrial remodeling process. Biochem Biophys Res Commun. 2009;378:518–523. doi: 10.1016/j.bbrc.2008.11.086. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, et al. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci USA. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mortiboys H, et al. Mitochondrial function and morphology are impaired in parkin-mutant fibroblasts. Ann Neurol. 2008;64:555–565. doi: 10.1002/ana.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morais VA, et al. Parkinson's disease mutations in PINK1 result in decreased Complex I activity and deficient synaptic function. EMBO Mol Med. 2009;1:99–111. doi: 10.1002/emmm.200900006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dagda RK, et al. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lutz AK, et al. Loss of parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J Biol Chem. 2009;284:22938–22951. doi: 10.1074/jbc.M109.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandebring A, et al. Mitochondrial alterations in PINK1 deficient cells are influenced by calcineurin-dependent dephosphorylation of dynamin-related protein 1. PLoS One. 2009;4:e5701. doi: 10.1371/journal.pone.0005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Twig G, Hyde B, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: The bioenergetic view. Biochim Biophys Acta. 2008;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatsuta T, Langer T. Quality control of mitochondria: Protection against neurodegeneration and ageing. EMBO J. 2008;27:306–314. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway: A mitochondrial quality control system? J Bioenerg Biomembr. 2009;41:499–503. doi: 10.1007/s10863-009-9253-3. [DOI] [PubMed] [Google Scholar]
- 23.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 24.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinbrink S, Boutros M. RNAi screening in cultured Drosophila cells. Methods Mol Biol. 2008;420:139–153. doi: 10.1007/978-1-59745-583-1_8. [DOI] [PubMed] [Google Scholar]
- 27.Whitworth AJ, et al. Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson's disease factors Pink1 and Parkin. Dis Model Mech. 2008;1:168–174. doi: 10.1242/dmm.000109. discussion 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gandhi S, et al. PINK1 protein in normal human brain and Parkinson's disease. Brain. 2006;129:1720–1731. doi: 10.1093/brain/awl114. [DOI] [PubMed] [Google Scholar]
- 29.Haque ME, et al. Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc Natl Acad Sci USA. 2008;105:1716–1721. doi: 10.1073/pnas.0705363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou C, et al. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci USA. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim Y, et al. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem Biophys Res Commun. 2008;377:975–980. doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 32.Sha D, Chin LS, Li L. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-kappaB signaling. Hum Mol Genet. 2010;19:352–363. doi: 10.1093/hmg/ddp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuda N, et al. Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J Biol Chem. 2006;281:3204–3209. doi: 10.1074/jbc.M510393200. [DOI] [PubMed] [Google Scholar]
- 34.Hampe C, Ardila-Osorio H, Fournier M, Brice A, Corti O. Biochemical analysis of Parkinson's disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum Mol Genet. 2006;15:2059–2075. doi: 10.1093/hmg/ddl131. [DOI] [PubMed] [Google Scholar]
- 35.Fallon L, et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat Cell Biol. 2006;8:834–842. doi: 10.1038/ncb1441. [DOI] [PubMed] [Google Scholar]
- 36.Joch M, et al. Parkin-mediated monoubiquitination of the PDZ protein PICK1 regulates the activity of acid-sensing ion channels. Mol Biol Cell. 2007;18:3105–3118. doi: 10.1091/mbc.E05-11-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore DJ, West AB, Dikeman DA, Dawson VL, Dawson TM. Parkin mediates the degradation-independent ubiquitination of Hsp70. J Neurochem. 2008;105:1806–1819. doi: 10.1111/j.1471-4159.2008.05261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim KL, Dawson VL, Dawson TM. Parkin-mediated lysine 63-linked polyubiquitination: A link to protein inclusions formation in Parkinson's and other conformational diseases? Neurobiol Aging. 2006;27:524–529. doi: 10.1016/j.neurobiolaging.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 39.Olzmann JA, et al. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuda N, Tanaka K. Does impairment of the ubiquitin-proteasome system or the autophagy-lysosome pathway predispose individuals to neurodegenerative disorders such as Parkinson's disease? J Alzheimers Dis. 2009;19:1–9. doi: 10.3233/JAD-2010-1231. [DOI] [PubMed] [Google Scholar]
- 41.Soubannier V, McBride HM. Positioning mitochondrial plasticity within cellular signaling cascades. Biochim Biophys Acta. 2009;1793:154–170. doi: 10.1016/j.bbamcr.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 42.Liesa M, Palacín M, Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiol Rev. 2009;89:799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- 43.Koshiba T, et al. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 44.Chu CT. Tickled PINK1: Mitochondrial homeostasis and autophagy in recessive Parkinsonism. Biochim Biophys Acta. 2010;1802:20–28. doi: 10.1016/j.bbadis.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Vos KJ, Sheetz MP. Visualization and quantification of mitochondrial dynamics in living animal cells. Methods Cell Biol. 2007;80:627–682. doi: 10.1016/S0091-679X(06)80030-0. [DOI] [PubMed] [Google Scholar]
- 46.Greene JC, et al. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci USA. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information